Abstract
Ribosomes are synthesized by large ribonucleoprotein complexes cleaving and properly assembling highly structured rRNAs with ribosomal proteins. Transcription and processing of pre-rRNAs are linked by the transcription-Utp sub-complex (t-Utps), a sub-complex of the small subunit (SSU) processome and prompted the investigations for the requirements of t-Utp formation and transition into the SSU processome. The rDNA promoter, the first 44 nucleotides of the 5΄ETS, and active transcription by pol I were sufficient to recruit the t-Utps to the rDNA. Pol5, accessory factor, dissociated as t-Utps matured into the UtpA complex which permitted later recruitment of the UtpB, U3 snoRNP and the Mpp10 complex into the SSU processome. The t-Utp complex associated with short RNAs 121 and 138 nucleotides long transcribed from the 5΄ETS. These transcripts were not present when pol II transcribed the rDNA or in nondividing cells. Depletion of a t-Utp, but not of other SSU processome components led to decreased levels of the short transcripts. However, ectopic expression of the short transcripts slowed the growth of yeast with impaired rDNA transcription. These results provide insight into how transcription of the rRNA primes the assemble of t-Utp complex with the pre-rRNA into the UtpA complex and the later association of SSU processome components.
Keywords: RNA polymerase I, rDNA transcription, ribosome biogenesis, SSU processome, small nucleolar RNA (snoRNA), U3 snoRNP, precursor ribosomal RNA (pre-rRNA), t-Utp complex, bhm-21
Cell growth requires the production of mature ribosomes and the assembly protein-RNA complex required for all cell growth occurs through the association of supcomplexes in a stepwise manner.
INTRODUCTION
A significant amount of cellular energy is invested in pre-rRNA transcription and processing to produce enough ribosomes to maintain cell growth (Warner 1999). In eukaryotes, the small subunit (SSU) processome forms co-transcriptionally at the 5΄ end of the pre-rRNA transcript to constitute the terminal knob of the Christmas trees (Dragon et al.2002; Gallagher et al.2004). The SSU processome is also referred to as the 90S pre-ribosome, though as originally used, this term would also include pre-60S processing factors (Trapman, Retel and Planta 1975). Recent structural studies have found that the large particle is composed of SSU processing factors assembled on the pre-rRNA containing the U3 snoRNA (Hunziker et al.2016; Kornprobst et al.2016; Zhang et al.2016; Barandun et al.2017; Chaker-Margot et al.2017; Sun et al.2017). The 35S pre-rRNA undergoes a series of endo and exonucleolytic cleavages that release the mature rRNAs from the external and internal spacers. The U3 snoRNA directly basepairs with the pre-rRNA and is required for the earliest cleavage events in the 5΄ETS (external transcribed spacer). The SSU processome forms around the U3 snoRNA and is required for the endonucleolytic cleavages that release the 20S pre-rRNA, the direct precursor to the 18S rRNA (reviewed by (Granneman and Baserga 2004; Raška, Shaw and Cmarko 2006; Henras et al.2008; Kressler, Hurt and Baßler 2010). The SSU processome, composed of subcomplexes, assembles as the pre-rRNA is folded and assembled with ribosomal proteins (Henras et al.2015; Kressler, Hurt and Baßler 2017; Peña, Hurt and Panse 2017; Tomecki, Sikorski and Zakrzewska-Placzek 2017).
Several subcomplexes have been found to constitute the SSU processome so far: t-Utp/UtpA, UtpB, UtpC, U3 snoRNP (small nucleolar ribonucleoprotein) and Mpp10-Imp3-Imp4 complex (Wehner, Gallagher and Baserga 2002; Dosil and Bustelo 2004; Gallagher et al.2004; Krogan et al.2004; Perez-Fernandez et al.2007; Kong et al.2011; Hunziker et al.2016; Yip et al.2016). These subcomplexes may represent assembly intermediates that can be seen when individual components are depleted or when smaller complexes are selectively purified. One of these, the transcription-Utp sub-complex (t-Utp), contains Utp4, 5, 8, 9, 10, 15 and Utp17/Nan1. In addition to its role in pre-rRNA processing as an SSU processome component, the t-Utps is associated with the rDNA and is required for optimal transcription of the rRNA in vivo (Gallagher et al.2004). Mapping studies found that the t-Utps directly interact with 5΄ETS at +40 from the transcriptional start site (TSS) and the second major site is located at +250 with a weaker site at +500 (Hunziker et al.2016; Kornprobst et al.2016). To differentiate the t-Utp complex that has not yet assembled into the processome and Utp4, 5, 8, 9, 10, 15 and Utp17/Nan1 assembled into the mature SSU processome, UtpA complex will be used to refer to these proteins within the SSU processome and t-Utp complex before other components of the SSU processome such as the U3 snoRNP, UtpB, and the Mpp10 complex have assembled. Cryo-EM shows that the N-terminus of Utp10 from the UtpA complex extends up from the bottom of the mature SSU processome and stabilizes the association of the UtpB complex within the larger complex (Kornprobst et al.2016; Cheng et al.2017). The t-Utp complex is conserved and similar effects on transcription have been observed in humans (Prieto and McStay 2007; Kong et al.2011). Therefore, the t-Utps provide the link between the transcription of the pre-rRNA and the later formation of the SSU processome.
The t-Utps associate with Pol5, the yeast homolog of Mybbp1a (Krogan et al.2004), which negatively regulates rDNA transcription in humans (Hochstatter et al.2012; Tan et al.2012). While in yeast, the t-Utp complex forms as pol I begins transcription (Gallagher et al.2004). As transcription progresses, Pol5 dissociates and the t-Utp complex becomes the UtpA. Pol5 directly associates with the rDNA and has not been detected in the mature SSU processome. Normally transcription of the rDNA is solely carried out by pol I (Reichel and Benecke 1984). In Saccharomycescerevisiae, the rDNA locus contains 100–200 repeats solely on chromosome XII. The regulation of transcription of the rDNA is the focus of multiple pathways. Because of the heavy investment of energy in ribosome biogenesis in unfavorable growth conditions, the activity of pol I is rapidly down-regulated but not completely (Kos-Braun, Jung and Koš 2017) while association Rrn3 to pol I promote loading onto the rDNA array and transcription (Torreira et al.2017). Deubiquitination by Ubp10, which physically interacts and stabilizes pol I, is required for optimal growth (Richardson et al.2012). Inhibiting ribosome biogenesis is an attractive chemotherapeutic target. Compounds, such as bmh-21 that interact with G-quadaplex DNA activating degradation by ubiquitination of stalled pol I (Wei et al.2018). Ubp10 interacts with several components of the SSU processome including Utp4 and Utp10, as well as Pol5 (Richardson et al.2012). Elongation mutants of pol I are particularly sensitive to bmh-21 and accumulate paused short 5΄ETS transcripts that are later elongated (Zhang et al.2010; Wei et al.2018).
Because of the head to tail arrangement of the rDNA genes, additional factors are required to maintain chromatin stability by blocking replication forks and relieving torsional stress from transcription (reviewed in Schneider 2012). Saccharomycescerevisiae maintains control over the number and homogeneity of the rDNA repeats by homologous recombination that is tightly linked to pol I transcription and starvation (Jack et al.2015). A screen for DNA sequences that increase recombination found that HOT1 is a hotspot of mitotic recombination (Keil and Roeder 1984). HOT1 is composed of the rDNA promoter and initiation site, known as the I element, and the enhancer and replication fork blocking site (RFB), known as the E element. (Fig. 1). The transcription by pol I in the direction of the recombination reporter is also required for mitotic recombination (Steven Huang and Keil 1995; Stewart and Roeder 1989; Lin and Keil 1991). E element functions to recruit pol I to the rDNA promoter outside the rDNA array (Wai et al.2001). The HOT1 element also contains the first 44 nucleotides of the 5΄ETS and previously it was shown that the first 22 nucleotides of the 5΄ETS are required for transcription (Musters et al.1990).
Figure 1.
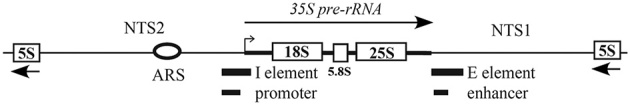
Schematic of the rDNA gene organization in Saccharomyces cerevisiae. The rDNA array encodes 100–200 repeats of the 5S and 35S rDNA genes separated by non-transcribed spacers (NTS). NTS2 contains the ARS (autonomous replicating sequence) and the I element of HOT1, which consists of the promoter and the transcriptional start. The end of the 35S pre-RNA contains the E element of the HOT1 sequence, which overlaps the 35S enhancer. The thick bar marks the transcribed spacer sequence. The 5΄ and 3΄ ends of the E and I elements are noted with the 5΄ end of the 35S pre-rRNA. The direction of transcription is depicted with an arrow above each gene. The 5΄ and 3΄ external transcribed spacers (ETS) are indicated.
Several short, sense, noncoding rRNA transcripts that begin at the pol I TSS were expressed only under optimal growth conditions. The short pol I transcripts were immunoprecipitabled by the t-Utp subcomplex proteins. Perturbations in growth decreased the levels of the short transcripts and affected the formation of the mature SSU processome. The short rDNA sequences within the HOT1 locus were sufficient to recruit the t-Utps co-immunoprecipitated to another chromosome outside the rDNA array, providing additional evidence of close association of the t-Utps with these sequences. Pol5, which was previously shown to have a role in maintaining DNA copy number (Shimizu et al.2002; Yang, Rogozin and Koonin 2003) and co-purified with the other t-Utps (Krogan et al.2004), shares some characteristics with the t-Utps. This work further details the steps in the formation of the earliest pre-rRNA processing complex that provides insights that link transcription and pre-rRNA processing.
RESULTS
t-Utps are associated with the rDNA at least at one site as seen with chromatin immunoprecipitation (ChIP: Gallagher et al.2004; Prieto and McStay 2007). To investigate this further, ChIP was carried out at three different sites in the rRNA: the rDNA promoter, the 5΄ ETS, and 25S regions of the rDNA sequence. All the tested t-Utps (Utp9, Utp15, Utp17) associated with the rDNA at all three sites (Fig. 2A). In addition, the common box C/D protein, Nop1 and Box H/ACA common protein, Gar1, immunoprecipitated the rDNA (Fig. 2A) as well as Rpa190 strongly associated with the promoter sequence, consistent with previous reports. To assess if the promoter region of the rDNA would be sufficient to recruit the t-Utps, the association of Utp8 (t-Utp) and Nop1 when these sequences were translocated to chromosome III was tested. The 5΄ end of the short rRNA transcripts overlaps with the previously described HOT1 sequence. HOT1 is a cis-acting sequence known to increase local recombination over 100-fold when placed outside the rDNA array (Keil and Roeder 1984). This and similar reporter strains have been previously used for searches for proteins that promote HOT1 recombination (Lin and Keil 1991; Kobayashi and Horiuchi 1996; Prusty and Keil 2004; Hepfer et al.2005). The rDNA sequences represented in HOT1 were further investigated to determine if they were sufficient for t-Utp:rDNA association. The HOT1 reporter construct contains the E and I sequences upstream of the his4 promoter, and the URA3 gene is integrated into a duplicated his4 gene with a mutation in the 3΄ segment in a strain called K3207 (Fig. 2B; Lin and Keil 1991). In contrast to ChIP performed on the many repeats of the rDNA array, there is only one HOT1 reporter per genome and thus quantitative PCR on purified chromatin was more sensitive. Quantitative ChIP of purified chromatin of Utp8-HA showed 40-fold enrichment at the HOT1-his4 locus compared to ACT1 used as the negative control, while ChIP with Nop1 and with the untagged parental strain showed no enrichment (Fig. 2C). Thus, the HOT1 sequences, comprising the rDNA promoter, the first 44 nucleotides of the 5΄ETS and the 3΄ end of the 35S pre-rRNA were sufficient to recruit the t-Utps outside the nucleolus to a nuclear locus.
