Abstract
The fate and activity of drugs are frequently dictated not only by the host per se but also by the microorganisms present in the gastrointestinal tract. The gut microbiome is known to, both directly and indirectly, affect drug metabolism. More evidence now hints at the effects that drugs can have on the function and composition of the gut microbiome. Both microbiota‐mediated alterations in drug metabolism and drug‐mediated alterations in the gut microbiome can have beneficial or detrimental effects on the host. Greater insights into the mechanisms driving these reciprocal drug–gut microbiota interactions are needed to guide the development of microbiome‐targeted dietary or pharmacological interventions, which may have the potential to enhance drug efficacy or reduce drug side effects. In this review, we explore the relationship between drugs and the gut microbiome, with a specific focus on potential mechanisms underpinning the drug‐mediated alterations on the gut microbiome and the potential implications for psychoactive drugs.
Linked Articles
This article is part of a themed section on When Pharmacology Meets the Microbiome: New Targets for Therapeutics? To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v175.24/issuetoc
Abbreviations
- B‐GOS
Bimuno™ galacto‐oligosaccharides
- CDI
Clostridium difficile infection
- CYP
cytochrome P450
- FMT
fecal microbiota transplantation
- GF
germ‐free
- GI
gastrointestinal
- HFD
high‐fat diet
- MAOI
MAO inhibitor
- MDD
major depressive disorder
- NSAID
non‐steroidal anti‐inflammatory drug
- PEG
polyethylene glycol
- PKPD
pharmacokinetics/pharmacodynamics
- PPI
proton pump inhibitor
- SCFA
short chain fatty acids
- SSRI
selective serotonin reuptake inhibitor
- TCA
tricyclic antidepressant
Introduction
Although not prominently featured in the classical teaching of pharmacokinetics and pharmacodynamics (PKPD), the fate and activity of drugs are frequently decided not only by the host per se but also by the collection of microorganisms present in the gastrointestinal (GI) tract. These microorganisms represent the gut microbiota, the trillions of microbes with more than 700–1000 different bacterial species which reside in the gut (Qin et al., 2010). The terms microbiota and microbiome are often used interchangeably in the literature but can be differentiated based on their more recent definitions. The term ‘microbiota’ refers to the collection of microorganisms, whereas the term ‘microbiome’ refers to the collective genomes of these microorganisms (Jandhyala et al., 2015; Marchesi et al., 2016). It is estimated that 10 different phyla are thought to contribute to the functional role of the gut microbiome; Firmicutes and Bacteroidetes are the most dominant phyla. Indeed, the ratio of Firmicutes to Bacteroidetes (F/B ratio) is often used as a surrogate indicator of bacterial shifts, although it may be an overly simplistic descriptor of variations in human gut microbiota composition (Mariat et al., 2009). Many factors have been identified that influence the biogeography and composition (both abundance and diversity) of bacteria along the GI tract including pH variation, diet, mucus, host immunity and environmental factors (Thursby and Juge, 2017). For example, the acidic environment of the stomach has a sparse microbiota (101 bacteria/g) compared to the estimated 103 bacteria/g found in the less acidic small intestine, which is the main site of drug absorption. The neutral pH or weakly basic environment found in the large intestine is the most densely colonized area of the GI tract, with an estimated 1012 bacteria/g (O'Hara and Shanahan, 2006).
The gut microbiome is implicated in the maturation of the immune system, nutrient absorption, energy homeostasis and protection against pathogens (Antunes et al., 2011; Jandhyala et al., 2015). It also plays a significant role in health, both in homeostasis and, likely, in the pathogenesis as well as treatment of disease. The initial composition and assembly of the gut microbiome are considered unique among individuals, and research has identified many factors which affect its composition including age, ethnicity, gender, environmental factors, diet and lifestyle (Quigley, 2017). For example, the composition of the gut microbiota in the newborn is dictated by the method of delivery, gestational age, infant antibiotic exposure and method of feeding (formula vs. breastfeeding) (Penders et al., 2006; Salazar et al., 2014). Though considered highly stable and resilient to change during adulthood, instability and variability in the gut microbiome become more prevalent during disease and at the extremes of life (e.g. early infancy and senescence) (Salazar et al., 2014). The young and the old are also considered to be the most vulnerable patient groups regarding the occurrence of adverse drug reactions and display the most variation in drug pharmacokinetic responses (Turnheim 2003).
The implication of the gut microbiome in drug metabolism is gaining more traction and has led to the emergence of a new term, pharmacomicrobiomics (Rizkallah et al., 2010). Pharmacomicrobiomics investigates the effect of variations of the gut microbiome on drugs, through the lens of PKPD. In this review, we explore the relationship between drugs and the gut microbiota, with a specific focus on the potential mechanisms underpinning the drug‐mediated alterations on the gut microbiota and the potential implications for the PKPD profile of drugs. With the increasing development of extended‐release drug formulations, especially in the field of neuropharmacology, which may increase the proportions of the drug reaching the bacterially dense large intestine, a higher number of psychoactive drugs, which are subject to microbiota‐mediated degradation, may be identified. While the gut microbiome may affect all classes of drugs, the purpose of this review is to focus specifically on psychoactive drugs given that most of these drugs display high lipophilicity to ensure CNS penetration (Alavijeh et al., 2005). In general, highly lipophilic drugs are at greater risk of poor drug absorption from the intestine; hence, any microbial‐mediated changes in intestinal absorption or metabolism merit further consideration. Developing a greater understanding of these important, but underappreciated, drug‐microbiota dynamics could help to identify drug–drug interactions and perhaps explain inter‐individual variation in drug efficacy and adverse effects.
The effects of drug therapy on the gut microbiome
Drug effects on the gut microbiome
Recently, two large‐scale human observational studies highlighted correlations between the gut microbiota and the intake of different classes of medication. Falony et al. (2016) compared the composition of the gut microbiota against the use of β‐lactam antibiotics, nitrofuran antibiotics (nitrofurantoin), an osmotic laxative, biological agents (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5074 inhibitor), disease modifying anti‐rheumatic drugs (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2700 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7120), oestrogen and progesterone hormones, benzodiazepines (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6963), antidepressants (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7321) and antihistamines. While 95% of samples had a similar core microbiome, medication exposure was identified as the factor causing greatest variability in the study cohort, with 63% of the detected covariate interactions driven by medication. A positive correlation, for example, was identified between the abundance of a species from the Eggerthella and Coprabacillus genera and medication use. In another metagenomic‐based study, Zhernakova et al. (2016) identified extrinsic and intrinsic factors correlating with microbial shifts in a Dutch population study. Forty‐four categories of drugs were tested in the analysis and antibiotics, gastric acid suppressants [proton pump inhibitors (PPIs)], lipid‐lowering medication (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=639 inhibitors or ‘statins’), anti‐hyperglycaemic (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4779) and laxatives were found to have a distinct effect on the gut microbiome. PPIs were, for example, associated with profound changes in 33 bacterial pathways, and metformin use positively correlated with Escherichia coli abundance. While a wide range of medication has been identified to interact with the gut microbiota, certain drug classes, including antibiotics, anti‐inflammatories, gastric acid suppressants, anti‐hyperglycaemics and psychotropics, have repeatedly been shown to have distinct gut microbiota signatures and will be discussed further in the review (see sections: Secondary effects of drugs on the gut microbiome and Effects of psychoactive drugs on the composition of the gut microbiota sections).
Combination of drugs and polypharmacy
The composition of the microbiota can vary depending on both the number and combination of medication ingested. A significant difference in microbiota composition was observed in patients taking a single drug on a long‐term basis in comparison to non‐drug taking patients (Rogers and Aronoff, 2016). Moreover, the combined use of non‐steroidal anti‐inflammatory drugs (NSAIDs) and PPIs differentially influenced the relative abundance of Bacteroides spp. and Erysipelotrichaceae spp., compared with NSAIDs alone. Similarly, Bacteroides spp. and a species belonging to the Ruminococcaceae family differentiated individuals who were concomitantly taking NSAIDs, antidepressants and laxatives from NSAID‐only users. Therefore, the composition of the gut microbiota may depend on an individual's drug use. In this study, the diversity of the gut microbiota was not significantly influenced by the number of medications taken, even though it was significantly influenced by the specific type of medication ingested by study participants. Of note, the median number of medications taken by the community‐dwelling subjects was 4, which falls below the threshold for polypharmacy. Once this threshold is reached (i.e. ≥5 concomitantly taken medications), an effect is frequently observed. A significant decrease in species richness, a measurement of the number of species present in an area and significant alterations in the relative abundance of 15 bacterial taxa have been reported in patients taking more than five concomitant medications (Ticinesi et al., 2017). Specifically, the relative abundance of the Helicobacter genus was significantly associated with polypharmacy, and an inverse relationship between polypharmacy and the abundance of the Lachnospiraceae and Succinivibrionaceae families was also found. The co‐administration of drugs may precipitate shifts in the composition of the microbiota to favour the abundance of microbial taxa that have metabolizing capacity for those drugs (Ticinesi et al., 2017). Not only is it important, therefore, to evaluate specific microbiota alterations induced by a single drug but it is also imperative to account for differences when multiple drugs are co‐administered. Further study of the effects of multiple co‐administered drugs on the gut microbiota is very relevant due to the increasing prevalence of polypharmacy in the ageing population (Dagli and Sharma, 2014). In a European study across eight countries, the prevalence of polypharmacy in 4023 nursing home residents, the mean age of whom was 83.5 (SD 9.3) years, was found to be almost 50%, while excessive polypharmacy (defined as ≥10 medication) was evident in an additional 24.3% of residents, which serves to highlight the potential for drug‐associated effects on the microbiota in a vulnerable population in which the gut microbiome is already compromised (Claesson et al., 2012; Onder et al., 2012).
