SlARF10 plays an important role in the chlorophyll accumulation and photosynthesis of tomato plants, and regulation of its expression affects the starch, fructose, and sucrose content of fruit.
Keywords: ARF10, auxin, chlorophyll, fruit, starch, sugar, tomato
Abstract
The photosynthesis of green tomatoes contributes to fruit growth and carbon economy. The tomato auxin response factor 10 (SlARF10) belongs to the ARF family and is located in nucleus. In this study, we found that SlARF10 was highly expressed in green fruit. Overexpression of SlARF10 in fruit produced a dark-green phenotype whilst knock-down by RNAi produced a light-green phenotype. Autofluorescence and chlorophyll content analyses confirmed the phenotypes, which indicated that SlARF10 plays an important role in chlorophyll accumulation. Overexpression of SlARF10 positively affected photosynthesis in both leaves and fruit. Furthermore, SlARF10-overexpression lines displayed improved accumulation of starch, fructose, and sucrose in fruit, whilst SlARF10-RNAi lines showed decreased accumulation of starch and sucrose. Regulation of SlARF10 expression altered the expression of AGPase starch biosynthesis genes. SlARF10 positively regulated the expression of SlGLK1, POR, CBP1, and CBP2, which are related to chlorophyll metabolism and regulation. Electrophoretic mobility shift assays confirmed that SlARF10 directly targets to the SlGLK1 promoter. Our results thus indicate that SlARF10 is involved in chlorophyll accumulation by transcriptional activation of SlGLK1 expression in tomato fruit, and provide insights into the link between auxin signaling, chloroplast activity, and sugar metabolism during tomato fruit development.
Introduction
Tomato (Solanum lycopersicum), a nutrient-rich multicarpellar berry with strong adaptability and high yield, has become the world’s second largest vegetable crop (Tanksley, 2004). It has also become the research model species for fleshy fruits, due to its short life cycle, self-pollination, and the ease with which mechanical crossing and genetic transformation can be conducted (Klee and Giovannoni, 2011).
Fruit development can be divided into three main stages (Ho and Hewitt, 1986). The first stage is characterized by intense mitotic activity, with an increase in cell number and starch accumulation (Ho, 1996). Cell enlargement associated with the degradation of starch into soluble sugars is characteristic of the second stage (Schaffer and Petreikov, 1997), whilst the third stage corresponds to fruit ripening, which is associated with the conversion of chloroplasts into chromoplasts, and the accumulation of carotenoids, sugars, organic acids, and volatile aromatic compounds in the cells (Klee and Giovannoni, 2011). The accumulation of soluble solids in ripening tomatoes is related to the starch level in immature and mature green fruit (Davies and Cocking, 1965). It has been reported that 10–15% of the total carbon and of the net sugar accumulation during fruit development comes from the photosynthetic activity in the fruit itself (Tanaka et al., 1974; Obiadalla-Ali et al., 2004). Hence, chloroplast development and the photosynthetic activity of green fruit affect the composition and quality of ripening tomatoes (Nadakuduti et al., 2014).
Several genes influence the development of fruit chloroplasts and the subsequent quality of ripening fruit in tomatoes. The DE-ETIOLATED 1/high pigment 2 (DET1/hp2) and UV-DAMAGED DNA-BINDING PROTEIN 1/high pigment 1 (DDB1/hp1) genes encode negative regulators of photomorphogenesis. Down-regulation of DET1/hp2 and DDB1/hp1 increases the number of chloroplasts and plastid compartment size, thereby leading to higher levels of chlorophyll and carotenoids in the fruit (Liu et al., 2004; Kolotilin et al., 2007; Rohrmann et al., 2011). GOLDEN2-LIKE (GLK) transcription factors are required for the regulation of chloroplast and chlorophyll levels (Waters et al., 2008). Tomato contains two GLKs, namely SlGLK1 and SlGLK2, which encode functionally similar peptides. Differential expression renders SlGLK1 more important in leaves and SlGLK2 more important in the fruit. The latitudinal gradient of SlGLK2 expression across the axis of the fruit results in the typical uneven coloration of green and ripe wild-type tomatoes (Nguyen et al., 2014). Tomato ARABIDOPSIS PSEUDO RESPONSE REGULATOR 2-LIKE (SlAPRR2-like) is the closest global relative of SlGLK2. Overexpression of the APRR2-like gene in tomatoes produces larger and more numerous chloroplasts, and consequently higher chlorophyll levels in green fruit and higher amounts of carotenoids in red ripening fruit (Pan et al., 2014). Two Class I KNOTTED1-LIKE HOMEOBOX (KNOX) proteins, namely TKN2 and TKN4, positively influence SlGLK2 and SlAPRR2-LIKE expression to promote chloroplast development in tomato fruit (Nadakuduti et al., 2014).
Phytohormones have been reported to be involved in chloroplast development and the quality of ripening fruit (Martineau et al., 1994; Galpaz et al., 2008; Sagar et al., 2013). Studies on the auxin signaling transduction pathway have indicated that auxin response factors (ARFs) are required for auxin-dependent transcriptional regulation in plants, and they can function as either transcriptional activators or repressors of auxin-responsive genes (Ren et al., 2011). Most ARF proteins contain an N-terminal DNA-binding domain (B3), which is involved in transcription of auxin-response genes, a middle region acting as an activation domain or repression domain, and a C-terminal dimerization domain that requires the formation of heterodimers or homodimers (Zouine et al., 2014). An increasing number of studies have demonstrated that ARFs play important roles in many developmental processes in tomato (Krogan et al., 2012; Wang et al., 2012; Guan et al., 2013; Ckurshumova et al., 2014; Liu et al., 2014a, 2014b; Zhang et al., 2015). SlARF7 acts as a negative regulator of fruit set and development in tomato (De Jong et al., 2009). ARF6 and ARF8 have important roles in controlling flower growth and development (Liu et al., 2014a). SlARF9 is required for the regulation of cell division during early tomato fruit development (De Jong et al., 2015), SlARF3 is involved in the formation of epidermal cells and trichomes (Zhang et al., 2015), and SlARF4 controls the accumulation of chlorophyll and starch in the fruit (Jones et al., 2002; Sagar et al., 2013). The influence of SlARF4 on fruit chlorophyll accumulation seems to be mediated through the transcriptional up-regulation of SlGLK1 in the fruit (Sagar et al., 2013).
Hendelman et al. (2012) reported that SlARF10 is post-transcriptionally regulated by Sl-miR160, and constitutive expression of mSlARF10 (Sl-miR160a-resistant version) produced narrow leaflet blades, sepals, and petals, and abnormally shaped fruit in tomato. Repression of SlARF10 expression by Sl-miR160 is essential for auxin-mediated blade outgrowth and early fruit development (Hendelman et al., 2012). In the present study, the functions of SlARF10 were studied in the development of tomato fruit, and it was found to be involved in chlorophyll and sugar accumulation. This study expands our understanding of the functions of ARFs during the development of tomato fruit, and provides new insights into the regulation mechanisms of chlorophyll and sugar accumulation.
Materials and methods
Plant material and growth conditions
Tomato (Solanum lycopersicum L. cv. Micro Tom) plants were grown in a culture chamber under a 16/8 h light/dark photoperiod at 25 ± 2/18 ± 2 °C and 80% relative humidity.
