Abstract
Nectar is one of the key rewards mediating plant–mutualist interactions. In addition to sugars, nectars often contain many other compounds with important biological functions, including proteins. This study was undertaken to assess the proteinaceous content of Brassica rapa nectar. SDS-PAGE analysis of raw B. rapa nectar revealed the presence of ~10 proteins, with a major band at ~10 kDa. This major band was found to contain a non-specific lipid transfer protein encoded by B. rapa locus Bra028980 and subsequently termed BrLTP2.1. Sequence analysis of BrLTP2.1 predicted the presence of a signal peptide required for secretion from the cell, eight cysteines, and a mature molecular mass of 7.3 kDa. Constitutively expressed BrLTP2.1–GFP in Arabidopsis displayed accumulation patterns consistent with secretion from nectary cells. BrLTP2.1 was also found to have relatively high sequence similarity to non-specific lipid-transfer proteins with known functions in plant defense, including Arabidopsis DIR1. Heterologously expressed and purified BrLTP2.1 was extremely heat stable and bound strongly to saturated free fatty acids, but not methyl jasmonate. Recombinant BrLTP2.1 also had direct antimicrobial activity against an extensive range of plant pathogens, being particularly effective against necrotrophic fungi. Taken together, these results suggest that BrLTP2.1 may function to prevent microbial growth in nectars.
Keywords: Arabidopsis thaliana, Brassica rapa, lipid transfer protein, LTP, nectar, nectaries, nectary
Brassica nectar contains a lipid-transfer protein with strong lipid-binding and antimicrobial activities in vitro. This is the first functional evaluation of a lipid-transfer protein in floral nectar.
Introduction
Floral nectar is one of the primary rewards plants offer to pollinators to enhance visitation. While simple sugars are the primary solutes found in nectars, approximately 10% of nectar dry weight is represented by many classes of non-sugar metabolites (Lüttge, 1977). Depending on the species, nectars may contain amino acids, organic acids, terpenes, alkaloids, flavonoids, glycosides, vitamins, phenolics, inorganic ions, free fatty acids, and proteins (Heil, 2011; Roy et al., 2017). These non-sugar compounds have been shown to perform a wide variety of functions, from acting as a deterrent to nectar robbers (Baker, 1978), to promoting pollination by attracting pollinators (Raguso and Pichersky, 1999; Carter et al., 2006).
The production of a nutrient-rich nectar may be a double-edged sword. While nectar does facilitate pollinator visitation, it can also serve as an excellent growth medium for microbes. Indeed, microbial infection of plants via the nectaries is known to occur in cotton, bean, squash, apple, pear, aucuba, banana, pineapple, hawthorn, and gourds (Trapp, 1936; Temkin-Gorodeiski and Chorin, 1971; Rohrbach, 1986; Elada, 1988; Wilson et al., 1990; Jaber and Vidal, 2009; Sasu et al., 2010). In one of the best-known examples, fireblight in apple and pear trees is caused by colonization of nectar by Erwinia amylovora and subsequent invasion of the floral vasculature through the nectary glands (Buban et al., 2003; Farkas et al., 2006). In addition to pathogens, some relatively benign bacteria and yeasts are also known to grow in some nectars, which can impact pollinator behavior (Kevan et al., 1988; Brysch-Herzberg, 2004; Vannette and Fukami, 2016).
It appears that some plants proactively limit microbial growth in nectar, usually via the secretion of antimicrobial proteins and secondary metabolites (Heil, 2011; Roy et al., 2017). For example, two proteins secreted into tobacco nectar, nectarins I and V, have superoxide dismutase and glucose oxidase activities, which both produce hydrogen peroxide of up to 4 mM levels in nectar (Carter and Thornburg, 2000, 2004a, c). This high accumulation of H2O2 was found to be antimicrobial to a wide range of plant pathogens (Carter et al., 2007). Many other examples of nectarins also exist in the literature (Heil, 2011; Roy et al., 2017).
Approximately 75% of all crop species benefit from animal-mediated pollination (Klein et al., 2007) and US pollinator-dependent crops alone have been estimated to have an annual value of nearly $29 billion (Calderone, 2012). For example, Brassica spp. are major worldwide crops, with varieties including canola, broccoli, cauliflower, turnip, and Chinese cabbage (bok choy) (Musgrave, 2000). Each year, over 23 million ha of canola and related varieties (e.g. oilseed rape) are planted globally, with up to 0.8 million ha planted in the USA alone. These species are largely self-incompatible and dependent on honeybees, wild bees, and flies to achieve full fecundity (Rahman, 1940; Downey, 1964; Downey et al., 1970). Poor pollinator visitation has been reported to reduce yields of Brassica and unrelated species by up to 50% (Louveaux and Verge, 1952). Since nectar composition can greatly impact the frequency of pollinator visitation (e.g. Silva and Dean, 2000), full knowledge of the chemical constituents of nectar may have broad implications, ranging from a better understanding of the co-evolution of plant–pollinator and plant–microbe interactions, to increasing yields in multiple crop species. As such, here is described the identification and characterization of a lipid-transfer family protein with antifungal activity secreted into the nectar of B. rapa.
Materials and methods
Plant materials, growth conditions, and nectar collection
Rapid-cycling B. rapa (CrGC 1–33), obtained from Wisconsin Fast Plants at the University of Wisconsin, was used for nectar collection for the identification of nectar proteins. Arabidopsis thaliana ecotype Columbia-0 was used for protein localization studies. Plants were grown in individual pots on a peat-based medium with vermiculite and perlite (Pro-Mix BX; Premier Horticulture, Rivière-du-Loup, Quebec, Canada). All plants were grown under a 16 h day/8 h night cycle, photosynthetic photon flux of 150 µmol m−2 s−1 and temperature of 22 °C. Nectar was collected from B. rapa flowers via microcapillary pipette as previously described (Bender et al., 2012).
Chemicals and reagents
Unless noted otherwise, all chemicals were obtained through Sigma-Aldrich Chemical Co. (St Louis, MO, USA) or Thermo Fisher Scientific (Waltham, MA, USA).
Nectar protein identification
Twenty-five microliters of raw B. rapa nectar was electrophoresed via standard one-dimensional 12% SDS-PAGE and silver stained with a method compatible for identification via liquid chromatography–tandem mass spectrometry (LC-MS/MS), as previously described (Shevchenko et al., 1996). The major protein band at ~10 kDa was excised, rinsed twice in 300 μl ddH2O, dehydrated in 300 μl of 100% acetonitrile for 10 min, and dried in a SpeedVac. The dried gel slice was submitted to the Center for Functional Proteomics at the University of Albany, NY, USA for identification via LC-MS/MS. Briefly, the gel piece was washed, reduced, alkylated, and in-gel tryptic digested. Proteolytic peptides were extracted from the gel. The peptide mixture was concentrated and reconstituted in 5% formic acid for LC-MS/MS analysis. An Ultimate HPLC (Dionex, USA) was used for peptide separation on a Magic C18 column (5 μm, 100 μm ID×150 mm, Michrom Bioresources, Auburn, CA, USA), with a gradient based on solvent A (5% acetonitrile, 0.1% formic acid, 0.01% trifluoroacetic acid) and solvent B (85% acetonitrile, 10% isopropanol, 5% H2O, 0.1% formic acid, 0.01% trifluoroacetic acid). The flow rate was held at 250 nl min−1 with a 100 minute linear gradient ranging from 10% to 100% solvent B. Parent and fragmented peptides were recorded via a QSTAR XL MS/MS (Applied Biosystems, USA).
An MS/MS peak list was created using an Analyst ‘script’, mascot.dll. The peak list files were used to query B. rapa gene sequences using MASCOT 2.51 from Matrix Science (London, UK) with the following parameters: peptide mass tolerance, 0.3 Da; MS/MS ion mass tolerance, 0.3 Da; allow up to 1 missed cleavages. Variable modifications included deamidation (N, Q), oxidation (M), and carbamidomethylation (C). This analysis identified the major B. rapa nectar protein as a lipid-transfer protein encoded by the locus Bra028980. We subsequently termed this protein ‘BrLTP2.1’.