Duration of drug treatment and recovery
In vitro evidence suggests that even a single dose of antibiotic or short‐term antibiotic courses, albeit at high doses, can significantly affect the microbiome (Maurice et al., 2013). Specifically, eight different antibiotics altered the fecal microbiota after a 4 hour incubation, and a significantly increased proportion of cells associated with loss of membrane integrity and altered polarity was observed. Most antibiotic‐induced alterations to the microbiome are reversed upon cessation of treatment. Some modifications can, however, persist after treatment (Jakobsson et al., 2010). For example, the combined treatment with the antibiotics clarithromycin and metronidazole and a PPI, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4279, induced alterations to macrolide resistance gene, ermB, which persisted in patients for up to 4 years after treatment.
Additionally, the antipsychotic, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=96, was associated with a reduction in the F/B ratio, which was evident at the 2–3 month follow‐up of treatment, but a further reduction in Bacteroidetes, and increased weight gain, was observed after prolonged use (Bahr et al., 2015a). The continued use of drugs may, therefore, strengthen microbiota alterations, which may be evident from initiation of treatment, but further comparisons between short‐term and long‐term users of medication are required.
Excipient effects
An excipient is a pharmacologically inert substance or non‐active ingredient that is added to a formulation to stabilize the active substance or enhance the function of the dosage form (Debotton and Dahan, 2017). Most excipients are not absorbed from the gut lumen. Moreover, polysaccharide‐based formulations are often utilized in the delivery of probiotics and colonic‐targeted drug dosage forms, for example, targeted local delivery of the anti‐inflammatory http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4840 (Kosaraju 2005; Prudhviraj et al., 2015). Recent research hints at the possibility that the excipients themselves might mediate microbiota changes, independent of changes induced by the drug. Recently, it was hypothesized that the presence of fermentable polysaccharides as drug formulation excipients may act as metabolic substrates for the gut microbiota, thereby promoting their growth (Ticinesi et al., 2017). Additionally, polyethylene glycol (PEG), a polymer used in drug delivery, is linked to changes in GI transit time and changes in the gut microbiota (Kashyap et al., 2013). Humanized mice [ex‐germ‐free (GF) mice colonized with human faecal microbiota] fed a standard diet supplemented with 15% PEG 3350 for 10 days, had significantly reduced abundance of the Peptococcaceae and Eubacteriaceae bacterial families and the Anaeroplasmataceae order. As the dosage used in this animal study was expressed as %w/w of the animal chow, it is, however, difficult to extrapolate this dose to that used in pharmaceutical formulations. PEGs are commonly included in dosage forms, with diverse excipient functions including as solubilizing agents, tablet binders, plasticizers in film coating, tablet lubricants and vehicles (D'souza and Shegokar, 2016). Additional studies are warranted to elucidate the potential effect of the different categories of PEG polymers used in drug formulations at pharmaceutically relevant concentrations. Excipient‐induced changes in the composition or functionality of the microbiome could potentially complicate the interpretation of drug–microbiota‐based observational studies, as it may be difficult to differentiate drug‐induced changes from excipient‐induced changes in the gut microbiome. More research is needed to evaluate the independent effects of different formulation excipients, and the possible unintended consequences that their inclusion in drug formulations could have on the gut microbiome.
Gut microbiome drug–drug interactions
Drug‐induced changes in the gut microbiome may alter the pharmacokinetics of concomitantly taken medication. An increased plasma level of the antiplatelet drug, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4139, was observed in rats who were treated with a β‐lactam antibiotic, ampicillin, 3 days previously (Kim et al., 2016). This enhanced bioavailability was attributed to the antibiotic‐induced suppression of the metabolic activity of the gut microbiome. Moreover, ampicillin treatment significantly prolonged bleeding time in aspirin‐treated rats, suggesting antibiotic treatment may potentiate its anti‐thrombotic effect. Similar microbiome‐mediated drug interactions have been demonstrated for both the lipid‐lowering drug, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2739, and the antihypertensive, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6981 (Yoo et al., 2014; Yoo et al., 2016).
Gut microbiota–drug interactions
The gut microbiome can both directly metabolize drugs and indirectly influence host metabolizing capacity of drugs (Haiser and Turnbaugh, 2013; Wilson and Nicholson, 2017). While many drugs undergo microbial biotransformation, the specific microorganisms involved are often unknown. In this review, we will mainly refer to bacterial drug‐metabolizing enzymes, and the effects of the gut microbiome on the metabolism of drugs by the liver, as the primary focus of the review, is the reciprocal effect drugs can have on the gut microbiome, a relatively unexplored and unappreciated area in the literature. Microbial drug metabolism has been extensively reviewed recently (Koppel et al., 2017; Wilson and Nicholson, 2017).
Drug metabolism by microbial enzymes
Microbial‐derived drug‐metabolizing enzymes have been implicated both in the activation and inactivation of drugs. For example, the activation of the prodrug sulfasalazine into its anti‐inflammatory active moiety mesalazine (5‐aminosalicyclic acid or 5‐ASA) is mediated by bacterial azoreductase (Peppercorn and Goldman, 1972). The direct metabolism of drugs by microbial‐derived enzymes has been shown for a variety of different drugs, examples of which are detailed in Table 1. In contrast to the oxidation and conjugation reactions characteristic of hepatic metabolism, reduction and hydrolysis reactions dominate gut microbiota‐mediated metabolic reactions (Haiser and Turnbaugh, 2013). β‐Glucuronidase is among the most studied bacterial drug‐metabolizing enzymes due to its role in the deconjugation of hepatically glucuronidated metabolites and the resultant enterohepatic recycling of drugs (Takasuna et al., 1998; Klaassen and Cui, 2015). Multiple different bacterial genera, including Clostridium, Streptococcus, Lactobacillus, Ruminococcus and Bifidobacterium, express β‐glucuronidase (Gloux et al., 2011).
Table 1.
The metabolism of drugs by bacterial drug‐metabolizing enzymes
| Microbiota‐derived enzyme | Hypothesized reaction mechanisms | Drug (or metabolite) substrate | References |
|---|---|---|---|
| β‐Glucuronidase | Remove glucuronic acid moiety from hepatic phase 2 metabolites |
http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6823 (SN‐38 glucuronide) NSAIDS, for example, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1909 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2714 |
Yamamoto et al. (2008) and Saitta et al. (2014) |
| Azoreductase | Reduction of azo or quinone bonds |
Azo‐containing drugs, for example, olsalazine (5‐ASA prodrug) Nitrofuran antibiotics, for example, nitrofurazone and nitrofurantoin Ester‐containing prodrugs |
Ryan (2017) |
| Carboxylesterase |
Hydrolyse ester, thioester, amide or carbamate containing drugs to corresponding free acids Hydrolyse esters to carboxylic acids |
Aspirin, ester‐containing prodrugs | Imai and Ohura (2010), Laizure et al. (2013) and Kim et al. (2016) |
| Nitroreductase | Reduction of nitro group |
Metronidazole Benzodiazepines |
Koch et al. (1979), Elmer and Remmel (1984) and Takeno et al. (1990) |
| N‐acetyltransferase | Transfer of acetyl group to nitrogen or oxygen atom of primary arylamines, hydrazines and N‐hydroxylated metabolites | 5‐Aminosalicylic acid | van Hogezand et al. (1992) and Deloménie et al. (2001) |
| β‐Lyase | Cleavage of C–S bond in hepatic‐production cysteine‐S‐conjugated metabolites |
Cysteine‐conjugated metabolites Bio‐activation of sulfur and selenocysteine derivatives |
Mikov (1994) |
| Sulfatases | Hydrolysis of sulfate esters utilizing formylglycine | Sulfate ester hepatic metabolites | Ulmer et al. (2014) and Koppel et al. (2017) |
The development of a chemically guided functional strategy to identify, quantify and assign functionality of enzymes associated with the gut microbiome is among the recent advances in this area. This strategy led to the identification of trans‐4‐hydroxy‐l‐proline dehydratase as a microbial enzyme implicated in the metabolism of a non‐proteinogenic amino acid, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4704 (Levin et al., 2017). Application of this strategy for the identification and characterization of microbial drug‐metabolizing enzymes holds much potential and could uncover previously unknown metabolic activities of the gut microbiome.