Figure 2.
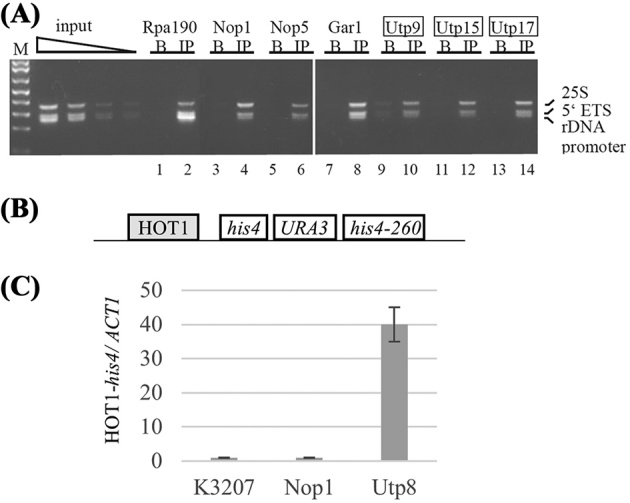
The t-Utps associate with rDNA sequences even outside of the rDNA repeats. (A) Semi-quantitative ChIPs of a subunit of RNA polymerase I (Rpa190), core box C/D snoRNP proteins (Nop1 and Nop5), core box H/ACA snoRNP proteins (Gar1), and t-Utps (Utp9, Utp15, Utp17/Nan1) tagged with HA in YPH499 were carried out. Probes used were to the rDNA promoter (primers –200 and 3΄ start), the 5΄ETS (primers +300 and oligo x) and the 25S rDNA (primers 5΄25S and oligo y; Table 1 for oligos). Lanes are marked B for beads alone in the immunoprecipitation; HA-indicates immunoprecipitation with the anti-HA antibody. (B) Schematic of the HOT1-his4 reporter from the K3207 strain. (C) Quantitative ChIP of HA-tagged Utp8 and Nop1 in K3207 using 5΄ start and 3΄ HIS4 primers that can only amplify the HOT1 sequence at the his4 locus but not at the rDNA. The signal was normalized to ACT1 and the standard deviation is indicated on the graph.
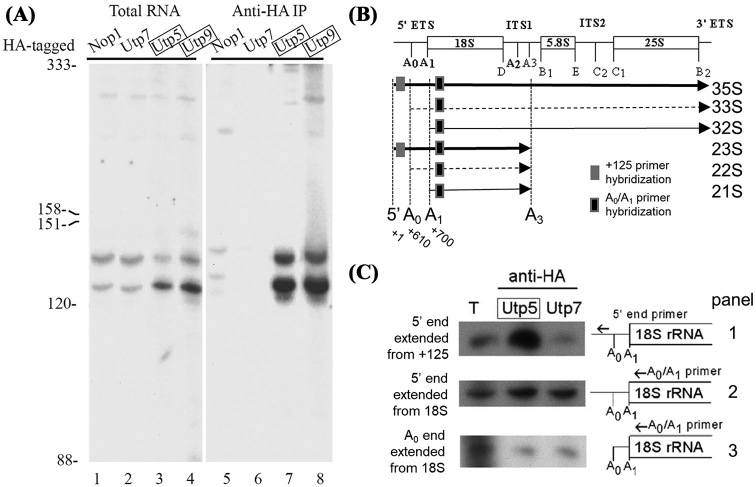
In an effort to further define the assembly of protein subcomplexes involved in ribosome biogenesis on the pre-rRNA, northern blots were probed to detect the presence of very short rRNA transcripts representing early products of transcription. RNA was extracted from early log-phase yeast and analyzed on a denaturing 8% polyacrylamide gel, followed by northern blotting with an oligonucleotide that is complementary to the first 24 nt of the pre-rRNA (Table 1). Several short rRNAs over 100 nt were detected from normally growing yeast (Fig. 3A, lanes 1–4), while RNAs smaller than 100 nt were not detected. Sequential hybridizations with probes to small stable RNAs of known size (5S, 5.8S, U3 snoRNA, tRNA-Tyr) were used to approximately size the two major short transcripts at 125 and 138 nt. Consistent with this sizing, an oligonucleotide probe complementary to nt 107–125 detected both short transcripts, while an oligonucleotide complementary to nt 120–138 detected only the longer transcript (data not shown). HA-tagging alone of the t-Utps shifted short transcripts to the shorter isoform. Similarly, neither an oligonucleotide complementary to the promoter in the NTS2 region (–24 to –47) detected the short transcripts. Therefore, the size heterogeneity most likely occurred at the 3΄ end of these transcripts and was not the result of polyadenylation as a poly-T oligonucleotide did not hybridize to them. In addition, only sense probes detected these RNAs, ruling out antisense transcription from this region of the rDNA (data not shown).
Table 1.
List of oligonucleotides.
| name | Nucleotide relative to +1 of 35S pre-rRNA | 5΄ or 3΄ of pre-rRNA | Genomic location | Primer sequence 5΄ to 3΄ |
|---|---|---|---|---|
| 5΄5S | −1249– −1269 | 5΄ | NTS2 | GGT AGA TAT GGC CGC AAC C |
| 3΄NTS2 | −956– −975 | 3΄ | NTS2 | CTT CAT AAC CTG TCA CCT TG |
| -200 | −242– −266 | 5΄ | promoter | GTG AGG AAC TGG GTT ACC CGG |
| -24 | −24– −47 | 3΄ | promoter | CTC ACA CTT GTA CTC CAT GAC |
| 3΄ start | 1–24 | 3΄ | 5΄ETS | GT CTT CAA CAA CTG CTT TCG CAT |
| 5΄ start | 1–24 | 5΄ | 5΄ETS | ATG CGA AAG CAG TTG AAG |
| +107/5’end | 107–125 | 3΄ | 5΄ETS | CTG ACG ATC ACC TAG CGA C |
| +120 | 120–138 | 3΄ | 5΄ETS | AGA CTA GGC AGA TCT GAC |
| +177 | 177–194 | 3΄ | 5΄ETS | AAT ACG ATC AAC CCA TG |
| +300 | 349–369 | 5΄ | 5΄ETS | GAA TAG CCG GTC GCA AGA CTG |
| oligo x | 611–632 | 3΄ | 5΄ETS | ACC TAT TCC CTC TTG CTA GAA G |
| oligo z | 688–704 | 3΄ | 5΄ETS | GAT AAC TAT CTT AAA AG |
| +923 | 924–953 | 5΄ | 18S | CAA TGT CTT CGG ACT CTT TG |
| 5΄ITS | 2656–2675 | 5΄ | ITS1 | ACGGTGAGAGATTTCTGTGC |
| 3΄ITS | 22826–2844 | 3΄ | ITS2 | CAAGAATTTTCGTAACTGG |
| 5.8S | 2886–2906 | 3΄ | 5.8S | GTT CTT CAT CGA TGC GAG AAC |
| 5΄25S | 5276–5296 | 5΄ | 25S | GAC TAC TTG CGT GCC TTG TTG |
| oligo y | 56511–5627 | 3΄ | 25S | CCG TTC CCT TGG CTG TG |
| 3΄HIS4 | 415–434 | 3΄ | HIS4 | GTA CGT ACT TCA CCA AGC AC |
| U3 | 3΄ | U3 snoRNA | GATCCTATGAAGTACGTCGAC |
Figure 3.
The short 5΄ ETS rRNA transcripts are associated with the t-Utps. (A) The indicated proteins were HA-tagged by chromosomal integration. Nop1 is a core box C/D snoRNA protein and Utp7 is not a t-Utp; Utp5 and Utp9 are t-Utps (boxed). RNAs co-immunoprecipitated by HA-tagged proteins were probed with an oligonucleotide complementary to nt 1–24 of the pre-rRNA (3΄ start oligo; Table 1). The blot was re-probed for tRNA-Tyr, 5S rRNA, 5.8S rRNA and the U3 snoRNA, and their sizes are indicated. Total RNA represents 10% of input from each immunoprecipitation (lanes 1–4). RNAs associating with Nop1-HA, Utp7-HA, Utp5-HA and Utp9-HA were analyzed in lanes 5–8. (B) Different 18S pre-rRNA species generated from different endonucleolytic events. U3 snoRNA dependent cleavages are in bold and the nucleotide is noted under. Internal transcribed spacer (ITS) 1 and 2 separate the mature rRNAs while the external transcribed spacers (ETS) flank the pre-rRNA. 5' ETS primer hybridization are noted in gray boxes while the A0/A1 primer hybridization is a black box. (C) Primer extensions of RNA immunoprecipitated from HA-tagged Utp7 and Utp5 and total RNA (T) were carried out with indicated primers.
Because the t-Utp complex is the earliest known SSU processome subcomplex associated with the pre-rRNA (Gallagher et al.2004) association of Utp5 and Utp9 proteins with the 5΄ short transcripts was tested. Nop1, Utp7, Utp5 and Utp9 with HA epitope-tagged at the C-terminus (Dragon et al.2002) were immunoprecipitated, RNA was extracted and analyzed by northern blotting. Both t-Utps strongly co-immunoprecipitated the short rRNA transcripts (Fig. 3A, lanes 7 and 8), while neither Nop1 nor Utp7 did so to an appreciable extent (Fig. 3A, lane 5–6). Western blots of these proteins did not show appreciable differences in immunoprecipitation (Supplemental Fig. 1) arguing against differences in protein levels accounting to differences in associating short transcripts. These proteins do associate with the U3 snoRNA and full-length 35S rRNA (Gallagher et al.2004). Thus, this analysis has uncovered previously unknown short 5΄ ETS pre-rRNA transcripts associated with a t-Utps.
