Abstract
Purpose
High mobility group box 3 (HMGB3) is associated with hepatocytes malignant transformation by our previous work. We continued to investigate the diagnostic and prognostic values of HMGB3 for hepatocellular carcinoma (HCC).
Patients and methods
Circulating HMGB3 levels were quantitatively detected in a cohort of 225 patients with chronic liver diseases by ELISA and compared with alpha-fetoprotein by the receiver operating characteristic curve. HMGB3 expression in tissues of 170 HCC was detected by tissue microarray and immunohistochemistry. Relationship between HMGB3 level and HCC prognosis was evaluated by the Kaplan–Meier curves and Cox regression model.
Results
The incidence of serum HMGB3 >2.0 ng/mL was 75.6% in HCC (96/127), 20.8% in liver cirrhosis (10/48), 16.0% in chronic hepatitis (8/50), and none in healthy controls (0/49). Significant difference (P<0.001) of circulating HMGB3 level was found between HCC and benign liver diseases. Total diagnostic sensitivity of serum HMGB3 plus alpha-fetoprotein was up to 89.0% for HCC. Higher HMGB3 expression was confirmed to be 73.5% in HCC tissues (125/170) >30.6% in their paracancerous tissues (52/170). HMGB3 expression was closely related to tumor size, TNM stage, poor survival, and high recurrence, suggesting an independent prognosis factor for HCC.
Conclusion
HMGB3 with aberrant expression could be a novel diagnostic and prognostic marker for HCC.
Keywords: hepatocellular carcinoma, HMGB3, diagnosis, prognosis, biomarker
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in China, particularly in the eastern and southern areas, including the inshore area of the Yangtze River.1 For decades, great progression has been made in HCC treatment, especially in new-generation sequencing, immune therapy, and molecular-targeted drugs. However, conventional therapies for advanced HCC have limited efficacy.2 Given the poor prognosis of HCC, early surveillance is of great importance. Although serum α-fetoprotein (AFP) level is a useful marker for HCC, the diagnostic value is far away from ideal due to its high false-negative rate at early stage and low incidence at advanced stage.3 New tumor markers, such as oncogenic secretory clusterin,4 insulin-like growth factor-I receptor,5 and insulin-like growth factor-II,6 have been developed to improve the sensitivity and specificity of diagnosis or prediction of prognosis; however, the overall outcome is still unsatisfying. Hence, searching novel and effective biomarkers for HCC diagnosis, prognosis, or targeted therapy is still of paramount clinical significance.
High mobility group box (HMGB) family is a group of highly conserved chromatin-associated proteins and consists of four members, including HMGB1, HMGB2, HMGB3, and HMGB4.7 It is widely expressed in mammalian cells and involved in various physiological process, such as DNA repairing, chromatin remodeling, and cell death.8,9 Among the family members, HMGB1 is known as a common secreted protein distributed in nuclear and cytoplasm and implicated in inflammation, neurodegeneration, cancer, and aging. According to recent studies, HMGB1 contributed to the genesis and progression of HCC, with the stimulation of hepatitis B virus X protein,10 prostate cancer-associated non-coding RNA transcript 1 (PCAT-1),11 or hypoxia condition.12 Similarly, HMGB2 was also related to aggressiveness and poor prognosis of HCC. Parallelly, our recent work for the first time indicated that HMGB3 might participate in the malignant transformation of hepatocytes.13 However, its possible clinical value in HCC remained unclear.
Thus, in this study, we detected HMGB3 expression in serum samples from 225 patients with chronic liver diseases to explore its diagnostic value of HCC. Additionally, hepatic HMGB3 expression and its correlation with HCC prognosis were also investigated in 170 HCC tissue samples. The current study aimed to evaluate HMGB3 as a prognostic and diagnostic biomarker for HCC.
Patients and methods
Serum samples
To evaluate the diagnostic value of serum HMGB3 levels, the current study conducted a cohort containing patients with HCC, liver cirrhosis (LC), chronic hepatitis (CH), and healthy control. One hundred twenty-seven HCC serum samples enrolled in this serological test were collected from HCC patients at Affiliated Hospital of Nantong University, China, from January 2015 to March 2017. Some basic information of these HCC sera cases was listed as follows: the patients’ age ranged from 37 to 86 years old (mean age: 60.54 years old) and included 93 men and 34 women. Among 127 HCC patients (105 primary and 22 recurrence), 86 cases were Child–Pugh classification A, 30 cases for B, and 11 cases for C; 55 cases had AFP values <20 ng/mL, 35 cases between 20 and 400 ng/mL, and 37 cases >400 ng/mL. One hundred one patients (80%) had a history of cirrhosis; 94 patients had positive hepatitis B surface antigen (HBsAg). Serum AFP level exceeding 20 ng/mL was defined as a positive result. HCC was diagnosed according to the criteria set by the 2010 Chinese National Collaborative Cancer Research Group.14 Other cases in this study included 48 patients with LC, 50 patients with CH, and 49 healthy volunteers as normal controls (NCs) with negative hepatitis B markers (HBsAg, HBcAb, and HBV-DNA) and normal serum alanine aminotransferase levels from the Nantong Central Blood Bank. The diagnosis of viral hepatitis was based on the criteria proposed at the Chinese National Viral Hepatitis Meeting.15 Clinical information was collected from medical records, including age, gender, tumor size, AFP value, cirrhosis, HBV infection, TNM stage, differentiation degree, and Child classification. The correlations of serum HMGB3 expression with clinical parameters above were also investigated in this study. Prior written informed consent was obtained from all patients according to the World Medical Association Declaration of Helsinki, and the study obtained approval from the Ethics Committee of Affiliated Hospital of Nantong University.