Sequence analysis
BLAST analysis was performed at the website http://www.ncbi.nlm.nih.gov/blast/. Protein domains were identified using the Pfam program (http://pfam.xfam.org). Alignment of protein sequences was performed with ClustalX version 2.1.
Expression patterns and qRT-PCR analysis
Expression patterns were analysed online according to the tomato gene expression database (http://tomexpress.toulouse.inra.fr/). The pericarp of fruit was used for total RNA isolation. Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen). Quantitative real-time (qRT)-PCR was carried out as previously described by Deng et al. (2012). The relative transcript abundance was monitored on a CFX96 real-time PCR detection system (Bio-Rad) using All-in-One™ qPCR Mix (GeneCopoeia). The relative expression for each gene of interest was calculated using the ΔΔCt values with ubiquitin as an internal standard. Primer sequences are listed in Supplementary Table S1 at JXB online. The following genes were examined: DDB1, UV damaged DNA binding protein 1 (Solyc02g021650); THY5, bZIP domain of plant elongated/long HY5-like transcription factors and similar proteins (Solyc08g061130); SlGLK1, golden2-like protein 1 (Solyc07g053630); SlGLK2, golden2-like protein 2 (Solyc10g008160); POR, protochlorophyllide oxidoreductase (Solyc10g006900) CBP1, chlorophyll binding protein 1 (Solyc02g070990) and CBP2, chlorophyll binding protein 2 (Solyc02g070950).
Subcellular localization of SlARF10
For the construction of the SlARF10-GFP fusion expression vector, the forward 5′-ATGAAGGAGGTTTTGGAGAAGTG-3′ and reverse 5′-CTATGCAAAGATGCTAAGAGGTC-3′ primers were used to amplify the sequence of SlARF10 coded frames. Protoplasts were obtained from suspension-cultured cells of tobacco (Nicotiana tabacum cv. Bright Yellow-2) and transfected with the SlARF10-GFP (green fluorescent protein) fusion expression vector. Transformation assays were performed as previously described by Chaabouni et al. (2009).
Generation of transgenic plants
The ORF sequence of SlARF10 was amplified using the forward 5′-TCCCCCGGGGATGAAGGAGGTTTTGGAGAA-3′ and reverse 5′-CGGGATCCCTATGCAAAGATGCTAAGAGGTC-3′ primers. The sequence was cloned into the plant binary vector pLP100, resulting in an overexpression vector. For the construction of the RNAi vector, 200-bp sequences of SlARF10 were amplified, and the PCR products were inserted around a spacer of the β-glucuronidase (GUS) gene in pCAMIBA2301 driven by a Cauliflower mosaic virus (CaMV) 35S promoter. Transgenic plants were generated via Agrobacterium tumefaciens-mediated transformation according to the methods described by Jones et al. (2002). All experiments were performed using homozygous lines of T3 generations.
Chlorophyll analysis in tomatoes
Chlorophyll content was determined in the fruit pericarp and leaves according to the methods described by Powell et al. (2012). The experiments were performed using 2-month-old plants and the leaf samples were selected from the fourth internode counting from the shoot tip. For the determination of chlorophyll autofluorescence, the pericarp was peeled off the fruit and observed under a TCS SP2 laser confocal microscope (Leica, Germany).
Measurement of chlorophyll fluorescence parameters
Chlorophyll fluorescence parameters were measured using a pulse-amplitude modulation fluorometer (PAM-2500, Walz, Germany). The experiments were performed using 2-month-old plants and the leaf samples were selected from the fourth internode counting from the shoot tip. The measurements were performed on leaves and green fruit, at 36 d post-anthesis (DPA), according to the methods described in detail by Maury et al. (1996).
Determination of photosynthetic substances
Samples of 1 g of tomato fruit pericarp were ground in liquid nitrogen and extracted three times with 10 ml of 80% ethanol at 80 °C for 30 min. After centrifugation at 5000 g for 5 min, the supernatant was used for measurement of glucose, fructose, and sucrose, and the pellet was used for starch analysis. The supernatant was completely evaporated under vacuum, and dissolved in 3 ml distilled water. Sub-samples of 1 ml were used to measure glucose, fructose, and sucrose using HPLC. For starch analysis, 4 ml of 0.2 M KOH was added to the pellet and heated at 100 °C for 30 min. Each sample was added to 1.48 ml of 1 M acetic acid (pH 4.5), hydrolysed with 7 Units of amyloglucosidase for 45 min, and dissolved in 10 ml distilled water. Sub-samples of 1 ml were then used to measure starch content via HPLC.
HPLC analysis was performed using an Agilent 1260 Series liquid chromatograph system (Agilent Technologies), equipped with a vacuum degasser, a binary pump, an auto-sampler, and a diode array detector, controlled by the Agilent ChemStation software. A Waters XBridge Amide column (4.6 × 150 mm i.d., 3.5 μm) together with a pre-column (Waters XBridge BEH Amide column, 3.9 × 5 mm i.d., 3.5 μm) were employed. Separation was achieved using an isocratic solvent system that consisted of solvent A (0.2% triethylamine water solution) and solvent B (acetonitrile) as mobile phases, eluting at 75% B for 15 min. The solvent flow rate was 0.6 ml min–1, the column temperature was kept at 38 °C, and the injection volume for all samples was 10 μl. The detection system was an ELSD 2000 (Alltech), with a drift tube temperature of 80 °C, using air as the carrier gas with flow rate of 2.2 l min–1. The contents of glucose, fructose, sucrose, and starch were calculated according to the methods described by Geigenberger et al. (1996).
Promoter analysis and dual-luciferase transient expression assays
The promoter sequences of SlGLK1 and SlGLK2 genes were analysed using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Transcription activity analyses of SlARF10 were conducted in leaves of tobacco (Nicotiana benthaminana). The full coding sequence of SlARF10 was amplified and cloned into pGreenII 62-SK (Hellens et al., 2005), which was used as an effector vector. The promoters of SlGLK1, POR, CBP1, and CBP2 were cloned into the pGreenII 0800-LUC (luciferase) double-reporter vector (Hellens et al., 2005). All primers used for construction of transient expression vectors are listed in Supplementary Table S1. The activities of LUC and Renilla (REN) luciferase were measured with a Luminoskan Ascent microplate luminometer (ThermoFisher Scientific) using a dual-luciferase assay kit (Promega) according to the manufacturer’s instructions. Six biological repeats were performed for each pair of vectors.
Protein expression and electrophoretic mobility shift assays (EMSAs)
The B3 domain sequence (amino acid position 1–366) of SlARF10 was fused to the glutathione S-transferase (GST) tag in GEX-4T-1 (GE Healthcare Life Science) and expressed in Escherichia coli strain BM Rosatta (DE3). The recombinant proteins were induced with 0.8 mM isopropyl-β-D-thiogalactopyranoside for 6 h at 30 °C and purified using a GST-Tagged Protein Purification Kit (Clontech). The recombinant proteins were used for the EMSA assays with a LightShift Chemiluminescent EMSA Kit (ThermoFisher Scientific) according to the procedure described by Han et al. (2016). The probe containing the TGTCTC sequence from the promoter of SlGLK1 was labelled with biotin using a Pierce Biotin 3′ End DNA Labeling Kit (ThermoFisher Scientific). An unlabeled DNA fragment of the same sequence was used as a competitor. The TGTCTC DNA fragment was modified into AAAAAA and used as a mutant probe. Biotin-labelled DNA was detected using a Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFisher Scientific) according to the manufacturer’s instructions. All the primers used in the EMSA assays are listed in Supplementary Table S1.
