Abstract
Background and Purpose
The nociceptin/orphanin FQ (N/OFQ) receptor (NOP) is a member of the opioid receptor family and is involved in a number of physiological responses, pain and immune regulation as examples. In this study, we conjugated a red fluorophore‐ATTO594 to the peptide ligand N/OFQ (N/OFQATTO594) for the NOP receptor and explored NOP receptor function at high (in recombinant systems) and low (on immune cells) expression.
Experimental Approach
We assessed N/OFQATTO594 receptor binding, selectivity and functional activity in recombinant (CHO) cell lines. Live cell N/OFQATTO594 binding was measured in (i) HEK cells expressing NOP and NOPGFP receptors, (ii) CHO cells expressing the hNOPGαqi5 chimera (to force coupling to measurable Ca2+ responses) and (iii) freshly isolated human polymorphonuclear cells (PMN).
Key Results
N/OFQATTO594 bound to NOP receptor with nM affinity and high selectivity. N/OFQATTO594 activated NOP receptor by reducing cAMP formation and increasing Ca2+ levels in CHOhNOPGαqi5 cells. N/OFQATTO594 was also able to visualize NOP receptors at low expression levels on PMN cells. In NOP‐GFP‐tagged receptors, N/OFQATTO594 was used in a FRET protocol where GFP emission activated ATTO, visualizing ligand–receptor interaction. When the NOPGFP receptor is activated by N/OFQATTO594, movement of ligand and receptor from the cell surface to the cytosol can be measured.
Conclusions and Implications
In the absence of validated NOP receptor antibodies and issues surrounding the use of radiolabels (especially in low expression systems), these data indicate the utility of N/OFQATTO594 to study a wide range of N/OFQ‐driven cellular responses.
Abbreviations
- DPN
diprenorphine
- N/OFQ
nociceptin/orphanin FQ
- NOP receptor
N/OFQ peptide receptor
- PMN
polymorphonuclear cells
- SB‐612111
7‐[[4‐(2,6‐dichlorophenyl)‐1‐piperidinyl]methyl]‐6,7,8,9‐tetrahydro‐1‐methyl‐5H‐benzocyclohepten‐5‐ol hydrochloride
Introduction
The nociceptin/orphanin FQ (N/OFQ) receptor (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=320) is the newest member of the opioid receptor family (Lambert, 2008). It exhibits similar signalling mechanisms to the classical opioid receptors (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=319, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=317 and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=318), through activation of a Gαi‐mediated G‐protein pathway (Meunier et al., 1995; Reinscheid et al., 1995). The NOP receptor, however, differs from the classical opioid receptors in a number of key areas. It has little to no affinity for the endogenous ligands of classical opioid receptors, nor does it have affinity for the opioid antagonist http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1638. Furthermore, its own endogenous ligand, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1681, is highly selective for the NOP receptor, displaying little or no affinity for the classical opioid receptors.
Like the classical opioid receptors, NOP receptor expression has been demonstrated throughout the pain pathway, with antinociceptive actions ascribed to activation (Mollereau and Mouledous, 2000; Schroder et al., 2014; Ding et al., 2015). At initial identification, in mice and rat studies, it was believed that the NOP receptor had pronociceptive activity supraspinally; however, work by Ding and colleagues in non‐human primates demonstrated antinociceptive activity throughout the pain pathway underscoring important interspecies differences (Ding et al., 2015). The NOP receptor is expressed in a range of other non‐neuronal tissues and presumed levels vary widely (Lambert, 2008). NOP‐eGFP knock‐in mice have been used to examine expression‐function but with a neuronal focus (Ozawa et al., 2015). Outside the nervous system, we and others have shown the presence of the NOP receptor in immune tissues, but due to presumed ultra‐low expression, this has been suggested through PCR and modulation of immune function (Kruger et al., 2006; Zhang et al., 2013). While it is possible to obtain acceptable amounts of cerebral tissue to measure opioid expression through radioligand binding, the quantities of peripheral tissue available are most often inadequate for such studies.
Assessment of NOP receptor expression outside the brain is hindered by several key issues. The quantity of sample collected (e.g. immune cells) is often insufficient to perform radioligand assays, the current gold standard. Therefore, as noted above, receptor identity is often confirmed by either PCR or antibody methods. PCR detects mRNA, and these levels do not necessarily translate to functional protein (Guo et al., 2008). Antibodies for GPCRs have been shown to lack the high levels of specificity required to provide evidence of expressed protein (Michel et al., 2009). Previous work in our laboratory and others has demonstrated high levels of non‐selectivity using commercially available antibodies (Scherrer et al., 2009; Niwa et al., 2012). In standard Western blotting, we demonstrated the presence of bands at the expected weight in CHO cells expressing opioid receptors as well as those that had not been transfected with the opioid receptor of interest (Niwa et al., 2012). The importance of selectivity is highlighted in a study of δ (DOP) receptors (Scherrer et al., 2009). In their study identifying regions of expression of δ receptors, Scherrer and colleagues demonstrated large differences between areas of expression determined by antibody when compared to δ‐eGFP expressing mice (Scherrer et al., 2009).
Because the NOP receptor has no affinity for classical opioid ligands and N/OFQ is highly selective for NOP receptors, labelling of N/OFQ is an exciting possibility as a replacement for radioprobes and antibodies. Here, we describe for the first time a method using N/OFQ conjugated to the highly fluorescent ATTO dye (594 nm) and show this probe has a remarkable similarity to the unlabelled peptide.
Methods
Cell culture and immune cell preparation
Hams F12 was used to culture CHO cells expressing recombinant human opioid receptors (μ, δ and κ), DMEM/F12 1:1 media were used for both CHOhNOP and CHOhNOPGαqi5 (from T Costa, Istituto Superiore di Sanità, Rome, Italy) and MEM was used for HEK cells expressing the recombinant NOP (HEKhNOP) receptor and HEKhNOP‐GFP. All media were supplemented with 10% FBS, 100 IU·mL−1 penicillin, 100 μg·mL−1 streptomycin and 2.5 μg·mL−1 fungizone. Cultures were maintained in selection media; for CHO cells containing the classical opioid receptors, 200 μg·mL−1 G418 was used. For HEKhNOP cells, 200 μg·mL−1 hygromycin B was used. For both CHOhNOP and CHOhNOPGαqi5 cells, 200 μg·mL−1 G418 and 200 μg·mL−1 hygromycin B were used. Recombinant cell lines were grown to confluency in T75 cm3 flasks in supplemented media at 37°C in 5%‐CO2/humidified air. Polymorphonuclear cells (PMN) were extracted from healthy volunteers as previously described (Thompson et al., 2013) and with University of Leicester volunteer research ethics committee approval. Up to 30 mL of blood was collected from each volunteer (mean age of 38; range 25–55, 4 male : 1 female) into Monovette blood collection tubes (Sardstedt, Germany) containing K‐EDTA (7.5 mL blood per tube, final EDTA concentration 1.6 mg·mL−1) and used within 1 h of venepuncture. PMN were extracted by centrifugation over an equal volume of Polymorphprep (Axis‐Shield, Dundee). Following extraction, PMN were cleared of any potential erythrocyte contamination by using a 1:1 dilution with BD PharmaLyse (Becton, Dickinson and Company, Oxford), resuspended in Krebs buffer (0.126 M NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 2.5 mM CaCl2) for counting and imaging. Extractions were carried out at room temperature, and the resulting cell suspension kept on ice until use (maximum 4 h). Viability and yields were quantified by Trypan Blue exclusion and counting using a haemocytometer (Strober, 2001).