Tissue specimen of HCC
HCC – and their self-matched paracancerous – tissues from 170 HCC patients who underwent hepatectomy in Affiliated Hospital of Nantong University from June 2007 to May 2011 were involved in this study. Basic information of these HCC tissue samples was described as follows: the patients’ age ranged from 29 to 84 years old (mean ages: 51.28 years old) and included 142 men and 28 women. Fifty cases had AFP values <20 ng/mL, 48 cases between 20 and 400 ng/mL, and 72 cases >400 ng/mL; 151 patients had a history of cirrhosis; 150 cases had positive HBsAg. All patients were histopathologically confirmed with HCC and underwent no treatment before the surgery. Clinical information on the patients was collected from medical records, including differentiation degree, TNM stage, tumor size, AFP value, cirrhosis, HBV infection, metastasis, and cirrhosis. Besides, each patient was followed up to obtain the information of survival and recurrence. The procedures were approved by the Ethics Committee of Affiliated Hospital of Nantong University.
Tissue microarray analysis
The tissue microarrays were constructed by the SuperChip Company (Shanghai, China), containing formalin-fixed and paraffin-embedded (FFPE) specimens from 170 HCC tissues and their self-matched paracancerous tissues. Cores from the selected areas were arrayed into a recipient block with unique tissue microarray location numbers.
Immunohistochemistry (IHC)
After deparaffinization and rehydration using xylene and graded alcohol, antigen of slides was retrieved by Tris– EDTA–citrate buffer for 5 minutes at 121°C. The slides were washed with Tris-buffered saline containing 0.05% Tween-2 and treated with 3% H2O2. After blocking with 10% goat serum for 2 hours, the slides were incubated with primary antibody (#MAB55071, R&D, Minneapolis, MN, USA) for 1 hour at room temperature. After washing for three times, the slides were treated with secondary antibody (#VC002-125, R&D) at 37°C for 30 minutes and stained with diaminobenzidine. Negative controls were performed by omitting the application of primary antibody.
IHC evaluation
Two experienced pathologists independently evaluated the immunohistochemical staining according to the H score system.16 The immunostaining intensity was indicated by four grades (0, negative; 1, weakly positive; 2, positive; and 3, strongly positive). Then, the H score was calculated by multiplication of staining intensities with percentage of positive cells. The final score ranged from 0 to 300. The score of 0–49 was considered negative, and the score of 50–300 was positive.
Enzyme-linked immunosorbent assay
Serum HMGB3 levels in four groups (HCC, LC, CH, and healthy control) were detected using the human HMGB3 ELISA kit (Cloud-Clone Corp, Wuhan, China) according to the manufacturer’s instructions. In brief, 100 µL sample or standard solution was added into each indicating well of 96-well plates and incubated at 37°C for 2 hours. After removing the supernatant, 100 µL HMGB3 biotin-conjugated antibody was added and incubated at 37°C for 1 hour. Subsequently, following washing three times, 100 µL avidin conjugated to horseradish peroxidase (HRP) was added to each well and incubated for 30 minutes at 37°C. Next, after incubation with 90 µL substrate solution for 30 minutes, each well was added 50 µL stop solution. Finally, value of each well was measured by a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) at a wave length of 450 nm. Each sample was detected repeatedly.
Statistical analysis
All statistical analyses were performed by SPSS19.0 or Stata14.0. The data are presented as mean ± SD. Student’s t-test (for quantitative data) and chi-square tests (for categorical data) were used to examine the differences between two groups. Overall survival (OS) curves and diseases-free survival (DFS) curves were calculated using the Kaplan–Meier method with the log-rank test. Univariate and multivariate analyses were performed to discover the independent factors for HCC prognosis. Receiver operating characteristic (ROC) curve was used to assess the diagnostic value of HCC. P-value <0.05 was considered statistically significant.
Results
Serological HMGB3 levels in chronic liver diseases
Quantitative analysis of serum average HMGB3 levels in a cohort of patients with malignant or benign chronic liver diseases is shown in Table 1. According to the results of HMGB3 levels in sera of the NCs, the upper limitation (mean ± 1.96 SD, 2.0 ng/mL) was defined as a cutoff value. The incidence of serum HMGB3 level was 75.6% (96/127) in HCC, 20.8% (10/48) in LC, 16.0% (8/50) in CH, and none (0/49) in NC. In comparison, the incidence of serum AFP (>20.0 ng/mL) was 56.7% (72/127) in HCC, 25.0% (12/48) in LC, 22.0% (11/50) in CH, and none (0/49) in NC. Both the serum HMGB3 and AFP levels are significantly higher (P<0.001) in the HCC group more than those in any group of patients with benign chronic liver diseases.
Table 1.