In silico characterization of BrLTP2.1
The translated sequence of BrLTP2.1 was analysed in silico via PSORT (http://psort1.hgc.jp/form.html) with default parameters for the presence of an N-terminal signal peptide. Structural prediction of BrLTP2.1 was conducted via iTasser (Yang et al., 2015; https://zhanglab.ccmb.med.umich.edu/I-TASSER/) using the predicted mature sequence (amino acids 27–98) via default parameters, with predicted models subsequently processed and viewed with DeepView/Swiss-PDBViewer v4.1.0.
Phylogenetic analysis of BrLTP2.1 was conducted via BLASTP (Altschul et al., 1990) to identify homologs of known or predicted function, as well as all paralogs encoded by the B. rapa genome (Supplementary Tables S1, S2 at JXB online). The relationship between these proteins was subsequently analysed in Geneious Pro 5.4.7 using the Geneious Tree Builder with the following tree alignment parameters: cost matrix: Blosum62; gap open penalty: 12; gap extension penalty: 3; alignment type: global alignment with free end gaps. Tree builder option parameters included: genetic distance model: Jukes-Cantor; tree build method: Neighbor-Joining.
BrLTP2.1 localization in Arabidopsis
Full-length BrLTP2.1, including the predicted signal peptide, was PCR amplified (primers in Supplementary Table S3) out of B. rapa genomic DNA and cloned into the XbaI and AscI sites of pMDC85 (Curtis and Grossniklaus, 2003), which placed it downstream of the constitutive 35S CaMV promoter and upstream and in-frame with green fluorescent protein (GFP) (35S::BrLTP2.1-GFP). This construct was transformed into Arabidopsis Col-0 via Agrobacterium-mediated infiltration (Clough and Bent, 1998) and selected on solid 0.5× Murashige and Skoog medium supplemented with hygromycin (50 μg ml−1). Ten independent Arabidopsis transformants with similar GFP accumulation patterns were obtained. Plants confirmed to carry the construct were observed with an Olympus BX53 compound fluorescence microscope mounted with a SPOT Insight 4 MP CCD color digital camera and configured for the detection of GFP fluorescence. Sample preparation simply consisted of sepal removal from flowers to expose the nectaries prior to imaging, or the detachment of rosette leaves with a razor blade.
Protein expression and purification
The predicted mature BrLTP2.1 (amino acids 27–98, minus signal peptide) was PCR amplified from genomic DNA and cloned into the BamHI and XhoI restriction sites of the protein expression vector pET21a(+) in frame with the N-terminal T7-tag and C-terminal His6 tag, to form pCH1, which was verified by Sanger sequencing at the University of Minnesota Genomics Center. Two additional constructs containing mutations in cysteine 69 (C69A and C69Y) were also synthesized and cloned into the BamHI and XhoI sites of pET21a(+) by GenScript Biotech (Piscataway, NJ, USA). Each expression construct was transformed into SHuffle® T7 Express lysY Escherichia coli (C3030; New England Biolabs, USA), which allows for expression of cytosolic proteins with disulfide bonds. Escherichia coli cultures for protein expression were grown at 30 °C until log phase was reached (OD600 of ~0.6) and induced for expression of T7-BrLTP2.1-His6 with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Induced cells grew for another 4 h at 30 °C and were subsequently harvested by centrifugation for protein purification. T7-BrLTP2.1-His6 was purified from E. coli with the HisPur™ Cobalt Purification Kit, 1 ml (Thermo Scientific™), according to the manufacturer’s instructions. Combined elution fractions were further purified by applying them to a 30K MWCO Amicon Ultra-4 Centrifugal Filter (Merk Millipore, Cork, Ireland), with the flow through containing BrLTP2.1 being collected. The flow through from the 30K MWCO filter was applied to a 3K MWCO Amicon Ultra-4 Centrifugal Filter to concentrate BrLTP2.1, which was lastly desalted with ~10 volumes of 25 mM NaPO4, pH 7.4. The concentration of the final purified BrLTP2.1 protein was determined by absorbance at 280 nm using predicted molecular masses and extinction coefficients. Purity was assessed by 4–20% SDS-PAGE and staining with PageBlue™ Protein Staining Solution (Thermo Fisher Scientific).
Lipid-binding assays
The ability and preference for recombinant BrLTP2.1 to bind lipids was assessed by fluorescence spectroscopy and a ‘protein lipid overlay assay’. The fluorescence assay was performed essentially as previously described (Buhot et al., 2004). In initial experiments, BrLTP2.1 concentration was kept constant at 2.5 μM in binding measurement buffer (BMB; 175 mM glucose, 0.5 mM K2SO4, 0.5 mM CaCl2, and 5 mM MES, pH 7.0), with the lipophilic fluorophore 2-p-toluidinonaphthalene-6-sulfonate (TNS) being added at final concentrations ranging from 0 to 10 μM. Blanks consisted of TNS in BMB without protein added. TNS binding to BrLTP2.1 was assessed through excitation at 320 nm and recording the emission at 437 nm. To test lipid binding specificity, free fatty acids and methyl jasmonate were added to a mixture of equimolar 2.5 μM BrLTP2.1 and 2.5 μM TNS in BMB, with TNS displacement from BrLTP2.1 being observed by a reduction in fluorescence at 437 nm.
A blot-based analysis of BrLTP2.1 binding to lipids consisted of a ‘protein lipid overlay assay’ as previously described (Dowler et al., 2002), with minor modifications. In this case, 1 µl aliquots of 500 μM lipids dissolved in a 2:1:0.8 solution of methanol:chloroform:water were spotted onto nitrocellulose and allowed to dry at room temperature for 1 h. The blot was first incubated in blocking buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20 (v/v), 2 mg ml−1 fatty acid-free BSA] for 1 h at room temperature. His-tagged BrLTP2.1 was then added to the membrane in blocking buffer at a final concentration of 10 nM and incubated overnight at 4 °C with gentle rocking. The membrane was then washed 10 times for 30 min in TBS-T [50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20 (v/v)] and then incubated for 1 h at room temperature in a 1:2000 dilution of anti-His rabbit polyclonal antibody (GenScript Biotech) in blocking buffer. The membrane was then again washed 10 times for 30 min in TBS-T. A 1:5000 dilution of alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Thermo Fisher Scientific) was incubated in blocking buffer with the membrane for 1 h and then washed 10 times for 30 min in TBS-T. After washing, BrLTP2.1 binding to lipids was detected by developing the membrane in substrate buffer (100 mM Tris-HCl, pH 9.5; 100 mM NaCl; 5 mM MgCl2) containing 150 μg ml−1 nitroblue tetrazolium chloride and 300 μg ml−1 5-bromo-4-chloro-3-indoxyl phosphate, p-toluidine salt.
In vitro antimicrobial assays
A microplate assay was used to monitor fungal growth inhibition adapted from the previously described assay (Broekaert et al., 1990). All fungal pathogen strains were retrieved from the University of Minnesota Mycological Culture Collection and were originally collected in Minnesota from commercial production fields. Fungal samples from Fusarium oxysporum, F. graminearum, Bipolaris oryzae, Trichoderma viride, Alternaria solani, and Colletotrichum trifolii were grown on potato dextrose agar plates for 1 week. Harvesting the spores was done by flooding the plates with sterile water and rubbing with a sterile rubber policeman. The spore suspensions were filtered, and spore densities were determined microscopically using a hemocytometer. Clear, flat bottom microplates were used with each well containing half-strength potato dextrose broth, approximately 2000 spores, and concentrations of BrLTP2.1 peptide up to 300 μg ml−1 in a total volume of 100 μl. The microplates were shaken on an orbital shaker and spores were allowed to sediment for 30 min before absorbance was measured. The absorbance of the wells was measured at 595 nm on a Synergy H1 microplate reader (BioTek, Winooski, VT, USA). Further absorbance measurements were carried out after 24-h and 48-h incubation periods. Absorbance values were calculated by subtracting the initial measurement from the final measurement. From these values, the amount of BrLTP2.1 needed to inhibit growth of the pathogens strains by 50% (IC50) was calculated.