Radioligand binding
Radioligand binding was quantified in CHO cells stably transfected with μ, δ, κ and NOP receptors. Twenty to forty micrograms of membrane protein were incubated in 0.5 mL wash buffer (50 mM Tris–HCL, pH 7.4 KOH with the addition of 0.5% BSA), ∼0.8 nM http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1612 (DPN; for classical opioid membranes) or ∼0.8 nM [3H]‐N/OFQ (for CHOhNOP membranes), varying concentrations (1 pM–10 μM) of the control ligand or N/OFQATTO594. Non‐specific binding was measured in the presence of 10 μM naloxone (μ, δ, κ receptors) or 1 μM of N/OFQ (NOP receptor). Samples were incubated at room temperature for 1 h. Reactions were terminated by vacuum filtration onto polyethylenimine‐soaked Whatman GF/B filters, using a Brandel harvester (Bird et al., 2015).
cAMP inhibition assay
HEKhNOP‐GFP cells were suspended in Krebs/HEPES buffer, pH 7.4 NaOH containing http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5190 (1 μM) and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=388 (1 mM). Both the control ligand (unlabelled N/OFQ) and the N/OFQATTO594 were included in a range of concentrations (0.1 pM–1 μM) and incubated at 37°C for 15 min. Reactions were terminated through the initial addition of 20 μL 10 M HCl, followed by 20 μL 10 M NaOH and 200 μL 1 M Tris–HCl (pH 7.4) to equilibrate the pH. Reactions were centrifuged at 16 000× g and supernatant collected. The supernatant was incubated with http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5096 and an in‐house prepared binding protein overnight at 4°C. Charcoal/BSA suspension was added and the reaction centrifuged at 16 000× g, following which, samples of the supernatant were counted as previously described (Kitayama et al., 2007).
Confocal microscopy
At confluence, NOP expressing cells were passaged onto ethanol‐sterilized 28 mm Menzel glaser #1‐coverslips (Thermo Scientific, Loughborough, UK) and incubated for 24 h before use. Cells were perfused with Krebs buffer, pH 7.4 at 4°C using a temperature controller and microincubator (PDMI‐2 and TC202A; Burleigh, Digitimer, Cambridge, UK) when studying ligand–receptor binding, while in functional studies (internalization of the ligand–receptor complex), cells were incubated at 37°C. Where used, immune cells were plated onto coverslips pretreated with Celltak™ (1 μg·mL−1) (Sigma, UK) and incubated at 37°C for 1 h, before being washed in ice‐cold Krebs buffer.
N/OFQATTO594 was injected onto coverslips, allowing for a range of concentrations to be measured (1 pM−100 nM, cumulatively), following which, images were captured using a Nikon Eclipse C1Si microscope (Surrey, UK), using an oil immersion 60× objective. N/OFQATTO594 was allowed to incubate for 5 min at 4°C before the coverslip was washed with ice‐cold Krebs buffer. Following this incubation period, HEKhNOP cells were imaged using the 594 nm wavelength laser, with images collected by the Nikon C1Si software. HEKhNOP‐GFP and CHOhNOPgαq/i5 cells were imaged sequentially using the 594 nm wavelength followed by 488 nm to assess N/OFQATTO594 and GFP/Fluo4‐AM respectively, 10 s per frame. In order to determine specificity of binding, the NOP antagonist http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1693 (1 μM) was pre‐incubated in desired cells for 15 min before addition of N/OFQATTO594. PMN were incubated with 100 nM N/OFQATTO594 and, following washing with ice‐cold Krebs, were imaged using the 594 nm wavelength laser. In all experiments, the laser and gain settings were maintained constant. For experiments with N/OFQATTO594 alone, settings were 37% 594 nm laser power, 7.10 gain‐red channel. In experiments including Fluo4‐AM stained cells (see below), 488 nm laser, an 18% power setting with 7.15 gain setting was used. Internalization studies were performed using 100 nM N/OFQATTO594 at 1 and 15 min time points (488 nm/594 nm wavelengths) in HEKhNOP‐GFP cells. FRET studies were undertaken by measuring the binding of N/OFQATTO594 to HEKhNOP‐GFP. NOP‐GFP receptors were stimulated using a 488 nm laser, and measurements were made using the RED (594 nm) filter channel (Wallrabe et al., 2003). In order to confirm FRET‐pairing, several additional controls were performed (Snapp and Hegde, 2006). N/OFQATTO594 (100 nM) was incubated on HEKhNOP cells (not expressing the GFP fluorophore) and measured using 488 nm laser. Furthermore, photobleaching of the ligand was performed, after binding to HEKhNOP‐GFP, by exposing it to 594 nm laser until fluorescence was undetectable, following which levels of GFP fluorescence intensity were measured. While GFP and ATTO594 may not be specifically designed as an optimum FRET pair, there is still significant crossover as demonstrated by spectra analyser data.
CHOhNOPGαqi5 functional assays
In experiments using CHOhNOPGαqi5 cells (Camarda and Calo, 2013), the cells were incubated with 1 μM Fluo4‐AM for 45 min in Krebs buffer, following which they were washed for 3 min in 4°C Krebs via the perfusion system. A series of experiments were also performed at the physiological temperature of 37°C. N/OFQATTO594 (100 nM) was perfused while the cells were monitored under the confocal microscope [imaging using both 488 nm laser (Fluo4‐AM) and 594 nm laser (N/OFQATTO594)].
Data analysis
All data are the mean of five experiments ± SEM as appropriate. Specimen confocal data sets are presented. All confocal images were analysed using ImageJ with resulting data analysed using GraphPad Prism‐v7. To measure corrected cell fluorescence, the formula, Corrected total cell fluorescence = Integrated density − (Area of selected cell × Mean fluorescence of background readings), was used to determine levels of N/OFQATTO594 as previously described (Burgess et al., 2010). In internalization studies, ImageJ tool plot profile was used to determine the position of N/OFQATTO594 and/or NOPGFP relative to the cell membrane (Wallrabe et al., 2003). All experiments were performed unblinded.
Materials
N/OFQATTO594, N/OFQ, dermorphin, Leu‐enkephalin and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1620 were synthesized in‐house at the University of Ferrara. Tritiated diprenorphine ([3H]‐DPN) and tritiated N/OFQ ([3H]‐N/OFQ) were purchased from Perkin Elmer (UK). Details of the synthesis of N/OFQATTO594 can be found in the Supporting Information Data S1 and Figures S1–S3.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017).