Indirect effects of the gut microbiome on host drug metabolism
It was traditionally assumed that only drugs reaching the colon are subject to microbiota‐associated alterations with relevance to drug PKPD. The concerted role of the gut bacteria and the liver in the enterohepatic recirculation of drugs is not a new concept (Takasuna et al., 1998). However, more recently, it was highlighted that the gut microbiome could also influence hepatic function, which may, in turn, precipitate changes in patient response to drugs.
Microbial‐derived metabolites can mimic and compete with drug intermediates of hepatic metabolic reactions and thereby interfere with host detoxification pathways. For example, P‐cresol is a microbial metabolite of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4791 or phenylalanine, which competes with http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5239 for the hepatic enzyme, sulfotransferase family 1A member 1 (SULT1A1) (Clayton et al., 2009). This competition for SULT1A1 induces a shift in the metabolism of paracetamol towards alternative host metabolic reactions leading to increased production of the paracetamol metabolite, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6299, whose accumulation has been associated with hepatotoxicity (Mitchell et al., 1973).
Moreover, the gut microbiome can alter the expression of hepatic enzymes or genes responsible for host metabolism. Claus et al. (2011) detected significantly reduced expression of hepatic cytochrome P450 (CYP) 2c29, CYP3a11 and CYP8b1 in GF mice in comparison to conventionally‐raised control mice. Increased expression of hepatic drug‐sensing transcription factors has also been observed in GF mice including the http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2951, a regulator of downstream CYP enzyme expression (Selwyn et al., 2015). However, the mechanisms responsible for the microbiome‐induced changes in the expression of hepatic enzymes requires further exploration. The accumulation of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4577, bile acids and steroid hormones in GF mice has been suggested as a potential mechanism mediating the altered activation of the http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=607, a ‘xenosensor’ nuclear receptor, and further implicates an indirect role for the microbiome in drug metabolism (Björkholm et al., 2009).
Effect of diet‐induced changes on the gut microbiome and drug pharmacokinetics
Diet is a factor that can shape the composition and function of the gut microbiome (Sandhu et al., 2016; Shanahan et al., 2017). It is plausible that in the future, dietary interventions may be utilized to augment drug efficacy or decrease toxicity. The amino acid http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4680 has been shown to inhibit the Eggerthella lenta‐encoding cgr operon, previously identified as essential for the microbiota‐mediated reduction of the cardiac glycoside, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4726 (Haiser et al., 2013). Following confirmation of high levels of cgr operon expression, E. lenta colonized GF mice were fed a high‐protein diet, and significantly increased serum and urine levels of digoxin (following a single 0.2 mg·mL−1 intra‐gastric digoxin dose) were identified. Interestingly, high‐protein diet conferred no effect on digoxin pharmacokinetics in GF mice colonized with a non‐digoxin reducing strain, FAA1‐3‐56, ruling out any indirect effect of host diet. Increased dietary protein, leading to increased arginine, could thus constitute a potential dietary intervention to improve digoxin efficacy. Recent data have also elucidated the role of diet‐induced changes in the gut microbiota on the oral bioavailability of the herbal supplement, berberine, used for the treatment of hyperlipidaemia and type 2 diabetes (Wang et al., 2017). The bioavailability of berberine was significantly increased in high‐fat diet (HFD)‐fed hamsters in comparison to hamsters fed a normal diet. The HFD significantly induced bacterial nitroreductase activity in hamster feces. The authors also observed higher blood concentrations of berberine in individuals with a higher fecal nitroreductase activity, which further corroborated the findings in their animal model.
Effect of probiotics on drug pharmacokinetics
Microbiota‐targeted interventions, including probiotics, prebiotics and antibiotics, may alter drug pharmacokinetics. There are no studies thus far, to our knowledge, exploring the effects of probiotics on psychoactive drug absorption and metabolism. The pharmacokinetics of an oral hypoglycaemic agent, gliclazide (administered as a single dose at 20 mg·kg−1), was significantly altered in diabetic rats pretreated with a cocktail of three probiotics (Lactobacillus acidophilus, Lactobacillus rhamnosus and Bifidobacterium lactis) for 3 days (Al‐Salami et al., 2008). Similarly, probiotic treatment significantly altered the absorption of the anti‐arrhythmic agent http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2566 (Matuskova et al., 2014). In this study, the probiotic E. coli strain Nissile (EcN) 1917 was administered to rats for 7 days, prior to a single p.o. dose of amiodarone hydrochloride (50 mg·kg−1). EcN significantly increased amiodarone bioavailability by 43% in comparison to saline‐treated control rats. No significant effect was observed with the treatment of a non‐probiotic E. coli strain, further indicating that the bacterial‐mediated changes in drug pharmacokinetics may be strain specific.
Probiotics may also affect drug pharmacokinetics by modulating the composition or metabolic activity of the gut microbiota. Recent data showed probiotic treatment significantly increased the microbiota‐mediated degradation of the antipyretic and analgesic, paracetamol; an effect suggested to be mediated by probiotic‐induced modulation of gut microbial enzyme activity (Kim et al., 2018). In this study, the probiotic, Lactobacillus reuteri, significantly increased both sulfatase and arylsulfate transferase and significantly decreased β‐glucuronidase activity during treatment, which are the bacterial enzymes involved in paracetamol metabolism. Following a 24 hour washout period after pretreatment with L. reuteri, a single dose of paracetamol [by i.v. (0.5 mg·kg−1) or p.o. gavage (10 mg·kg−1)] was administered to mice. L. reuteri significantly reduced the paracetamol plasma concentration to 68.4% in comparison to control mice. Similarly, administration of a probiotic cocktail consisting of L. acidophilus, B. lactis and Streptococcus salivarius to rats for 3 days significantly increased azoreductase activity in ex vivo colon contents (Lee et al., 2012). The ex vivo incubation of sulfasalazine with colon contents significantly increased the metabolism of the drug. No clinical significance was observed, however, in the in vivo setting when a single dose of sulfasalazine (100 mg·kg−1) was administered to rats following pretreatment with the probiotic cocktail for 3 days.
Drug–microbiome interactions
In this section, the mechanisms underpinning the drug‐mediated changes to the composition and function of the gut microbiome will be explored.
Antibacterial effects of drugs on the gut microbiota
Direct disruption of the gut microbiota, through bacteriostatic or bactericidal activity, can alter the metabolic capability of the microbiota and have long‐term effects on host functions and health. Such ‘antibiotic’‐induced changes have been well studied. Antibiotic therapy in neonates can disrupt the bacterial colonization of the intestine and can have long‐term health implications with links to the development of eczema, allergic rhinitis and inflammatory bowel disease in later life (Rodríguez et al., 2015). Antimicrobial activity, linked to the depletion of ‘good’ commensal gut microbiota, provides an impetus for the overgrowth of hazardous commensal bacteria (Antunes et al., 2011). For example, antibiotics can decrease host resistance to the growth of the opportunistic pathogen, Clostridium difficile (Theriot et al., 2016).
Non‐antibiotic drugs have also been shown to exert antimicrobial activity. In vitro studies have detected antimicrobial activity, against at least one bacterial strain, for a wide range of non‐antibiotic drug classes including NSAIDs, mucolytic agents, bisphosphonates, PPIs, antihistamines, statins, cytostatic agents and psychotropics (Kruszewska et al., 2000; Kruszewska et al., 2002). Sulfasalazine and the bacterial metabolite, N‐acetyl 5‐ASA, inhibited the in vitro growth of fecal anaerobic strains including C. difficile (Sandberg‐Gertzen et al., 1985; Deloménie et al., 2001). The anti‐neoplastic drug, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4789 (5‐FU), has also shown bactericidal effects against clinical isolates of Staphylococcus aureus even at lower absorption concentrations than would be expected after i.v. administration of 5‐FU (Bodet et al., 1985). Similarly, the antimetabolite and antifolate drug, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4815, displayed strong in vitro antibacterial activity only against S. aureus following a surveillance study of the drug against a variety of microbial strains including E. coli, Pseudomonas aeruginosa and Candida albicans (Kruszewska et al., 2000). Non‐antibiotic drugs, chiefly psychotropics, can also affect the gut microbiome by acting synergistically with a co‐administered antibacterial or by modulating the activity or pathogenicity of microbes, which will be discussed later in the review (Effects of Psychoactive Drugs on the Composition of the Gut Microbiota section) (Martins et al., 2008).