A spread-plate method was used to quantify antibacterial activity of the BrLTP2.1. Bacterial lawns of Pseudomonas syringae pv. tomato were grown on LB plates for 2 days. The plates were flooded with sterile water, and a bacterial cell suspension was made by rubbing the plate with a sterile rubber policeman. Cultures were diluted with sterile water to an OD600 value of 0.1. In microcentrifuge tubes, 200 μl of bacteria was incubated with shaking for 3 h with concentrations of BrLTP2.1 up to 300 μg ml−1. After the peptide treatment, the bacteria were serially diluted, and 100 μl was plated in triplicate onto nutrient broth yeast extract (NBY) plates. After 48 h of incubation, the bacterial colonies were counted. From these values, the amount of BrLTP2.1 needed to inhibit growth of bacterial strains by 50% (IC50) was calculated.
BrLTP2.1 heat stability assays and immunodetection
To test the heat stability of BrLTP2.1, 1 ml of E. coli cell culture was harvested 4 h after induction of BrLTP2.1 expression with IPTG and directly boiled for 5, 10, 30, and 60 min. Boiled samples were incubated on ice for 15 min and centrifuged at 17000 g for 10 min to precipitate denatured protein and cell debris. Remaining soluble proteins were evaluated by 4–20% SDS-PAGE as described above. In some cases, the boiled supernatant was further purified via affinity chromatography as described above. Western blot analysis of boiled and purified BrLTP2.1 was assessed with polyclonal rabbit anti-His (GenScript Biotech A00174) and alkaline-phosphatase-conjugated goat anti-rabbit (Thermo Fisher Scientific 31346) antibodies as previously described above in the protein lipid overlay assay.
Results
The major B. rapa nectar protein is a lipid-transfer family protein
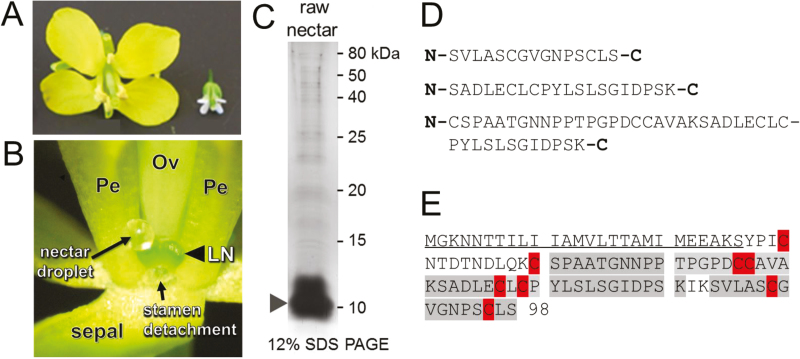
Brassica rapa and Arabidopsis are close relatives that have similar floral and nectary structures (Fig. 1A; Davis et al., 1998). As with Arabidopsis, the bulk of nectar in B. rapa is secreted by nectaries located at the base of short stamens (Fig. 1B; Davis et al., 1998). Approximately 10 protein bands were observed in raw B. rapa nectar via SDS-PAGE, with a major band accumulating at ~10 kDa (Fig. 1C, arrowhead). LC-MS/MS analysis of the trypsinized gel slice containing this major band identified three peptides (Fig. 1D) that mapped to a predicted non-specific lipid-transfer protein (nsLTP) encoded by the B. rapa locus Bra028980.
Fig. 1.
A lipid transfer protein (LTP) is the major protein in Brassica rapa nectar. (A) Whole B. rapa flower (left) beside one from its close relative, Arabidopsis. (B) Example of a nectar droplet collected from B. rapa flowers for protein identification. LN, lateral nectary; Ov, ovary; Pe, petal. A short stamen was removed from the flower to visualize the nectar droplet. (C) Protein profile of raw B. rapa nectar after separation by 12% SDS-PAGE and silver staining. The major protein band (arrowhead) was excised from the gel and processed for protein identification. (D) Peptides identified from the major protein band [arrowhead in (C)] by LC-MS/MS. (E) BLAST searches identified the major protein band as Bra028980, a putative lipid-transfer protein. Peptides identified by MS/MS are shaded in gray, cysteines are highlighted, and a putative signal peptide required for secretion from the cell is underlined.
Bra028980 encodes a full-length protein of 98 amino acids, which places it in the Type 2 class of nsLTPs. Following a nomenclature system recently outlined (Salminen et al., 2016), we subsequently named the gene encoded by the Bra028980 locus as ‘BrLTP2.1’. The three peptides from the MS/MS analysis covered 57 of 98 amino acids (58%) of the predicted full-length coding region of BrLTP2.1 (shaded in gray in Fig. 1E). However, the online localization prediction tool PSORT predicted that BrLTP2.1 is secreted from cells via the presence of a 26 amino acid long N-terminal signal peptide. Thus, the peptides identified by LC-MS/MS covered 80% (57 out of 71 amino acids) of the mature, secreted protein (Fig. 1E). Like other nsLTPs, mature BrLTP2.1 contains eight cysteine residues (highlighted in red in Fig. 1E), which likely form disulfide bonds. Other physiochemical properties of BrLTP2.1 include a predicted mature molecular mass of 7.3 kDa and an acidic isoelectric point (pI) of 4.46.
Heterologously expressed BrLTP2.1 is secreted from Arabidopsis cells
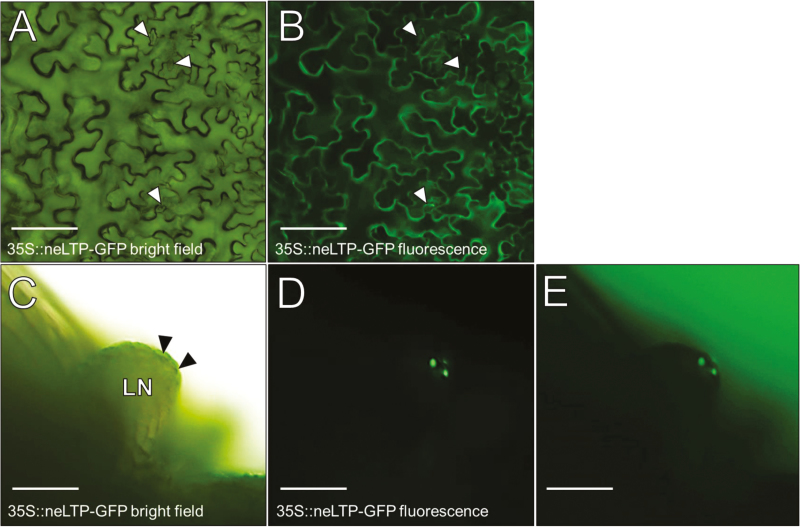
To confirm that BrLTP2.1 is indeed secreted from cells, a 35S::BrLTP2.1-GFP construct was transformed into Arabidopsis. BrLTP2.1–GFP clearly outlined the pavement cells of rosette leaves (Fig. 2A, B), which is consistent with extracellular accumulation. Perhaps more strikingly, BrLTP2.1–GFP preferentially accumulated in the stomatal pores of lateral nectaries (Fig. 2C–E, arrowheads), which are the presumed locations of nectar secretion (Davis et al., 1986, 1998). Note that the entire nectary fluoresces in 35S::BrLTP2.1-GFP lines and the images shown represent a relatively short exposure time when compared with the rosette leaf images (Fig. 2A, B), which is indicative of very high accumulation. Moreover, a similar pattern of GFP accumulation in stomatal pores was not observed in rosette leaves (arrowheads in Fig. 2A, B). The small size of the Arabidopsis nectary and associated nectar volume make it technically difficult to unambiguously determine if BrLTP2.1–GFP reaches the nectar itself. Therefore, while not conclusive, these results cumulatively suggest that heterologously expressed BrLTP2.1 is secreted into Arabidopsis nectar.
Fig. 2.
Constitutively expressed BrLTP2.1–GFP is secreted from Arabidopsis cells. (A, B) Full-length BrLTP2.1–GFP driven under control of the 35S-CaMV promoter leads to secretion from rosette leaf pavement cells. Arrowheads in (A) and (B) point to stomata. (C–E) BrLTP2.1–GFP preferentially accumulates in the stoma formed by guard cells in a lateral nectary (LN). Note that a similar accumulation of BrLTP2.1–GFP is not observed in the stomatal pores of rosette leaves (arrowheads).