Results
Binding selectivity and affinity
Selectivity for N/OFQATTO594 was measured in CHO cells expressing human μ, δ, κ and NOP receptors. In CHOhNOP cells, N/OFQATTO594 displaced [3H]‐N/OFQ in a concentration dependent and saturable manner (pKi: 9.21), which was not significantly (Student's t‐test) different from unlabelled N/OFQ (9.37) (Figure 1). N/OFQATTO594 failed to displace [3H]‐DPN in cells expressing μ, δ or κ receptors (Figure 1). In HEKhNOP cells, N/OFQATTO594 also displaced [3H]‐N/OFQ in a concentration dependent and saturable manner (pKi 9.14 ± 0.13; n = 5), which was not significantly (Student's t‐test) different from unlabelled N/OFQ (pKi: 9.34 ± 0.08; n = 5). A range of concentrations of N/OFQATTO594 were added to HEK cells expressing the NOP receptor and binding measured using 594 nm laser on a confocal microscope (Figure 2). N/OFQATTO594 bound in a concentration‐dependent and saturable manner, producing a pKd of 9.51 ± 0.18 (n = 5; Figure 2). In HEK cells expressing GFP‐tagged NOP (HEKhNOP‐GFP) cells, N/OFQATTO594 bound to NOP with an affinity (pKi) of 8.98 ± 0.06 in displacement binding assays using [3H]‐N/OFQ as the radiolabel (Figure 3). When binding affinity was assessed directly using confocal microscopy, N/OFQATTO594 produced a binding affinity (pKd) of 8.53 ± 0.34, which was not significantly (Student's t‐test) different from that determined by displacement (Figure 3A–I, J). In confocal experiments, pre‐incubation with SB‐612111 abolished N/OFQATTO594 binding.
Figure 1.
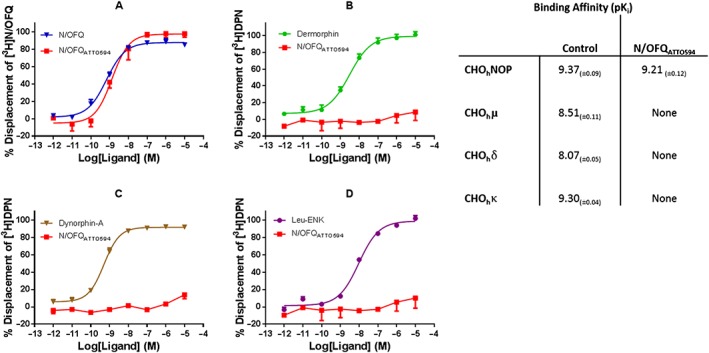
Displacement of [3H]‐N/OFQ in CHOhNOP cell membranes by a range of concentrations of N/OFQ and N/OFQATTO594 (A). Displacement of [3H]‐DPN in CHOhμ cell membranes by a range of concentrations of dermorphin and N/OFQATTO594 (B). Displacement of [3H]‐DPN in CHOhκ cell membranes by a range of concentrations of dynorphin‐A and N/OFQATTO594 (C). Displacement of [3H]‐DPN in CHOhδ cell membranes by a range of concentrations of Leu‐enkephalin and N/OFQATTO594 (D). Displacement binding affinities for N/OFQATTO594 and control ligands are summarised in the table (NOP: N/OFQ; μ: dermorphin; δ: Leu‐enkephalin; κ: dynorphin‐A). Data are the mean ± SEM of five experiments.
Figure 2.
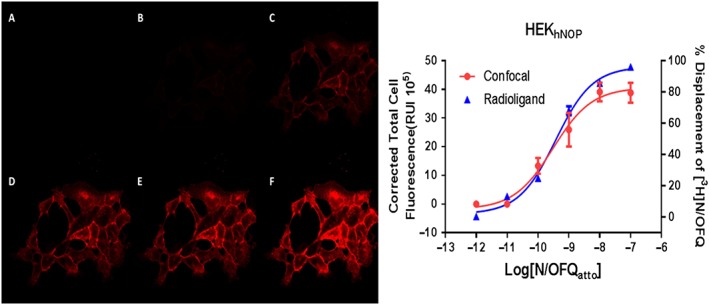
Concentration‐dependent binding of N/OFQATTO594 to HEKhNOP cells using confocal microscopy and stimulation with 594 nm laser and [3H]‐N/OFQ. On the left representative images of the binding of various concentrations (A: 1 pM; B: 10 pM; C: 100 pM; D: 1 nM; E: 10 nM; F: 100 nM) of N/OFQATTO594 using confocal microscopy. On the right are the binding curves analysed as in methods overlayed with data from a [3H]‐N/OFQ displacement analysis; data are mean ± SEM for five experiments. Confocal experiments were performed at 4°C while radioligand binding assays were performed at room temperature.
Figure 3.
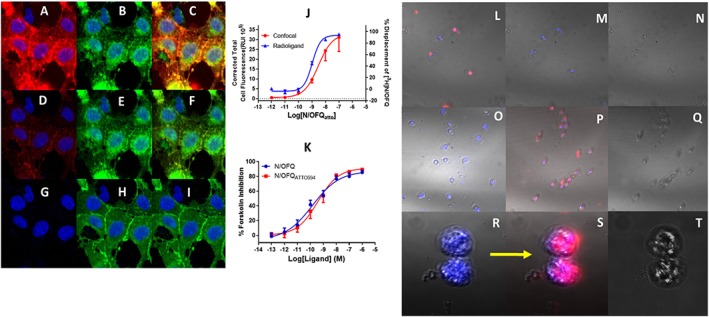
A–C: Binding of 100 nM N/OFQATTO594 in HEKhNOP‐GFP split into the red channel (A) with N/OFQATTO594 binding, (B) the green channel to identify NOPGFP tagged receptors and (C) a composite image demonstrating the interaction between N/OFQATTO594 and NOPGFP. (D–F) Binding of 1 nM of N/OFQATTO594 split into the red channel (D) showing N/OFQATTO594 binding, (E) the green channel identify NOPGFP tagged receptors and (F) a composite image demonstrating overlapping binding of N/OFQATTO594 with NOPGFP. (G–I) Binding of 1 pM of N/OFQATTO594 showing split into the red channel (G) showing N/OFQATTO594 binding, (H) the green channel identify NOPGFP tagged receptors and (I) a composite image demonstrating no binding of N/OFQATTO594 to NOPGFP. Nucleus is blue with DAPI stain. (J) Comparison between a radioligand displacement binding curve (using 3H‐N/OFQ at room temperature) and saturation curve measurement obtained from confocal microscopy. Data are mean ± SEM for five experiments. (K) Functional analysis of N/OFQatto activity (red) in a cAMP inhibition assay when compared to unlabelled N/OFQ. Data are mean ± SEM for five experiments performed at 37°C. (L) The ability of 100 nM N/OFQATTO594 to bind and visualize low expression systems is demonstrated in human PMN at 4°C. This binding can be disrupted by both unlabelled N/OFQ (1 μM) (M) and SB‐612111 (1 μM) (O). Furthermore, the classical opioid antagonist naloxone (1 μM) is unable to inhibit N/OFQATTO594 binding (P). In the enlarged image, unlabelled N/OFQ is pre‐incubated with PMN cells to occupy NOP receptors, before addition of 100 nM N/OFQATTO594 (R) [and as in M]), again inhibiting binding of the fluorescent ligand. After washing in Krebs to ‘clear’ the receptor for 10 min, 100 nM N/OFQATTO594 is able to bind to the surface of the PMN (S) [and as in L]). Bright field images for the accompanying experiments are shown in (N), (Q) and (T). All PNM data have been repeated for a total of five experiments. All confocal experiments were undertaken at 4°C.