Recently, Maier et al. (2018) further elucidated the extensive effects of non‐antibiotic drugs on the gut microbiota. The authors explored the antibacterial effects of 362 anti‐infective and 835 host‐targeted drugs (all drugs screened at 20 μM; a concentration deemed representative of the predicted concentration of the drugs reaching the bacteria‐dense ileum and colon) in vitro against 40 bacteria found to colonize the GI tract. About 24% of host‐targeted drugs had antibacterial activity against at least one bacterial strain, with 40 drugs inhibiting the growth of up to 10 different bacteria. Antipsychotics, chemotherapeutic agents and antihypertensives were among the host‐targeted drugs exhibiting the greatest antibacterial activity. Certain bacteria, previously identified as being highly abundant in healthy individuals such as Prevotella copri and Eubacterium rectale, were found to be most susceptible to the host‐targeted drugs. Using available data from previously published metagenomic‐based human studies, the authors validated the antibacterial effects of the drugs identified in their high‐throughput drug screens and suggested that the use of some host‐targeted drugs may increase patient susceptibility to antibiotic resistance. This study reaffirms the importance of accounting for drug‐induced changes in the gut microbiota as a potential confounder in assessing compositional changes in microbiota‐related studies.
Secondary effects of drugs on the gut microbiome
Drugs can directly alter the GI tract environment (e.g. pH and transit time), mucosa integrity, host and bacterial metabolic activity and the production of microbial metabolites. These drug‐induced changes could, in turn, have secondary effects on the microbiome with the potential to cause drug–drug interactions.
As the gradient of pH along the GI tract is known to influence bacterial abundance and diversity, drug‐induced changes in gastric and intestinal pH may shape the gut microbiome (Walker et al., 2005; Krajmalnik‐Brown et al., 2012). Imhann et al. (2017) reported changes in 20% of bacterial taxa and a significant decrease in alpha diversity in PPI users (211 participants) compared with non‐users (1604 participants). Alpha diversity refers to the within habitat or sample diversity (a single sample value for average species diversity at an individual site). It usually includes species richness but also accounts for the abundance of the species present in the sample (Gotelli and Colwell, 2001). A PPI‐mediated increase in the pH of the stomach and upper small intestine precipitated the growth of specific taxa (Enterococcaceae and Streptococcaceae) and increased the susceptibility of enteric mucosa to NSAID‐induced damage (Freedberg et al., 2015). Increased abundance of gastric and fecal Streptococcus in PPI users has been linked to the suppression of gastric acid production and associated with increased risk of C. difficile infection (CDI). Moreover, PPI use has also been linked to increases in expression of bacterial invasion genes, which may also mediate the predisposition to CDI in PPI users (Freedberg et al., 2015).
Additionally, GI transit time can dictate the length of exposure of the microbiota to the gut environment. The anti‐diarrhoeal, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7215, administered to humanized GF mice in drinking water (0.1%) for 10 days significantly increased GI transit time and altered the composition of the distal gut microbiota with an increased F/B ratio and a significant reduction in abundance of the Lachnospiraceae family (Kashyap et al., 2013). These changes were reversible upon cessation of loperamide administration and normalized gut transit time.
Drugs may also challenge both the integrity and permeability of the intestinal mucosa. The anti‐hyperglycaemic drug, metformin, has been linked with mucosal modifications that influenced the bacterial growth of Akkermansia spp. (Forslund et al., 2015). An enrichment of virulence factors and gas metabolism genes, mediated by the associated increase in Escherichia, are thought to contribute to the GI disturbances, bloating and increased flatulence, associated with metformin use.
The liver is continually exposed to gut microbiota‐derived metabolites, including secondary bile acids and short chain fatty acids (SCFAs), as it receives an estimated 70% of its blood supply from the intestine (Marchesi et al., 2016). Recent data showed bile acids were involved in the solubilization and absorption of lipophilic drugs (Enright et al., 2017). Drug treatment can, however, alter the gut microbiota production of bile salts, which may subsequently affect the absorption and metabolism of co‐administered medication. Cefoperazone, vancomycin and clindamycin were identified as antibiotics associated with changes in the gut microbiota composition and caused decreased levels of secondary bile acids precipitating the growth and spore germination of C. difficile (Theriot et al., 2016). Studies have found a correlation between the reduced levels of SCFAs in the proximal to distal colon with the corresponding increase in pH from the caecum to rectum (den Besten et al., 2013). Drug‐induced changes in the production of SCFAs could thus indirectly alter GI pH, which as mentioned previously may precipitate changes in the microbiome. For example, increased levels of SCFAs were linked with metformin, which could instigate the microbiota modifications associated with this drug (Zhernakova et al., 2016).
Secondary effects of drugs on the gut microbiome could also arise from drug‐induced modification of genes or enzymes involved in drug metabolism or drug transport. Metabolomic studies have been employed to investigate the effects of antibiotics on the gut microbiome before and after treatment (Antunes et al., 2011). High‐dose treatment of mice with an aminoglycoside antibiotic (20 mg streptomycin via p.o. gavage) altered 87% of all detected intestinal metabolite features and affected many host metabolic pathways involving the metabolism of bile acids, sugars, amino acids and fatty acids with the most significant effect on the steroidal metabolic pathway. Additionally, a marginally increased level of CYP‐metabolizing enzymes was observed after antibiotic treatment. Meta‐transcriptomic approaches have also been employed to analyse gut microbiome samples exposed to various drugs and have identified many candidate genes for their microbial metabolism (Maurice et al., 2013). This ex vivo study investigated the effects of short‐term exposure of human faeces to various non‐antibiotic drugs (10 mg·mL−1 concentration used for all the following drugs) including cardiac glycosides (digoxin and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6782), an anthelmintic (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7210), gastric acid suppressant (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7248), an analgesic (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7402) and sulfasalazine. Even though these drugs did not directly alter microbial physiology (i.e. membrane integrity and polarity), even at very high concentrations, they all significantly changed the expression of microbial genes linked to drug import and metabolism. For example, sulfasalazine induced the expression of thioredoxins and nitrate reductases, while nizatidine, subject to bacterially mediated N‐oxide bond cleavage, up‐regulated the expression of drug enzymes and transporters acting on nitrogen bonds (Maurice et al., 2013). This finding supports the earlier hypothesis that drugs may shift the microbiota to favour the abundance of taxa involved in its metabolism. Furthermore, this altered metabolic capacity of the microbiota could consequently affect not only the pharmacokinetics of subsequent doses of the drug itself (a phenomenon referred to as autoinduction) but also the pharmacokinetics of co‐administered medication that are substrates of the same metabolic pathway or transporter.
Implications of the drug–gut microbiome relationship for neuropharmacology
Effect of antibiotics on inflammation and brain–gut axis
Recent studies have explored the effects of antibiotics on the inflammasome‐gut microbiota regulation of brain function and behaviour. Wong et al. (2016) investigated whether the caspase‐1 antagonist and tetracycline antibiotic, minocycline, has a protective effect on the stress response by modulating the microbiota–gut–brain axis. Minocycline treatment (5 mg·kg−1 via i.p. injection for 21 days) increased the abundance of Akkermansia spp., which is consistent with attenuation of inflammation and ameliorated the stress‐induced depressive‐like behaviour in wild‐type mice. This suggests that the behavioural effects of minocycline may be mediated via the microbiota and not just via its effects on the activation of CNS microglia (Inta et al., 2017).
Psychoactive drugs and the gut–brain axis: effects on intestinal barrier function
Several psychoactive drugs have marked effects on the intestinal barrier (e.g. permeability and mucus secretion) and alter GI transit. Serotonergic psychoactive drugs have been utilized as a treatment in functional GI diseases; the selective serotonin reuptake inhibitor (SSRI), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4790, increases the motility rate of the small intestine, while tricyclic antidepressants (TCAs) are known to delay gastric emptying (Grover and Camilleri, 2013). Concurrently, the gut microbiota modulates intestinal permeability with consequential effects on the absorption and metabolism of psychoactive drugs. Maintenance of intestinal mucosal integrity is important for appropriate absorption of psychoactive drugs in the small intestine. The gut microbiota challenges the intestinal barrier function via induced alterations in intestinal pH and effects on intestinal epithelial integrity (Cani et al., 2008; Carvalho et al., 2012). The presence of the bacterium Helicobacter pylori decreases the absorption of the anti‐Parkinson's drug and dopamine precursor, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=3639. It was hypothesized that H. pylori might alter gastric motility, disrupt the duodenal mucosa or produce ROS, which may inactivate levodopa (Hamlet et al., 1999; Miyaji et al., 1999). More recently, eradication of H. pylori has been shown to significantly improve clinical symptoms of patients with Parkinson's disease and improve levodopa efficacy (Hashim et al., 2014). Additionally, several neuropsychiatric disorders have been associated with a dysfunctional intestinal barrier; acute stress is associated with expression of the tight junction proteins, zonula occludens‐1 and occludin, in the duodenal mucosa of rats subjected to water immersion restraint stress (Lee et al., 2013). Moreover, http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=19 mediate the colonic barrier dysfunction observed in response to maternal separation‐induced mild stress (Söderholm et al., 2002). Hence, both the gut microbiota and psychiatric disorders modulate intestinal permeability and barrier function, which in turn may precipitate changes in the absorption of psychoactive drugs (Kelly et al., 2015).