Phylogenetic analysis of BrLTP2.1
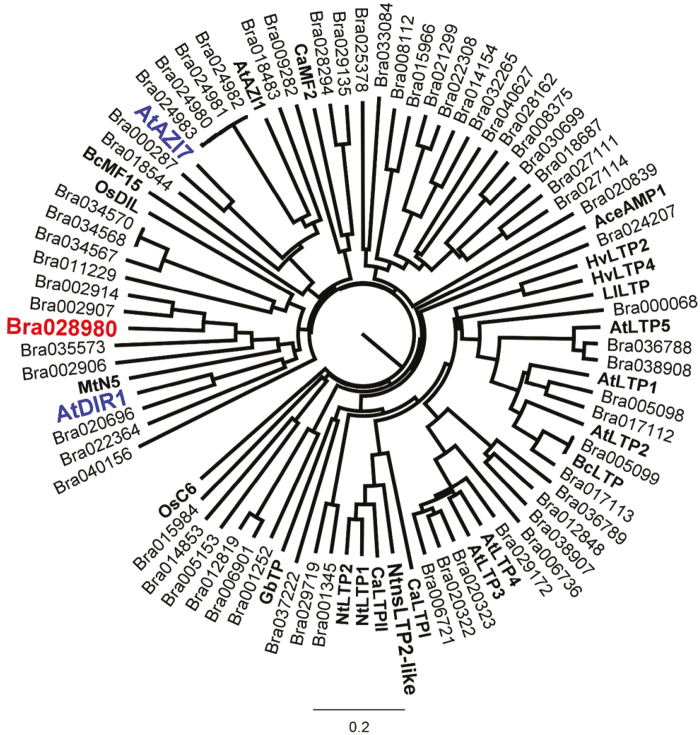
In order to identify a potential biological function for BrLTP2.1, BLASTP and literature searches were used to identify close homologs, as well as nsLTPs with known or implicated functions (Supplementary Table S1). Included in this analysis were all 63 nsLTPs encoded by the B. rapa genome (Supplementary Table S2, contains sequences of all nsLTPs used for analysis). Of the nsLTPs with known functions, BrLTP2.1 was most closely related to Arabidopsis DIR1 (Fig. 3; 35% identity, 50% similarity), a protein involved in mediating systemic acquired resistance to pathogens (Maldonado et al., 2002; Champigny et al., 2013). In a previous report, both DIR1 and BrLTP2.1 fell into the same clade and were classified as ‘Type IV’ nsLTPs via a classification system based on sequence rather than functional similarity. Interestingly, BrLTP2.1 was also closely related to Arabidopsis AtAZI7, which we previously showed to have strong nectary-enriched expression profiles by microarray and RT-PCR analysis (Kram et al., 2009). The biological function of AtAZI7 is currently unknown; however, its close paralog AtAZI1 was implicated in the long-distance priming of defense responses mediated by azelaic acid in Arabidopsis (Cecchini et al., 2015). Azelaic acid, a saturated 7-carbon dicarboxylic acid, is produced in response to local infections and moves throughout the plant to promote systemic acquired resistance (SAR). Interestingly, azelaic acid production is dependent on the SAR regulator ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) (Wittek et al., 2014).
Fig. 3.
Phylogenetic analysis of Bra028980 (BrLTP2.1). The protein sequence of Bra02890 (BrLTP2.1) was subjected to CLUSTAL Omega multiple sequence alignment and tree analysis with all members of LTP family encoded by B. rapa genome, as well as select LTPs from other species with known or implicated biological function (in bold, see Supplementary Table S1 for detailed list). One of the nearest Arabidopsis LTPs with implicated function, AtAZI7, was also previously found to have enriched expression in nectaries by microarray analyses, suggesting conservation of BrLTP2.1 function, at least within the Brassicaceae.
In silico structural analysis of BrLTP2.1
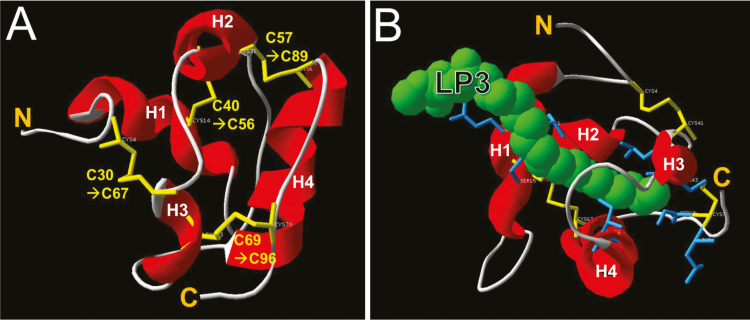
The mature amino acid sequence of BrLTP2.1, without the signal peptide, was subjected to structural modeling at i-TASSER by using the crystal structure of DIR1 (2rknA; Lascombe et al., 2008) as the threading template. Not surprisingly, and like most nsLTPs, BrLTP2.1 was predicted to have four α-helices and four disulfide bonds formed by eight cysteine residues (Fig. 4A). Moreover, a hydrophobic binding pocket complexed with a lipid ligand (1-stearoyl-sn-glycero-3-phosphocholine) was predicted based on the ligand-bound DIR1 crystal structure (Fig. 4B).
Fig. 4.
Structural prediction of BrLTP2.1. (A) AtDIR1, a close homolog to BrLTP2.1 involved in plant defense responses, was used as a template to model BrLTP2.1 structure. This analysis predicted the presence of four α-helices (labeled H1–H4 from N- to C-terminus) and four disulfide bonds. (B) Model of BrLTP2.1 with the lipid LP3 (1-stearoyl-sn-glycero-3-phosphocholine) bound. The specific sidechains predicted to coordinate lipid binding are Leu37, Gln38, Cys40, Ser15, Leu65, Leu68, Cys69, Ile82, Ser95, and Leu97. The models shown in both (A) and (B) were predicted by iTASSER using Arabidopsis DIR1 (2rknA) as a threading template. The sequence of the predicted mature BrLTP2.1 without signal peptide was used as the input, but the amino acid numbering includes the predicted 26 amino acid signal peptide.
BrLTP2.1 has lipid-binding activity
The structural analysis of BrLTP2.1, along with the fact that nsLTPs are known to bind a number of different lipids, led us to examine the binding activity of BrLTP2.1 in vitro. His-tagged BrLTP2.1 was heterologously expressed in E. coli and purified with a combination of affinity chromatography and size filtration (Fig. 5A). Importantly, we used a strain of E. coli, NEB SHuffle® T7 Express lysY, which allows for the cytosolic formation of disulfide bonds, which are likely required for structural integrity. Therefore, we also purified two mutant versions of BrLTP2.1, both containing mutations in cysteine 69 (BrLTP2.1C69A and BrLTP2.1C69Y) as negative controls.
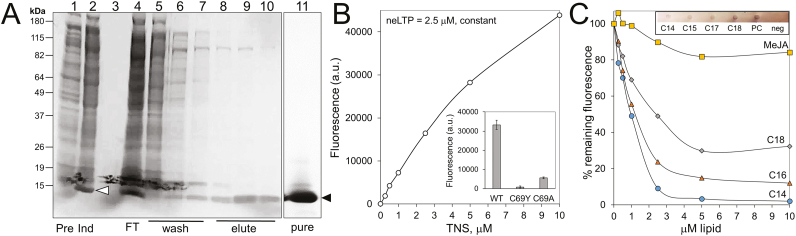
Fig. 5.