Comparative analysis of serum HMGB3 or AFP level in patients with different liver diseases
| Group | n | Mean ± SD | t value | P-value | Positive n (%) | χ2 value | P-value |
|---|---|---|---|---|---|---|---|
| HMGB3 | ng/mL | >2.0 ng/mL | |||||
| HCC | 127 | 2.7±1.3 | 96 (75.6) | ||||
| LC | 48 | 1.6±0.8 | 6.805 | <0.001 | 10 (20.8) | 43.733 | <0.001 |
| CH | 50 | 1.5±0.7 | 8.438 | <0.001 | 8 (16.0) | 52.571 | <0.001 |
| NC | 49 | 1.2±0.4 | 11.576 | <0.001 | 0 (0.0) | 81.487 | <0.001 |
| AFP | ng/mL | >20.0 ng/mL | |||||
| HCC | 127 | 2505.0±5634.4 | 72 (56.7) | ||||
| LC | 48 | 78.5±200.8 | 4.845 | <0.001 | 12 (25.0) | 14.018 | 0.001 |
| CH | 50 | 107.1±287.9 | 4.780 | <0.001 | 11 (22.0) | 17.339 | <0.001 |
| NC | 49 | 3.4±1.6 | 5.003 | <0.001 | 0 (0.0) | 47.012 | <0.001 |
Abbreviations: AFP, alpha-fetoprotein; CH, chronic hepatitis; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3; LC, liver cirrhosis; NC, normal control.
Circulating HMGB3 as a diagnostic marker for HCC
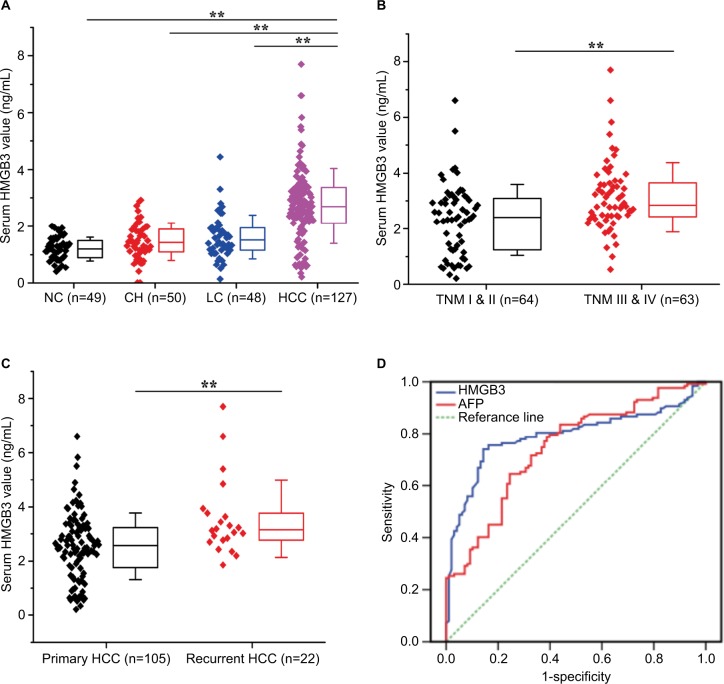
Distribution of serum HMGB3 expression and its diagnostic values in HCC are shown in Figure 1. Serum HMGB3 levels increased from benign liver diseases to HCC according to the distribution in different groups. The mean concentration of circulating HMGB3 in the HCC group was significantly higher (P<0.001) than any other groups (Figure 1A). Among the HCC patients, significant difference (P<0.001) in serum HMGB3 expression was found between patients at high and low TNM stage (Figure 1B) or patients with and without recurrence (Figure 1C). The diagnostic value of HMGB3 level compared with AFP for HCC was evaluated by the ROC curve (Figure 1D). The area under the curve was 0.791 in HMGB3 (CI: 0.730–0.853, P<0.001) and 0.743 in AFP (CI: 0.679–0.808, P<0.001).
Figure 1.
Distribution of serum HMGB3 expression and diagnostic values in HCC. The levels of circulating HMGB3 expression in the HCC, LC, CH, and NC group were detected by ELISA.
Notes: (A) The distribution of serum HMGB3 level in different groups; (B) serum HMGB3 expression in HCC patients at high or low TNM stages; (C) serum HMGB3 expression in HCC patients with or without recurrence; (D) the diagnostic value of HMGB3 level for HCC evaluated by the ROC curve. Area under curve of HMGB3 was 0.791 (CI: 0.730–0.853, P<0.001) and that of AFP was 0.743 (CI: 0.679–0.808, P<0.001). Bar represents the mean value of each group. **P<0.01.
Abbreviations: AFP, alpha-fetoprotein; CH, chronic hepatitis; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3; LC, liver cirrhosis; NC, normal controls; ROC, receiver operating characteristic curve.
The sensitivity, specificity, accuracy, positive predictive value, and negative predictive value of serum HMGB3 or AFP level (Table 2) for HCC diagnosis were 75.6% or 56.7%, 81.6% or 76.5%, 78.2% or 65.3%, 84.2% or 73.5%, and 72.1% or 57.7%, respectively. Notably, the sensitivity was up to 89.0% by combining detection of them. Interestingly, for the HCC patients with small tumor size (≤3 cm), the diagnostic sensitivity of serum HMGB3 or AFP level alone was 55.3% (21/38) or 39.5% (15/38), while it was up to 71.1% (27/38) in combining detection.
Table 2.