Results
The SlARF10 gene is highly expressed in tomato fruit and the SlARF10 protein is located in the nucleus
Amino-acid sequence analysis determined that SlARF10 had the domains B3-DNA, ARF, and AUX/IAA, which indicated that it had the typical conserved ARF domains (Supplementary Fig. S1).
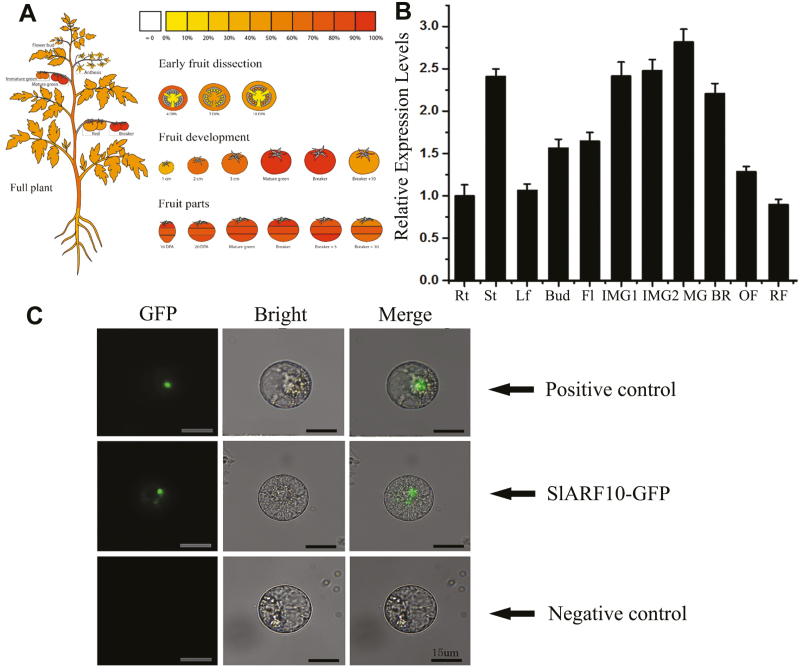
The expression profiles of SlARF10 in tomato were analysed via an online database and qRT-PCR. The database analysis revealed that it was expressed in all tissues, including roots, stems, leaves, flowers, and fruit. The expression level was high in fruit, especially in immature green, mature green, and breaker fruit (Fig. 1A). The qRT-PCR analysis also showed a similar expression profile (Fig. 1B). These results suggested that the SlARF10 gene may be involved in the development of tomato fruit.
Fig. 1.
Expression patterns of the SlARF10 gene and subcellular localization analysis of the SlARF10 protein in tomato plants. (A) Online analysis of SlARF10 in tomato plants (see Methods). The degree of shading indicates the gene expression level. (B) qRT-PCR analysis of SlARF10 expression levels; the housekeeping gene ubiquitin was used as the reference. The data are means (±SD) of three biological replicates. Rt, root; St, shoot; Lf, leaf; Fl, flower; IMG, immature green fruit (IMG1, 4 d post-anthesis, DPA; IMG2, 18 DPA); MG, mature green fruit (35 DPA); BR, breaker fruit (40 DPA); OF, orange fruit (43 DPA); RF, red fruit (46 DPA). The fruit pericarp was used for total RNA isolation. (C) Subcellular localization analysis of the SlARF10 protein. PCX-DG-GFP was the negative control. Scale bars are 15 μm.
Amino-acid sequence analysis showed that SlARF10 had a nuclear-localization signal peptide. In order to verify the location in the nucleus, SlARF10-GFP fusion protein vectors were constructed and transferred into tobacco protoplasts. The green fluorescence of the fusion protein indicated that SlARF10 was located in the nucleus (Fig. 1C).
SlARF10 is involved in chlorophyll accumulation in tomato fruit
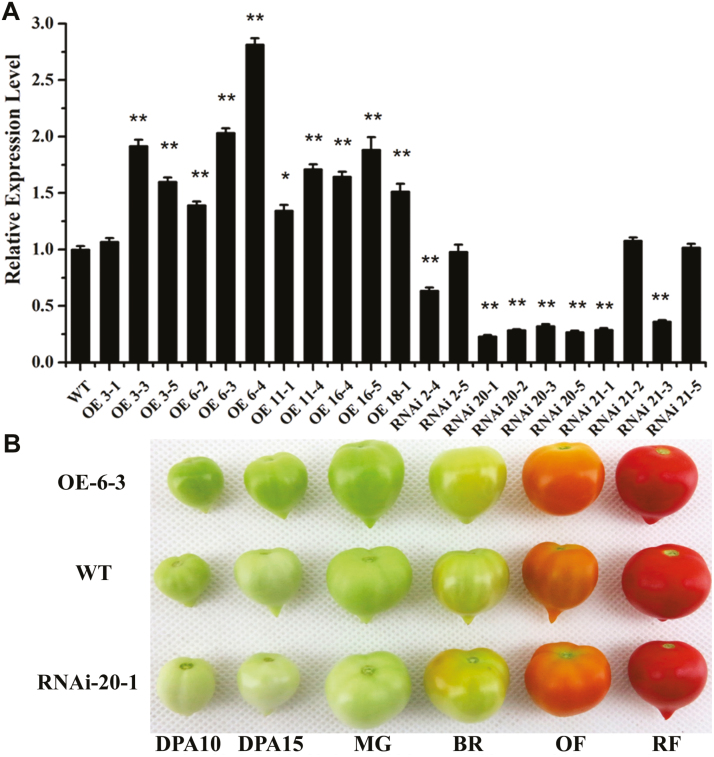
In order to examine its functions in tomato fruit development, transgenic techniques were used to up- and down-regulate SlARF10. Ten homozygous down-regulated transgenic lines (SlARF10-RNAi) and 11 homozygous up-regulated transgenic lines (SlARF10-OE) were generated, corresponding to independent transformation events. The T3 generation plants of the RNAi 2-4 and RNAi 20-1 lines and the OE 6-3 and OE 11-4 lines that exhibited lower and higher accumulation of SlARF10 transcripts, respectively, were selected for further study (Fig. 2A). At the green fruit stage, the two OE lines produced fruit that were darker green than the wild-type (WT) plants whilst the two RNAi lines produced fruit that were lighter green (Fig. 2B). However, there were no visual differences in the fruit colors between the transgenic and WT plants at the subsequent breaker, orange, and red-ripe stages.
Fig. 2.
SlARF10 transgenic plants and their fruit phenotypes. (A) qRT-PCR analysis of the expression of SlARF10 in the transgenic lines; the housekeeping gene ubiquitin was used as the reference. (B) Fruit phenotypes. WT, wild-type plants; OE, SlARF10-overexpression lines; RNAi, SlARF10-RNAi lines. DPA, days post-anthesis. MG, mature green fruit; BR, breaker fruit; OF, orange fruit (2 d post-breaker stage); R, red fruit (5 d post-breaker stage). The fruit pericarp was used for total RNA isolation. Data are means (±SD) of three biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01.