To further map the 5΄ end of the short transcripts, primer extensions were carried out on immunoprecipitated RNAs associated with the Utps. The primer hybridizes the RNA and RT polymerase makes a cDNA copy until it falls off the 5΄ end allowing precise mapping when a dideoxy sequencing reaction is carried out in parallel. Two primers were used: one upstream of the A0 cleavage (within the 5΄ETS between +107 and 125) and one in the 18S rRNA (+924-932 from the beginning of transcript). Primer extensions can detect multiple RNAs with the same 5΄ end, if the target RNAs can hybrid to the primer. 35S and 23S pre-rRNAs have the same 5΄ end (A0) which would be detected by the A0/A1 primer but have different 3΄ ends (Fig. 3B gray boxes). While a primer to the 5΄ end primer (+107 nucleotide) should only detect the 5΄ of the 35S pre-rRNA. The proportion of pre-rRNA with the A1 end as measured by the A0/A1 primer would equal for both types of Utps. The 5΄ ends of RNAs associated with Utps were mapped by primer extensions of immunoprecipitations of the HA-tagged Utp5 and Utp7. Using the A0/A1 primer, two 5΄ ends of RNAs were detected associated with Utp5 and Utp7. The 5΄ ends mapped back to the TSS (5΄ end of 35S rRNA) and the A0 site (5΄ end of the 33S pre-rRNA) (Fig. 3C). Using the 5΄ end primer which hybridizes upstream of the A0 site, only the 5΄ end of the 35S rRNA can be detected. Unlike mapping with the A0/A1 primer, mapping the 5΄ with the end start primer more RNA with the 5΄ end at the TSS was detected associated with Utp5 compared to Utp7 (Fig. 3C). If the short transcripts were a result of cleavage, then the signal from primer extensions should be the same whether the A0/A1 primer or the 5΄ end primer was used. Additionally, if there was an endonucleolytic cleavage event, then there should be a corresponding signal from the hypothetical 3΄ cleavage product at 125 and 138 nucleotides but no transcript with that corresponding 5΄ end mapping to 126 and 139 nt within the 5΄ETS was detected. Thus, by size, sequential hybridization and primer extension, these transcripts corresponded to the first 125–138 nt of the 5΄ETS containing helix I and part of II (Chaker-Margot et al.2017).
Transcriptional run-on analysis can determine whether the short 5΄ ETS rRNA transcripts resulted from premature transcription termination/ pausing with exonucleolytic trimming or endonucleolytic cleavage from the processing of longer pre-rRNA transcripts. Yeast were permeabilized with sarkosyl detergent and then incubated with 32P labeled UTP. Transcriptional run-ons only allow bound polymerases to continue transcription; this allows a snapshot of active transcription that occurs during the 10 minute in vivo labeling period. The radiolabeled RNA is extracted and used to detect different regions of transcription. An increase in transcripts detected with a probe complementary to the short 5΄ ETS rRNAs would indicate that they likely result from premature transcription termination/ pausing of pol I or exonucleolytic trimming of the 5΄ETS region. Probes were generated to span the length of the 5΄ETS and the NTS2 (nontranscribed spacer) of the rDNA (Fig. 4A). Analysis of the transcripts generated in the run-on assay indicated that transcripts corresponding to the 5΄ end of the pre-rRNA, where transcription starts, were about 35% more abundant than the longer pre-rRNAs (Figs. 4B and C). The amounts of transcription detected with the two downstream probes (middle and end of the 5΄ETS) were approximately equal and are likely detecting 35S and 33S with the middle probe and only 33S and 18S with the end probe. A little signal was seen by hybridization to the NTS2 probe. These results are consistent with the short 5΄ rRNA transcripts, which can only be detected with the start probe, were active transcribed by pol I and either premature termination or pausing and trimming which allowed accumulation of the short transcripts compared to 18S rRNA.
Figure 4.
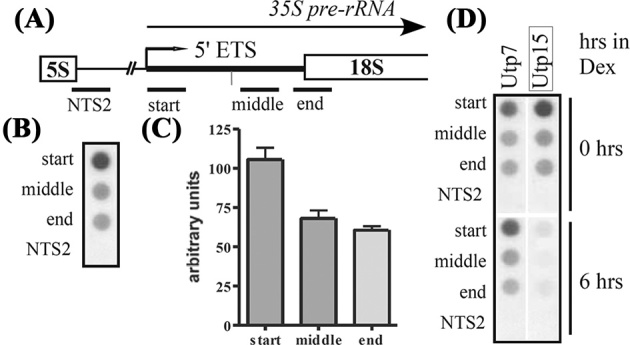
The short 5΄ ETS rRNA transcripts are actively transcribed and are dependent on t-Utps. (A) Underlines indicate probes used in the transcriptional run-on assays. The non-transcribed spacer (NTS2) probe is approximately 1 kb upstream from the pre-rRNA transcription start site. The start probe spans from 1 to 194 nucleotides of the 5΄ external transcribed spacer (ETS) pre-rRNA. The middle probe spans 350–631 nucleotides of the 5΄ETS pre-rRNA. The end probe spans 687–945 nucleotides, from within the 5΄ETS pre-rRNA and extending into the 18S rRNA. The A0 cleavage site is marked with a vertical line. (B) Radiolabeled RNA transcripts from transcriptional run-on assay were used to hybridize to membranes containing the four probes to the rDNA. (C) Quantitation of transcriptional run-on assay in Figure 3B. The signal was normalized to NTS2 and standard error is shown from three independent assays. (D) Transcriptional run-on assays were carried out on GAL: HA-UTP strains. Radiolabeled RNA transcripts from cells depleted of Utp7 and Utp15 for zero and six hours by growth in dextrose-containing media were used to probe blots containing fragments of rDNA.
Are the t-Utp subcomplex proteins required for the transcriptional events detected in the transcriptional run-on? As the t-Utps are depleted transcription of the full-length 35S decreased when measured with probes to 351–609 nt in the 5΄ETS and 5270–5670 nt within the 25S coding region (Gallagher et al.2004). Transcriptional run-on assays were carried out on yeast strains where UTP7 and UTP15 are under the control of a GAL promoter. Like most proteins involved in ribosome biogenesis, Utp15 and Utp7 are essential for growth and were depleted in dextrose for six hours, during which the doubling time does not change (Gallagher et al.2004). Transcription of all of these rRNA transcripts, including the short 5΄ ETS rRNA transcripts (start probe), was affected by depletion of Utp15, the t-Utp, but not by depletion of Utp7 (Fig. 4D). Thus, the t-Utps are required for optimal transcription of both the short and longer transcripts from the rDNA. From the primer extension there was no 5΄ end that would correspond to an endonucleolytic cleavage at nucleotide 125 or 138 from a longer rRNA as well as an increased short rRNAs actively transcribed hybridize to the start probe compared to the middle or end 5΄ETS probes further supports that the short transcripts result from increased transcription over the first 125–138 nucleotides of 5΄ETS.
To investigate whether the formation of the SSU processome affects the nature of the short rRNA transcripts and assembly of the t-Utp complex, U3 snoRNA mutants were expressed in yeast. Briefly, one copy of the gene encoding the U3 snoRNA is deleted from the genome while the other copy is under the GAL promoter. Exogenous U3 snoRNA is expressed from a plasmid (Fig. 5A; Wehner, Gallagher and Baserga 2002). When cells are shifted to glucose-containing media, chromosomal wild-type U3 snoRNA is repressed so that the sole source of U3 snoRNA is from plasmids expressed from these strains (Wehner, Gallagher and Baserga 2002). The t-Utp complex can form in the absence of the U3 snoRNA and depletion of U3 snoRNA does not affect the formation of the t-Utp complex or transcription of the rRNA (Gallagher et al.2004). The U3 snoRNA directly basepairs at three locations in the 5΄ETS of the pre-rRNA and two regions within the mature 18S rRNA using two different regions of the U3 snoRNA called 5΄ETS and Box A/A’(Fig. 5B; Yeh and Lee 1992; Sharma and Tollervey 1999; Dutca, Gallagher and Baserga 2011). The first region of complementarity is in the 5΄ETS which corresponds to nucleotides 281 to 290 and the second is at 469 to 479 of the pre-rRNA. The U3 snoRNA Box A/A' directly basepairs to the 5΄ end of the 18S rRNA which is 701 to 725 nucleotides from the 5΄ end of the 35S pre-rRNA and at a second site within the 18S rRNA. Unexpectedly, a mutation in the first 5΄ETS:U3 snoRNA interaction abolished expression of both the long and short forms of the 5΄ETS transcripts (Fig. 5C top lanes 1, 4, 7 and 10). In contrast, expression of the Box A mutant U3 snoRNA shifted the two short transcripts to primarily the longer form, while the distribution of short transcripts associated with other Utps, Utp5 and Utp9 did not change (Fig. 5C bottom lanes 8, 9, 11 and 12). Therefore, when present, the t-Utps associated with the short transcripts that were stably expressed. Mutations interrupting the first U3:5΄ETS at +281 of the 5΄ETS but Box A U3:5΄ETS basepairing interaction affected levels of the short transcripts.
Figure 5.
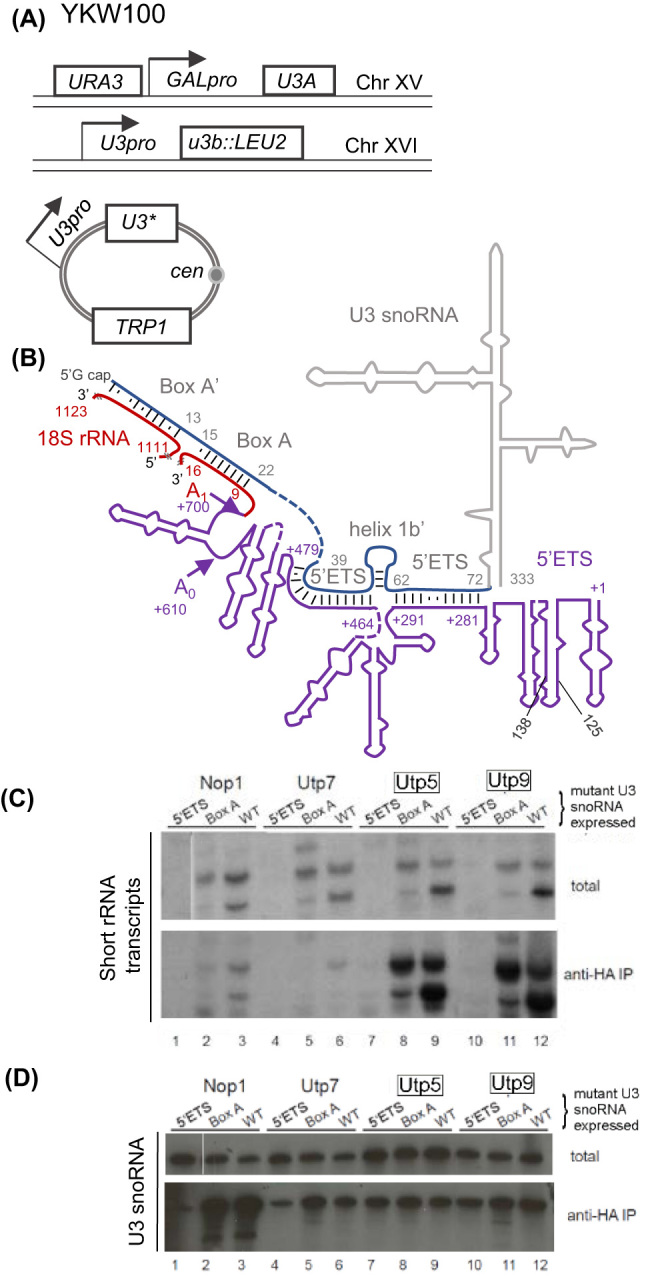
Early 5΄ETS pre-rRNA-U3 snoRNA interaction is required for the production of short transcripts and association with the other Utps with the U3 snoRNA. (A) Schematic of yeast strain YKW100 containing U3B on chromosome XVI replaced with LEU2, U3A placed under the control of the GAL promoter. Different plasmids containing U3 mutants (U3*) are expressed under the endogenous U3 promoter. (B) Schematic of the U3 snoRNA and 5’ETS of the rRNA basepairing. Sequences comprising the mature 18S rRNA are red while the 5΄ETS is in purple and relevant nucleotides are in matching colors numbering begins at the first nucleotide of that region (ETS starts at +1). Canonical basepairing is noted a black dash and other nucleotide interactions are a dot. Regions of the U3 snoRNA interacting with the pre-rRNA are blue and the body of the U3snoRNA not directly interacting with the pre-rRNA is grey. Cleavage sites are noted with arrows. (C) RNA was co-immunoprecipitated from yeast with C-terminal HA-tagged Nop1, Utp7, Utp5 and Utp9 expressing mutant U3 snoRNA (5΄ETS or Box A binding sites) or wild-type U3 snoRNA (WT). Total RNA represented 5% of lysate immunoprecipitated. Northern blots were probed with the start oligo complementary the 5΄ end of the pre-rRNA. t-Utps were boxed. (D) Northern blot in part C was reprobed to detect co-immunoprecipitated U3 snoRNA expressed from a plasmid, extracted yeast expressing C-terminal tagged Nop1, Utp7, Utp 5 and Utp9. t-Utps are boxed.