Assessment of diagnostic validity of serum HMGB3/AFP level for HCC
| HMGB3 >2.0 ng/mL (%) | AFP >20.0 ng/mL (%) | HMGB3 + AFP (%) | |
|---|---|---|---|
| Sensitivity (%) | 75.6 | 56.7 | 89.0 |
| Specificity (%) | 81.6 | 76.5 | 63.3 |
| Accuracy (%) | 78.2 | 65.3 | 77.8 |
| Positive predictive value (%) | 84.2 | 73.5 | 75.8 |
| Negative predictive value (%) | 72.1 | 57.7 | 81.6 |
Abbreviations: AFP, alpha-fetoprotein; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
Clinicopathologic features of serum HMGB3 expression
The clinicopathologic characteristics of circulating HMGB3 expression in 127 HCC patients are shown in Table 3. The HMGB3 expression in sera of HCC patients was closely associated with portal vein invasion (χ2=7.519, P=0.006), tumor size (χ2=18.357, P=0.019), differentiation degree (χ2=15.175, P<0.001), TNM stage (χ2=9.292, P<0.001), and recurrence (χ2=5.690, P=0.017). However, no significant relationship was found between HMGB3 expression and age, gender, cirrhosis, HBV infection, AFP level, gross classification, or Child–Pugh degree of HCC patients.
Table 3.
Clinicopathologic features of serum HMGB3 expression in 127 HCC patients
| Group | n | Mean ± SD (ng/mL) | t value | P-value | Positive, n (%) | χ2value | P-value |
|---|---|---|---|---|---|---|---|
| Age (years) | |||||||
| ≤50 | 65 | 2.8±1.4 | 0.677 | 0.500 | 48 (73.85) | 0.220 | 0.639 |
| >50 | 62 | 2.6±1.2 | 48 (77.42) | ||||
| Gender | |||||||
| Male | 93 | 2.6±1.3 | 1.018 | 0.311 | 67 (72.04) | 2.369 | 0.124 |
| Female | 34 | 2.9±1.3 | 29 (85.29) | ||||
| AFP (ng/mL) | |||||||
| ≤20 | 55 | 2.5±1.2 | 1.387 | 0.168 | 40 (72.73) | 0.431 | 0.511 |
| >20 | 72 | 2.9±1.4 | 56 (77.78) | ||||
| HBsAg | |||||||
| Negative | 33 | 2.6±0.8 | 0.475 | 0.636 | 29 (87.88) | 3.649 | 0.056 |
| Positive | 94 | 2.8±1.5 | 67 (71.28) | ||||
| Liver cirrhosis | |||||||
| With | 89 | 2.6±1.3 | 1.460 | 0.147 | 64 (71.91) | 2.184 | 0.139 |
| Without | 38 | 3.0±1.3 | 32 (84.21) | ||||
| Portal vein invasion | |||||||
| With | 37 | 3.3±1.2 | 3.523 | 0.001 | 34 (91.89) | 7.519 | 0.006 |
| Without | 90 | 2.5±1.3 | 62 (68.89) | ||||
| Tumor size | |||||||
| ≤5 cm | 60 | 2.3±1.3 | 3.472 | 0.001 | 35 (58.33) | 18.357 | <0.001 |
| >5 cm | 67 | 3.1±1.3 | 61 (91.04) | ||||
| Metastasis | |||||||
| With | 63 | 2.9±1.3 | 1.199 | 0.233 | 50 (79.37) | 0.965 | 0.326 |
| Without | 64 | 2.6±1.3 | 46 (71.88) | ||||
| Differentiation | |||||||
| Well | 57 | 2.3±1.4 | 3.594 | <0.001 | 34 (59.65) | 15.175 | <0.001 |
| Moderate and poor | 70 | 3.1±1.2 | 62 (88.57) | ||||
| Gross classification | |||||||
| Unifocal | 92 | 2.7±1.3 | 0.760 | 0.448 | 68 (73.91) | 0.509 | 0.476 |
| Multifocal | 35 | 2.8±1.3 | 28 (80.00) | ||||
| TNM | |||||||
| I–II | 64 | 2.3±1.3 | 3.639 | <0.001 | 41 (64.06) | 9.292 | 0.002 |
| III–IV | 63 | 3.1±1.2 | 55 (87.30) | ||||
| Child classification | |||||||
| A | 82 | 2.7±1.3 | 0.762 | 0.448 | 61 (74.39) | 0.181 | 0.671 |
| B and C | 45 | 2.8±1.4 | 35 (77.78) | ||||
| Recurrence | |||||||
| Yes | 22 | 3.6±1.2 | 3.426 | 0.001 | 21 (95.45) | 5.690 | 0.017 |
| No | 105 | 2.5±1.4 | 75 (71.42) | ||||
Note: Bold indicates P<0.05.
Abbreviations: AFP, alpha-fetoprotein; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
Distribution of HMGB3 in HCC tissues
The immunohistochemical staining of HMGB3 expression in a cohort of 170 self-matched human cancerous and paracancerous tissues is shown in Figure 2. Human cancerous and paracancerous tissues were examined by the histopathology with H&E staining. Representative HMGB3 staining was mostly located in cell nucleus and cytoplasm. Moreover, the HMGB3 staining exhibited stronger intensity and more nuclear-positive cells in the cancerous tissues with advanced stages than that in the paracancerous tissues or early stages (Figure 2A). As presented in Figure 2B, increased score of HMGB3 expression was observed in the cancerous tissues with advanced stage compared with that in the paracancerous tissues or HCC tissues with early stage by the semiquantitative analysis of IHC staining. The positive HMGB3 expression (score of 50–300) detected in 73.53% of the cancerous tissues (125/170) was significantly higher (χ2=62.801, P<0.001) than that in the paracancerous tissues (30.59%, 52/170).