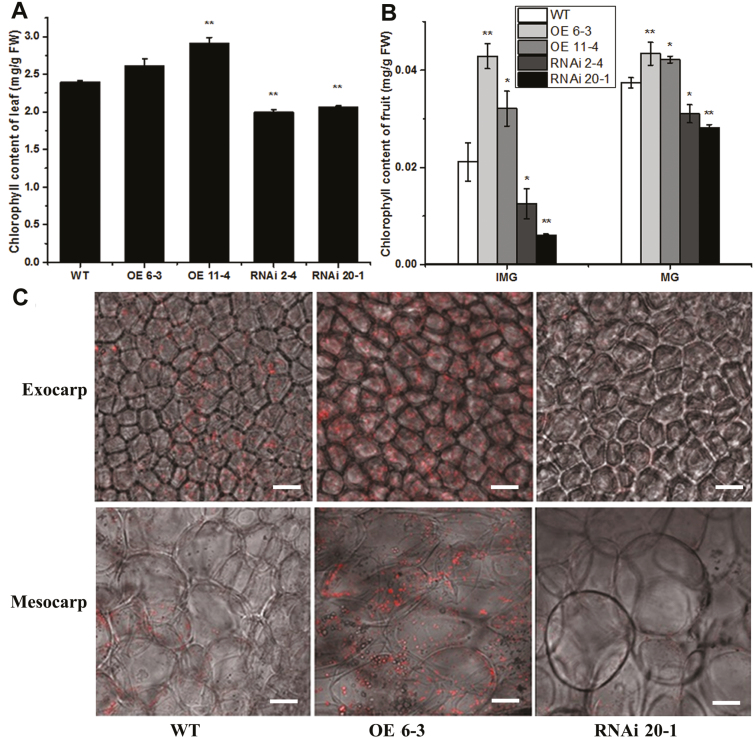
To verify these observations, the chlorophyll contents of green fruit and leaves were determined in the plants. Compared with the WT, the RNAi lines showed lower accumulation of chlorophyll and the OE lines showed higher accumulation (Fig. 3A, B). Confocal laser-scanning microscopy was used to detect chlorophyll autofluorescence in the mesocarp (middle layer of the pericarp) and exocarp (outer layer) of the fruit. The OE 6-3 line had stronger chlorophyll autofluorescence compared with the WT plants whilst the RNAi 20-1 line had weaker autofluorescence (Fig. 3C). These results indicated that SlARF10 is involved in chlorophyll accumulation, and that its regulation could therefore control the chlorophyll content in tomato fruit.
Fig. 3.
Chlorophyll accumulation in SlARF10 transgenic plants. (A, B) Chlorophyll contents in leaves and fruit of SlARF10-overexpression (OE) and SlARF10-RNAi lines Compared with the wild-type (WT). Data are means (±SD) of three biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01. (C) Autofluorescence of chlorophyll in the pericarp of tomato fruit, as determined by confocal laser scanning microscopy for the SlARF10-OE line 6-3 and SlARF10-RNAi line 20-1, compared with the WT. Scale bars are 10 μm.
Regulation of SlARF10 expression affects the photochemical efficiency and synthesis of photosynthetic substances in tomato fruit
Increased chlorophyll content may potentially confer higher photosynthetic performance in the transgenic plants. To assess this hypothesis, the photosynthetic performance was examined in the fruit and leaves of the selected RNAi and OE lines. Relative to the WT, the OE lines had increased photochemical potential in the fruits and leaves whilst the RNAi lines had decreased potential (Fig. 4A, B). Similar results were found for the effective photochemical quantum yield of PSII (Fig. 4C, D). Thus, regulation of SlARF10 gene expression positively affected photosynthesis in the plants.
Fig. 4.
Photochemical potential in SlARF10 transgenic plants. Photochemical potential in (A) the fruit and (B) the leaves of SlARF10-overexpression (OE) and SlARF10-RNAi plants compared with the wild-type (WT). Effective photochemical quantum yield of PSII in (C) the leaves and (D) the fruit. Data are means (±SD) of three biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01.
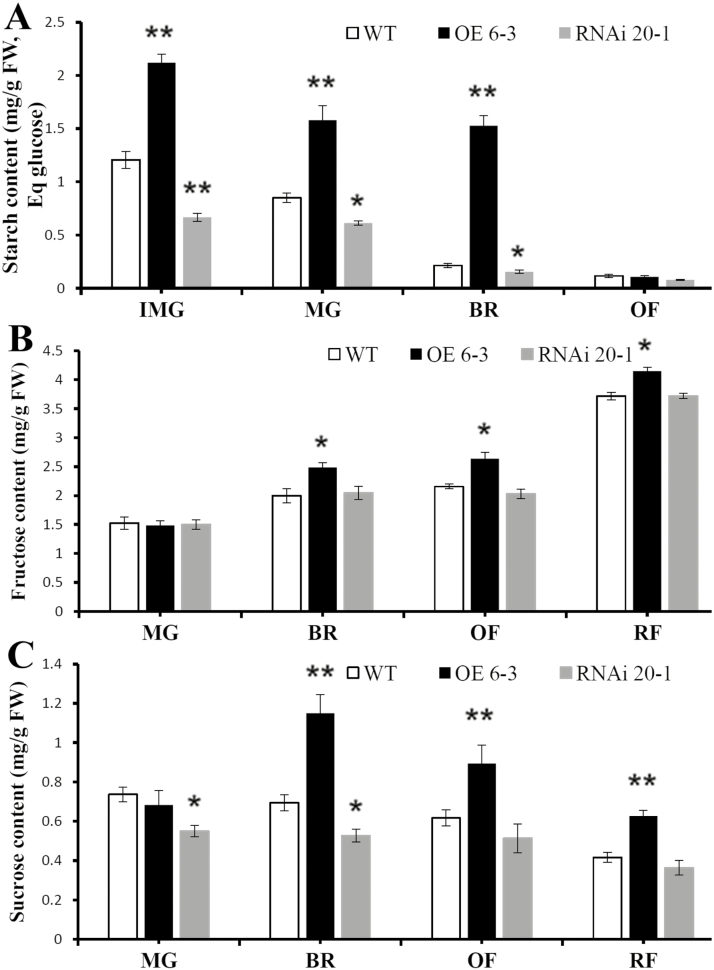
Sugars are the main products of chloroplast activity and photosynthesis, and therefore their accumulation was determined in the plants using HPLC. Starch accumulated at the immature green stage of fruit development and had decreased dramatically by the orange stage (Fig. 5A). Up-regulation of SlARF10 increased starch accumulation in the fruit at the immature green, mature green, and breaker stages compared with the WT plants, whilst down-regulation of SlARF10 decreased accumulation. These results indicated that regulation of SlARF10 gene expression controlled starch synthesis during early stages of fruit development.
Fig. 5.
Accumulation of photosynthetic substances in the fruit of SlARF10 transgenic plants. Contents of (A) starch, (B) fructose, and (C) sucrose in SlARF10-overexpression (OE) and SlARF10-RNAi lines compared with the wild-type (WT). Data are means (±SD) of three biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01.
Starch degradation is the main source of soluble sugars and therefore we assessed the impact of SlARF10 regulation on fructose, glucose, and sucrose in the fruit. The OE 6-3 line had significantly higher fructose contents than the WT at the breaker, orange-, and red-fruit stages, whilst the RNAi 20-1 line showed no significant differences compared with the WT plants (Fig. 5B). There were no significant differences in glucose content between the WT, OE 6-3, and RNAi 20-1 plants (data not shown). The sucrose content in the OE 6-3 line was significantly higher than in the WT at the breaker, orange-, and red-fruit stages, whilst in the RNAi 20-1 line the content was lower at the mature green and breaker stages (Fig. 5C).