Effects of CNS‐related disorders on the composition of the gut microbiota
In addition to drug‐induced changes in the gut microbiome, alterations in the composition of the gut microbiota have also been associated with neurological dysfunction and psychiatric disorders (Cenit et al., 2017). Kelly et al. (2016) observed decreased richness and diversity of the gut microbiota in depressed patients, and numerous studies have identified an altered microbiota composition in children with autism spectrum disorder; increased abundance of Lactobacillus and Desulfovibrio spp. has correlated with autism severity (Finegold, 2011; Tomova et al., 2015). The gut microbiota has also been identified as a potential regulator of other neuropsychiatric disorders, for example, schizophrenia (Dinan et al., 2014; Shen et al., 2018). It is yet to be established whether these disease‐associated changes to the gut microbiota are involved in the initiation of disease pathogenesis (altered microbiota state precipitating changes in brain development or function) or occur because of the disease (i.e. whether changes in gut microbiota arise following alterations in brain development or function). Alternatively, the microbiota changes could also occur as a result of dietary patterns or medication (Mayer et al., 2015). As both the CNS‐related disease itself and the pharmacological treatment of the disease can alter the composition of the microbiota, it is thus essential to account for independent effects of the disease or drug treatment in both animal and human studies.
Microbial metabolism of psychoactive drugs
Few studies have offered mechanistic insight into the microbiota‐mediated metabolism of psychoactive drugs; however, those that have are summarized in Table 2. In most cases, the specific microbial species, or microbial‐derived enzymes, responsible for the metabolism of psychoactive drugs by the gut microbiome are unknown.
Table 2.
Psychoactive drugs are subject to direct metabolism by the gut microbiota
| Drug | Drug class | Nature of study; animal species/human | Suggested implicated microbial species | Microbial‐mediated mechanism | References |
|---|---|---|---|---|---|
| Risperidone | Anti‐psychotic | In vivo; rats and dogs | Unknown | Isoxazole scission of benzisoxazole ring system | Meuldermans et al. (1994) |
| Nitrazepam and clonazepam | Hypnotic | In vivo; rats | Unknown | Nitroreductase‐mediated reduction | Takeno et al. (1990) |
| http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4803 | Psychostimulant | In vitro | Lactobacilli, Enterococci and Clostridia | N‐demethylation | Caldwell and Hawksworth (1973) |
| Levodopa | Dopamine precursor | In vivo; rats | Unknown | Decarboxylation and p‐dehydroxylation‐mediated conversion to m‐tyramine and subsequent oxidation to m‐hydroxyphenylacetic acid | Goldin et al. (1973) |
There is evidence to suggest that benzodiazepines are subject to microbiota‐mediated drug metabolism. Older studies have demonstrated the microbiota‐mediated nitro‐reduction of the hypnotics, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6963 and nitrazepam, to their corresponding 7‐amino metabolite derivatives. Elmer and Remmel (1984) administered radiolabelled clonazepam and quantified the production of the nitro‐reduced clonazepam metabolites in GF rats before and after colonization with faecal microbiota. The production of these metabolites increased significantly from 15% in GF mice to 77% after microbial colonization. Similarly, the reduction of the hypnotic, nitrazepam, to 7‐aminonitrazepam and 7‐acetylaminonitrazepam was shown to be mediated by several anaerobic bacteria isolated from human gut isolates including species from the Clostridium, Bacteroides and Eubacterium genera. The authors identified the bacterium, Clostridium leptum, as having highly specific nitroreductase activity (Rafii et al., 1997). A previous study highlighted antibiotics can deplete the activity of this microbial enzyme (Takeno and Sakai, 1991). The production of nitro‐reduced nitrazepam metabolites in pregnant rats, following a p.o. dose of nitrazepam 300 mg·kg−1, was quantified before and after antibiotic treatment, which was given to diminish nitroreductase enzymatic activity. The levels of 7‐aminonitrazepam and 7‐acetylaminonitrazepam decreased from 30% pretreatment to 2% after antibiotic treatment.
Effects of psychoactive drugs on the composition of the gut microbiota
Antidepressants and antipsychotics are associated with distinct gut microbiota signatures. Gastric acid suppressants, antipsychotics and antidepressants have repeatedly been confirmed as the three non‐antibiotic drug classes most associated with the abundance of single taxa. Antipsychotics, for example, were associated with the abundance of Prevotella, an unclassified member of the Desulfovibrionaceae family and Victivallis genus in an observation‐based study in an elderly (≥65 years) hospitalized cohort (Ticinesi et al., 2017). On the other hand, antidepressant use in this cohort significantly correlated with the increased abundance of five specific bacterial taxa belonging to the Helicobacter, Asteroleplasma and Marinilactibacillus genera and unclassified members of both the Bacillus class and Succinivibrionaceae family. In an observation‐based study in community‐dwelling patients (mean age 52 years), the SSRI antidepressant, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7547, was associated with a significantly increased relative abundance of the Enterobacteriaceae family in comparison to patients not taking any medication (Rogers and Aronoff, 2016). The specific use of the antipsychotics, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4790 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=96, causes a shift in the gut microbiota composition to a state previously shown to be associated with obesity in both rat and human [12.2 (SD 2.5) years] in vivo studies (Davey et al., 2012; Bahr et al., 2015a). Chronic olanzapine treatment of rats (2–4 mg·kg−1·day−1) and mice (50 mg·kg−1 HFD) was associated with an increase in Firmicutes, a decrease in Bacteroidetes and an overall reduction in biodiversity (Davey et al., 2012; Morgan et al., 2014). Similarly, chronic risperidone treatment (approximately 80 μg risperidone per day for 58 days) resulted in a 22.4% decrease in Bacteroidetes and a reciprocal 32.6% increase in Firmicutes in the risperidone‐treated mice relative to control mice (Bahr et al., 2015b).
The mechanisms underpinning these changes in bacterial abundance or diversity induced by psychoactive drugs are not fully understood, but both these drug classes have shown in vitro antimicrobial activity. There is preliminary evidence to suggest that olanzapine has antimicrobial activity. Morgan et al. (2014) found olanzapine inhibited the in vitro growth of two commensal strains, E. coli and Enterococcus faecalis. The dose investigated, however, was above the recommended dosage range of 5–20 mg·day−1, and therefore, whether clinically relevant doses of olanzapine would still possess antimicrobial activity remains to be tested. Similarly, the phenothiazine group of antipsychotics exerted antimicrobial effects at higher than clinically relevant drug doses (Amaral et al., 2004). The phenothiazine antipsychotic, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=83, was the first psychoactive drug associated with antibacterial properties. S. aureus, Mycobacterium and some gram‐negative rods such as Shigella spp. have been identified as the bacteria most susceptible to the antibacterial activity of phenothiazine antipsychotics (Amaral et al., 2004). Evidence suggests phenothiazine antipsychotics may mediate their effects on microbial growth through the alteration of bacterial morphology (phenothiazines cause filamentation of E. coli) or inhibition of bacterial adherence to epithelial cells (phenothiazines reduce E. coli adherence to urinary epithelium) (Amaral et al., 2004).
Similarly, antidepressant‐induced changes in gut microbiota may be mediated by direct antimicrobial activity. SSRIs (including paroxetine, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4798 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=203) are associated with a broad‐spectrum of antibacterial activity, including activity against strains of Staphylococcus, Enterococcus, Clostridium, Pseudomonas and Citrobacter (Munoz‐Bellido et al., 2000). Sertraline can also influence microbial growth by acting synergistically with other antibiotics. Sertraline increased the efficacy of co‐administered tetracycline and fluoroquinolone antibiotics against, a pathogenic strain associated with urinary tract infections, Corynebacterium urealyticum (Munoz‐Bellido et al., 1996). The inhibition of efflux pumps in bacterial cells by sertraline is hypothesized as one causative mechanism. SSRIs may also interfere with the biosynthesis of the slime layer on bacteria (Munoz‐Bellido et al., 2000); the disruption of the slime layer may also act to increase bacterial susceptibility to co‐administered antibiotics. Apart from SSRIs, other antidepressants have antimicrobial effects, including MAO inhibitors (MAOIs) and TCAs. The inhibition of cell wall synthesis and anti‐plasmid activity has been proposed as mechanisms underlying the antimicrobial activity of the MAOI, iproniazid and the TCA drug class respectively (Molnar, 1988; Macedo et al., 2017).