Figure 2.
Immunohistochemical analysis of HMGB3 expression in HCC tissues.
Notes: Distribution of hepatic HMGB3 expression in HCC and paracancerous tissues was detected by tissue microarray with immunohistochemical staining. (A) The staining of H&E or HMGB3 immunohistochemistry in paracancerous tissues and HCC tissues with different TNM stage (original magnification ×40 or ×400). (B) The scores of hepatic HMGB3 staining were demonstrated as scatter and box plots. Horizontal lines represent the median score of the two group; the bottom and top lines indicated the 10th and 90th percentiles, respectively; vertical bars indicated the range of data; **P<0.01.
Abbreviations: HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
Clinical features of hepatic HMGB3 expression
The correlations of hepatic HMGB3 expression with clinicopathologic parameters in HCC patients are elucidated in Table 4. Abnormality of HMGB3 expression was significantly associated with tumor size (χ2=7.502, P=0.006), advanced TNM stages (χ2=5.801, P=0.016), and high recurrence ratio (χ2=26.176, P<0.001). No significant difference was observed between HMGB3 expression and age, gender, AFP, HBV infection, LC, degree of differentiation, or gross classification.
Table 4.
Clinicopathologic features of HMGB3 expression in 170 HCC tissues
| Group | n | Pos. n (%) | χ2 value | P-value | |
|---|---|---|---|---|---|
| Age (years) | |||||
| ≤50 | 82 | 58 (70.73) | 0.637 | 0.425 | |
| >50 | 88 | 67 (76.14) | |||
| Gender | |||||
| Female | 28 | 17 (60.71) | 2.828 | 0.093 | |
| Male | 142 | 108 (76.06) | |||
| AFP (ng/mL) | |||||
| ≤20 | 50 | 35 (70.00) | 0.453 | 0.501 | |
| >20 | 120 | 90 (75.00) | |||
| HBsAg | |||||
| Negative | 20 | 16 (80.00) | 0.488 | 0.485 | |
| Positive | 150 | 109 (72.67) | |||
| Tumor size (cm) | |||||
| ≤5 | 87 | 53 (60.92) | 7.502 | 0.006 | |
| >5 | 83 | 72 (86.75) | |||
| Liver cirrhosis | |||||
| Without | 19 | 12 (63.16) | 1.182 | 0.277 | |
| With | 151 | 113 (74.83) | |||
| Differentiation | |||||
| Well | 72 | 50 (69.44) | 1.071 | 0.301 | |
| Moderate and poor | 98 | 75 (76.53) | |||
| Gross classification | |||||
| Unifocal | 139 | 99 (71.22) | 2.083 | 0.149 | |
| Multifocal | 31 | 26 (83.87) | |||
| TNM | |||||
| I and II | 149 | 105 (70.47) | 5.801 | 0.016 | |
| III and IV | 21 | 20 (95.24) | |||
| Metastasis | |||||
| Without | 160 | 115 (71.88) | 3.825 | 0.050 | |
| With | 10 | 10 (100.00) | |||
| Recurrence | |||||
| No | 88 | 50 (56.82) | 26.176 | <0.001 | |
| Yes | 82 | 75 (91.46) | |||
Note: Bold indicates P<0.05.
Abbreviations: AFP, alpha-fetoprotein; HBsAg, hepatitis B surface antigen; HMGB3, high mobility group box 3.
Prognostic value of HMGB3 level for HCC
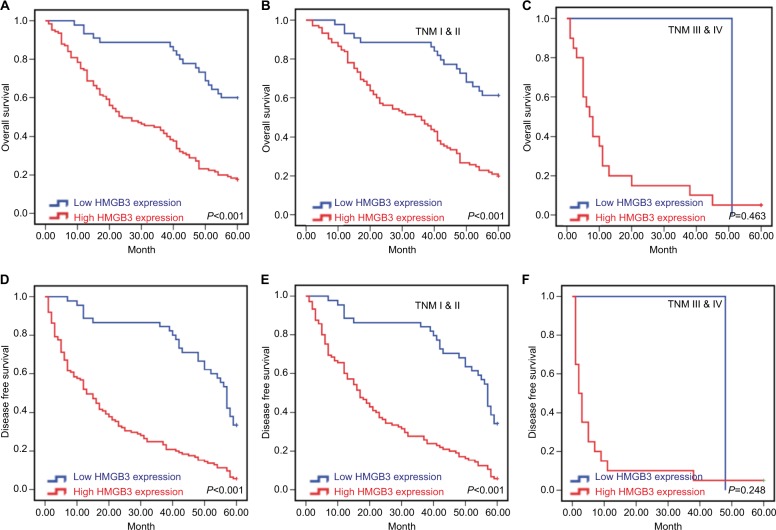
The Kaplan–Meier analysis to discover the prognostic significance of HMGB3 level in HCC patients is shown in Figure 3. HCC patients with high HMGB3 expression had significantly poorer OS (χ2=11.348, P<0.001, Figure 3A) and DFS (χ2=14.947, P<0.001, Figure 3D), in contrast to the cases with low HMGB3 expression. Besides, for the patients in early stage (TNM I and II, Figures 3B, E), high HMGB3 also indicated poor OS and DFS, while it had no statistical significance for patients with advanced stages (TNM III and IV, Figures 3C, F).