Regulation of SlARF10 expression alters the expression of starch biosynthesis genes
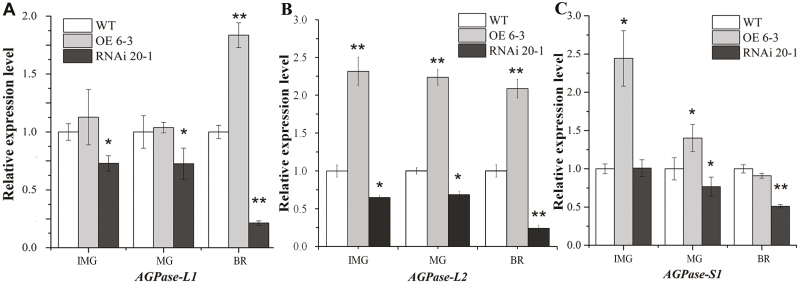
To gain more insight into the mechanism of sugar metabolism in the SlARF10 transgenic plants, we examined the expression patterns of starch biosynthesis genes. AGPase genes, with four subtypes (AGPase-L1, AGPase-L2, AGPase-L3, and AGPase-S1), encode the most important enzymes in starch synthesis that catalyse the first step of the reaction. Relative to the WT, the expression of AGPase-L1 was increased at the breaker stage in the OE 6-3 fruit and decreased in the immature green, mature green, and breaker stages of the RNAi 20-1 fruit (Fig. 6A). AGPase-L2 showed increased expression at each of the three stages in the OE 6-3 fruit and decreased expression in the RNAi 20-1 fruit (Fig. 6B). AGPase-S1 showed increased expression at the immature green and mature green stages in the OE 6-3 fruit and decreased expression at the mature green and breaker stages of the RNAi 20-1 fruit (Fig. 6C). The AGPase-L3 gene showed no altered expression in the SlARF10 transgenic lines compared with the WT plants (data not shown). Analysis of the promoter sequences in the AGPase genes showed that the ARF binding site, the TGTCTC box, existed in the promoters of AGPase-L1 and AGPase-L2 (Supplementary Table S2). The direct binding of the SlARF10 protein to the SlGLK1 promoter was verified by EMSAs. The purified recombinant truncated SlARF10 and glutathione S-transferase (GST) fusion protein (GST-tSlARF10) were successfully obtained (Supplementary Fig. S3), but the EMSA results showed that there was no specific binding between SlARF10 and the promoters of AGPase-L1 and AGPase-L2, (data not shown).
Fig. 6.
The expression of AGPase genes in SlARF10 transgenic plants. The levels of transcripts were assessed in tomato fruit by qRT-PCR for (A) AGPase-L1, (B) AGPase-L2, and (C) AGPase-S1 in SlARF10-overexpression (OE) and SlARF10-RNAi lines compared with the wild-type (WT). The housekeeping gene ubiquitin was used as the reference. IMG, immature green stage; MG, mature green stage; BR, breaker stage. Data are means (±SD) of three biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01.
SlARF10 positively regulates the expression of SlGLK1, POR, CBP1, and CBP2
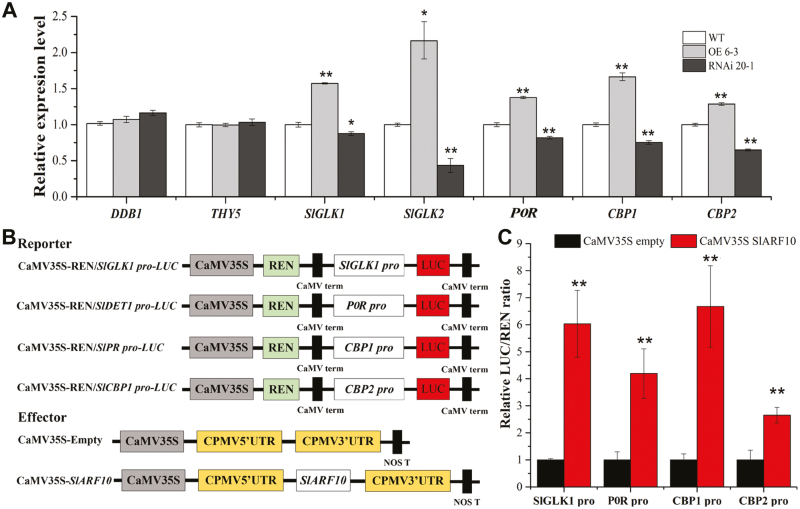
The chlorophyll and starch phenotypes of the SlARF10-OE plants were reminiscent of those described in transgenic SlGLK overexpression plants. The expression levels of two GLK genes, SlGLK1 and SlGLK2, were therefore examined in the OE 6-3 and RNAi 20-1 lines. qRT-PCR analysis demonstrated that there was enhanced accumulation of SlGLK1 and SlGLK2 transcripts in the fruit of the OE 6-3 plants and reduced accumulation of transcripts in the fruit of RNAi 20-1 plants relative to the WT (Fig. 7A). The qRT-PCR results also showed that there were no significant differences between the WT and transgenic plants in the expression levels of DDB1 and THY5, which indicated that the effect of SlARF10 on chlorophyll accumulation may have acted independently or downstream of the DDB1 pathway. The expression levels of POR, CBP1, and CBP2 were also examined and increased accumulation of transcripts were found in the fruit of OE 6-3 plants, whilst decreased accumulation was found in the fruit of RNAi 20-1 plants. No differences in expression of SlARF4 were found in the OE and RNAi lines relative to the WT (Supplementary Fig. S2).
Fig. 7.
SlARF10 activates the expression of genes related to chlorophyll metabolism and regulation in tomato fruit. (A) Expression profiles of genes related to chlorophyll metabolism and regulation in SlARF10-overexpression (OE) and SlARF10-RNAi lines compared with the wild-type (WT). The housekeeping gene ubiquitin was used as the reference. Data are means (±SD) of three biological replicates. (B) Diagrams of the reporter and effector constructions used in the dual-luciferase reporter assay. (C) In vivo interactions of SlARF10 with the promoters obtained from transient assays in tobacco leaves. The ratio of LUC/REN of the empty vector plus promoter was used as a calibrator (set as 1). Data are means (±SD) of six biological replicates. Significant differences between transgenic and WT plants were determined using Student’s t-test: *P<0.05, **P<0.01.
Transient expression assays were performed using the dual-luciferase reporter system to investigate whether SlARF10 could directly activate the expression of SlGLK1, POR, CBP1, and CBP2. Tobacco leaves were co-transformed with LUC reporter plasmids under the control of the promoters of SlGLK1, POR, CBP1, and CBP2 together with an overexpression vector carrying SlARF10 under the control of the CaMV35S promoter (Fig 7B). As shown in Fig. 7C, the activities of all the promoters were significantly increased in the presence of SlARF10, with relatively higher LUC/REN ratios being observed compared to the controls.