Pharmacological activity
In order to determine whether conjugation of the fluorescent tag had any effect on functional activity, cAMP inhibition assays were performed (Figure 3K). The control ligand, unlabelled N/OFQ produced a pEC50 of 10.23 ± 0.25 and Emax of 85.43 ± 2.70% inhibition of forskolin‐stimulated cAMP formation. N/OFQATTO594 produced a pEC50 of 9.73 ± 0.29 and Emax of 89.30 ± 2.04%. There was no significant (Student's t‐test) difference in functional activity of N/OFQATTO594 when compared to N/OFQ (Figure 3K).
In experiments performed at 4°C, 100 nM N/OFQATTO594 was added to CHOhNOPGαqi5 cells loaded with the calcium indicator, Fluo4‐AM (Figure 4A–D). An increase in red fluorescence can be seen around the membrane after addition of N/OFQATTO594, following which an increase in green fluorescence can be seen indicating activation of the Gαqi5‐coupled NOP receptor and subsequent increase in cytosolic calcium (Figure 4A–D and Supporting Information Video S1). The kinetics of the rise in Ca2+ is slow as the experiment was performed at 4°C. Binding of N/OFQATTO594 at 37°C led to a more rapid increase of Ca2+ (Figure 4E–G).
Figure 4.
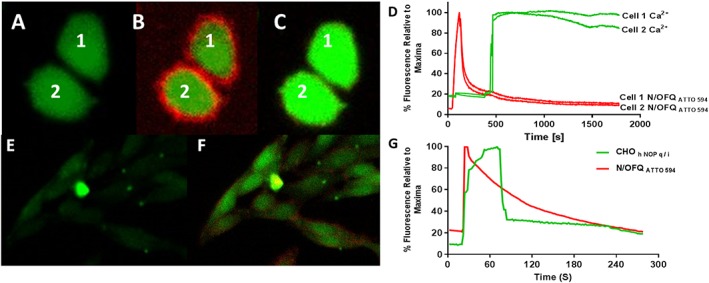
Live cell binding of N/OFQATTO594 and subsequent stimulation in CHOhNOPGαqi5 cells. Fluo4‐AM loaded CHOhNOPGαqi5 cells at 4°C (A) are incubated with 100 nM N/OFQATTO594 after 30 s (B) leading to release of calcium (cells labelled 1 and 2) (B and C). The process is demonstrated in the Supporting Information Video S1. This video has been created through ImageJ (eight frames per second), with N/OFQATTO594 added after 15 s, with the entirety of the video covering approximately 4.5 min. (D) A representative figure demonstrating N/OFQATTO594 binding to CHOhNOPGαqi5 cells at 4°C and the increase in Ca2+. Note: red is N/OFQATTO594 binding and green is Ca2+. Fluo4‐AM loaded CHOhNOPGαqi5 cells at 37°C (E) are incubated with 100 nM N/OFQATTO594 after 30 s, which leads to a prompt increase in binding and calcium (small field of cells; F). (G) A representative figure demonstrating binding of N/OFQATTO594 at 37°C and increase in Ca2+. Note: red is N/OFQATTO594 binding and green is Ca2+. These data are representative of n = 5.
Low expression systems
Immune cells are known to express NOP mRNA and do not bind radiolabelled N/OFQ, but their function is modulated in response to N/OFQ. We next used N/OFQATTO594 to determine whether we could detect NOP at low levels of expression. Human PMN cells separated from healthy volunteers were seeded onto cover slips. At a temperature of 4°C, N/OFQATTO594 (100 nM) binding was observed (Figure 3L), indicating the presence of NOP receptors. Furthermore, pre‐incubation with unlabelled N/OFQ (1 μM; Figure 3M) or the selective NOP antagonist SB‐612111 (1 μM; Figure 3O) blocked the binding of N/OFQATTO594. The addition of naloxone (1 μM) had no effect on N/OFQATTO594 binding (Figure 3P); this is consistent with the selectivity of binding of N/OFQATTO594. In a further experiment to confirm NOP expression, PMN cells were pre‐incubated with N/OFQ before the addition of 100 nM N/OFQATTO594 (Figure 3R), demonstrating no binding of N/OFQatto594. Cells were washed for several minutes in ice‐cold Krebs, following which 100 nM N N/OFQATTO594 was added for a second time. In this instance, binding was restored (Figure 3S). Brightfield images are shown in panels N, Q and T.
NOP receptor internalization
We then used N/OFQATTO594 to track NOP receptor internalization. Binding was initially measured at 4°C using 100 nM of N/OFQATTO594 in HEKhNOP‐GFP cells (Figure 5A, C and E) clearly demonstrating a localization of N/OFQATTO594 (Figure 5C) on the cellular membrane with GFP‐tagged NOP (A and overlay E). Following a temperature increase to 37°C, N/OFQATTO594 is seen to mobilize within the cell, away from the cell membrane (D). These pools of N/OFQATTO594 are shown to co‐localize with internalized NOP‐GFP (B and overlay F). Using ImageJ line analysis (dotted line in A–F), it is possible to assess the change in fluorescence at different time points across the cell after binding of N/OFQATTO594 (Figure 5G) and the subsequent localization of NOP‐GFP (Figure 5H).
Figure 5.
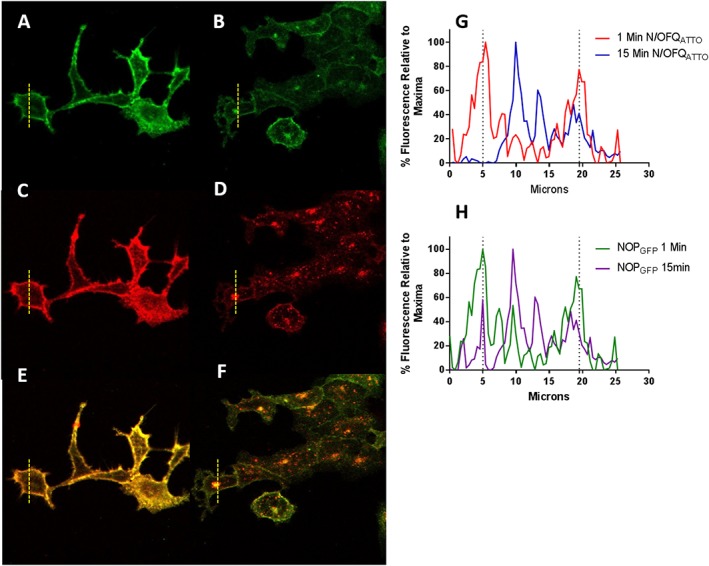
Use of N/OFQATTO594 and NOPGFP to examine cell surface receptor expression. All images depict HEKhNOP‐GFP cells. Panels A, C and E are at 4°C and measured 1 min after ligand addition while panels B, D and F are at 37°C, 15 min after ligand addition. Green channel for NOPGFP is in A and B; red channel for N/OFQATTO594 is in C and D, and the overlap is in E and F. Panel G shows a representative graph of the initial binding of N/OFQATTO594 (red; 1 min after adding N/OFQATTO594) and subsequent change in localization (blue; 15 min after adding N/OFQATTO594). Panel H shows a representative graph of initial location of NOPGFP (green; 1 min after adding N/OFQATTO594) and subsequent change in localization (purple; 15 min after adding N/OFQATTO594) In (G and H), the dotted lines are used to indicate the position of the membrane with the cell interior between the two.