Figure 3.
Kaplan–Meier survival curves of 170 HCC patients.
Notes: The Kaplan–Meier survival curves were generated after follow-up of 170 HCC patients. (A) The overall survival (OS) curves were calculated according to high or low HMGB3 expression (P<0.001); (B) OS curves of patients in TNM I and II stages according to HMGB3 expression (P<0.001); (C) OS curves of patients in TNM III and IV stages according to HMGB3 expression(P=0.463). (D) The disease-free survival (DFS) curves were performed according to high or low HMGB3 expression (P<0.001); (E) DFS curves of patients in TNM I and II stages according to HMGB3 expression (P<0.001); (F) DFS curves of patients in TNM III and IV stages according to HMGB3 expression (P=0.248).
Abbreviations: HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
The univariate Cox regression analysis of OS or DFS in HCC patients (Table 5) demonstrated that tumor size, TNM stage, differentiation, tumor number, metastasis, and HMGB3 expression were potential factors affecting OS or DFS in HCC patients. Then, multivariate Cox regression analysis indicated that high HMGB3 expression (HR =3.042, P<0.001), differentiation (HR =1.538, P=0.034), and TNM stage (HR =2.471, P=0.003) were independent indicators of OS in HCC patients (Table 6). On the contrary, for the DFS in HCC patients, HMGB3 expression (HR=3.227, P<0.001), TNM stage (HR=1.926, P=0.031), differentiation (HR=1.635, P=0.006), and tumor number (HR=1.804, P=0.007) should be independent prognostic factors for HCC recurrence.
Table 5.
Univariate analysis to identify the risk factors of HCC
| Group | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
| HR | P-value | 95% CI | HR | P-value | 95% CI | |
| Gender | ||||||
| Male vs female | 1.432 | 0.160 | 0.868–2.364 | 1.492 | 0.079 | 0.955–2.330 |
| Age (years) | ||||||
| ≤50 vs >50 | 0.910 | 0.605 | 0.637–1.301 | 0.800 | 0.175 | 0.579–1.104 |
| Tumor diameter (cm) | ||||||
| ≤5 vs >5 | 1.863 | 0.001 | 1.300–2.671 | 1.617 | 0.004 | 1.169–2.236 |
| Differentiation | ||||||
| Well vs moderate and poor | 1.868 | 0.001 | 1.285–2.713 | 1.872 | <0.001 | 1.338–2.617 |
| AFP (ng/mL) | ||||||
| ≤50 vs >50 | 1.294 | 0.207 | 0.867–1.931 | 1.396 | 0.068 | 0.976–1.997 |
| Liver cirrhosis | ||||||
| Yes vs no | 0.935 | 0.812 | 0.535–1.633 | 0.903 | 0.692 | 0.545–1.497 |
| Gross classification | ||||||
| Multifocal vs unifocal | 1.584 | 0.041 | 1.018–2.466 | 1.778 | 0.006 | 1.177–2.687 |
| HBsAg | ||||||
| Yes vs no | 0.827 | 0.777 | 0.547–1.569 | 1.071 | 0.786 | 0.654–1.753 |
| TNM | ||||||
| I–II vs III–IV | 3.870 | <0.001 | 2.373–6.309 | 3.295 | <0.001 | 2.039–5.324 |
| Metastasis | ||||||
| Yes vs no | 3.844 | <0.001 | 1.990–7.425 | 3.509 | <0.001 | 1.827–6.737 |
| HMGB3 expression | ||||||
| High vs low | 3.658 | <0.001 | 2.208–6.061 | 3.382 | <0.001 | 2.243–5.098 |
Note: Bold indicates P<0.05.
Abbreviations: AFP, alpha-fetoprotein; HBsAg, hepatitis B surface antigen; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
Table 6.
Multivariate analysis (adjusted for age and sex) to identify the risk factors of HCC
| Group | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
| HR | P-value | 95% CI | HR | P-value | 95% CI | |
| Tumor diameter (cm) | ||||||
| ≤5 vs >5 | 1.400 | 0.077 | 0.964–2.034 | 1.202 | 0.296 | 0.851–1.700 |
| Differentiation | ||||||
| Well vs moderate and poor | 1.538 | 0.034 | 1.033–2.288 | 1.635 | 0.006 | 1.150–2.323 |
| Gross classification | ||||||
| Multifocal vs unifocal | 1.550 | 0.059 | 0.984–2.444 | 1.804 | 0.007 | 1.171–2.779 |
| TNM | ||||||
| I–II vs III–IV | 2.471 | 0.003 | 1.350–4.525 | 1.926 | 0.031 | 1.063–3.478 |
| Metastasis | ||||||
| Yes vs no | 1.302 | 0.520 | 0.582–2.913 | 1.305 | 0.500 | 0.602–2.826 |
| HMGB3 expression | ||||||
| High vs low | 3.042 | <0.001 | 1.809–5.115 | 3.227 | <0.001 | 2.098–4.963 |
Note: Bold indicates P<0.05.
Abbreviations: AFP, alpha-fetoprotein; HCC, hepatocellular carcinoma; HMGB3, high mobility group box 3.