While the Nop1, Utp7, Utp5 and Utp9 stably associated with both 5΄ETS and Box A mutant U3 snoRNAs (Fig. 5D), there were changes in protein-protein interaction when mutant 5΄ETS U3 snoRNA was expressed but not the Box A mutant. The larger SSU processome around the 5΄ETS U3 snoRNA mutant did not form properly as shown by the loss of Nop1, Utp7 and t-Utps association with Mpp10 protein component of the SSU processome (Supplemental Fig. 2 lane 1, 4, 7, 10 and 14). Protein levels of Nop1, Utp7, 5, 8 and 9 in the strains expressing the U3 snoRNA mutants did not change, and the 5΄ETS interaction was required for the Utps to associate with Mpp10 (Supplemental Fig. 2).
Transcription of the rDNA in yeast can be driven by either pol I or RNA polymerase II (pol II) (Nogi, Yano and Nomura 1991). To determine whether the short 5΄ ETS rRNA transcripts were dependent on the type of polymerase transcribing the rDNA, RNA from yeast that contained an inactivated pol I or when a pol II promoter drove transcription was analyzed. The chromosomal rDNA was deleted from the genome and rDNA encoded on a plasmid was introduced (Wai et al.2001) and RNA from yeast expressing rDNA from different plasmids bearing either pol I or pol II promoters was examined by northern blot (Fig. 6A). Levels were quantitated and compared to the levels of short transcripts expressed in the YPH499 strain or to the levels of 5.8S rRNA or the U3 snoRNA. Only when the plasmid rDNA was transcribed by pol I (NOY908) did the short 5΄ ETS rRNA transcripts stably accumulate, although less than short transcripts from the YPH499 strain containing chromosomal rDNA (Fig. 6A, lane 1 and 3). In contrast, when the GAL promoter (pol II) drove the expression of the rDNA from a plasmid, did not exhibit the short 5΄ ETS rRNA transcripts (Fig. 6A, lane 2). To control for the possibility that the chromosomal context of the rDNA locus itself was important, the presence of the short transcripts was assessed in yeast where the rDNA was transcribed by a cryptic pol II promoter in the rDNA repeat in the presence of deleted large subunit of pol I (Oakes et al.1999). Yeast that have switched to using this pol II promoter are called PSW (promoter switch) strains. To determine if the short 5΄ ETS rRNA transcripts could be detected in a promoter-switched strain, PSW/NOY878. Northern blots indicate that the PSW strain, driven by pol II, also did not transcribe the short 5΄ ETS rRNA transcripts (Fig. 6A, lane 4). The short transcripts were not detected when two different pol II constructs drove transcription of the rDNA or when the rDNA was encoded on a plasmid or at its endogenous chromosomal location. As a control, Rpa43, a pol I component, was also found at the rDNA and its association did not require the presence of the t-Utps. (Supplemental Fig. 3A). Likewise, Rpa43 did not associate with the short 5΄ETS transcripts, the t-Utps or the U3 snoRNA (Supplemental Fig. 3B-D). Thus, the t-Utp and snoRNP core protein association with the rDNA was distinct from that of RNA polymerase I. Thus, the short 5΄ ETS rRNA transcripts were apparent only when the rDNA is transcribed by pol I, although to a lower level when expressed from a plasmid.
Figure 6.
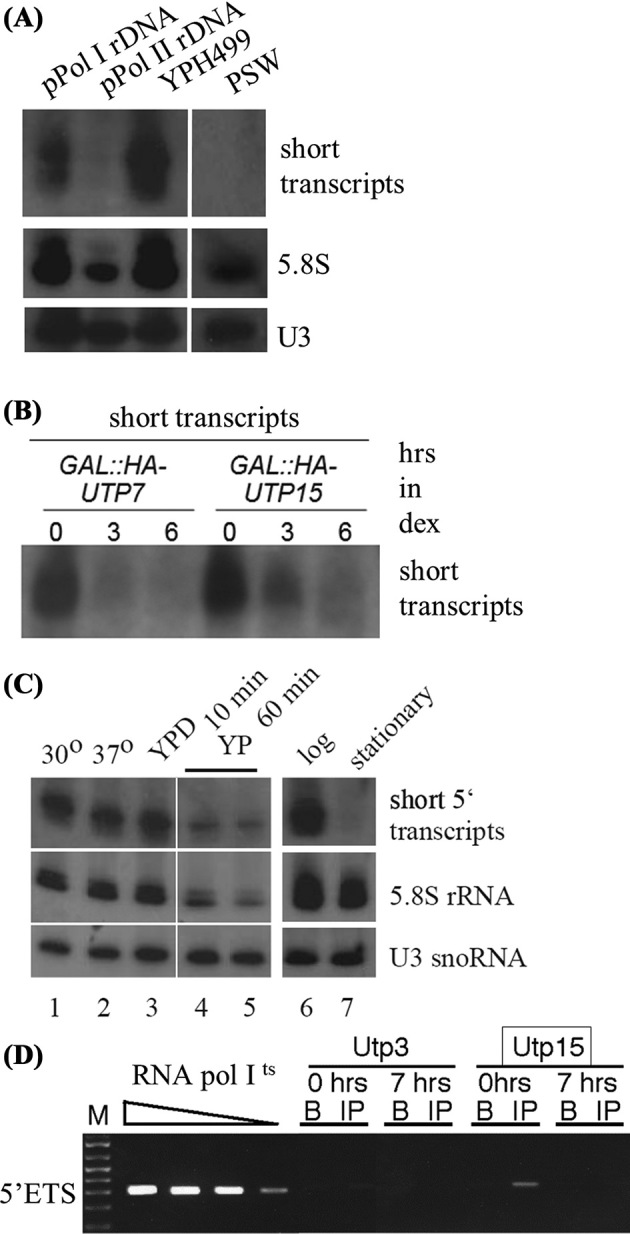
The short 5΄ ETS rRNA transcripts are not transcribed by RNA polymerase II. (A) Northern blot of total RNA from cells expressing the rDNA from a cryptic RNA pol II promoter (PSW/ NOY878) or from plasmid-encoded pPol I rDNA (NOY908) and pPol II rDNA (NOY891). The blot was probed with radiolabeled +107 oligonucleotide. (B) Northern blot of total RNA from yeast grown in galactose for 0, 3 and 6 hours to deplete UTP7 and UTP15 and hybridized to detect the short transcripts. (C) Northern blot of total RNA from YPH499 cells grown at 30°C in YPD then shifted to 37°C or media with no dextrose (YP) or grown to stationary phase (OD600 5.5). (D) ChIPs with 5΄ETS primers of Utp3 and Utp15-HA in NOY504, carrying a temperature sensitive RNA pol I were shifted to 37°C for 7 hours to repress rDNA transcription. Input DNA was diluted 2.5-fold.
The requirement of the pol I promoter and the specific association of the t-Utps with the short 5΄ETS rRNA transcripts led us to investigate if the t-Utps were specifically required for the accumulation of the short transcripts. Utp7 and Utp15 (t-Utp) were depleted and the levels of the short transcripts were assessed at three and 6 hours after transcription of these genes were repressed (Gallagher et al.2004). Surprisingly, depletion of both proteins reduced the levels of the short 5΄ETS rDNA transcripts (Fig. 6B), indicating that normal levels of these proteins are required for their accumulation.
Various stress conditions are known to decrease transcription of the rDNA in yeast, including starvation and growth to stationary phase (reviewed; Warner 1999). The accumulation of the short 5΄ ETS rRNA transcripts in these stress conditions was tested. Yeast were grown at 30°C and then shifted to 37°C for heat shock, shifted to a medium lacking dextrose to achieve starvation conditions and grown to an OD600 of 5.5 to achieve stationary phase. RNA was isolated from stressed cells and then analyzed for the presence of the short 5΄ ETS rRNA transcripts. After six hours of heat shock, the steady-state levels of the short 5΄ ETS rRNA transcripts were not altered (Fig. 6C lanes 1 and 2). In contrast, inducing both starvation and growth to stationary phase decreased the levels of the short 5΄ ETS rRNA transcripts (Figure 6C lanes 4, 5 and 7). Under several conditions where rDNA transcription is decreased, the stable accumulation of short 5΄ ETS rRNA transcripts was reduced, again providing support that the expression of the 5΄ ETS short transcripts is related to pol I transcription.
To determine if functional pol I was required for association of the t-Utps to the rDNA, Utp5 (t-Utp) was HA-tagged in a temperature sensitive yeast strain containing a deletion of Rrn3, a nonessential subunit of pol I, that reduces the stability of the complex (NOY504 (Nogi et al.1993)). At the nonpermissive temperature, pol I transcription is greatly reduced to less than 5% of levels at the permissive temperature (Gallagher et al.2004). ChIP of Utp5 and of a non-t-Utp, Utp3, was carried out. When shifted to the nonpermissive temperature for 7 hours, Utp15 no longer associated with the rDNA (Fig. 6D, compare IP at 0 vs. 7 hours). As published previously, Utp3 was not associated with the rDNA under any conditions (Gallagher et al.2004). Under conditions where pol I is not expressed, the t-Utps no longer associated with the rDNA.