Because of the close proximity formed between ligand–receptor complexes (Figure 6M), a FRET stimulation was performed in HEKhNOP‐GFP cells using N/OFQatto594. When the receptor complex was formed, the 488 nm laser was used (Figure 6A–D), while filters in the red channel were viewed. In the absence of N/OFQATTO594, no image was obtained (Figure 6B). After administration of 100 nM N/OFQATTO594, a clear image demonstrating areas of interaction of NOPGFP‐N/OFQATTO594 was seen (Figure 6C), indicating FRET. The overlap of NOPGFP and N/OFQATTO594 can be seen in an overlaid composite image (Figure 6D). In order to confirm FRET‐pairing, 100 nM N/OFQATTO594 was incubated with HEKhNOP cells (not expressing GFP), following which detection of binding was attempted using the 488 nm wavelength laser (Figure 6E). N/OFQATTO594 was not stimulated by the laser in this environment. Binding of N/OFQATTO594 to the surface was confirmed by stimulation with 594 nm laser (Figure 6F). A second control experiment was performed, whereby 100 nM N/OFQATTO594 was incubated with HEKhNOP‐GFP cells and exposed to prolonged stimulation through 594 nm laser wavelengths (Figure 6G–L). This produced photobleaching of the ligand (Figure 6J). The level of GFP fluorescence was taken before and after photobleaching and demonstrated that the corrected total cell fluorescence of GFP had a statistically significant increase from 0.78 ± 0.11 to 1.11 ± 0.17 (Student's t‐test; P < 0.05) (Figure 6N).
Figure 6.
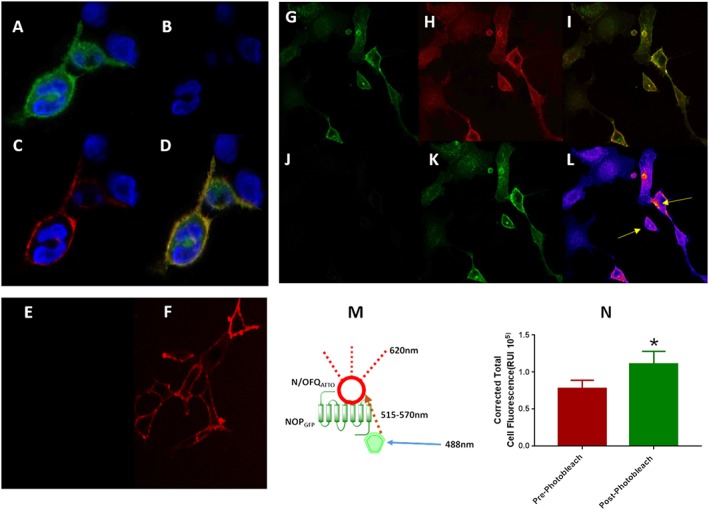
N/OFQATTO594 can be activated when in close proximity to GFP attached to the NOP receptor (FRET), as described in the cartoon in the middle of the figure (M). (A) Visualizes the NOP‐GFP receptors stimulated by the 488 nm laser in the green filter window. (B) Demonstrates no ‘leak’ of fluorescence into the red window when using the green laser in the absence of N/OFQATTO594. Upon addition of 100 nM N/OFQATTO594, the ligand is stimulated through FRET pairing (C) to fluoresce when in close proximity to the NOPGFP receptors. Signals are overlapped in (D). (E) In order to confirm FRET pairing, the 488 nm laser was used in conjunction with HEKhNOP cells (no fluorescent linker present) to again demonstrate lack of activation of N/OFQATTO594. (F) Binding of N/OFQATTO594 was confirmed using 594 nm laser. Photobleaching of the acceptor molecule is a further method to confirm FRET pairing. HEKhNOPGFP cells (G) were labelled with 100 nM N/OFQATTO594 (H) with binding of the receptor‐ligand complex shown as a composite image (I). N/OFQATTO was exposed to 594 nm light until photobleaching was achieved (J) at which point changes in NOPGFP fluorescence were measured (K) with the heatmap indicating increases of fluorescence shown and highlighted by arrows (blue: low and red: high) (L). (N) Average increase in NOPGFP fluorescence after photobleaching of N/OFQATTO594; P < 0.05 Students t‐test. Data are the mean of eight experiments.
Discussion
In this study, we report the synthesis and use of a novel fluorescent probe for the NOP receptor, N/OFQATTO594. We have conjugated ATTO594 to the highly selective endogenous NOP ligand N/OFQ, and this new ligand retains high NOP selectivity (over classical opioid receptors) and full agonist activity in (i) cAMP inhibition experiments performed in cells expressing NOP receptors (Kitayama et al., 2007), (ii) Ca2+ mobilisation experiments in cells co‐expressing NOP and chimeric G proteins (Camarda and Calo, 2013) and (iii) receptor internalization studies (Arttamangkul et al., 2000). N/OFQATTO594 was visibly detected over a range of concentrations when studied using confocal microscopy with sufficient sensitivity to determine a pKd; this did not differ from values obtained using the standard [3H]‐N/OFQ binding protocol. The nM affinity reported here, coupled with the high intrinsic brightness of ATTO594 allows for the visualization of NOP receptors in low expression systems, PMN. Use of cells expressing NOP receptors coupled to GFP allows further use of N/OFQATTO594 in a FRET‐based assay to document ligand–receptor interaction. Finally, this new ligand is well suited to studies of receptor internalization, and when coupled with a GFP tagged receptor, it should be possible to track both receptor and ligand fate following binding and activation and unbinding.
Because N/OFQ is an agonist for the NOP receptor, we have measured binding in confocal experiments at 4°C. At the more conventional 37°C, there would be substantial activation and loss of cell surface receptors (see below) as we have shown previously (Hashimoto et al., 2002; Barnes et al., 2007). The assessment of binding affinity would not be affected by temperature (Ahmadi et al., 2014) provided the ligand–receptor interaction was allowed to reach equilibrium, and we have a consistent assessment of affinity in HEKNOP and HEKhNOP‐GFP. These values were not different from the native peptide, but coupling of NOP to GFP has reduced binding affinity. At 4°C, there is the potential for non‐equilibrium and an ideal solution would be to confirm affinity using association/dissociation time courses. In this paradigm, (i) the chamber floods with ‘free’ label making it impossible to determine bound ligand (i.e. separate from free) and (ii) experiments would be very long as the label is added cumulatively with the potential for internalization. We are reassured that our estimate of K D is sensible as the values overlap with [3H]‐N/OFQ binding in membranes, and cognisant of the issues, we used two cell lines to confirm that all values are in good agreement. Measuring binding at the lower temperature followed by rapid warming allows the kinetics of internalization to be assessed. Despite measuring binding at 4°C, it is still possible to track receptor activation, at least at the level of Ca2+ in cells co‐expressing NOP and Gαqi5 chimeric G‐protein. A similar shaped but more rapid response is also seen at 37°C. The kinetics at the lower temperature are substantially slower than we have previously reported for experiments performed at the higher temperature in population measurements, but the shape of the response is similar at all temperatures (Camarda and Calo, 2013).