Discussion
HMGB family consists of four members with multiple physiology and pathology features. HMGB3 is an X-linked member of this family and mainly distributed in nucleus, chromosome, and cytoplasm. Due to the high homology among HMGB family members, HMGB3 may share some similar properties with HMGB1 and HMGB2.17 Recently, the abnormal expression of HMGB1 and HMGB2 in this family were associated with the carcinogenesis and progression of HCC.12,18 Although our previous work showed its roles in HCC progression in vitro and in vivo, its clinical values have not been investigated. Thus, the current study discovered the relationships between HMGB3 expression and HCC progression in sera and tissues, attempting to evaluate HMGB3 as a novel molecular marker for HCC diagnosis and prognosis.
Early diagnosis of HCC is of utmost significance.19 Although serum AFP has been routinely tested for HCC diagnosis, it exhibits a low sensitivity and specificity iñ40% of negative patients.20 Circulating soluble HMGB1 was examined as a useful predictor for advanced HCC patients treated with sorafenib and hepatic arterial infusion chemotherapy.21 However, no significant difference in serum HMGB1 level between HCC patients and healthy controls was observed in another cohort.22 Apart from that, other HMGB family members have not been evaluated as serum marker for HCC. In this study, for the first time, serum HMGB3 level was detected in a cohort of patients with chronic liver diseases. Abnormal HMGB3 levels were significantly higher in HCC patients than those in cases with cirrhosis or CH. According to the sensitivity and the ROC curve, circulating HMGB3 showed superiority over AFP for HCC diagnosis, especially in cases with tumor size <3 cm. Moreover, combining detection of AFP and HMGB3 presented supplementary value in improving sensitivity, indicating that serum HMGB3 level might be a novel useful biomarker for HCC diagnosis.
However, HMGB3 was also first evaluated as an indicator for HCC prognosis. Indeed, elevated HMGB3 has been associated with poor survival of patients in several kinds of cancer types, including gastric cancer,23 lung adenocarcinoma,24 esophageal squamous cell carcinoma,25 and bladder cancer.26 Besides, it could contribute to tumor progression by enhancing proliferation,27 migration,28 and chemoresistance29 of cancer cells. In the current study, HMGB3 overexpression was observed in HCC tissues with advanced TNM stages rather than paracancerous tissues or HCC tissues with early stages. In addition, upregulation of HMGB3 was associated with larger tumor size, higher recurrence ratio, and lower survival rate. Univariate or multivariate assays recommended as an independent factor for prognosis of HCC patients.
Given its overexpression with clinical malignant features in HCC, HMGB3 might contribute to the HCC progression. As expected, elevated HMGB3 levels were observed during hepatocarcinogenesis in a rat model by our recent work.13 In addition, in vitro evidence showed that HMGB3 promoted proliferation and tumor growth of HCC cells by regulating cell cycle and DNA replication pathways, which was further confirmed by Gene Set Enrichment Analysis in bioinformatic databases. While HMGB3 was also implicated in classic tumor-related pathways such as Wnt and MAPK pathways in other cancer types, the exact regulatory and interaction mechanisms remain to be elucidated. Therefore, it needs more mechanism investigations for further research.
Conclusion
To the best of our knowledge, this study is the first to investigate HMGB3 expression, a little-known member of HMGB family, in cancerous tissues and sera of HCC patients. Although some limitations in this study should be solved in further study, including sample size of cancerous tissues, comprehensively functional exploration, and interaction with HCC-associated signal pathway, this work sheds light on the promising clinical application of HMGB3 as a novel biomarker for HCC diagnosis or prognosis. Future studies should clarify the exact molecular mechanisms relating to the upregulation of HMGB3 expression and its important role in hepatocarcinogenesis or potentially therapeutic value.30
Acknowledgments
The study was supported by National Natural Science Foundation (81673241, 81702419), Projects of Jiangsu Medical Science (BE2016698), and Graduate innovation (KYCX17_1934), Nantong HFPC (WQ2016083) of China.