To investigate whether the short 5΄ transcripts can function in trans, the 18S, 5.8S and 25S were deleted from plasmid-encoded rDNA using the pol I promoter (pNOY373) in a high expression plasmid so that the entire 5΄ETS could be expressed without the mature rRNAs. Three different deletions of pNOY373 were constructed. The long deletion has the entire 5΄ETS, the beginning of 18S, the 3΄ ETS and the transcriptional terminator (Fig. 7A). The next deletion deleted most of the 3’ETS but maintained the terminator stop (medium deletion), the short deletion plasmid deleted everything after the 129th nucleotide of the 5’ETS (short). The deletion plasmids was transformed into pPol II strain (NOY891) from Fig. 6A that did not express the short transcripts. NOY891 was also transformed with an empty plasmid and full-length pPol I rDNA (pNOY373) (Fig. 7A). Yeast were grown overnight in selective media containing galactose and an equal number of cells were serially diluted onto media that would maintain the plasmids (Fig. 7B). The pol II promoter driving the rDNA is the GAL promoter and the yeast grow slower than yeast with pol I transcribing the rDNA. Surprisingly, ectopic expression of the short transcripts reduced the growth of yeast that otherwise did not express the short transcripts. The effect was extubated with the shortest deletion. The negative effect of growth was detected in the overnight cultures because yeast carrying the long deletion plasmid grew10-fold less dense (an average OD600 0.14 after 24 hours of growth) than yeast with pPol II or pPol I rDNA plasmids (OD600 1.24 and OD600 1.73, respectively). The dominant negative effect may be a result of sequestering the t-Utps from SSU processome assembly. To determine if there was a dosage effect from expression of the deletion plasmids, rDNA segment from the medium deletion plasmid was cloned onto a centromeric plasmid (Fig. 7C). The growth defect was less severe when the single copy plasmid was expressed but not as decreased as an empty plasmid.
Figure 7.
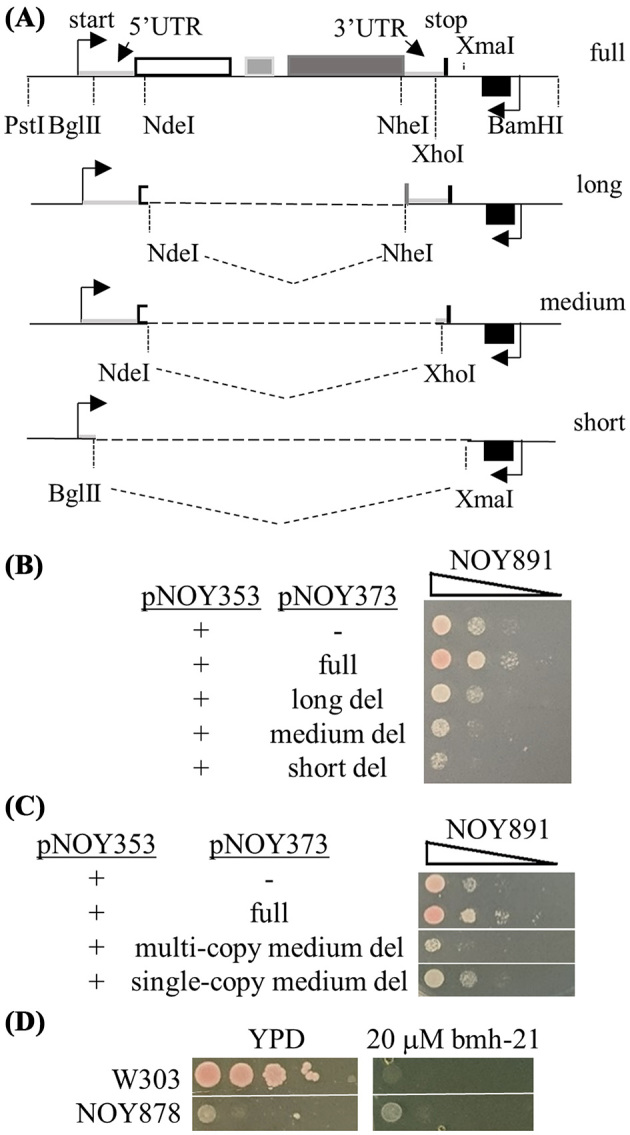
Ectopic expression of short 5΄ETS slows growth of impaired yeast. (A) Deletion schematic of the 35S rDNA from plasmid pNOY373. The 9.1Kb rDNA repeat was cut with restriction enzymes and religated to generate the long, medium and short deletions. The 5΄ and 3΄ UTR is light grey line, the 18S is white box, 5.8S is a grey box, 26S is a dark grey box and the 5S is a black box. The transcriptional starts are noted with arrows and the transcriptional stop is a thick black vertical line. (B) Serial dilution of yeast with rDNA expressed from the plasmid and the short transcripts (the rDNA promoter and 5΄ETS) expressed from high copy plasmid. NOY891 carrying pNOY353 (GAL-35S rDNA) was transformed with an empty plasmid (-), pNOY373 (full) or pNOY373 that had the 18S, 5.8S and 25S deleted (long, medium or short). Yeast were grown to saturation and an equal number of cells diluted ten-fold, spotted onto selective media (YM+AU) containing galactose, and grown for three days. (C) Serial dilution of yeast with rDNA expressed from the plasmid and the short transcripts (medium deletion) expressed from high or low copy plasmid. (D) Wild-type and PSW yeast growth in response to bmh-21 on YPD.
To determine if the expression of the short rRNAs, yeast were exposed to bhm-21 which inhibits elongation of pol I by stabilizing G-quadaplex structures. However, bhm21 did not inhibit yeast growth under the conditions required to maintain the plasmids (Supplemental Fig. 5). It is not uncommon for differences in drug sensitivity in minimal media compared to rich media because biochemical target pathways may not be active (Rong-Mullins et al.2017) or the transporter may not be expressed (Webster and Dickson 1983; Cheng et al.2000). Using the PSW yeast strain that has pol I mutated (NOY878) and the genomic rDNA is expressed using a cryptic pol II promoter present in the repeats, the effect of a pol I inhibitor was tested. At same concentrations that fail to affect yeast growth in minimal media, wild-type yeast growth was completely inhibited in YPD (Fig. 7D) while the PSW yeast were not affected by bmh-21.
Several recent cyro-EM structures have placed the Utp4, 5, 8, 9, 10 and 17 on the 5΄ETS (Tulha et al.2010; Kornprobst et al.2016; Calviño et al.2017; Chaker-Margot et al.2017) and in vivo crosslinking (CRAC) has precisely mapped UtpA component interactions on both the 5’ETS and U3 snoRNA (Baßler et al.2017; Calviño et al.2017). Previous work had found Pol5 stably associated with proteins of the UtpA complex (Krogan et al.2004) but behaves differently than the other components. To determine if Pol5 shared known characteristics of t-Utps by testing association with other t-Utps and SSU processome components, rDNA and the short 5’ ETS transcripts. By co-immunoprecipitation Pol5 associated with the t-Utp, Utp17 (Fig. 8A lane 3), but not with the non-t-Utp, SSU processome component, Mpp10 (Fig. 8A lane 3). In contrast, Utp5 co-immunoprecipitated both Mpp10 and Utp17 (Fig. 8A lane 4), as do all t-Utps (Gallagher et al.2004). This suggests that Pol5 should not be considered a component of the mature SSU processome, but a t-Utp accessory component which dissociates before the SSU processome is formed. Similar to depletion of a t-Utp, Pol5 depletion blocked the SSU processome dependent pre-RNA cleavages (Supplemental Fig. 5A and B). Consistent with previously published results (Shimizu et al.2002), Pol5 was enriched four-fold by ChIP to the 5΄ETS compared to mock ChIP (Fig. 8B).
Figure 8.
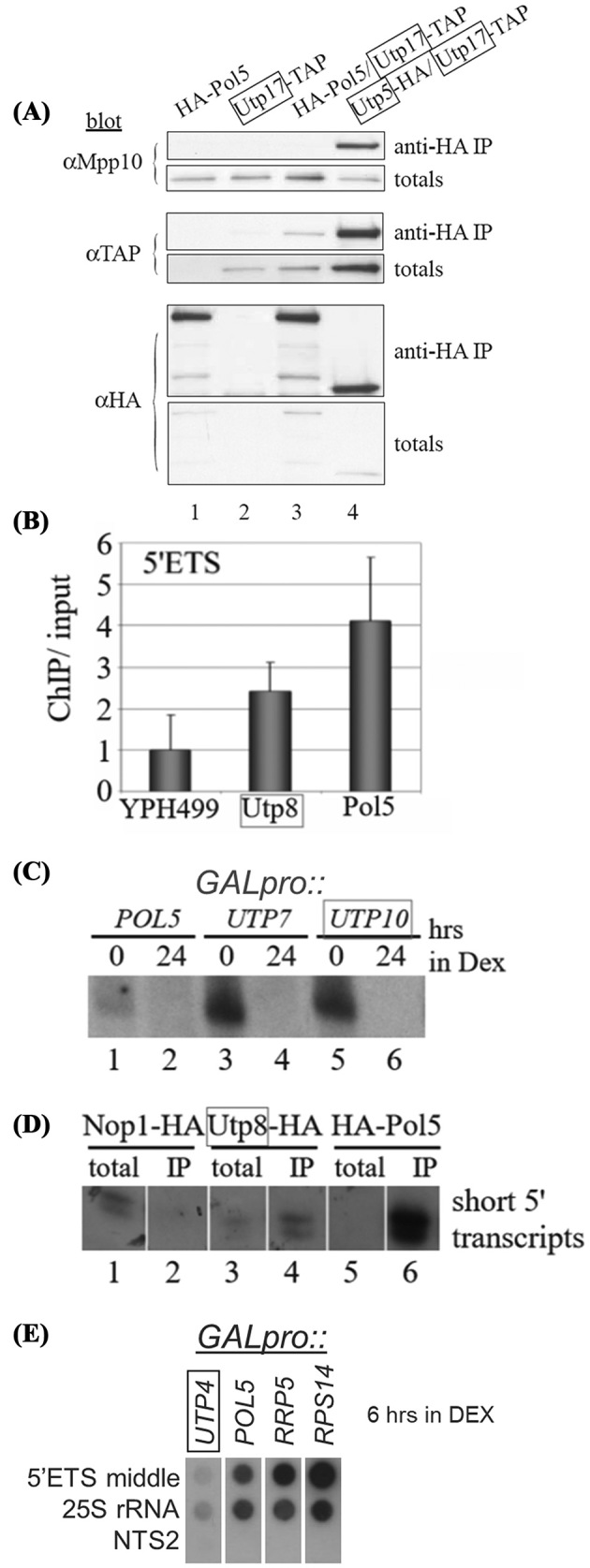
Pol5 association with the t-Utps and the SSU processome. (A) Anti-HA immunoprecipitation from yeast containing HA-Pol5 or Utp5-HA and Utp17-TAP proteins. Proteins were tagged at either the C-terminus or the N-terminus in YPH499. Western blots of co-immunoprecipitation were sequentially blotted with antibodies to HA tag, TAP tag and Mpp10 and are labeled to the left of the blots. (B) Quantitative PCR of ChIP from yeast expressing HA-tagged Utp8 and
Figure 8.