Heterologously expressed BrLTP2.1 has lipid binding activity. (A) Heterologously expressed and purified BrLTP2.1 from E. coli. Lane 1: pre-induction E. coli lysate; lane 2: 4 h post-induction lysate; lane 3: cell media; lane 4: flow-through from Co2+ affinity column; lanes 5–7: column washes; lanes 8–10: elutions with 300 mM imidazole; lane 11: pure protein after concentration and desalting. (B) TNS, a lipophilic fluorophore binds to BrLTP2.1. TNS concentration ranged from 0 to 10 μM, while BrLTP2.1 concentration was held constant at 2.5 μM, with excitation at 320 nm and emission recorded at 437 nm. Inset: TNS-dependent fluorescence in wild-type and two mutant versions of BrLTP2.1 (protein and TNS both at 2.5 μM). (C) Lipids present in Brassica nectars competitively displaced TNS from BrLTP2.1. TNS and BrLTP2.1 concentration were each held constant at 2.5 μM, while myristic acid (C14), palmitic acid (C16), stearic acid (C18), and methyl jasmonate (MeJA) ranged from 0 to 10 μM. Inset: Dot blot analysis of BrLTP2.1 binding to myristic (C14), pentadecanoic (C15), heptadecanoic (C17), and stearic (C18) acid, as well as phosphatidylcholine (PC). A 2:1:0.8 solution of methanol:chloroform:water, the solvent for all lipids, was used as a negative control (neg). BrLTP2.1 binding was detected with anti-His antibodies.
We used the lipophilic fluorescent dye TNS to assess lipid binding to BrLTP2.1. TNS is weakly fluorescent in aqueous solution, but fluoresces brightly when bound to hydrophobic regions of proteins (Buhot et al., 2004). In an initial test, BrLTP2.1 was held constant at 2.5 μM, with TNS added from 0 to 10 μM. Blanks containing TNS at the indicated concentrations in binding buffer (without BrLTP2.1) were subtracted from the observed fluorescence intensities from the test samples containing 2.5 μM BrLTP2.1. Total fluorescence was positively correlated to TNS concentration (Fig. 5B). Conversely, the two mutant versions, BrLTP2.1C69A and BrLTP2.1C69Y, displayed minimal binding to TNS relative to the wild-type version of the protein (inset of Fig. 5B).
We previously identified free fatty acids in Brassica nectars accumulating at near millimolar levels, all of which were saturated (Bender et al., 2013). To test if these lipids may bind BrLTP2.1, saturated free fatty acids with chain lengths of 14, 16, and 18 carbons (myristic, palmitic, and stearic acids) were added to solutions containing equimolar BrLTP2.1 and TNS (2.5 μM each). If BrLTP2.1 had a stronger binding affinity for a given lipid over TNS, then the observed fluorescence would decrease due to displacement of TNS via competitive binding. Each of the three free fatty acids tested strongly reduced TNS-dependent fluorescence (Fig. 5C), indicating that they displaced TNS from the BrLTP2.1 binding pocket. Moreover, BrLTP2.1 displayed a preference for myristic acid (C14) over the longer chain free fatty acids (based on the much larger decrease in fluorescence at lower concentrations). It is also important to note that maximal displacement of TNS from BrLTP2.1 occurred at 2.5 μM myristic acid, suggesting a 1:1 stoichiometry between myristic acid and BrLTP2.1 (which was held constant at 2.5 μM). Some LTPs also bind jasmonates (Bakan et al., 2006), which are important in regulating nectary function (Radhika et al., 2010; Stitz et al., 2014; Wang et al., 2014). Therefore, we tested the ability of methyl jasmonate (MeJA) to displace TNS from BrLTP2.1. MeJA did slightly reduce TNS-dependent fluorescence, though not nearly as much as the saturated free fatty acids tested. Lastly, a blot-based protein lipid overlay assay confirmed the binding of BrLTP2.1 to saturated free fatty acids, as well as phosphatidylcholine (inset of Fig. 5C).
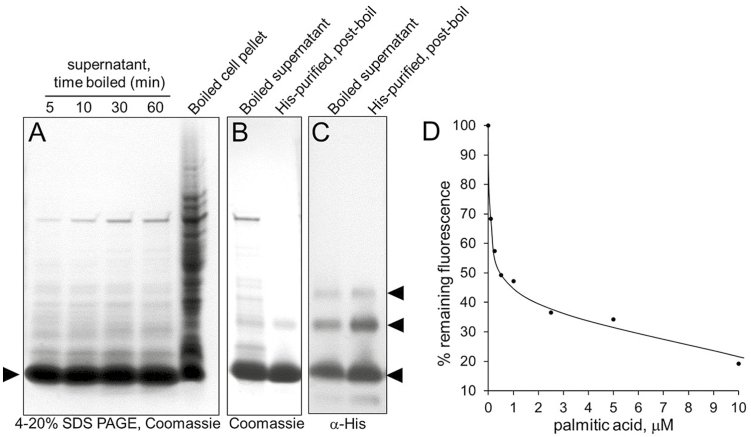
Due to its small size and four disulfide bonds, BrLTP2.1 can be predicted to have significant heat stability, as has been shown for a number of other LTPs (Salminen et al., 2016). Indeed, induced cultures of E. coli could be boiled for at least 60 min with no apparent loss or denaturation of BrLTP2.1 (Fig. 6A). The boiled protein was further purified via affinity chromatography (Fig. 6B) and analysed via Western blot (Fig. 6C). Interestingly, this analysis identified several isoforms of BrLTP2.1 (arrowheads in Fig. 6C), in spite of being boiled in Laemli loading buffer, which contains SDS and β-mercaptoethanol. This boiled and purified protein was also verified to still be able to bind free fatty acids, as determined by TNS displacement (Fig. 6D).
Fig. 6.
BrLTP2.1 is extremely heat stable. (A) One milliliter of E. coli cell culture was harvested 4 h after induction of BrLTP2.1 expression with IPTG and directly boiled for the indicated times. Boiled samples were incubated on ice for 15 min and centrifuged at 17000 g for 10 min to precipitate denatured protein and cell debris. Remaining proteins were evaluated by SDS-PAGE. (B) SDS-PAGE analysis of the clarified supernatant of cell cultures boiled for 15 min (left lane) and further purified BrLTP2.1 via Co2+ affinity chromatography (right lane). (C) Western blot analysis of boiled and purified BrLTP2.1 from (B) as detected by anti-His-tag antibodies. Arrowheads indicate the multiple bands corresponding to BrLTP2.1 post-boil and affinity purification. (D) Boiled BrLTP2.1 retains lipid-binding activity after purification [from (B)], as determined by displacement of TNS with palmitic acid.
BrLTP2.1 has broad antimicrobial activity in vitro
A number of nsLTPs display antimicrobial activity in vitro (Salminen et al., 2016). To test a potential role for BrLTP2.1 in limiting microbial growth, the recombinant protein was tested against a battery of fungal and bacterial plant pathogens. BrLTP2.1 displayed strong antimicrobial activity, particularly against necrotrophic fungi (Fig. 7; Table 1). The IC50 of BrLTP2.1 (the concentration at which microbe growth was reduced by half) was in the high nanomolar to low micromolar range against most fungal pathogens (Table 1). To verify that the observed antifungal activity was due to the purified BrLTP2.1, the mutant protein BrLTP2.1C69Y was tested against two of these plant pathogens. Indeed, BrLTP2.1C69Y displayed no activity against Fusarium oxysporum and ~30-fold lower activity against Trichoderma viride than the wild-type protein (Table 1). BrLTP2.1 also displayed activity against the single bacterial pathogen tested, Pseudomonas syringae pv. tomato, though the IC50 at ~35 μM was somewhat higher than for most fungal pathogens.
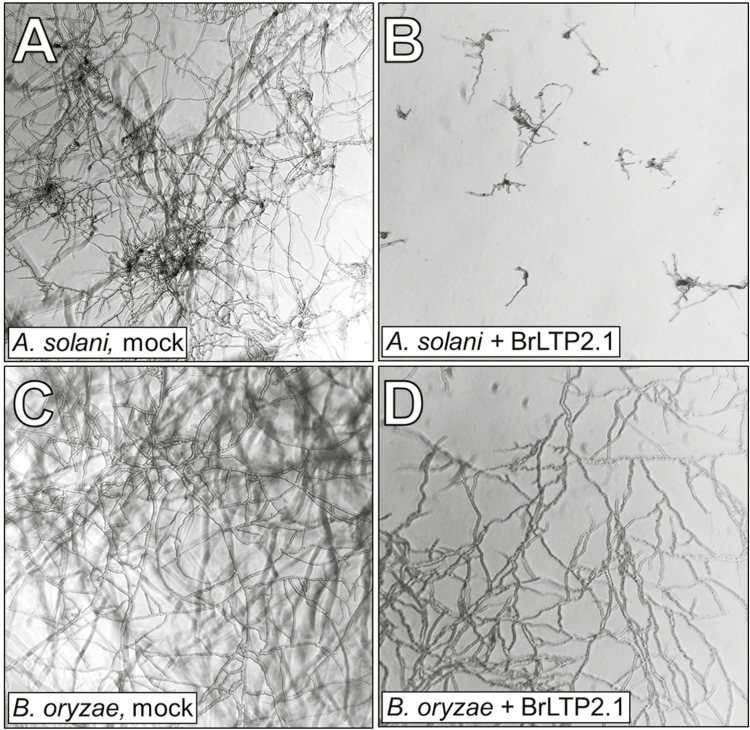
Fig. 7.