Footnotes
Author contributions
WZ, JY, and ZD contributed equally to this work, conceived the idea, and designed the experiments; MF and WW collected and processed clinical data; LW analyzed data; MY and DY are the guarantors and supervised the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen JG, Zhu J, Zhang YH, et al. Cancer survival in Qidong between 1972 and 2011: a population-based analysis. Mol Clin Oncol. 2017;6(6):944–954. doi: 10.3892/mco.2017.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn RS, Zhu AX, Farah W, et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: a systematic review and meta-analysis. Hepatology. 2018;67(1):422–435. doi: 10.1002/hep.29486. [DOI] [PubMed] [Google Scholar]
- 3.Yao D, Jiang D, Huang Z, et al. Abnormal expression of hepatoma specific gamma-glutamyl transferase and alteration of gamma-glutamyl transferase gene methylation status in patients with hepatocellular carcinoma. Cancer. 2000;88(4):761–769. [PubMed] [Google Scholar]
- 4.Zheng W, Yao M, Qian Q, et al. Oncogenic secretory clusterin in hepatocellular carcinoma: expression at early staging and emerging molecular target. Oncotarget. 2017;8(32):52321–52332. doi: 10.18632/oncotarget.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan XD, Yao M, Wang L, et al. Overexpression of insulin-like growth factor-I receptor as a pertinent biomarker for hepatocytes malignant transformation. World J Gastroenterol. 2013;19(36):6084–6092. doi: 10.3748/wjg.v19.i36.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian J, Yao D, Dong Z, et al. Characteristics of hepatic IGF-II expression and monitored levels of circulating IGF-II mRNA in metastasis of hepatocellular carcinoma. Am J Clin Pathol. 2010;134(5):799–806. doi: 10.1309/AJCPTFDSE2V3LCZP. [DOI] [PubMed] [Google Scholar]
- 7.Reeves R. High mobility group (HMG) proteins: modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair. 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799(1–2):101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Gerlitz G. HMGNs, DNA repair and cancer. Biochim Biophys Acta. 2010;1799(1–2):80–85. doi: 10.1016/j.bbagrm.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Dong Z, Yang P, et al. Hepatitis B virus X protein stimulates high mobility group box 1 secretion and enhances hepatocellular carcinoma metastasis. Cancer Lett. 2017;394:22–32. doi: 10.1016/j.canlet.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhang D, Cao J, Zhong Q, et al. Long noncoding RNA PCAT-1 promotes invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway in hepatocellular carcinoma. Biomed Pharmacother. 2017;95:1187–1193. doi: 10.1016/j.biopha.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 12.Tohme S, Yazdani HO, Liu Y, et al. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth through HMGB1 and TLR9 interaction. Hepatology. 2017;66(1):182–197. doi: 10.1002/hep.29184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng WJ, Yao M, Fang M, Wang L, Dong ZZ, Yao DF. Abnormal expression of HMGB-3 is significantly associated with malignant transformation of hepatocytes. World J Gastroenterol. 2018;24(32):3650–3662. doi: 10.3748/wjg.v24.i32.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ministry of Health of the People’s Republic of China Updated standards for the diagnosis and treatment of primary liver cancer. Zhonghua Gan Zang Bing Za Zhi. 2012;20(6):419–426. Chinese [with English abstract] [PubMed] [Google Scholar]
- 15.O Y. Management of clinical diagnosis, and antiviral therapy for HBV-related cirrhosis. Zhonghua Gan Zang Bing Za Zhi. 2014;22(5):327–335. [PubMed] [Google Scholar]
- 16.Specht E, Kaemmerer D, Sänger J, et al. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology. 2015;67(3):368–377. doi: 10.1111/his.12662. [DOI] [PubMed] [Google Scholar]
- 17.Yanai H, Ban T, Taniguchi T. High-mobility group box family of proteins: ligand and sensor for innate immunity. Trends Immunol. 2012;33(12):633–640. doi: 10.1016/j.it.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Kwon JH, Kim J, Park JY, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16(22):5511–5521. doi: 10.1158/1078-0432.CCR-10-0825. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Yao M, Dong Z, Zhang Y, Yao D. Circulating specific biomarkers in diagnosis of hepatocellular carcinoma and its metastasis monitoring. Tumour Biol. 2014;35(1):9–20. doi: 10.1007/s13277-013-1141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roskilly A, Rowe IA. Surveillance for hepatocellular cancer. Clin Med. 2018;18(Suppl 2):s66–s69. doi: 10.7861/clinmedicine.18-2s-s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda K, Ono A, Aikata H, et al. Serum HMGB1 concentrations at 4 weeks is a useful predictor of extreme poor prognosis for advanced hepatocellular carcinoma treated with sorafenib and hepatic arterial infusion chemotherapy. J Gastroenterol. 2018;53(1):107–118. doi: 10.1007/s00535-017-1348-8. [DOI] [PubMed] [Google Scholar]
- 22.Sadeghi M, Lahdou I, Oweira H, et al. Serum levels of chemokines CCL4 and CCL5 in cirrhotic patients indicate the presence of hepatocellular carcinoma. Br J Cancer. 2015;113(5):756–762. doi: 10.1038/bjc.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang HR, Luo XQ, Xu G, et al. High mobility group-box 3 overexpression is associated with poor prognosis of resected gastric adenocarcinoma. World J Gastroenterol. 2012;18(48):7319–7326. doi: 10.3748/wjg.v18.i48.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song N, Liu B, Wu JL, et al. Prognostic value of HMGB3 expression in patients with non-small cell lung cancer. Tumour Biol. 2013;34(5):2599–2603. doi: 10.1007/s13277-013-0807-y. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Zou Z, Gao J, et al. Increased expression of HMGB3: a novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8(1):345–352. [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Cai Y, Zhao H, et al. Overexpression of HMGB3 protein promotes cell proliferation, migration and is associated with poor prognosis in urinary bladder cancer patients. Tumour Biol. 2015;36(6):4785–4792. doi: 10.1007/s13277-015-3130-y. [DOI] [PubMed] [Google Scholar]
- 27.Guo S, Wang Y, Gao Y, et al. Knockdown of high mobility group-box 3 (HMGB3) expression inhibits proliferation, reduces migration, and affects chemosensitivity in gastric cancer cells. Med Sci Monit. 2016;22:3951–3960. doi: 10.12659/MSM.900880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Chang Y, Zhang J, et al. HMGB3 promotes growth and migration in colorectal cancer by regulating WNT/β-catenin pathway. PLoS One. 2017;12(7):e0179741. doi: 10.1371/journal.pone.0179741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Wu Y, Liu A, Tang X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB3. Biochem Biophys Res Commun. 2016;477(4):768–773. doi: 10.1016/j.bbrc.2016.06.133. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68(3):526–549. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]