Pol5. Primers to the 5΄ETS amplified chromatin isolated from each tagged strain, normalized to ACT1 and standard deviation noted. (C) Northern blot of short RNAs when Pol5, Utp7 and Utp10 were depleted for 24 hours. (D) Northern blot of RNA extracted from anti-HA immunoprecipitations. HA-tagged proteins were immunoprecipitated and RNA was separated on an 8% polyacrylamide gel. Oligonucleotide +107 was radiolabeled and blotted to detect the short 5΄ ETS pre-rRNA transcripts.
To determine whether Pol5 was required for accumulation of the 5’ETS short rRNA transcripts, GAL promoter was placed upstream of POL5 gene and northern blots were performed as in Fig 2A. Interestingly, the levels of these rRNAs were lower than that in the GAL:UTP7 or GAL:UTP10 strains before depletion (Fig. 8C). This may be due to over-expression of Pol5 similar to what has been observed for Mybbp1a (Tan et al.2012). However, depletion of Pol5 increased the levels of the 35S pre-RNA in contrast to depletion of t-Utp components which demonstrated reduced transcription of the rDNA (Supplemental Fig. 6A). Before depletion of Pol5, the abundance of pre-rRNA precursors was different compared to the wild-type control. Depletion of Pol5 also resulted in a mild decrease of 5.8S levels (Supplemental Fig. 6B). The levels of short rRNAs immunoprecipitated were much higher by Pol5 than Utp8 (Fig. 8D). Transcriptional run-on from yeast depleted of Pol5 showed that levels of pre-rRNAs complementary to the middle of 5΄ETS and 25S rRNA did not reduce to the levels when Utp4 was depleted (Fig. 8E). Pol5 is, therefore, is a t-Utp complex accessory factor because it associated with other t-Utps, the rDNA, the short 5΄ rRNA transcripts but was not found associated with components of the later SSU processome.
DISCUSSION
While investigating the factors required for the procession of the t-Utps into the mature SSU processome, the existence of short stable transcripts comprising the first 125–138 nt of the 35S pre-rRNA was uncovered. These transcripts were associated with the t-Utps and the protein, Pol5, but not with other components required for SSU biogenesis. Overexpression of the short transcripts impaired growth which was dose-dependent. The 5΄ETS-U3 snoRNA interaction was required for the stability of these transcripts as well as the transcription by pol I. The nucleolar run-on experiments suggest that these transcripts resulted from increased pol I dependent transcription within the rDNA 5΄ETS. Depletion of a t-Utp led to both a decrease in transcription of the rRNA and of the short 5΄ETS rRNA transcripts. The t-Utps associated with the rDNA at several sites and were even associated with the rDNA sequences were translocated (HOT1).
What is the nature of the short 5΄ ETS rRNA transcripts? These results suggest that they specifically a pol I products that pause or stop early in the 5΄ETS. If they are degradation products then, there may be an exonuclease recruited by pol I like splicing factors and the CTD of pol II or an RNA modification that permits specific cleavage of the pre-rRNA that is not present when the pre-rRNA is transcribed by pol II, stress, mutations in the U3 snoRNA, or depletion of t-Utps or Pol5. However, the lack of the corresponding 3΄ cleavage product argues against this explanation. If the short transcripts are a result of a pause site, one has not yet been posited for pol I but has been observed for pol II where so-called abortive transcripts of 20 nt are often evident (Wade, Hall and Struhl 2004). The short stable transcripts have not yet been identified for pol I before perhaps because most studies were based on reporter constructs or have used probes that would not detect them in yeast (Tschochner 1996; Keener et al.1998; Oakes et al.1999). The 5’ end of the short transcripts map to the known pol I TSS for the 35S pre-rRNA ruling out heterogeneous TSSs. However, it is unknown whether the 5΄ ETS transcripts result from a single pause event followed by variable exonucleolytic trimming of the 138 nucleotide RNA to 125 nucleotide RNA or from multiple pause events or are further extended. The 5’ ETS is extensively structured (Yeh and Lee 1992; Tulha et al.2010; Kornprobst et al.2016; Calviño et al.2017; Chaker-Margot et al.2017) and the 138 and 125 positions are nearly opposite of each other in the second stem loop of the 5’ETS (Fig. 5B). The 5’ETS is bound and remodeled directly by RNA helicases (Sardana et al.2015). Ectopic expression of a series of deletions of the 5’ETS decreased the growth of yeast with impaired rDNA transcription but not normal yeast. The drug bmh-21 blocks pol I elongation which induces ubiquitintation and degradation of pol I. In vitro transcription assays with pol I elongation mutants show pausing at an undetermined site in the 5’ETS (Viktorovskaya et al.2013) are approximately the size of the short transcripts described here. These transcripts are later extended to then end of the reporter. Bmh-21 also causes pausing (Wei et al.2018) and yeast that use pol II to transcribe the rDNA were not sensitive. Because bmh-21 was not effective on minimal media suggesting that the transporter is not expressed, the combination of the ectopic expression of the short transcripts and bmh-21 could not be directly tested. If ectopic short transcripts also blocked elongation then pol I would be ubiquitintated. The physical interaction of Ubp10 and the t-Utps, Pol5, and other components of the SSU processome (Richardson et al.2012) support this as an important checkpoint of ribosome biogenesis as previously suggested (Wei et al.2018). The lack of growth inhibition in wild-type yeast points to the robustness of the system and only when the rDNA was expressed from outside it's normal repetitive genomic location did expression of the short transcripts alone from a plasmid negatively affects the growth of yeast, suggesting that excess short transcripts or when expressed ectopically may sequester processing factors from productive rRNA transcripts by preventing recycling of factors.
An intriguing question remains of how the 5’ETS and U3 snoRNA interaction, which is 3’ of the short transcripts affects the levels of these short rRNA transcripts. Without the 5΄ETS-U3 snoRNA base-pairing interaction, no stable 5'ETS transcripts were detected despite the first 5΄ETS-U3 snoRNA base-pairing being located 143 nucleotides downstream of the longest short rRNA transcript of 138 nucleotides. Based on the secondary structure of the RNA (Chaker-Margot et al.2017), the first helix and most of the second helix of the 5’ETS comprise the short 5’ ETS rRNA transcripts. However, the secondary structure based on full-length 5’ETS and does not rule out conformational changes that may occur as the SSU processome matures as there are no current structures of the t-Utp complex alone. The second interaction between the U3 snoRNA and the 5’ETS is 71 nucleotides upstream of the A0 cleavage site (nt 551) in the 5’ETS of the pre-rRNA and may represent a checkpoint for the cells to degrade pre-rRNAs that will not be properly processed by an incompletely assembled SSU processome. The reporter constructed used to measure pauses in elongation by pol I notes several sites of pausing at the approximate size of the short transcripts described here (Wei et al.2018). The U3 snoRNA is required for the A0, A1 and A2 cleavages, and without it, the SSU processome fails to form. The t-Utp complex formation is independent of the U3 snoRNA and unlike other Utps, the t-Utps did not require the 5΄ETS or helix 1b' sequences of the U3 snoRNA for association with each other or the pre-rRNA. The U3 snoRNA is at the core of the SSU processome and basepairs directly to regions in the 18S rRNA, preventing the formation of the central pseudoknot.
Studies in human cells have not detected any short transcripts from the 5’ ETS (Kuhn and Grummt 1992; Stefanovsky et al.2006; Moss et al.2007), while others have argued on the basis of kinetics that there might be (Panov, Friedrich and Zomerdijk 2001; Panov et al.2006). Notably, short RNAs (snPI RNAs) transcribed by pol I from the start site of transcription have been observed in HeLa and other metazoan cells (Benecke and Penman 1977; Reichel et al.1982; Reichel and Benecke 1984). It would not be unexpected to find short 5’ETS transcripts in mammalian cells stabilized by binding to the t-Utps, as there are orthologous t-Utps in human cells with similar functions in pol I transcription and pre-rRNA processing (Prieto and McStay 2007).
However, polyadenylated RNAs transcribed from the 5’ETS have been described (Schneider et al.2007). These RNAs were observed in the presence of an elongation defective pol I and are subsequently degraded by the TRAMP exosome (Hage et al.2010). These aberrant RNAs start at the transcription start site and end as a result of cleavage approximately 270 nt from the 5’ end of the 18S rRNA, and therefore are much longer than the ones that were described here and are polyadenylated (600 to 2000 nt vs 125–138 nt). In contrast, the short 5΄ ETS rRNA transcripts described here were neither polyadenylated nor did they immunoprecipitate subunits of the TRAMP complex (data not shown). Thus, the previously described longer 5’ polyadenylated 5’ETS RNAs are likely distinct from the short 5’ ETS rRNA transcripts described here.
The association of the t-Utps with the short transcripts signify the earliest complex. Detailed structures of the SSU processome published to date are a static picture of the pre-ribosome assembly process (reviewed; Barandun, Hunziker and Klinge 2018). These large ribonucleoprotein complexes have been purified using two differentially tagged proteins each from the UtpA and UtpB complexes and therefore represent a later step in the process of ribosome assembly. Additionally, as the presence of the short rRNA transcripts appears to signal active growth, purification of the SSU processome after early-log phase (defined here as 0.2–0.5 OD600) would not be optimal for identifying the earliest processing complexes. Using tagged RNAs would also not have identified the short 5’ETS RNAs because the smallest reporter/ probes are longer than the short transcripts (Schneider et al.2007; Hunziker et al.2016; Zhang et al.2016; Barandun et al.2017; Chaker-Margot et al.2017). Intriguingly, in vivo crosslinking (Hunziker et al.2016) found that the likely order of the t-Utps loading on to the pre-RNA is Utp9, Utp8 and Utp17 first then Utp4, Utp15 and Utp10 and Utp5, if 5’ to 3’ position on the 5’ETS is reflective of their order of association. Utp10 has an N-terminal domain that reaches up into the SSU processome that may set up the loading of UtpB complex (Hunziker et al.2016). All these t-Utp interactions can account for interactions with the 125–138 nucleotide short RNAs seen here. These proteins also crosslink to the U3 snoRNA centering on helix 1b’ and helix 3 (Hunziker et al.2016). Utp10 also interacts at further 3’ nucleotides of the U3 snoRNA and may act as the bridge tethering the UtpA, UtpB and U3 snoRNA complexes (Hunziker et al.2016; Chaker-Margot et al.2017).