Psychotropic drugs can also modulate the activity and pathogenicity of microbes, which can precipitate changes in the gut microbiome. Sertraline can alter the in vitro pathogenicity of fungi, for example, by significantly affecting the virulence properties associated with Candida including decreased hyphal elongation and reduced secretion of aspartyl proteinases, thereby preventing fungal adherence and tissue invasion of Candida spp. (Lass‐Florl et al., 2003).
The effects of psychoactive drug–microbiota interactions: implications for drug response and toxicity
The growing evidence of the complex relationship between drugs and the gut microbiome, illustrated in Figure 1, underscores the importance of considering the gut microbiota as an additional factor contributing to the inter‐individual variation observed in drug metabolism and response.
Figure 1.
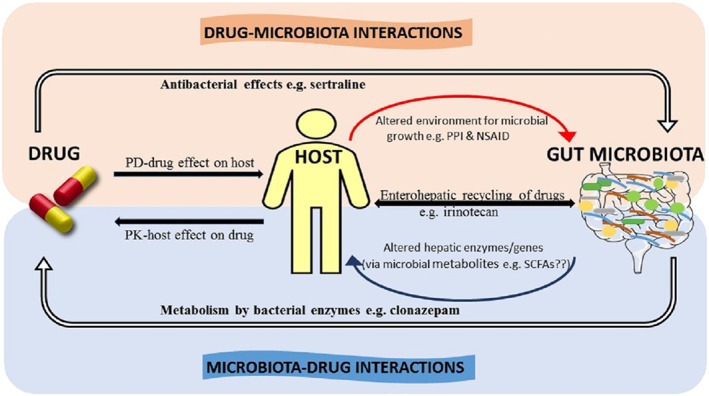
The complex interplay between drugs and the gut microbiota. The reciprocal relationship between drugs and the gut microbiome is composed of both microbiota‐mediated alterations to drug pharmacokinetics and drug‐mediated alterations to the function and composition of the gut microbiome. These interactions can occur by both direct (illustrated by solid white arrows with black outline) and indirect or secondary (illustrated by curved line arrows) mechanisms. ‘Microbiota–Drug Interactions’: the microbiota can directly metabolize drugs through bacterial‐derived enzymes (e.g. nitroreductase‐mediated metabolism of clonazepam) but can also indirectly affect drug metabolism through the alteration of the hosts capacity to metabolize drugs (curved‐up line arrow), potentially altering drug PK profile. Microbial‐derived metabolites (e.g. SCFAs and secondary bile acids) may be potential mediators of this effect. The interactions between the host and gut microbiome are responsible for the enterohepatic recirculation of drugs. For example, the hepatic‐glucuronidated irinotecan metabolite is deconjugated by β‐glucuronidase enzymes expressed by the gut microbiota. ‘Drug–Microbiota Interactions’: the mechanisms underpinning the drug‐mediated changes to the function and composition the gut microbiome are yet to be fully elucidated. Drugs can have antibacterial properties that directly affect the composition of the gut microbiota (e.g. sertraline). Drugs can also alter the physiological properties or functions of host organs (i.e. PD effect) (e.g. PPI‐mediated alterations to gastric acid production and pH, NSAID‐induced changes to mucosal integrity), which may, in turn, precipitate secondary effects on the composition of the gut microbiota (illustrated by the curved‐down line arrow).
The interaction of psychoactive drugs with the gut microbiota has implications for drug absorption and bioavailability. As mentioned previously, levodopa is one of the most studied psychoactive drugs that interact with the gut microbiome. Research illustrating the metabolism of this drug by the gut microbiota‐mediated dehydroxylation dates back to the 1970s (Goldin et al., 1973). Follow‐on studies found decreased plasma levels of the drug with the presence of H. pylori, attributed to levodopa‐mediated interaction with the bacterial surface adhesions (Niehues and Hensel, 2009).
Antidepressants are associated with considerable inter‐individual variation in drug response and a lack of efficacy; it is estimated that antidepressants are only 20–30% more effective than placebo (Arroll et al., 2005). Pharmacogenetics, although still in its relative infancy, has thus far proven unsuccessful in identifying and optimizing factors that may hinder antidepressant efficacy. The pathophysiology of major depression disorder (MDD) has, however, been linked to alterations in the gut microbiota composition. MDD is associated with increased levels of Bacteroidetes and Enterobacteriaceae and decreased levels of Firmicutes. This altered microbiota state has been linked to increased gut permeability, a factor that can dictate intestinal drug transport and absorption (Jiang et al., 2015). Continued metabolomic and metagenomic analysis of the microbiome and further pharmacomicrobiomic based‐studies may thus offer some additional insight.
The gut microbiota has also been implicated in the propagation of the side effects and toxicity of psychoactive drugs. With patient adherence to psychotropic therapy estimated to be only 30%, poor compliance has been linked to the unfavourable side‐effect profile associated with these drugs (Weich et al., 2007). As previously highlighted, antibiotic treatment of rats attenuated both the microbial metabolism and the teratogenicity‐associated adverse effect of nitrazepam and its metabolites (Elmer and Remmel, 1984). More recently, antipsychotic‐induced metabolic dysfunction has been linked to shifts in the composition of the gut microbiota. The role of the gut microbiome in the development of olanzapine‐induced weight gain was elucidated when olanzapine‐treated GF mice did not gain weight but did so upon colonization with caecal microbiota (Morgan et al., 2014). Furthermore, an antibiotic cocktail co‐administered with olanzapine, to chemically deplete the gut microbiota, attenuated the metabolic side effects associated with the drug treatment alone in rats; increased weight gain, increased uterine fat deposition and increased plasma free fatty acid levels were side effects attenuated in microbiota‐depleted rats (Davey et al., 2013).
A recent follow‐on study by Kao et al. (2018) investigated whether treatment with a prebiotic, BimunoTM galacto‐oligosaccharides (B‐GOS), modified the olanzapine‐induced weight gain. Female rats were treated with B‐GOS (0.5 g·kg−1·day−1) for 21 days, in conjunction with administration of olanzapine (10 mg·kg−1 once daily via i.p. injection) or saline on days 8–21. B‐GOS treatment significantly attenuated the drug‐induced weight gain without altering the antagonism of central serotoninergic receptors necessary for drug efficacy. B‐GOS treatment alone altered the microbiota composition; differences in the abundance of Bifidobacterium spp. and Firmicutes spp. were evident in B‐GOS‐only‐treated rats in comparison to both water‐only and B‐GOS‐olanzapine‐treated rats. Contrary to the previous findings by Davey et al. (2013) and Morgan et al. (2014) discussed above, administration of olanzapine, however, did not significantly alter the composition of the faecal microbiota, although different dosage regimens were employed in the respective studies. Furthermore, the authors deduced, contrary to their hypothesis, that elevated levels of the SCFA, acetate, may mediate the olanzapine‐induced weight gain. Considering the risks associated with antibiotic resistance, prebiotic treatment may constitute a more suitable adjunctive therapy in comparison to long‐term antibiotic use to improve patient tolerance to olanzapine.
Both olanzapine‐induced and risperidone‐induced weight gain correlated with an altered composition of gut microbiota (Davey et al., 2012; Bahr et al., 2015b). Suppressed energy expenditure, induced by the gut microbiota, was found to cause the observed weight gain in risperidone‐treated mice. Furthermore, fecal microbiota transplantation (FMT) from risperidone‐treated mice to treatment‐naive mice induced weight gain and suppressed energy expenditure in the FMT‐recipient mice (Bahr et al., 2015b). On the premise that some antidepressants (e.g. SSRIs and TCAs) are associated with similar effects to these antipsychotics, that is, changes in the gut microbiota and drug‐induced weight gain, it may be timely to extend these studies to antidepressants.
Conclusion and future directions
Research has identified many clinically relevant drugs, including psychotropics, which are metabolized by the microbiome, often via as yet unknown mechanisms. There remains a need to establish the microorganism responsible for metabolism and identify the molecular mechanisms involved. While medication use can precipitate changes in the abundance and diversity of a wide array of different bacterial phyla, the resultant effects of these compositional changes on drug response and patient outcomes requires further study. Matching specific microbial genes or enzymes, which may dictate drug response, to specific bacterial phyla could lead to greater understanding of the consequences of disturbances in the microbiome on patient outcomes and provide additional impetus to explore the contribution of these important, but underappreciated, drug–microbiome interactions to the inter‐individual variability observed in drug response.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org,thecommon portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a,b).
Conflict of interest
N.P.H. has acted as a consultant for Johnson & Johnson Consumer Inc., Janssen Research & Development and Steigerwald Arzneimittelwerk GmbH (Bayer) and has received research support from Marigot Ltd.
Acknowledgements
The APC Microbiome Institute is a research institute funded by the Science Foundation Ireland (SFI) through the Irish Government's National Development Plan (grant no. SFI/12/RC/2273).