Studies with GPCRs are, in general, hampered by lack of good antibodies for use in Western blotting (Hamdani and van der Velden, 2009; Jensen et al., 2009; Pradidarcheep et al., 2009; Berahovich et al., 2010; Cecyre et al., 2014; Talmont and Mouledous, 2014). This is likely due to the high degree of structural conservation both between families and more specifically in members of the same family, for example, the opioid family. Use of knockout tissue controls is often lacking, and as such, the validity of work using these probes could be questioned; this is not the case with our highly selective N/OFQATTO594. A more conventional strategy is to use radiolabelled probes. For NOP, this includes [3H]‐N/OFQ (McDonald et al., 2003), [3H]‐N/OFQ(1–13)‐NH2 (Hashiba et al., 2002), [125I]‐N/OFQ (Singh et al., 2013) and [3H]‐UFP‐101 [(Bird et al., 2016); their use is best employed where there is favourable access to tissue or where radioligand specific activity is high (e.g. with 125I). There are a series of elegant studies using radioligands for NOP in autoradiographic protocols (Slowe et al., 2001), but these retain the same issues regarding expression. Radioligands are in general not suited to use in tissues with presumed low expression, for example, vas deferens (Guerrini et al., 1998) and on immune cells (see below).
A further approach to track receptor fate is to use fluorescently labelled receptors, as we have done here with NOPGFP receptors. An elegant example of how this can be used comes from the work of Evans et al. (2010). In this study, the authors produced fluorescently labelled isoforms of μ, δ and κ along with NOP receptors. Using a confocal paradigm, they went on to address receptor dimerization. They measured the degree of colour overlap in a manner similar to our data set examining overlap of GFP and ATTO to visualize ligand–receptor interaction either in the ‘conventional’ sense or in a FRET protocol. They conclusively showed that NOP was capable of hetero‐dimerization with all members of the opioid family (Evans et al., 2010).
In this study, as controls for FRET, we ensured that 488 nm wavelength alone did not stimulate ATTO (HEKNOP cells without GFP tag) and in the presence of the GFP tag SB‐612111 (NOP antagonist) inhibits any FRET response. This indicates (i) ATTO is being stimulated by the emission spectra of GFP not through the laser, and (ii) it must be bound to the receptor to see the response, that is, locality which is the basis of FRET. Furthermore, photobleaching of the acceptor (N/OFQATTO594) leads to increased fluorescence measurements of GFP, a positive indicator of FRET pairing (Snapp and Hegde, 2006).
It has been known for many years that opioids are immune modulators (Vallejo et al., 2004), recently reviewed by Plein and Ritter (2018). The site of immune modulation is contentious, and we have reviewed this topic recently (Al‐Hashimi et al., 2013). Opioids could interact directly with the immune cell, modulate the activity of the hypothalamic–pituitary–adrenal axis and/or exert central actions involving glia (Hutchinson et al., 2011). The expression of opioid receptors on immune cells is the most contentious. There is well known modulation of immune cell activity (migration and cytokine release) (Liang et al., 2016), but we have failed to detect classical opioid (μ, δ and κ) receptor mRNA in any peripheral circulating immune cells (Al‐Hashimi et al., 2016). This is not consistent with the modulation of function, and in this area particularly, the lack of reliable antibodies for Western blot is a major disadvantage. It is possible that opioids could be working via a non‐opioid mechanism such as TLR4 receptors (Franchi et al., 2012), or there could be differences in circulating and resident immune cells. There is no evidence for the latter in the area of opioid pharmacology. What is clear is that all circulating immune cells that we have examined to date expressed mRNA for NOP receptors, and we and others have been able to report modulation of immune function (Singh et al., 2016). We have attempted to measure [125I]‐N/OFQ and [3H]‐N/OFQ binding to membranes from circulating mixed human immune cells (predominantly polymorphs). These experiments failed despite the use of relatively large amounts of membrane tissue, and we infer this is due to ultra‐low expression. After careful characterization in high expressing recombinant systems, in the present study, we went on to use N/OFQATTO594 in polymorphs from human volunteers and were able to detect binding. The small size of these immune cells (relative to the recombinants) and resolution of the microscope are limiting factors in pictorially demonstrating membrane location (see Supporting Information Figure S4). However, we were able to detect binding that could be blocked by pre‐occupying NOP with N/OFQ or the selective NOP antagonist SB‐612111 indicating both selectivity of binding to NOP and membrane location. Moreover, unlabelled N/OFQ could be effectively washed and replaced with N/OFQATTO594. These results demonstrate that NOP receptor mRNA measured in PCR experiments is effectively translated into protein capable of binding N/OFQ and, unlike classical opioid receptors, provides a target to explain the observed immune modulation.
Upon activation opioid receptors are internalized in an arrestin‐driven fashion (Williams et al., 2013). This is a fairly standard mechanism utilized by a number of GPCRs (Peterson and Luttrell, 2017) and leads to reduced cellular responsiveness or desensitization. For the NOP receptor, we have used a BRET protocol (using Renilla luciferase‐NOP and Renilla GFP on arrestin) to show efficient arrestin coupling (Malfacini et al., 2015). We have used radioligand binding to measure loss of cell surface NOP receptors following a desensitizing challenge. These latter studies were in high expressing (Hashimoto et al., 2002) or inducible expressing (Barnes et al., 2007) systems. The issues with these types of experiments is complete removal of the desensitizing challenge as any residual receptor occupancy would effectively reduce apparent density leading to an erroneous conclusion as to loss of receptors. The fluorescent probe we have designed, as an agonist itself, overcomes these problems. In this study, we show that receptors on the cell surface (GFP tagged) with N/OFQ (ATTO) bound leave the cell surface together (line analysis); ligand and receptor appear to co‐localize inside the cell. Use of N/OFQATTO594 to study internalization and the role of arrestins is particularly apposite here as there is now compelling evidence to suggest that agonists biased away from arrestin recruitment produce analgesia without the development of tolerance, a game changer in the development of analgesics for chronic pain.
Moreover, it is also possible to measure the ligand receptor interaction process in a FRET‐based assay coupling the ATTO‐labelled peptide with a GFP‐labelled receptor in cell lines. This will also be possible in NOP‐eGFP knock‐in mice (Ozawa et al., 2015).