The t-Utps do not interact directly with pol I but because of their close association with the rDNA and their binding to the first nucleotides of the 5’ETS, they are in a prime location to contribute to regulation of elongation or provide an important checkpoint for ribosome biogenesis to proceed. Indeed, translocation of the rDNA promoter contained in the HOT1 sequence to another chromosome was sufficient for ectopic recruitment of the t-Utps to chromatin. The transcription of rDNA forms the nucleolus and has extensive intra rDNA interactions and notably between NTS and the 35S gene (Mayan and Aragón 2010) that may explain the diffuse t-Utp association across the repeat and possible recycling of components linked to the head to tail arrangement of the rDNA repeats. Purification of proteins in the t-Utp have found a mixture of complexes containing these proteins (Kornprobst et al.2016) and may represent the t-Utp complex conversion into the UtpA as transcription of the 5’ETS progresses. Purification of the pol I deubiquintase, Ubp10, identified Utp4, Utp10 and Pol5 as highly enriched (Richardson et al.2012) but none of these proteins have been identified as ubiquitinated. All published structures of the SSU processome to date have not identified Pol5, supporting that these structures represent a fully formed SSU processome and not the t-Utp complex, as defined here. As transcription of the 5’ETS, other subcomplexes are incorporated and leave as the 18S rRNA matures and the ribosomal proteins replace processing proteins (Bernstein et al.2004; Jakob et al.2012).
The short rRNA transcripts described here are markers of exponentially growing cells. The short transcripts were not detected in cells starved either by glucose depletion, grown to stationary phase or depletion of ribosome biogenesis factors. Starvation can be mimicked by the addition of rapamycin to inhibit mTOR. Within 10 minutes of starvation, the Rrn3 dependent association of pol I to the promoter drops to 30% (Torreira et al.2017) as do the short transcripts as shown here. The downregulation is dependent on Paf1, which stimulates in vitro pol I transcription (Zhang et al.2009). Rapamycin-induced rapid downregulation of ribosome biogenesis including phosphorylation. Pol5 and several Utps are rapidly phosphorylated upon the addition of rapamycin (Oliveira et al.2015). In particular, serines 789 and 800 of Pol5 are phosphorylated and are conserved across yeast but not in humans. However, the mouse Pol5 ortholog, Mybbp1a is phosphorylated in response to rapamycin at the C-terminal end (Yu et al.2011) and highlights the conservation of the essential process of ribosome biogenesis. Mutations in the human homolog of Utp4, a t-Utp, cause Indian Childhood Liver (Zhao et al.2014; Sondalle, Baserga and Yelick 2016). Other diseases of ribosome biogenesis and upregulation of ribosome biogenesis in cancer point to the allocation of resources as a critical point of regulation in cell growth and apoptosis.
MATERIALS AND METHODS
Strains, media and plasmids
Yeast strains expressing triple HA carboxyl-tagged proteins (KanR) in YPH499 and YJV100 were constructed by homologous recombination in the genome as previously described (Longtine et al.1998). The UTP genes were placed under the control of the GAL promoter to make GAL:HA-UTP strains (Longtine et al.1998). YPH499 containing Utp8-HA or Nop1-HA were first backcrossed with YPH500 and then crossed to K3207 (Lin and Keil 1991), sporulated, and KanR, URA+, HIS+ strains were used for further study. Unless otherwise stated, cells were grown at 30°C in YPD. YJV100 was described in (Venema et al.1995). YJV100 cells were grown in YPG/R (2% yeast extract, 1% peptone, 2% galactose and 2% raffinose) and then shifted to YPD (2% yeast extract, 1% peptone and 2% dextrose) for three to 6 hours. GAL:HA strains were grown in the same way as YJV100. NOY504 (Nogi et al.1993), NOY878(Oakes et al.1999), NOY891 and NOY908 (Wai et al.2001) strains were grown as described by others. YKW100 were grown in SC-Gal/Raf-Trp and then shifted to SC-glucose for 24 hours to deplete endogenous U3 snoRNA (Wehner, Gallagher and Baserga 2002). Mutant U3 snoRNA was expressed from plasmids (Lee and Baserga 1997; Wormsley et al.2001). The pNOY373 containing the rDNA repeat with the pol I promoter was cut with NdeI and NheI, the overhangs were filled in the Klenow and religated. The long deletion plasmid contained the rDNA promoter 206 nucleotides upstream of the TSS, all 699 nucleotides of the 5’ETS, 30 nucleotides the mature 18S, 174 of the 3’ end of the 25S region. The medium deletion was cut at NdeI and XhoI, filled in and religated. The short deletion plasmid was cut at BglII and XmaI, filled in and religated. The medium deletion plasmid was cut with PstI and BamHI and cloned into pRS315. All plasmids were confirmed by sequencing. The pNOY373 deletion plasmids were transformed into NOY891 and selected on yeast minimal media supplemented with adenine and uracil with galactose as the sole carbon source. 20 μM of bhm-21 was added to solid media and used within one day. Yeast were serially diluted as previously described (Rong-Mullins, et al.2017). All yeast strains are listed in Table 2.
Table 2.
Strain list.
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 | MATa ura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1 | (Sikorski and Hieter 1989) |
| YPH500 | MATα ura3-52 lys2-801_amber ade2-101_ochre trp1-Δ63 his3-Δ200 leu2-Δ1 | (Sikorski and Hieter 1989) |
| YPH499 HA | C-terminal 3xHA tag | (Dragon et al.2002) |
| YPH499 TAP | C-terminal TAP tag (TRP1) | (Gallagher et al.2004) |
| YKW100 | MATa, ura3-52, his3-Δ, leu2 lys2-801 amber, trp1-Δ6,3 u3aΔ UAS GALpro:U3A::URA3 u3b::LEU2 | (Wehner et al.2002) |
| YJV100 | MATα, ade3-1, his3-11, leu2-3,112, trp1-1, ura3-1 can100, rpa135::LEU2, GAL7-35rDNA::TRP1d | (Venema et al.1995) |
| K3207 | MATa his4-260::URA3, his4−Δ ade2-1, ade5 ura3-52, HOT1::leu2::ADE5,7::leu2-3,112, trp1HIII, lys2-BX::CAN1::LYS2, can1 | (Lin and Keil 1991) |
| YPH499 GAL:HA | GAL1 driven N-terminal 3xHA tag (KANR) | (Dragon et al.2002) |
| NOY504 | MATα, ade2-101, ura3-1, trp1-1, leu2-3,112,his3-11, can1-100, rrn4::LEU2, (pol I temperature sensitive) | (Nogi et al.1993) |
| NOY878 | MATa, ade2-101, ura3-1, trp1-1, leu2-3,112, can1-100, rrn9::HIS3, PSW (35S rDNA promoter switch from pol I to pol II) (GAL7-35S rDNA, 5S rDNA, URA3, 2 μm (pNOY103)) | (Oakes et al.1999) |
| NOY891 | MATa ade2-101, ura3-1, trp1-1, leu2-3,112, can1-100, rdnΔΔ::HIS3 carrying pNOY353 (GAL7-35S rDNA, 5S rDNA, TRP1, 2 μm (pNOY353)) | (Oakes et al.1999) |
| NOY908 | MATa ade2-101, ura3-1, trp1-1, leu2-3,112, can1-100, rdnΔΔ::HIS3 carrying pNOY373 (pol I rDNA LEU2, 2 μm (pNOY373) | (Wai et al.2001) |
| pPol II rDNA | NOY891 with pRS315 | This study |
| pPol I rDNA | NOY891 with pNOY373 | This study |
| pdeletions+ pPol II rDNA | NOY891 with pNOY373 with different length deletions of the 5΄ETS (long, medium or short 5’ETS transcripts) | This study |
RNA manipulations
Aliquots of YJV100 were collected 0 or 6 hours after the shift from galactose-containing media to glucose-containing media while keeping the OD600 of each culture below 0.5 unless otherwise noted. Total RNA was extracted with hot phenol (Ausubel et al.1995). RNA was separated on 8% polyacrylamide and transferred to Hybond N+ membrane (GE Healthcare). Hybridizations were done with 32P-labeled oligonucleotides complementary to the pre-rRNA, 5.8S and U3 snoRNA, as previously described (Dunbar et al.1997). Blots were serially hybridized with probes to RNAs of known length and used to determine the length of the short rRNAs. The sequences of all oligonucleotides used in this study are shown in Table 1. Primer extensions were performed as previously described (Dragon et al.2002).
Transcriptional run-on assays
Transcriptional run-on assays were performed with YPH499 and GAL:HA strains as previously published (Gallagher et al.2004). PCR fragments corresponding to segments of the rDNA transcription unit were cloned into the Invitrogen pCR21.1 TOPO TA cloning system and spotted onto Hybond N+ membrane (GE Healthcare). The sequences for the NTS2 and middle (5΄ ETS) probes were previously described (Gallagher et al.2004). Primers used for the 5΄ start and 3΄ +177 and for the end probe are 5΄ +923 and 3΄oligo z (Table 1 (Lee and Baserga 1997)). Plasmids containing the various regions of the rDNA were spotted on the membrane in excess of in vivo radiolabeled RNA that had been extracted from yeast. The dot blot hybridization signals were quantitated using Bio-Rad Multi-Analyst Version 1.0.2. To obtain the corrected values numbers, the background (the NTS2 dot) was subtracted from the start, middle, and end 5’ ETS dots. The amount of signal was determined in arbitrary units, and three independent transcription run-on assays were averaged together, with standard error indicated.
Protein manipulations
Co-immunoprecipitations were carried out as previously described (Gallagher et al.2004). Proteins were detected by incubation with 1:10,000 dilution of anti-Mpp10 antibody (Dunbar et al.1997) or 1:500 dilution of anti-HA antibody or 1:5000 dilution of PAP (to detect TAP from Sigma Aldrich), for an hour at room temperature after being transferred to ImmobilonTM-P. Secondary antibodies conjugated to HRP (anti-rabbit or anti-mouse) diluted 1:10,000 were then added for 15 minutes. ECLTM from GE Healthcare was used to detect the immunoreactive bands. Twenty-five μl of the extract was taken as 1/20th of the total and co-immunoprecipitation were carried out as previously described (Gallagher et al.2004). Co-immunoprecipitated RNA was extracted by phenol-chloroform and analyzed by northern blotting.
Chromatin immunoprecipitations
Semi-quantitative ChIP was carried out as described in (Gallagher et al.2004) for sequences in the repetitive rDNA. The sequences for the primers are found in Table 1. The primers to the rDNA promoter are 5΄ –200 and 3΄ –47. Primers to amplify the start 5’ETS were 5’start and +177 while primers for mid 5΄ETS were +300 and oligo x. Primers to the 25S rRNA coding sequence are 5΄25S and oligo y. Quantitative ChIP of single gene localizations such as the HOT1 reporter and the gene was carried out as described in (Kuras et al.2000; Gallagher et al.2014; Lefrançois, Gallagher and Snyder 2014) and the primers used are 5΄ start and 3΄ HIS4. The signal from the HOT1-HIS4 PCR was normalized to that from the ACT1 gene. Quantitative ChIP was carried out in biological triplicate and standard deviation noted. While Quantitative ChIP of the rDNA repeat cannot be normalized to a single nonnucleolar locus and instead was normalized to input.
Supplementary Material
Acknowledgements
These studies would not have been possible without the support of Susan Baserga and the Baserga lab. Strains and plasmids were gifts from Masayasu Nomura and Ralf Keil. John Woolford and Susan Baserga provided insightful comments. Sequencing of deletions was carried out by the WVU Genomics Core. We would like to acknowledge the WVU Genomics Core Facility, Morgantown WV for support provided to help make this publication possible. JEGG was supported by a pre-doctoral fellowship from the National Institutes of Health [GM20905].