Heterologously expressed BrLTP2.1 has direct antimicrobial activity. Spores harvested from a battery of fungal plant pathogens were incubated with BrLTP2.1 from 0 to 300 μg ml−1 and monitored for growth over 48 h. In the examples shown, Alternaria solani (A, B) and Bipolaris oryzae (C, D) were either mock treated (A, C) or incubated with 50 μg ml−1 BrLTP2.1 (~5 μM; B, D). Summarized data are provided in Table 1.
Table 1.
BrLTP2.1 has direct antimicrobial activity against plant pathogensa
| Microbe | Wild-type IC50 | C69Y IC50 | ||
|---|---|---|---|---|
| µg ml−1 | µM | µg ml−1 | µM | |
| Alternaria solani | 36.0 | 3.73 | ND | ND |
| Colletotrichum trifolii | 29.0 | 2.98 | ND | ND |
| Fusarium oxysporum | 140.2 | 14.4 | No activityb | No activityb |
| Fusarium graminearum | 248.3 | 25.5 | ND | ND |
| Trichoderma viride | 7.7 | 0.79 | 225.4 | 23.4 |
| Bipolaris oryzae | 16.7 | 1.72 | ND | ND |
| Pseudomonas syringae pv. tomato | 338.5 | 34.8 | ND | ND |
a Fungal spores or diluted bacterial cultures were incubated with BrLTP2.1 (0–300 μg ml−1) and assessed for growth after 48 h.
b Activity was tested at levels up to 300 μg ml−1.
ND, not determined.
Discussion
Nectars often contain only a few major proteins (Roy et al., 2017), therefore the finding of a single major protein, BrLTP2.1, in Brassica nectar (Fig. 1C) is not surprising. In a prior transcriptomic study we previously reported that BrLTP2.1 displays enriched expression in nectaries (Hampton et al., 2010), which is consistent with the expression patterns of most nectarins (Carter et al., 1999; Carter and Thornburg, 2003; Carter and Thornburg, 2004b,c; Seo et al., 2013). Several nsLTPs from other species are also highly expressed in nectaries, including AZI7 in Arabidopsis (Kram et al., 2009) and NaLTP1 and NaLTP2 in Nicotiana attenuata (Seo et al., 2013). Based on sequence similarity alone, it does not appear that BrLTP2.1 is a direct ortholog of these other nectary-enriched nsLTPs, as other Arabidopsis nsLTPs share much closer sequence similarity to BrLTP2.1 at the amino acid level than does AZI7. Similarly, NaLTP1 and NaLTP2 do accumulate in N. attenuata nectar, but neither has been functionally characterized and they share only ~16% identity with BrLTP2.1. Therefore, it is difficult to extrapolate functional or biochemical conservation of nsLTPs with nectary-enriched expression.
Heterologously expressed and purified BrLTP2.1 has strong antimicrobial activity, particularly against fungal plant pathogens, with IC50 values in the high nanomolar to low micromolar range. This antifungal activity is on a par with other cysteine-rich peptides involved in defense, such as plant defensins (Lacerda et al., 2014). These IC50 values, coupled with the apparently high concentration of BrLTP2.1 in nectar, strongly suggests that BrLTP2.1 is actively secreted into nectar as a broad-spectrum agent to limit fungal growth. Moreover, while the IC50 value for BrLTP2.1 against the bacterium P. syringae is on the high side, it is very rare for an nsLTP to have both antifungal and antibacterial activities – an onion nsLTP appears to be the only other known example (Cammue et al., 1995). Finally, it is possible that an unidentified antimicrobial molecule from E. coli co-purified with BrLTP2.1, though several pieces of evidence strongly suggest otherwise: (i) the purified protein was extensively dialysed during the purification process; (ii) the fluorophore TNS readily binds at roughly equimolar concentrations to the same purified protein preparations used for the antimicrobial assays, suggesting the presence of an empty binding pocket; (iii) TNS itself is easily competitively displaced from BrLTP2.1 by free fatty acids at equimolar concentrations (Fig. 5), suggesting that TNS weakly binds BrLTP2.1 and that it is unlikely to displace other potential co-purifying molecules, if present; and (iv) related nsLTPs isolated from both natural and recombinant sources display similar antimicrobial activities (reviewed in Salminen et al., 2016). These facts cumulatively indicate that it is highly unlikely that a small antimicrobial molecule from E. coli co-purifies with BrLTP2.1 and strongly suggest that the protein itself has direct activity. In a distinct but related point, the purified BrLTP2.1C69Y mutant protein displayed greatly diminished antifungal activity relative to the wild-type protein (Table 1).
Since BrLTP2.1 is expressed in flowers prior to any apparent challenge by pathogens, this could indicate that this broad-spectrum antimicrobial activity contributes to non-host resistance. However, BrLTP2.1 also falls into a clade of nsLTPs that contains Arabidopsis DIR1 (Fig. 3; Li et al., 2014). dir1 plants exhibit no change in local responses to bacterial pathogens, but are defective in the development of systemic acquired resistance (SAR) to virulent strains (Maldonado et al., 2002; Champigny et al., 2013). AtAZI1 is another nsLTP that has been implicated in the azelaic acid-dependent development of SAR (Cecchini et al., 2015). As mentioned above, AtAZI7, a close paralog to AZI1, is highly expressed in nectaries (Kram et al., 2009). The relatively close relationship between BrLTP2.1, AtDIR1, and AtAZI7/AtAZI1 suggests an alternative role for BrLTP2.1 in defense signaling in B. rapa. Direct antimicrobial activity has not yet been reported for AtDIR1 or the AtAZI1 family of nsLTPs. Future studies will test if overexpression of BrLTP2.1 in plants leads to enhanced disease resistance.
The molecular mechanism through which nsLTPs inhibit microbial growth are currently unknown, but it is clear that nsLTPs can increase cell permeability, possibly through disruption of membrane structure (Sun et al., 2008). A clue to the potential mechanism through which nsLTPs act may come from plant defensins. Defensins are small, cysteine-rich, antimicrobial proteins, like nsLTPs, that form large, multimeric pores in target membranes (De Coninck et al., 2013). Intriguingly, nsLTPs are also known to form multimers in vitro (Pokoj et al., 2010), and thus their antimicrobial activity may depend on a similar mechanism to that of some defensins.
An exhaustive study of potential ligands was not performed in our study, but BrLTP2.1 displayed an apparent preference for binding to shorter chain free fatty acids over longer ones, as well as over methyl jasmonate (Fig. 5C). These findings, along with the fact that the mutant BrLTP2.1C69A and BrLTP2.1C69Y did not bind TNS (Fig. 5C), suggests some degree of lipid ligand specificity by BrLTP2.1. Future studies will need to more thoroughly address the biochemical nature of BrLTP2.1 and its interactions with microbial membranes. While the physiological importance of the extreme heat stability of BrLTP2.1 (Fig. 6) is unknown, this characteristic could be very useful for purifying large amounts of the protein. It should also be noted that the extreme heat stability of BrLTP2.1 is not unique among nsLTPs (Salminen et al., 2016), though a biological function for such stability has yet to be demonstrated.