In summary, we describe a novel ligand for use in the study of live‐cell ligand–receptor interaction and tracking movement of liganded cell surface receptors. Use of higher resolution confocal technologies will facilitate a more detailed study of the ‘unbinding’ and recycling process. Interesting further possibilities for this new ligand include its use in (i) brain sections and (ii) receptor binding protocols where the fluorescent label totally replaces the radioligand; this would revolutionize binding methodology for this receptor and potentially other members of the opioid family.
Author contributions
M.F.B. collected and analysed the data and wrote the paper. R.G. designed and synthesised the ligand and wrote the paper. J.M.W. and J.P.T. wrote the paper and obtained funding. G.C. designed the ligand and wrote the paper. D.G.L. analysed the data, wrote the paper and obtained funding.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This http://onlinelibrary.wiley.com/doi/10.1111/bph.13405/abstract acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Data S1 Chemistry.
Figure S1 Analytical HPLC profile of the [Cys(ATTO 594)18]N/OFQ‐NH2 reaction mixture.
Figure S2 Analytical HPLC profile of the purified [Cys(ATTO 594)18]N/OFQ‐NH2.
Figure S3 Mass spectrum of the purified [Cys(ATTO 594)18]N/OFQ‐NH2.
Figure S4 A limited image z‐series stack for 100 nM N/OFQATTO594 binding to PMN is presented, panel A. In panel B, representative ‘slices’ at top, middle and bottom are shown.
Video S1 N/OFQATTO594 binding and increased Ca2+ at 4°C.
Acknowledgements
This study is funded by Biotechnology and Biological Sciences Research Council (BB/N000188/1). We thank the Advanced Imaging Facility (Dr K Straatman) at the University of Leicester for their support.
Bird, M. F. , Guerrini, R. , Willets, J. M. , Thompson, J. P. , Caló, G. , and Lambert, D. G. (2018) Nociceptin/Orphanin FQ (N/OFQ) conjugated to ATTO594: a novel fluorescent probe for the N/OFQ (NOP) receptor. British Journal of Pharmacology, 175: 4496–4506. 10.1111/bph.14504.
References
- Ahmadi F, Dabirian S, Faizi M, Tabatabai SA, Beiki D, Shahhosseini S (2014). Optimum conditions of radioligand receptor binding assay of ligands of benzodiazepine receptors. Iran J Pharm Res: IJPR 13: 79–86. [PMC free article] [PubMed] [Google Scholar]
- Al‐Hashimi M, Scott SWM, Thompson JP, Lambert DG (2013). Opioids and immune modulation: more questions than answers. BJA: Br J Anaesth 111: 80–88. [DOI] [PubMed] [Google Scholar]
- Al‐Hashimi M, McDonald J, Thompson JP, Lambert DG (2016). Evidence for nociceptin/orphanin FQ (NOP) but not μ (MOP), δ (DOP) or κ (KOP) opioid receptor mRNA in whole human blood. BJA: Br J Anaesth 116: 423–429. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Marrion NV, Peters JA et al (2017). The concise guide to pharmacology 2017/18: G protein‐coupled receptors. Br J Pharmacol 174: S17–S129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arttamangkul S, Alvarez‐Maubecin V, Thomas G, Williams JT, Grandy DK (2000). Binding and internalization of fluorescent opioid peptide conjugates in living cells. Mol Pharmacol 58: 1570–1580. [DOI] [PubMed] [Google Scholar]
- Barnes TA, McDonald J, Rowbotham DJ, Duarte TL, Lambert DG (2007). Effects of receptor density on nociceptin/orphanin FQ peptide receptor desensitisation: studies using the ecdysone inducible expression system. Naunyn Schmiedebergs Arch Pharmacol 376: 217–225. [DOI] [PubMed] [Google Scholar]
- Berahovich RD, Penfold ME, Schall TJ (2010). Nonspecific CXCR7 antibodies. Immunol Lett 133: 112–114. [DOI] [PubMed] [Google Scholar]
- Bird MF, Vardanyan RS, Hruby VJ, Calo G, Guerrini R, Salvadori S et al (2015). Development and characterisation of novel fentanyl‐δ opioid receptor antagonist based bivalent ligands. Br J Anaesth 114: 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MF, Cerlesi MC, Brown M, Malfacini D, Vezzi V, Molinari P et al (2016). Characterisation of the novel mixed μ‐NOP peptide ligand dermorphin‐N/OFQ (DeNo). PLoS One 11: e0156897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A (2010). Loss of human greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B‐Cdc2/PP2A balance. Proc Natl Acad Sci U S A 107: 12564–12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarda V, Calo G (2013). Chimeric G proteins in fluorimetric calcium assays: experience with opioid receptors. Methods Mol Biol (Clifton, NJ) 937: 293–306. [DOI] [PubMed] [Google Scholar]
- Cecyre B, Thomas S, Ptito M, Casanova C, Bouchard JF (2014). Evaluation of the specificity of antibodies raised against cannabinoid receptor type 2 in the mouse retina. Naunyn Schmiedebergs Arch Pharmacol 387: 175–184. [DOI] [PubMed] [Google Scholar]
- Ding H, Hayashida K, Suto T, Sukhtankar DD, Kimura M, Mendenhall V et al (2015). Supraspinal actions of nociceptin/orphanin FQ, morphine and substance P in regulating pain and itch in non‐human primates. Br J Pharmacol 172: 3302–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, You H, Hameed S, Altier C, Mezghrani A, Bourinet E et al (2010). Heterodimerization of ORL1 and opioid receptors and its consequences for N‐type calcium channel regulation. J Biol Chem 285: 1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi S, Moretti S, Castelli M, Lattuada D, Scavullo C, Panerai AE et al (2012). Mu opioid receptor activation modulates Toll like receptor 4 in murine macrophages. Brain Behav Immun 26: 480–488. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Calo G, Rizzi A, Bigoni R, Bianchi C, Salvadori S et al (1998). A new selective antagonist of the nociceptin receptor. Br J Pharmacol 123: 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y et al (2008). How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim Biophys Sin 40: 426–436. [DOI] [PubMed] [Google Scholar]
- Hamdani N, van der Velden J (2009). Lack of specificity of antibodies directed against human β‐adrenergic receptors. Naunyn Schmiedebergs Arch Pharmacol 379: 403–407. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS guide to pharmacology in 2018: updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiba E, Lambert DG, Farkas J, Toth G, Smith G (2002). Comparison of the binding of [3H]nociceptin/orphaninFQ(1–13)NH2, [3H]nociceptin/orphaninFQ(1–17) OH and [125I]Tyr14nociceptin/orphaninFQ(1–17) OH to recombinant human and native rat cerebrocortical nociceptin/orphanin FQ receptors. Neurosci Lett 328: 5–8. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Calo G, Guerrini R, Smith G, Lambert DG (2002). Effects of chronic nociceptin/orphanin FQ exposure on cAMP accumulation and receptor density in Chinese hamster ovary cells expressing human nociceptin/orphanin FQ receptors. Eur J Pharmacol 449: 17–22. [DOI] [PubMed] [Google Scholar]