Conflict of interest. None declared.
REFERENCES
- Ausubel F, Brent R, Kingston RE et al. Short protocols in molecular biology 1995. Wiley, New York. [Google Scholar]
- Barandun J, Chaker-Margot M, Hunziker M et al. The complete structure of the small-subunit processome. Nat Struct Mol Biol 2017;24:944–53. [DOI] [PubMed] [Google Scholar]
- Barandun J, Hunziker M, Klinge S. Assembly and structure of the SSU processome — a nucleolar precursor of the small ribosomal subunit. Curr Opin Struct Biol 2018;49:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Ahmed YL, Kallas M et al. Interaction network of the ribosome assembly machinery from a eukaryotic thermophile. Protein Sci 2017;26:327–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benecke B-J, Penman S. A new class of small nuclear RNA molecules synthesized by a type I RNA polymerase in HeLa cells. Cell 1977;12:939–46. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gallagher JE, Mitchell BM et al. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell 2004; Baßler JAhmed YL, Kallas M:1619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviño FR, Kornprobst M, Schermann G et al. Structural basis for 5΄-ETS recognition by Utp4 at the early stages of ribosome biogenesis. (Granneman S. Ed.). PLoS One 2017;12:e0178752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Barandun J, Hunziker M et al. Architecture of the yeast small subunit processome. Science (80) 2017;355:eaal1880. [DOI] [PubMed] [Google Scholar]
- Cheng TH, Chang CR, Joy P et al. Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res 2000;28:108e–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kellner N, Berninghausen O et al. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat Struct Mol Biol 2017;24:954–64. [DOI] [PubMed] [Google Scholar]
- Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem 2004;279:37385–97. [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JEG, Compagnone-Post PA et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002;417:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar DA, Wormsley S, Agentis TM et al. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast.. Mol Cell Biol 1997;17:5803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutca LM, Gallagher JEG, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res 2011;39:5164–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JEG, Dunbar DA, Granneman S et al. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 2004;18:2506–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JEG, Zheng W, Rong X et al. Divergence in a master variator generates distinct phenotypes and transcriptional responses. Genes Dev 2014;28:409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 2004;296:43–50. [DOI] [PubMed] [Google Scholar]
- Hage AEl, French SL, Beyer AL et al. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson-Chastang C, O’Donohue M-F et al. An overview of pre-ribosomal RNA processing in eukaryotes. WIREs RNA 2015;6:225–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 2008;65:2334–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepfer CE, Arnold-Croop S, Fogell H et al. DEG1, encoding the tRNA:pseudouridine synthase Pus3p, impacts HOT1-stimulated recombination in Saccharomyces cerevisiae. Mol Genet Genomics 2005;274:528–38. [DOI] [PubMed] [Google Scholar]
- Hochstatter J, Holzel M, Rohrmoser M et al. Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J. Biol Chem 2012;287:24365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker M, Barandun J, Petfalski E et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Comms 2016;7:12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CV, Cruz C, Hull RM et al. Regulation of ribosomal DNA amplification by the TOR pathway. Proc Natl Acad Sci USA 2015;112:9674–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob S, Ohmayer U, Neueder A et al. Interrelationships between Yeast Ribosomal Protein Assembly Events and Transient Ribosome Biogenesis Factors Interactions in Early Pre-Ribosomes (Kudla G, Ed.). PLoS One 2012;7:e32552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Josaitis CA, Dodd JA et al. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem 1998;273:33795–802. [DOI] [PubMed] [Google Scholar]
- Keil RL, Roeder GS. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 1984;39:377–86. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1996;1:465–74. [DOI] [PubMed] [Google Scholar]
- Kong R, Zhang L, Hu L et al. hALP, a novel transcriptional U three protein (t-UTP), activates RNA polymerase I transcription by binding and acetylating the upstream binding factor (UBF). J Biol Chem 2011;286:7139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell 2016;166:380–93. [DOI] [PubMed] [Google Scholar]
- Kos-Braun IC, Jung I, Koš M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner (Misteli T. Ed.). PLoS Biol 2017;15:e2000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem Sci 2017;42:640–54. [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim Biophys Acta 2010;1803:673–83. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng W-T, Cagney G et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell 2004;13:225–39. [DOI] [PubMed] [Google Scholar]
- Kuhn A, Grummt I. Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor.. Proc Natl Acad Sci 1992;89:7340–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M et al. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 2000;288:1244–48. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Baserga SJ. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci 1997;94:13536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançois P, Gallagher JEG, Snyder M. Global Analysis of Transcription Factor-Binding Sites in Yeast Using ChIP-Seq (Smith JS, Burke DJ Eds.). Yeast Genet 2014;1205:231–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Keil RL. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics 1991;127:31–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998;14:953–61. [DOI] [PubMed] [Google Scholar]
- Mayan M, Aragón L. Cis-interactions between non-coding ribosomal spacers dependent on RNAP-II separate RNAP-I and RNAP-III transcription domains. Cell Cycle 2010;9:4328–37. [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T et al. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol life Sci 2007;64:29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W, Boon K, van der Sande CA et al. Functional analysis of transcribed spacers of yeast ribosomal DNA. EMBO J 1990;9:3989–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Dodd J et al. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol 1993;13:114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA 1991;88:3962–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M, Siddiqi I, Vu L et al. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol 1999;19:8559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AP, Ludwig C, Zampieri M et al. Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci Signal 2015;8:rs4–rs4. [DOI] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J et al. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J 2006;25:3310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Zomerdijk JC. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol 2001;21:2641–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C, Hurt E, Panse VG. Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 2017;24:689–99. [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, Las Rivas JD et al. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 2007;27:5414–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 2007;21:2041–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty R, Keil RL. SCH9, a putative protein kinase from Saccharomyces cerevisiae, affects HOT1-stimulated recombination. Mol Genet Genomics 2004;272:264–74. [DOI] [PubMed] [Google Scholar]
- Raška I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 2006;18:325–34. [DOI] [PubMed] [Google Scholar]
- Reichel R, Benecke BJ. Localization of small nuclear polymerase I RNA sequences at the 5’ end of the human rDNA transcription unit. EMBO J 1984;3:473–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel R, Monstein HJ, Jansen HW et al. Small nuclear RNAs are encoded in the nontranscribed region of ribosomal spacer DNA. Proc Natl Acad Sci USA 1982;79:3106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson LA, Reed BJ, Charette JM et al. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep 2012;2:372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong-Mullins X, Ravishankar A, McNeal KA et al. Genetic variation in Dip5, an amino acid permease, and Pdr5, a multiple drug transporter, regulates glyphosate resistance in S. cerevisiae (Louis EJ, Ed.). PLoS One 2017;12:e0187522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong-Mullins X, Winans MJ, Lee JB et al. Proteomic and genetic analysis of S. cerevisiae response to soluble copper leads to improvement of antimicrobial function of cellulosic copper nanoparticles. Metallomics 2017;9:1304–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Liu X, Granneman S et al. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol 2015;13:e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene 2012;493:176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Michel A, Sikes ML et al. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell 2007;26:217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5’ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol 1999;19:6012–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Kawasaki Y, Hiraga S et al. The fifth essential DNA polymerase phi in Saccharomyces cerevisiae is localized to the nucleolus and plays an important role in synthesis of rRNA. Proc Natl Acad Sci USA 2002;99:9133–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989;122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondalle SB, Baserga SJ, Yelick PC. The Contributions of the Ribosome Biogenesis Protein Utp5/WDR43 to Craniofacial Development. J. Dent Res 2016;95:1214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T et al. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell 2006;21:629–39. [DOI] [PubMed] [Google Scholar]
- Steven Huang G, Keil RL. Requirements for activity of the yeast mitotic recombination hotspot HOTI: RNA polymerase I and multiple &-acting sequences. Genetics 1995;141:845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Roeder GS. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol 1989;9:3464–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife 2017;6:e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B, Yang C-C, Hsieh C-L et al. Epigeneitc silencing of ribosomal RNA genes by Mybbp1a. J Biomed Sci 2012;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska-Placzek M. Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett 2017;591:1801–50. [DOI] [PubMed] [Google Scholar]
- Torreira E, Louro JA, Pazos I et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife 2017;6:e20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J, Retel J, Planta RJ. Ribosomal precursor particles from yeast. Exp Cell Res 1975;90:95–104. [DOI] [PubMed] [Google Scholar]
- Tschochner H. A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3’ end. Proc Natl Acad Sci USA 1996;93:12914–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulha J, Lima A, Lucas C et al. Saccharomyces cerevisiae glycerol/H+ symporter Stl1p is essential for cold/near-freeze and freeze stress adaptation. A simple recipe with high biotechnological potential is given. Microb Cell Fact 2010;9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Dirks‐Mulder A, Faber AW et al. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast 1995;11:145–56. [DOI] [PubMed] [Google Scholar]
- Viktorovskaya OV, Engel KL, French SL et al. Divergent Contributions of Conserved Active Site Residues to Transcription by Eukaryotic RNA Polymerases I and II. Cell Rep 2013;4:974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 2004;432:1054–58. [DOI] [PubMed] [Google Scholar]
- Wai H, Johzuka K, Vu L et al. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol Cell Biol 2001;21:5541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 1999;24:437–40. [DOI] [PubMed] [Google Scholar]
- Webster TD, Dickson RC. Direct selection of Saccharomyces cerevisiae resistant to the antibiotic G418 following transformation with a DNA vector carrying the kanamycin-resistance gene of Tn903. Gene 1983;26:243–52. [DOI] [PubMed] [Google Scholar]
- Wehner KA, Gallagher JE, Baserga SJ. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol Cell Biol 2002;22:7258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Najmi SM, Liu H et al. Small-molecule targeting of RNA polymerase I activates a conserved transcription elongation checkpoint. Cell Rep 2018;23:404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormsley S, Samarsky DA, Fournier MJ et al. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA 2001;7:904–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Rogozin IB, Koonin EV. Yeast POL5 is an evolutionarily conserved regulator of rDNA transcription unrelated to any known DNA polymerases. Cell Cycle 2003;2:120–2. [DOI] [PubMed] [Google Scholar]
- Yeh L-CC, Lee JC. Structure analysis of the 5΄ external transcribed spacer of the precursor ribosomal RNA from Saccharomyces cerevisiae. J Mol Biol 1992;228:827–39. [DOI] [PubMed] [Google Scholar]
- Yip WSV, Shigematsu H, Taylor DW et al. Box C/D sRNA stem ends act as stabilizing anchors for box C/D di-sRNPs. Nucleic Acids Res 2016;44:8976–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon S-O, Poulogiannis G et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science (80-.). 2011;332:1322–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL et al. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci USA 2009;106:2153–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Smith AD, Renfrow MB et al. The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J Biol Chem 2010;285:14152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G et al. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev 2016;30:718–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Andreeva V, Gibert Y et al. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genet 2014;10:e1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.