Nectar is an inherently excellent growth medium for microbes, and therefore it is unsurprising that plants would secrete antimicrobial proteins, like BrLTP2.1, into nectar as a defense mechanism. For example, microbial infection of plants via the nectaries has been well documented in many species (Trapp, 1936; Temkin-Gorodeiski and Chorin, 1971; Rohrbach, 1986; Elada, 1988; Wilson et al., 1990; Jaber and Vidal, 2009; Sasu et al., 2010). Furthermore, yeast and bacterial communities inhabit nectars of a wide variety of plant species (Kevan et al., 1988; Brysch-Herzberg, 2004; Vannette and Fukami, 2016). There is also growing evidence that these microbes can shape nectar composition in such a way that could deter pollinators through the consumption and metabolism of nectar compounds (Herrera et al., 2008; Canto and Herrera, 2012; de Vega and Herrera, 2012; Vannette et al., 2013; Good et al., 2014). As such, nectar proteins are known to serve in a defensive capacity against nectar inhabiting microbes in both extrafloral (González-Teuber et al., 2009, 2010; Heil, 2011) and floral nectars (Carter and Thornburg, 2004a; Roy et al., 2017). In the constitutively secreted extrafloral nectar of two Acacia species, three pathogenesis-related enzymes were identified (chitinase, β-1,3-glucanase, and peroxidase), whose activities reduced fungal growth (González-Teuber et al., 2010). Since BrLTP2.1 is the major protein secreted in B. rapa nectar (Fig. 1C) and has strong in vitro antifungal activity (Table 1; Fig. 7A–D), we suggest that the major role of this protein is to limit the growth of potentially deleterious microbial communities. This protective service may be necessary to maintain the integrity and quality of the nectar in order to effectively manipulate pollinators to achieve successful pollination. Future studies should aim to determine whether the presence or absence of this protein can indeed alter microbial growth in nectar and if this alteration in growth impacts nectar composition and pollinator preference.
Lastly, free fatty acids have been reported to accumulate to near millimolar levels in a few nectars (Kram et al., 2008; Bender et al., 2012), but how they are secreted is still unknown. Intriguingly, BrLTP2.1 appears to have strong affinity for the same saturated fatty acids (palmitic and myristic acids; Fig. 5C) that accumulate to high levels in B. rapa nectar (Bender et al., 2012). Thus, a somewhat more speculative function of BrLTP2.1 is it may be involved in the transport of free fatty acids into nectars, which would not be dissimilar to the role that NtLTP1 plays in transporting lipids into glandular trichome secretions in tobacco (Choi et al., 2012).
In summary, we have identified and partially characterized a B. rapa nectar protein, BrLTP2.1, with strong lipid-binding and antifungal activities in vitro. Future studies addressing its true biological role will depend on the evaluation of null mutant and overexpression plants. Such an analysis would determine if BrLTP2.1 limits microbial growth in vivo or if it is involved in the movement of lipids into nectar. Biochemical and biophysical approaches to probe the mechanism through which BrLTP2.1 binds lipids and prevents pathogen growth will also be beneficial to a general understanding nsLTP structure and function.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of nsLTPs with known or putative biological functions used for phylogenetic analysis
Table S2. Sequences of nsLTPs used for tree building
Table S3. Oligonucleotides used in this study
Acknowledgements
The authors thank Dr Min Ni, University of Minnesota, for use of his fluorescence microscope. This work was supported by National Science Foundation grants 0820730 and 1339246 to CJC. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Glossary
Abbreviations:
- BrLTP2.1
Brassica rapa lipid-transfer protein 2.1
- nsLTP
non-specific lipid-transfer protein
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bakan B, Hamberg M, Perrocheau L, Maume D, Rogniaux H, Tranquet O, Rondeau C, Blein JP, Ponchet M, Marion D. 2006. Specific adduction of plant lipid transfer protein by an allene oxide generated by 9-lipoxygenase and allene oxide synthase. The Journal of Biological Chemistry 281, 38981–38988. [DOI] [PubMed] [Google Scholar]
- Baker HG. 1978. Chemical aspects of the pollination of woody plants in the tropics. In: Tomlinson PB, Zimmerman M, eds. Tropical trees as living systems. New York: Cambridge University Press, 57–82. [Google Scholar]
- Bender RL, Fekete ML, Klinkenberg PM, et al. 2013. PIN6 is required for nectary auxin response and short stamen development. The Plant Journal 74, 893–904. [DOI] [PubMed] [Google Scholar]
- Bender R, Klinkenberg P, Jiang Z, Bauer B, Karypis G, Nguyen N, Perera MADN, Nikolau BJ, Carter CJ. 2012. Functional genomics of nectar production in the Brassicaceae. Flora 207, 491–496. [Google Scholar]
- Broekaert W, Terras F, Cammue B, Vanderleyden J. 1990. An automated quantitative assay for fungal growth inhibition. FEMS Microbiology Letters 1–2, 55–59. [Google Scholar]
- Brysch-Herzberg M. 2004. Ecology of yeasts in plant-bumblebee mutualism in Central Europe. FEMS Microbiology Ecology 50, 87–100. [DOI] [PubMed] [Google Scholar]
- Buban T, Orosz-Kovacs Z, Farkas A. 2003. The nectary as the primary site of infection by Erwinia amylovora (Burr.) Winslow et al.: a mini review. Plant Systematics and Evolution 238, 183–194. [Google Scholar]
- Buhot N, Gomès E, Milat ML, Ponchet M, Marion D, Lequeu J, Delrot S, Coutos-Thévenot P, Blein JP. 2004. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Molecular Biology of the Cell 15, 5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone NW. 2012. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992-2009. PLoS One 7, e37235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue BP, Thevissen K, Hendriks M, Eggermont K, Goderis IJ, Proost P, Van Damme J, Osborn RW, Guerbette F, Kader JC. 1995. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiology 109, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto A, Herrera CM. 2012. Micro-organisms behind the pollination scenes: microbial imprint on floral nectar sugar variation in a tropical plant community. Annals of Botany 110, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW. 1999. Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Molecular Biology 41, 207–216. [DOI] [PubMed] [Google Scholar]
- Carter C, Healy R, O’Tool NM, Naqvi SM, Ren G, Park S, Beattie GA, Horner HT, Thornburg RW. 2007. Tobacco nectaries express a novel NADPH oxidase implicated in the defense of floral reproductive tissues against microorganisms. Plant Physiology 143, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Shafir S, Yehonatan L, Palmer RG, Thornburg R. 2006. A novel role for proline in plant floral nectars. Die Naturwissenschaften 93, 72–79. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. 2000. Tobacco nectarin I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. The Journal of Biological Chemistry 275, 36726–36733. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. 2003. The nectary-specific pattern of expression of the tobacco Nectarin I promoter is regulated by multiple promoter elements. Plant Molecular Biology 51, 451–457. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. 2004a Is the nectar redox cycle a floral defense against microbial attack?Trends in Plant Science 9, 320–324. [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. 2004b Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Molecular Biology 54, 415–425. [DOI] [PubMed] [Google Scholar]
- Carter CJ, Thornburg RW. 2004c Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiology 134, 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini NM, Steffes K, Schläppi MR, Gifford AN, Greenberg JT. 2015. Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nature Communications 6, 7658. [DOI] [PubMed] [Google Scholar]
- Champigny MJ, Isaacs M, Carella P, Faubert J, Fobert PR, Cameron RK. 2013. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Frontiers in Plant Science 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YE, Lim S, Kim HJ, Han JY, Lee MH, Yang Y, Kim JA, Kim YS. 2012. Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. The Plant Journal 70, 480–491. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Peterson RL, Shuel RW. 1986. Anatomy and vasculature of the floral nectaries of Brassica napus (Brassicaceae). Canadian Journal of Botany 64, 2508–2516. [Google Scholar]
- Davis AR, Pylatuik JD, Paradis JC, Low NH. 1998. Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae. Planta 205, 305–318. [DOI] [PubMed] [Google Scholar]