SlARF10 targets the promoter of SlGLK1
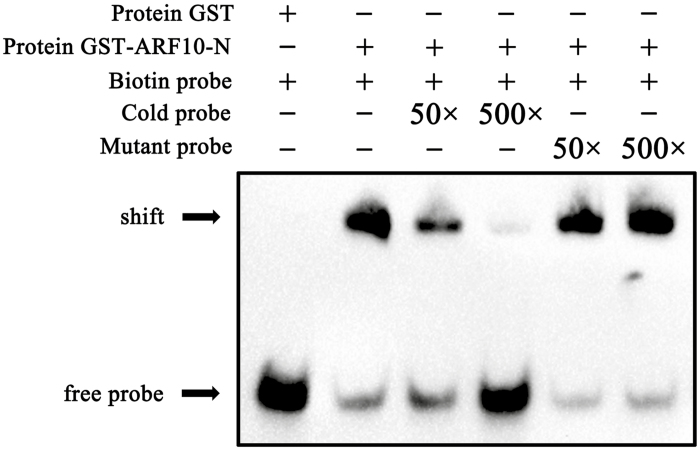
Analysis of the promoter sequence in the SlGLK1 gene showed two conserved ARF binding sites, the TGTCTC box. The direct binding of the SlARF10 protein to the SlGLK1 promoter was verified by EMSA. The GST-tSlARF10 fusion protein bound to the biotin-labeled probes containing the TGTCTC motif from the SlGLK1 promoter and caused mobility shifts. The mobility shift was effectively abolished in a dose-dependent manner by the addition of increasing amounts of unlabeled competitors with the same sequence, but not by the mutated probes (Fig. 8). The mobility shift was also not observed when biotin-labeled probes were incubated with GST only, indicating the specific binding between SlARF10 and the SlGLK1 promoter.
Fig. 8.
SlARF10 targets the promoters of SlGLK1 genes. Electrophoretic mobility shift assay showing the binding of SlARF10 to the SlGLK1 promoter. Biotin-labeled DNA probes from native promoter or mutants were incubated with the GST-SlARF10 protein, and the DNA–protein complexes were separated on 6% native polyacrylamide gels.
Discussion
SlARF10 is involved in chlorophyll accumulation in tomato fruit
Chlorophyll content, a critical feature of unripe fruit, affects the nutritional components and flavor of ripe fruit. Moreover, the link between chlorophyll content and photosynthesis (or photosynthate metabolism in fruit tissues) has been highlighted by a number of studies (Lopez-Juez and Pyke, 2005; Powell et al., 2012; Nadakuduti et al., 2014). However, the regulatory mechanisms by which chlorophyll impacts on photosynthetic capacity, as well as on photosynthate accumulation and thus fruit quality, remain unclear. Auxin plays a pivotal role in the initiation of fleshy fruit development and determination of final fruit size through the control and expansion of cell division (Devoghalaere et al., 2012; Sagar et al., 2013). Consequently, auxin impacts on an array of crucial regulators (such as ethylene, ABA, and the ripening regulator Rin) and vital effectors (such as genes for β-xanthophyll, lycopene biosynthesis, and chlorophyll degradation) (Su et al., 2015; Manoharan et al., 2017). Studies have also suggested that roots of Arabidopsis treated with auxin show increased chlorophyll accumulation and chloroplast development after detachment from shoots, and analyses of mutants indicated that auxin transported from the shoot represses chlorophyll accumulation via the function of ARF7, ARF19, and IAA14 (Kobayashi et al., 2012). A hypothesis can be drawn based on these findings in which auxin, a critical phytohormone, regulates chlorophyll accumulation and degradation via the function of ARFs during fruit setting and development.
Given that IAA14 and ARF7/19 mediate the auxin signaling pathway to repress chlorophyll biosynthetic genes in Arabidopsis (Kobayashi et al., 2012), we speculated that auxin is likely to regulate chlorophyll biosynthesis and accumulation in tomato via activated or repressed transcriptional functioning of ARFs. Previous work has shown that DR12/ARF4, a member of the tomato ARF gene family, influences the regulation of fruit development, such that transgenic tomato plants with down-regulated SlARF4 expression levels bear dark-green fruit at immature stages, with a significant increase in chlorophyll content, and accumulate more starch during the colour transition stages and more sugar at the ripening stages of fruit development. SlARF4 may function through the transcriptional repression of GLK1 expression in tomato fruit (Jones et al., 2002; Sagar et al., 2013). In contrast, in our current study, up-regulation of SlARF10, another transcriptional repressor, resulted in enhanced chlorophyll accumulation in tomato fruit (Fig. 3). Furthermore, overexpression of SlARF10 increased the accumulation of SlGLK1 and SlGLK2 transcripts in the fruit, whereas down-regulation had the opposite effect (Fig. 7). We also found that SlARF10 could positively regulate the expression of SlGLK1 and directly target its promoter (Fig. 8). Overexpression of SlGLK1 and SlGLK2 is known to produce dark-green fruit (Nguyen et al., 2014), which is similar what we observed in plants with SlARF10 overexpression. Although SlGLK2 expression was increased in the SlARF10 overexpression lines and decreased in the down-regulated lines, this would not account for the dark-green phenotypes as the Micro Tom variety that we used possesses two null alleles of SlGLK2 (Powell et al., 2012). Our results indicated that SlARF10 controls chlorophyll accumulation by regulating the expression of SlGLK1, with SlARF10 functioning as a transcriptional activator of SlGLK1. SlGLK1 and SlGLK2 have similar functions, but different expression patterns effectively restrict SlGLK1 mostly to leaf functions and SlGLK2 mostly to fruit functions in tomato plants (Nguyen et al., 2014). Our results indicated that SlGLK1 may have important functions in chlorophyll accumulation not only in tomato leaves, but also in the fruit, and in future studies knock-out of SlGLK1 in the fruit could be performed to further elucidate its roles in chlorophyll accumulation in tomato plants. It was interesting that the expression levels of SlARF4 were not changed in the SlARF10-OE and SlARF10-RNAi lines (Supplementary Fig. S2). Thus, the data suggest that regulation of SlGLK1 expression by SlARF10 may act independently of SlARF4.
Our results showed no significant differences between the WT and transgenic plants in the expression levels of DDB1 and THY5 (Fig. 7). In the dark, active DDB1 causes the degradation of photomorphogenesis-promoting transcription factors such as THY5. Exposure to light represses DDB1 and activates THY5, leading to transcriptional activation of photomorphogenesis and pigment genes (Cocaliadis et al., 2014). The regulation of chlorophyll accumulation by SlARF10 may act independently or downstream of the DDB1 pathway. Chlorophyll a is initially synthesized from glutamyl-tRNAglu, and chlorophyll b is synthesized at the final step of chlorophyll biosynthesis. Analysis of the complete genome of Arabidopsis has determined that there are 15 enzymes encoded by 27 genes for chlorophyll biosynthesis (Beale, 1999). Our study showed that SlARF10 positively regulated the expression of POR (Fig. 7), which encodes a protochlorophyllide oxidoreductase that catalyses protochlorophyllide into chlorophyllide, a key step in chlorophyll biosynthesis (Cahoon and Timko, 2000). Thus, SlARF10 may directly regulate the expression of this key gene, thereby influencing chlorophyll biosynthesis at the developmental stages of tomato fruit. We also found that SlARF10 positively regulated the expression of CBP1 and CBP2. Chloroplast membranes contain many kinds of chlorophyll binding proteins and they are associated with chlorophyll and xanthophylls, which absorb sunlight and transfer the excitation energy to the PSII core complexes to drive photosynthetic electron transport (Hiller et al., 1995). The increased expression of CBP1 and CBP2 may explain the improved photochemical efficiency in the SlARF10 overexpression lines (Fig. 4).