Walsh, J. , Griffin, B. T. , Clarke, G. , and Hyland, N. P. (2018) Drug–gut microbiota interactions: implications for neuropharmacology. British Journal of Pharmacology, 175: 4415–4429. 10.1111/bph.14366.
References
- Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017a). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Cidlowski JA, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Nuclear hormone receptors. Br J Pharmacol 174: S208–S224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Salami H, Butt G, Fawcett JP, Tucker IG, Golocorbin‐Kon S, Mikov M (2008). Probiotic treatment reduces blood glucose levels and increases systemic absorption of gliclazide in diabetic rats. Eur J Drug Metab Pharmacokinet 33: 101–106. [DOI] [PubMed] [Google Scholar]
- Alavijeh MS, Chishty M, Qaiser MZ, Palmer AM (2005). Drug metabolism and pharmacokinetics, the blood‐brain barrier, and central nervous system drug discovery. NeuroRx 2: 554–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral L, Viveiros M, Molnar J (2004). Antimicrobial activity of phenothiazines. In Vivo 18: 725–731. [PubMed] [Google Scholar]
- Antunes LCM, Han J, Ferreira RBR, Lolić P, Borchers CH, Finlay BB (2011). Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroll B, Macgillivray S, Ogston S, Reid I, Sullivan F, Williams B et al (2005). Efficacy and tolerability of tricyclic antidepressants and SSRIs compared with placebo for treatment of depression in primary care: a meta‐analysis. Ann Fam Med 3: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr SM, Tyler BC, Wooldridge N, Butcher BD, Burns TL, Teesch LM et al (2015a). Use of the second‐generation antipsychotic, risperidone, and secondary weight gain are associated with an altered gut microbiota in children. Transl Psychiatry 5: e652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr SM, Weidemann BJ, Castro AN, Walsh JW, de Leon O, Burnett CML et al (2015b). Risperidone‐induced weight gain is mediated through shifts in the gut microbiome and suppression of energy expenditure. EBioMed 2: 1725–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S (2009). Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4: e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodet CA, Jorgensen JH, Drutz DJ (1985). Antibacterial activities of antineoplastic agents. Antimicrob Agents Chemother 28: 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J, Hawksworth GM (1973). The demethylation of methamphetamine by intestinal microflora. J Pharm Pharmacol 25: 422–424. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM et al (2008). Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes 57: 1470–1481. [DOI] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf‐Filho P, Vassallo J et al (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high‐fat fed mice. Diabetologia 55: 2823–2834. [DOI] [PubMed] [Google Scholar]
- Cenit MC, Sanz Y, Codoñer‐Franch P (2017). Influence of gut microbiota on neuropsychiatric disorders. World J Gastroenterol 23: 5486–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S et al (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J et al (2011). Colonization‐induced host‐gut microbial metabolic interaction. MBio 2: e00271–e00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK (2009). Pharmacometabonomic identification of a significant host‐microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 106: 14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli RJ, Sharma A (2014). Polypharmacy: a global risk factor for elderly people. J Int Oral Health: JIOH 6: i–ii. [PMC free article] [PubMed] [Google Scholar]
- Davey KJ, Cotter PD, O'Sullivan O, Crispie F, Dinan TG, Cryan JF et al (2013). Antipsychotics and the gut microbiome: olanzapine‐induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Transl Psychiatry 3: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey KJ, O'Mahony SM, Schellekens H, O'Sullivan O, Bienenstock J, Cotter PD et al (2012). Gender‐dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology (Berl) 221: 155–169. [DOI] [PubMed] [Google Scholar]
- Debotton N, Dahan A (2017). Applications of polymers as pharmaceutical excipients in solid oral dosage forms. Med Res Rev 37: 52–97. [DOI] [PubMed] [Google Scholar]
- Deloménie C, Fouix S, Longuemaux S, Brahimi N, Bizet C, Picard B et al (2001). Identification and functional characterization of arylamine N‐acetyltransferases in eubacteria: evidence for highly selective acetylation of 5‐aminosalicylic acid. J Bacteriol 183: 3417–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D‐J, Bakker BM (2013). The role of short‐chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, Borre YE, Cryan JF (2014). Genomics of schizophrenia: time to consider the gut microbiome? Mol Psychiatry 19: 1252–1257. [DOI] [PubMed] [Google Scholar]
- D'souza AA, Shegokar R (2016). Polyethylene glycol (PEG): a versatile polymer for pharmaceutical applications. Expert Opin Drug Deliv 13: 1257–1275. [DOI] [PubMed] [Google Scholar]
- Elmer GW, Remmel RP (1984). Role of the intestinal microflora in clonazepam metabolism in the rat. Xenobiotica 14: 829–840. [DOI] [PubMed] [Google Scholar]
- Enright EF, Joyce SA, Gahan CGM, Griffin BT (2017). Impact of gut microbiota‐mediated bile acid metabolism on the solubilization capacity of bile salt micelles and drug solubility. Mol Pharm 14: 1251–1263. [DOI] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira‐Silva S, Wang J, Darzi Y, Faust K et al (2016). Population‐level analysis of gut microbiome variation. Science 352: 560–564. [DOI] [PubMed] [Google Scholar]
- Finegold SM (2011). Desulfovibrio species are potentially important in regressive autism. Med Hypotheses 77: 270–274. [DOI] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S et al (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC et al (2015). Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 149: 883–885.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloux K, Berteau O, El oumami H, Béguet F, Leclerc M, Doré J (2011). A metagenomic β‐glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci U S A 108: 4539–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin BR, Peppercorn MA, Goldman P (1973). Contributions of host and intestinal microflora in the metabolism of L‐dopa by the rat. J Pharmacol Exp Ther 186: 160–166. [PubMed] [Google Scholar]
- Gotelli NJ, Colwell RK (2001). Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett 4: 379–391. [Google Scholar]
- Grover M, Camilleri M (2013). Effects on gastrointestinal functions and symptoms of serotonergic psychoactive agents used in functional gastrointestinal diseases. J Gastroenterol 48: 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science (New York, N.Y.) 341: 295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Turnbaugh PJ (2013). Developing a metagenomic view of xenobiotic metabolism. Pharmacological research: the official journal of the Italian Pharmacological Society 69: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlet A, Thoreson AC, Nilsson O, Svennerholm AM, Olbe L (1999). Duodenal Helicobacter pylori infection differs in cagA genotype between asymptomatic subjects and patients with duodenal ulcers. Gastroenterology 116: 259–268. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim H, Azmin S, Razlan H, Yahya NW, Tan HJ, Manaf MRA et al (2014). Eradication of Helicobacter pylori infection improves levodopa action, clinical symptoms and quality of life in patients with Parkinson's disease. PLoS One 9: e112330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Ohura K (2010). The role of intestinal carboxylesterase in the oral absorption of prodrugs. Curr Drug Metab 11: 793–805. [DOI] [PubMed] [Google Scholar]
- Imhann F, Vich Vila A, Bonder MJ, Lopez Manosalva AG, Koonen DPY, Fu J et al (2017). The influence of proton pump inhibitors and other commonly used medication on the gut microbiota. Gut Microbes 8: 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inta D, Lang UE, Borgwardt S, Meyer‐Lindenberg A, Gass P (2017). Microglia activation and schizophrenia: lessons from the effects of minocycline on postnatal neurogenesis, neuronal survival and synaptic pruning. Schizophr Bull 43: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Jernberg C, Andersson AF, Sjölund‐Karlsson M, Jansson JK, Engstrand L (2010). Short‐term antibiotic treatment has differing long‐term impacts on the human throat and gut microbiome. PLoS One 5: e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN (2015). Role of the normal gut microbiota. World J Gastroenterol: WJG 21: 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y et al (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 48 (Supplement C): 186–194. [DOI] [PubMed] [Google Scholar]
- Kao AC‐C, Spitzer S, Anthony DC, Lennox B, Burnet PWJ (2018). Prebiotic attenuation of olanzapine‐induced weight gain in rats: analysis of central and peripheral biomarkers and gut microbiota. Transl Psychiatry 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA et al (2013). Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 144: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JR, Borre Y, O'Brien C, Patterson E, El Aidy S, Deane J et al (2016). Transferring the blues: depression‐associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res 82: 109–118. [DOI] [PubMed] [Google Scholar]