- Hutchinson MR, Shavit Y, Grace PM, Rice KC, Maier SF, Watkins LR (2011). Exploring the neuroimmunopharmacology of opioids: an integrative review of mechanisms of central immune signaling and their implications for opioid analgesia. Pharmacol Rev 63: 772–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, Swigart PM, Simpson PC (2009). Ten commercial antibodies for alpha‐1‐adrenergic receptor subtypes are nonspecific. Naunyn Schmiedebergs Arch Pharmacol 379: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama M, McDonald J, Barnes TA, Calo G, Guerrini R, Rowbotham DJ et al (2007). In vitro pharmacological characterisation of a novel cyclic nociceptin/orphanin FQ analogue c [Cys(7,10)]N/OFQ(1‐13) NH (2). Naunyn Schmiedebergs Arch Pharmacol 375: 369–376. [DOI] [PubMed] [Google Scholar]
- Kruger C, Kothe L, Struppert A, Pietruck C, Simm A, Grond S (2006). Expression und function of the ORL‐1 receptor on human leukocytes. Schmerz (Berlin, Germany) 20: 509–518. [DOI] [PubMed] [Google Scholar]
- Lambert DG (2008). The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat Rev Drug Discov 7: 694–710. [DOI] [PubMed] [Google Scholar]
- Liang X, Liu R, Chen C, Ji F, Li T (2016). Opioid system modulates the immune function: a review. Transl Perioper Pain Med 1: 5–13. [PMC free article] [PubMed] [Google Scholar]
- Malfacini D, Ambrosio C, Gro’ MC, Sbraccia M, Trapella C, Guerrini R et al (2015). Pharmacological profile of nociceptin/orphanin FQ receptors interacting with G‐proteins and β‐arrestins 2. PLoS One 10: e0132865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J, Barnes TA, Okawa H, Williams J, Calo G, Rowbotham DJ et al (2003). Partial agonist behaviour depends upon the level of nociceptin/orphanin FQ receptor expression: studies using the ecdysone‐inducible mammalian expression system. Br J Pharmacol 140: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier JC, Mollereau C, Toll L, Suaudeau C, Moisand C, Alvinerie P et al (1995). Isolation and structure of the endogenous agonist of opioid receptor‐like ORL1 receptor. Nature 377: 532–535. [DOI] [PubMed] [Google Scholar]
- Michel MC, Wieland T, Tsujimoto G (2009). How reliable are G‐protein‐coupled receptor antibodies? Naunyn Schmiedebergs Arch Pharmacol 379: 385–388. [DOI] [PubMed] [Google Scholar]
- Mollereau C, Mouledous L (2000). Tissue distribution of the opioid receptor‐like (ORL1) receptor. Peptides 21: 907–917. [DOI] [PubMed] [Google Scholar]
- Niwa H, Rowbotham DJ, Lambert DG (2012). Evaluation of primary opioid receptor antibodies for use in western blotting. Br J Anaesth 108: 530–532. [DOI] [PubMed] [Google Scholar]
- Ozawa A, Brunori G, Mercatelli D, Wu J, Cippitelli A, Zou B et al (2015). Knock‐in mice with NOP‐eGFP receptors identify receptor cellular and regional localization. J Neurosci 35: 11682–11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson YK, Luttrell LM (2017). The diverse roles of arrestin scaffolds in G protein‐coupled receptor signaling. Pharmacol Rev 69: 256–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plein LM, Rittner HL (2018). Opioids and the immune system – friend or foe. Br J Pharmacol 175: 2717–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradidarcheep W, Stallen J, Labruyere WT, Dabhoiwala NF, Michel MC, Lamers WH (2009). Lack of specificity of commercially available antisera against muscarinergic and adrenergic receptors. Naunyn Schmiedebergs Arch Pharmacol 379: 397–402. [DOI] [PubMed] [Google Scholar]
- Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR et al (1995). Orphanin FQ: a neuropeptide that activates an opioid like G protein‐coupled receptor. Science 270: 792–794. [DOI] [PubMed] [Google Scholar]
- Scherrer G, Imamachi N, Cao YQ, Contet C, Mennicken F, O'Donnell D et al (2009). Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell 137: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder W, Lambert DG, Ko MC, Koch T (2014). Functional plasticity of the N/OFQ‐NOP receptor system determines analgesic properties of NOP receptor agonists. Br J Pharmacol 171: 3777–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Sullo N, Bradding P, Agostino B, Brightling C, Lambert D (2013). Role of nociceptin orphanin FQ peptide – receptor system in mast cell migration. Eur Respir J 42. [Google Scholar]
- Singh SR, Sullo N, Matteis M, Spaziano G, McDonald J, Saunders R et al (2016). Nociceptin/orphanin FQ (N/OFQ) modulates immunopathology and airway hyperresponsiveness representing a novel target for the treatment of asthma. Br J Pharmacol 173: 1286–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowe SJ, Clarke S, Lena I, Goody RJ, Lattanzi R, Negri L et al (2001). Autoradiographic mapping of the opioid receptor‐like 1 (ORL1) receptor in the brains of μ‐, δ‐ or κ‐opioid receptor knockout mice. Neuroscience 106: 469–480. [DOI] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS (2006). Rational design and evaluation of FRET experiments to measure protein proximities in cells. Curr Protoc Cell Biol Chapter 17: Unit 17 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W (2001). Trypan blue exclusion test of cell viability. Curr Protoc Immunol Appendix 3: Appendix 3B. [DOI] [PubMed] [Google Scholar]
- Talmont F, Mouledous L (2014). Evaluation of commercial antibodies against human sphingosine‐1‐phosphate receptor 1. Naunyn Schmiedebergs Arch Pharmacol 387: 427–431. [DOI] [PubMed] [Google Scholar]
- Thompson JP, Serrano‐Gomez A, McDonald J, Ladak N, Bowrey S, Lambert DG (2013). The nociceptin/orphanin FQ system is modulated in patients admitted to ICU with sepsis and after cardiopulmonary bypass. PloS One 8: e76682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R, de Leon‐Casasola O, Benyamin R (2004). Opioid therapy and immunosuppression: a review. Am J Ther 11: 354–365. [DOI] [PubMed] [Google Scholar]
- Wallrabe H, Elangovan M, Burchard A, Periasamy A, Barroso M (2003). Confocal FRET microscopy to measure clustering of ligand‐receptor complexes in endocytic membranes. Biophys J 85: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S et al (2013). Regulation of mu‐opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol Rev 65: 223–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Stuber F, Stamer UM (2013). Inflammatory mediators influence the expression of nociceptin and its receptor in human whole blood cultures. PloS One 8: e74138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Chemistry.
Figure S1 Analytical HPLC profile of the [Cys(ATTO 594)18]N/OFQ‐NH2 reaction mixture.
Figure S2 Analytical HPLC profile of the purified [Cys(ATTO 594)18]N/OFQ‐NH2.
Figure S3 Mass spectrum of the purified [Cys(ATTO 594)18]N/OFQ‐NH2.
Figure S4 A limited image z‐series stack for 100 nM N/OFQATTO594 binding to PMN is presented, panel A. In panel B, representative ‘slices’ at top, middle and bottom are shown.
Video S1 N/OFQATTO594 binding and increased Ca2+ at 4°C.