Altering SlARF10 expression affects accumulation of photosynthates in tomato fruit
SlARF10 up-regulated lines display dark-green fruit phenotypes similar to those seen in SlARF4 down-regulated lines with enhanced chlorophyll content (Sagar et al., 2013). In contrast to SlARF4 down-regulated plants where the dark-green phenotype is restricted to immature fruit, significantly higher chlorophyll content was detected in leaf and fruit tissues of the SlARF10-OE lines (Fig. 3). This indicated that, in contrast to SlARF4, SlARF10 control of chlorophyll accumulation was not fruit-specific. Furthermore, the higher chlorophyll content in the SlARF10-OE lines correlated with higher photochemical efficiency compared with the WT plants (Fig. 4), and resulted in elevated starch and sugar contents in the transgenic fruit (Fig. 5). Starch is not only a significant carbohydrate reserve in the majority of plants but also a major factor in defining fruit nutrition and flavor. In starch synthesis, the first regulatory step (the synthesis of ADP-glucose) is catalysed by AGPase from glucose-1-phosphate and ATP (Stark et al., 1992; Yin et al., 2010). In potato (Solanum tuberosum) tubers, it has been shown that this critical catalytic reaction is also the limiting step during starch biosynthesis (Tiessen et al., 2002). Auxin regulates expression of SlAGPase genes (Miyazawa et al., 1999). Down-regulation of SlARF4 increases both starch content and the expression of essential genes involved in starch biosynthesis in tomato fruit, particularly genes coding for AGPase (Sagar et al., 2013). In our study, the improved starch content in the SlARF10-OE lines correlated well with increased expression of AGPase genes in starch biosynthesis (Fig. 6). The promoter regions of AGPase S1 and AGPase S2 have auxin-responsive motifs (Supplementary Table S2); however, the EMSA data showed that SlARF10 did not directly bind to the promoters of these genes, indicating that SlARF10 regulates starch accumulation by indirectly controlling the expression of SlAGPase genes.
Many studies of tomato have found that the vast majority of photoassimilates in the fruit are supplied by the leaves rather than produced de novo in the fruit (Do et al., 2010; Ruan et al., 2012): more than 80% of the total carbon of the fruit has been estimated to result from photosynthetic activity in the leaves (Cocaliadis et al., 2014). In this study, it is possible that the enhanced leaf photosynthesis observed in the SlARF10-OE lines improved the transportation of photoassimilates into the fruit, resulting in the increased starch content. Wang et al. (2005) reported that down-regulation of SlIAA9 alters auxin sensitivity and facilitates the development of vascular bundles, thereby probably increasing sink strength as well as the supply of assimilates to the fruit. Up-regulation of SlARF10 also resulted in higher fructose content at various stages of fruit development (Fig. 5), probably due to the increased starch content, which was available to be degraded into soluble sugars This finding was in accordance with previous studies demonstrating that starch content in the early stages of development determines the soluble solid content in mature fruit (Schaffer et al., 2000; Baxter et al., 2005). In our study, up-regulation of SlARF10 produced higher sucrose content, whereas down-regulation lines displayed decreased sucrose accumulation at some stages of tomato fruit development (Fig. 5). Sucrose is produced in photosynthetically active tissues (sources) and translocated to non-photosynthetic tissues (sinks) (Bihmidine et al., 2013). Li et al. (2002) reported that sucrose significantly elevates the expression of AGPase genes in tomato leaves and fruit. The increased starch accumulation in the SlARF10-OE lines may also be explained by the sucrose-enhanced expression of AGPase genes in the fruit.
It has been reported that miR160 targets and represses the SlARF10 gene; however, it has very low expression in fruit at the mature green, breaker, and red stages, and in leaves (Hendelman et al., 2012). In our study, SlARF10 driven by the 35S promoter in mature green fruit had higher expression levels compared with the WT plants (Fig. 7). The effects of SlARF10 overexpression on the accumulation of chlorophyll and sugars in the fruit and leaves may also demonstrate the low activity of miR160 during their development.
In summary, our study demonstrates that SlARF10 plays a significant role in chlorophyll accumulation during fruit development in tomato plants. Our results also shed some light on the ability of auxin to regulate starch accumulation during fruit development by altering the expression of SlARF10. However, auxin regulation of carbohydrate accumulation, especially its connection with other regulatory mechanism, remains to be determined. Future studies could focus on examining the auxin regulatory network for chlorophyll and starch biosynthesis, and determining the gene functions of relevant transcriptional factors.
Supplementary data
Supplementary data are available at JXB online.
Fig. 1. Sequence comparison of SlARF10 and AtARF10.
Fig. 2. Relative expression levels of SlARF4 in SlARF10 transgenic lines.
Fig. 3. SDS-PAGE gel demonstrating affinity purification of the recombinant GST-SlARF10 protein used for EMSAs.
Table S1. Primers used for qRT-PCR, vector construction and EMSA.
Table S2. ARF binding element (TGTCTC) in AGPase genes.
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFD0400100), the Project of Chongqing Science and Technology Commission (CSTC2015JCYJA80018), the Graduate Scientific Research and Innovation Foundation of Chongqing, China (CYS16013), the National Natural Science Foundation of China (31272165), and the National Basic Research Program of China (2013CB127106). We greatly appreciate the help of Prof. Wangjin Lu, Prof. Jianye Chen, and Prof. Jianfei Kuang in providing experimental instruments and instructions.