- De Coninck B, Cammue BPA, Thevissen K. 2013. Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biology Reviews 26, 109–120. [Google Scholar]
- de Vega C, Herrera CM. 2012. Relationships among nectar-dwelling yeasts, flowers and ants: patterns and incidence on nectar traits. Oikos 121, 1878–1888. [Google Scholar]
- Dowler S, Kular G, Alessi DR. 2002. Protein lipid overlay assay. Science’s STKE 2002, pl6. [DOI] [PubMed] [Google Scholar]
- Downey RK. 1964. Effect of bees on seed yields of Arlo rapeseed. Forage Notes 10, 1. [Google Scholar]
- Downey RK, Pawlowski SH, McAnsh J. 1970. Rapeseed – Canada’s “Cinderella” Crop. Winnipeg: Rapeseed Association of Canada. [Google Scholar]
- Elada Y. 1988. Scanning electron microscopy of parasitism of Botrytis cinerea on flowers and fruits of cucumber. Transactions of the British Mycologocial Society 91, 185–190. [Google Scholar]
- Farkas Á, Orosz-Kovács Z, Bubán T. 2006. Nectary structure of pear cultivars and its relation to fire blight susceptibility. Acta Horticulturae 704, 131–138. [Google Scholar]
- González-Teuber M, Eilmus S, Muck A, Svatos A, Heil M. 2009. Pathogenesis-related proteins protect extrafloral nectar from microbial infestation. The Plant Journal 58, 464–473. [DOI] [PubMed] [Google Scholar]
- González-Teuber M, Pozo MJ, Muck A, Svatos A, Adame-Alvarez RM, Heil M. 2010. Glucanases and chitinases as causal agents in the protection of Acacia extrafloral nectar from infestation by phytopathogens. Plant Physiology 152, 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AP, Gauthier MP, Vannette RL, Fukami T. 2014. Honey bees avoid nectar colonized by three bacterial species, but not by a yeast species, isolated from the bee gut. PLoS One 9, e86494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M, Xu WW, Kram BW, Chambers EM, Ehrnriter JS, Gralewski JH, Joyal T, Carter CJ. 2010. Identification of differential gene expression in Brassica rapa nectaries through expressed sequence tag analysis. PLoS One 5, e8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M. 2011. Nectar: generation, regulation and ecological functions. Trends in Plant Science 16, 191–200. [DOI] [PubMed] [Google Scholar]
- Herrera CM, García IM, Pérez R. 2008. Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89, 2369–2376. [DOI] [PubMed] [Google Scholar]
- Jaber LR, Vidal S. 2009. Interactions between an endophytic fungus, aphids and extrafloral nectaries: do endophytes induce extrafloral-mediated defences in Vicia faba?Functional Ecology 23, 707–714. [Google Scholar]
- Kevan PG, Eisikowitch D, Fowle S, Thomas K. 1988. Yeast-contaminated nectar and its effects on bee foraging. Journal of Apicultural Research 27, 26–29. [Google Scholar]
- Klein AM, Vaissiere BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T. 2007. Importance of pollinators in changing landscapes for world crops. Proceedings of the Royal Society B, Biological Sciences 274, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram BW, Bainbridge EA, Perera MA, Carter C. 2008. Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Molecular Biology 68, 173–183. [DOI] [PubMed] [Google Scholar]
- Kram BW, Xu WW, Carter CJ. 2009. Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biology 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda AF, Vasconcelos EA, Pelegrini PB, Grossi de Sa MF. 2014. Antifungal defensins and their role in plant defense. Frontiers in Microbiology 5, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascombe MB, Bakan B, Buhot N, Marion D, Blein JP, Larue V, Lamb C, Prangé T. 2008. The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Science 17, 1522–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gao G, Xu K, Chen B, Yan G, Li F, Qiao J, Zhang T, Wu X. 2014. Genome-wide survey and expression analysis of the putative non-specific lipid transfer proteins in Brassica rapa L. PLoS One 9, e84556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveaux J, Verge J. 1952. Research on the pollination of winter rape. Apiculteur 96, 15–18. [Google Scholar]
- Lüttge U. 1977. Nectar composition and membrane transport of sugars and amino acids: a review on the present state of nectar research. Apidologie 8, 305–319. [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK. 2002. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. [DOI] [PubMed] [Google Scholar]
- Musgrave ME. 2000. Realizing the potential of rapid-cycling Brassica as a model system for use in plant biology research. Journal of Plant Growth Regulation 19, 314–325. [DOI] [PubMed] [Google Scholar]
- Pokoj S, Lauer I, Fötisch K, Himly M, Mari A, Enrique E, Miguel-Moncin Mdel M, Lidholm J, Vieths S, Scheurer S. 2010. Pichia pastoris is superior to E. coli for the production of recombinant allergenic non-specific lipid-transfer proteins. Protein Expression and Purification 69, 68–75. [DOI] [PubMed] [Google Scholar]
- Radhika V, Kost C, Boland W, Heil M. 2010. The role of jasmonates in floral nectar secretion. PLoS One 5, e9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguso RA, Pichersky E. 1999. A day in the life of a linalool molecule: chemical communication in a plant-pollinator system. Part 1: Linalool biosynthesis in flowering plants. Plant Species Biology 14, 95–120. [Google Scholar]
- Rahman KA. 1940. Insect pollinators of toria (Brassica napus Linn., var. dichotoma prain) and sarson (B. campestris Linn., var. sarson prain) at Lyallpur. Indian Journal of Agricultural Science 10, 422–447. [Google Scholar]
- Rohrbach K. 1986. Unusual tropical fruit diseases with extended latent periods. Plant Disease 73, 607–609. [Google Scholar]
- Roy R, Schmitt AJ, Thomas JB, Carter CJ. 2017. Review: Nectar biology: From molecules to ecosystems. Plant Science 262, 148–164. [DOI] [PubMed] [Google Scholar]
- Salminen TA, Blomqvist K, Edqvist J. 2016. Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244, 971–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasu MA, Seidl-Adams I, Wall K, Winsor JA, Stephenson AG. 2010. Floral transmission of Erwinia tracheiphila by cucumber beetles in a wild Cucurbita pepo. Environmental Entomology 39, 140–148. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Wielsch N, Kessler D, Svatos A, Park CM, Baldwin IT, Kim SG. 2013. Natural variation in floral nectar proteins of two Nicotiana attenuata accessions. BMC Plant Biology 13, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Silva EM, Dean BB. 2000. Effect of nectar composition and nectar concentration on honey bee (Hymenoptera: Apidae) visitations to hybrid onion flowers. Journal of Economic Entomology 93, 1216–1221. [DOI] [PubMed] [Google Scholar]
- Stitz M, Hartl M, Baldwin IT, Gaquerel E. 2014. Jasmonoyl-L-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). The Plant Cell 26, 3964–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JY, Gaudet DA, Lu ZX, Frick M, Puchalski B, Laroche A. 2008. Characterization and antifungal properties of wheat nonspecific lipid transfer proteins. Molecular Plant-Microbe Interactions 21, 346–360. [DOI] [PubMed] [Google Scholar]
- Temkin-Gorodeiski N, Chorin M. 1971. The role of the nectary in the development of black heart disease of the Cavendish banana. Israel Journal of Botany 20, 91–95. [Google Scholar]
- Trapp G. 1936. The parasitism of Botrytis cinerea Pers. on Aucuba japonica thunb. Transactions of the British Mycological Society 20, 299–303. [Google Scholar]
- Vannette RL, Gauthier MPL, Fukami T. 2013. Nectar bacteria, but not yeast, weaken a plant–pollinator mutualism. Proceedings of the Royal Society B, Biological Sciences 280, 20122601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannette RL, Fukami T. 2016. Nectar microbes can reduce secondary metabolites in nectar and alter effects on nectar consumption by pollinators. Ecology 97, 1410–1419. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu G, Niu H, Timko MP, Zhang H. 2014. The F-box protein COI1 functions upstream of MYB305 to regulate primary carbohydrate metabolism in tobacco (Nicotiana tabacum L. cv. TN90). Journal of Experimental Botany 65, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Sigee D, Epton H. 1990. Erwinia amylovora infection of hawthorn blossom: III. The nectary. Journal of Phytopathology 128, 62–74. [Google Scholar]
- Wittek F, Hoffmann T, Kanawati B, Bichlmeier M, Knappe C, Wenig M, Schmitt-Kopplin P, Parker JE, Schwab W, Vlot AC. 2014. Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistance via azelaic acid and its precursor 9-oxo nonanoic acid. Journal of Experimental Botany 65, 5919–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.