Glossary
Abbreviations:
- ARF
auxin response factor
- B3
B3 DNA-binding domain
- CBP1
chlorophyll binding protein 1
- CBP2
chlorophyll binding protein 2
- DDB1
UV-DAMAGED DNA-BINDING PROTEIN 1
- GLK
GOLDEN2-LIKE
- qRT-PCR
quantitative real-time PCR
- WT
wild-type
References
- Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick PW, Fernie AR, Sweetlove LJ. 2005. Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant & Cell Physiology 46, 425–437. [DOI] [PubMed] [Google Scholar]
- Beale SI. 1999. Enzymes of chlorophyll biosynthesis. Photosynthesis Research 60, 43–73. [Google Scholar]
- Bihmidine S, Hunter CT III, Johns CE, Koch KE, Braun DM. 2013. Regulation of assimilate import into sink organs: update on molecular drivers of sink strength. Frontiers in Plant Science 4, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon AB, Timko MP. 2000. yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. The Plant Cell 12, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latché A, Pech JC, Bouzayen M. 2009. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. Journal of Experimental Botany 60, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ckurshumova W, Smirnova T, Marcos D, Zayed Y, Berleth T. 2014. Irrepressible MONOPTEROS/ARF5 promotes de novo shoot formation. New Phytologist 204, 556–566. [DOI] [PubMed] [Google Scholar]
- Cocaliadis MF, Fernández-Muñoz R, Pons C, Orzaez D, Granell A. 2014. Increasing tomato fruit quality by enhancing fruit chloroplast function. A double-edged sword?Journal of Experimental Botany 65, 4589–4598. [DOI] [PubMed] [Google Scholar]
- Davies JN, Cocking EC. 1965. Changes in carbohydrate, proteins and nucleic acids during cellular development in tomato fruit locule tissue. Planta 67, 242–253. [Google Scholar]
- De Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. 2009. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. The Plant Journal 57, 160–170. [DOI] [PubMed] [Google Scholar]
- De Jong M, Wolters-Arts M, Schimmel BC, et al.. 2015. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. Journal of Experimental Botany 66, 3405–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, Song H, Hu Y, Bouzayen M, Li Z. 2012. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytologist 194, 379–390. [DOI] [PubMed] [Google Scholar]
- Devoghalaere F, Doucen T, Guitton B, et al.. 2012. A genomics approach to understanding the role of auxin in apple (Malus × domestica) fruit size control. BMC Plant Biology 12, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR. 2010. The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiology 154, 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. 2008. Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. The Plant Journal 53, 717–730. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Lerchi J, Stitt M, Sonnewald U. 1996. Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant, Cell & Environment 19, 43–55. [Google Scholar]
- Guan XX, Xu T, Gao S, Qi MF, Wang YL, Liu X, Li TL. 2013. Temporal and spatial distribution of auxin response factor genes during tomato flower abscission. Journal of Plant Growth Regulation 33, 17–327. [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ. 2016. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and Expansins during fruit ripening. Plant Physiology 171, 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendelman A, Buxdorf K, Stav R, Kravchik M, Arazi T. 2012. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Molecular Biology 78, 561–576. [DOI] [PubMed] [Google Scholar]
- Hiller RG, Wrench PM, Sharples FP. 1995. The light-harvesting chlorophyll a–c-binding protein of dinoflagellates: a putative polyprotein. FEBS Letters 363, 175–178. [DOI] [PubMed] [Google Scholar]
- Ho LC. 1996. Tomato. In: Zamski E, Scheffer AA, eds. Photoassimilate distribution plants and crops. Source–sink relationships. New York: Marcel Dekker, 709–727. [Google Scholar]
- Ho LC, Hewitt JD. 1986. Fruit development. In: Atherton JG, Rudich J, eds. The tomato crop. London: Chapman and Hall, 201–240. [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M. 2002. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. The Plant Journal 32, 603–613. [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual Review of Genetics 45, 41–59. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, et al.. 2012. Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. The Plant Cell 24, 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I. 2007. Transcriptional profiling of high pigment-2dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiology 145, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T. 2012. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytologist 194, 391–401. [DOI] [PubMed] [Google Scholar]
- Li X, Xing J, Gianfagna TJ, Janes HW. 2002. Sucrose regulation of ADP-glucose pyrophosphorylase subunit genes transcript levels in leaves and fruits. Plant Sciences 162, 239–244. [DOI] [PubMed] [Google Scholar]
- Liu N, Wu S, Van Houten J, Wang Y, Ding B, Fei Z, Clarke TH, Reed JW, van der Knaap E. 2014a. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. Journal of Experimental Botany 65, 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dinh TT, Li D, Shi B, Li Y, Cao X, Guo L, Pan Y, Jiao Y, Chen X. 2014b. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. The Plant Journal 80, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. 2004. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proceedings of the National Academy of Sciences, USA 101, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. 2005. Plastids unleashed: their development and their integration in plant development. The International Journal of Developmental Biology 49, 557–577. [DOI] [PubMed] [Google Scholar]
- Manoharan RK, Jung HJ, Hwang I, Jeong N, Kho KH, Chung MY, Nou IS. 2017. Molecular breeding of a novel orange-brown tomato fruit with enhanced beta-carotene and chlorophyll accumulation. Hereditas 154, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau B, Houck CM, Sheehy E, Hiat WR. 1994. Fruit specific expression of the A. tumefaciens isopentenyl transferase gene in tomato: effects on fruit ripening and defense-related gene expression in leaves. The Plant Journal 5, 11–19. [Google Scholar]
- Maury P, Mojayad F, Berger M, Planchon C. 1996. Photochemical response to drought acclimation in two sunflower genotypes. Physiologia Plantarum 98, 57–66. [Google Scholar]
- Miyazawa Y, Sakai A, Miyagishima S, Takano H, Kawano S, Kuroiwa T. 1999. Auxin and cytokinin have opposite effects on amyloplast development and the expression of starch synthesis genes in cultured Bright Yellow-2 tobacco cells. Plant Physiology 121, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Holdsworth WL, Klein CL, Barry CS. 2014. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. The Plant Journal 78, 1022–1033. [DOI] [PubMed] [Google Scholar]
- Nguyen CV, Vrebalov JT, Gapper NE, Zheng Y, Zhong S, Fei Z, Giovannoni JJ. 2014. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. The Plant Cell 26, 585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiadalla-Ali H, Fernie AR, Lytovchenko A, Kossmann J, Lloyd JR. 2004. Inhibition of chloroplastic fructose 1,6-bisphosphatase in tomato fruits leads to decreased fruit size, but only small changes in carbohydrate metabolism. Planta 219, 533–540. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bradley G, Pyke K, et al.. 2014. Network inference analysis identifies an aprr2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiology 161, 1476–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, et al.. 2012. uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715. [DOI] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. 2011. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. Journal of Experimental Botany 62, 2815–2826. [DOI] [PubMed] [Google Scholar]
- Rohrmann J, Tohge T, Alba R, et al.. 2011. Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. The Plant Journal 68, 999–1013. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Patrick JW, Bouzayen M, Osorio S, Fernie AR. 2012. Molecular regulation of seed and fruit set. Trends in Plant Science 17, 656–665. [DOI] [PubMed] [Google Scholar]
- Sagar M, Chervin C, Mila I, et al.. 2013. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiology 161, 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Levin I, Oguz I, Petreikov M, Cincarevsky F, Yeselson Y, Shen S, Gilboa N, Bar M. 2000. ADP glucose pyrophosphorylase activity and starch accumulation in immature tomato fruit: the effect of a Lycopersicon hirsutum-derived introgression encoding for the large subunit. Plant Science 152, 135–144. [Google Scholar]
- Schaffer AA, Petreikov M. 1997. Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiology 113, 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM. 1992. Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258, 287–292. [DOI] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan JP, Bouzayen M, Giuliano G, Chervin C. 2015. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin–ethylene balance. BMC Plant Biology 15, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Fujita K, Kikuchi K. 1974. Nutrio-physiological studies on the tomato plant. III. Photosynthetic rate on individual leaves in relation to dry matter production of plants. Soil Science and Plant Nutrition 20, 173–183. [Google Scholar]
- Tanksley SD. 2004. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. The Plant Cell 16, S181–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JH, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P. 2002. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. The Plant Cell 14, 2191–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jones B, Li ZG, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech J, Bouzayen M. 2005. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. The Plant Cell 17, 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Deng D, Shi Y, Miao N, Bian Y, Yin Z. 2012. Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Molecular Biology Reports 39, 2401–2415. [DOI] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. 2008. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. The Plant Journal 56, 432–444. [DOI] [PubMed] [Google Scholar]
- Yin YG, Kobayashi Y, Sanuki A, Kondo S, Fukuda N, Ezura H, Sugaya S, Matsukura C. 2010. Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ‘Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. Journal of Experimental Botany 61, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yan F, Tang Y, Yuan Y, Deng W, Li Z. 2015. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant & Cell Physiology 56, 2110–2124. [DOI] [PubMed] [Google Scholar]
- Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, Audran C, Roustan JP, Bouzayen M. 2014. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE 9, e84203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.