- Kelly JR, Kennedy PJ, Cryan JF, Dinan TG, Clarke G, Hyland NP (2015). Breaking down the barriers: the gut microbiome, intestinal permeability and stress‐related psychiatric disorders. Front Cell Neurosci 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Yoo DH, Jung IH, Lim S, Jeong JJ, Kim KA et al (2016). Reduced metabolic activity of gut microbiota by antibiotics can potentiate the antithrombotic effect of aspirin. Biochem Pharmacol 122: 72–79. [DOI] [PubMed] [Google Scholar]
- Kim J‐K, Choi MS, Jeong J‐J, Lim S‐M, Kim IS, Yoo HH et al (2018). Effect of probiotics on pharmacokinetics of orally administered acetaminophen in mice. Drug Metab Dispos 46: 122–130. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY (2015). Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos 43: 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch RL, Chrystal EJT, Beaulieu BB, Goldman P (1979). Acetamide—a metabolite of metronidazole formed by the intestinal flora. Biochem Pharmacol 28: 3611–3615. [DOI] [PubMed] [Google Scholar]
- Koppel N, Maini Rekdal V, Balskus EP (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science 356 10.1126/science.aag2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaraju SL (2005). Colon targeted delivery systems: review of polysaccharides for encapsulation and delivery. Crit Rev Food Sci Nutr 45: 251–258. [DOI] [PubMed] [Google Scholar]
- Krajmalnik‐Brown R, Ilhan Z‐E, Kang D‐W, DiBaise JK (2012). Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract: official publication of the American Society for Parenteral and Enteral Nutrition 27: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszewska H, Zareba T, Tyski S (2000). Antimicrobial activity of selected non‐antibiotics–activity of methotrexate against Staphylococcus aureus strains. Acta Pol Pharm 57 Suppl: 117–119. [PubMed] [Google Scholar]
- Kruszewska H, Zareba T, Tyski S (2002). Search of antimicrobial activity of selected non‐antibiotic drugs. Acta Pol Pharm 59: 436–439. [PubMed] [Google Scholar]
- Laizure SC, Herring V, Hu Z, Witbrodt K, Parker RB (2013). The role of human carboxylesterases in drug metabolism: have we overlooked their importance? Pharmacotherapy 33: 210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass‐Florl C, Ledochowski M, Fuchs D, Speth C, Kacani L, Dierich MP et al (2003). Interaction of sertraline with Candida species selectively attenuates fungal virulence in vitro. FEMS Immunol Med Microbiol 35: 11–15. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Zhang H, Orlovich DA, Fawcett JP (2012). The influence of probiotic treatment on sulfasalazine metabolism in rat. Xenobiotica 42: 791–797. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim D‐K, Kim YB, Lee KJ (2013). Effect of acute stress on immune cell counts and the expression of tight junction proteins in the duodenal mucosa of rats. Gut and Liver 7: 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BJ, Huang YY, Peck SC, Wei Y, AM‐d C, Marks JA et al (2017). A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans‐4‐hydroxy‐L‐proline. Science 355 10.1126/science.%20aai8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo D, Filho AJMC, Soares de Sousa CN, Quevedo J, Barichello T, Júnior HVN et al (2017). Antidepressants, antimicrobials or both? Gut microbiota dysbiosis in depression and possible implications of the antimicrobial effects of antidepressant drugs for antidepressant effectiveness. J Affect Disord 208 (Supplement C): 22–32. [DOI] [PubMed] [Google Scholar]
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE et al (2018). Extensive impact of non‐antibiotic drugs on human gut bacteria. Nature 555: 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Adams DH, Fava F, Hermes GDA, Hirschfield GM, Hold G et al (2016). The gut microbiota and host health: a new clinical frontier. Gut 65: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J et al (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M, Dastidar SG, Fanning S, Kristiansen JE, Molnar J, Pagès J‐M et al (2008). Potential role of non‐antibiotics (helper compounds) in the treatment of multidrug‐resistant Gram‐negative infections: mechanisms for their direct and indirect activities. Int J Antimicrob Agents 31 (3): 198–208. [DOI] [PubMed] [Google Scholar]
- Matuskova Z, Anzenbacherova E, Vecera R, Tlaskalova‐Hogenova H, Kolar M, Anzenbacher P (2014). Administration of a probiotic can change drug pharmacokinetics: effect of E. coli Nissle 1917 on amidarone absorption in rats. PLoS One 9: e87150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA, Tillisch K, Gupta A (2015). Gut/brain axis and the microbiota. J Clin Invest 125: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuldermans W, Hendrickx J, Mannens G, Lavrijsen K, Janssen C, Bracke J et al (1994). The metabolism and excretion of risperidone after oral administration in rats and dogs. Drug Metab Dispos 22: 129–138. [PubMed] [Google Scholar]
- Mikov M (1994). The metabolism of drugs by the gut flora. Eur J Drug Metab Pharmacokinet 19: 201–207. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Jollow DJ, Potter WZ, Davis DC, Gillette JR, Brodie BB (1973). Acetaminophen‐induced hepatic necrosis. I. Role of drug metabolism. J Pharmacol Exp Ther 187: 185–194. [PubMed] [Google Scholar]
- Miyaji H, Azuma T, Ito S, Abe Y, Ono H, Suto H et al (1999). The effect of Helicobacter pylori eradication therapy on gastric antral myoelectrical activity and gastric emptying in patients with non‐ulcer dyspepsia. Aliment Pharmacol Ther 13: 1473–1480. [DOI] [PubMed] [Google Scholar]
- Molnar J (1988). Antiplasmid activity of tricyclic compounds. Methods Find Exp Clin Pharmacol 10: 467–474. [PubMed] [Google Scholar]
- Morgan AP, Crowley JJ, Nonneman RJ, Quackenbush CR, Miller CN, Ryan AK et al (2014). The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One 9: e115225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz‐Bellido JL, Munoz‐Criado S, Garcia‐Rodriguez JA (1996). In‐vitro activity of psychiatric drugs against Corynebacterium urealyticum (Corynebacterium group D2). J Antimicrob Chemother 37: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Munoz‐Bellido JL, Munoz‐Criado S, Garcia‐Rodriguez JA (2000). Antimicrobial activity of psychotropic drugs: selective serotonin reuptake inhibitors. Int J Antimicrob Agents 14: 177–180. [DOI] [PubMed] [Google Scholar]
- Niehues M, Hensel A (2009). In‐vitro interaction of L‐dopa with bacterial adhesins of Helicobacter pylori: an explanation for clinicial differences in bioavailability? J Pharm Pharmacol 61: 1303–1307. [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F (2006). The gut flora as a forgotten organ. EMBO Rep 7: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G, Liperoti R, Fialova D, Topinkova E, Tosato M, Danese P et al (2012). Polypharmacy in nursing home in Europe: results from the SHELTER study. J Gerontol A Biol Sci Med Sci 67: 698–704. [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I et al (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118: 511–521. [DOI] [PubMed] [Google Scholar]
- Peppercorn MA, Goldman P (1972). The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther 181: 555–562. [PubMed] [Google Scholar]
- Prudhviraj G, Vaidya Y, Singh SK, Yadav AK, Kaur P, Gulati M et al (2015). Effect of co‐administration of probiotics with polysaccharide based colon targeted delivery systems to optimize site specific drug release. Eur J Pharm Biopharm 97: 164–172. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley EMM (2017). Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet? Nature Rev Gastroenterology Hepatol 14: 315–320. [DOI] [PubMed] [Google Scholar]
- Rafii F, Sutherland JB, Hansen JEB, Cerniglia CE (1997). Reduction of nitrazepam by Clostridium leptum, a nitroreductase‐producing bacterium isolated from the Human intestinal tract. Clin Infect Dis 25 (Supplement_2): S121–S122. [DOI] [PubMed] [Google Scholar]
- Rizkallah MR, Saad R, Aziz RK (2010). The human microbiome project, personalized medicine and the birth of pharmacomicrobiomics. Curr PPharmacogenomics Pers Med 8: 182–193. [Google Scholar]
- Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N et al (2015). The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26: 26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MAM, Aronoff DM (2016). The influence of non‐steroidal anti‐inflammatory drugs on the gut microbiome. Clin Microbiol Infect 22: 178.e1–178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A (2017). Azoreductases in drug metabolism. Br J Pharmacol 174: 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta KS, Zhang C, Lee KK, Fujimoto K, Redinbo MR, Boelsterli UA (2014). Bacterial beta‐glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica 44: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar N, Arboleya S, Valdés L, Stanton C, Ross P, Ruiz L et al (2014). The human intestinal microbiome at extreme ages of life. Dietary intervention as a way to counteract alterations. Front Genet 5: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg‐Gertzen H, Kjellander J, Sundberg‐Gilla B, Jarnerot G (1985). In vitro effects of sulphasalazine, azodisal sodium, and their metabolites on Clostridium difficile and some other faecal bacteria. Scand J Gastroenterol 20: 607–612. [DOI] [PubMed] [Google Scholar]
- Sandhu KV, Sherwin E, Schellekens H, Stanton C, Dinan TG, Cryan JF (2016). Feeding the microbiota‐gut‐brain axis: diet, microbiome, and neuropsychiatry. Transl Res 179: 223–244. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cui JY, Klaassen CD (2015). RNA‐seq quantification of hepatic drug processing genes in germ‐free mice. Drug Metab Dispos 43: 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F, van Sinderen D, O'Toole PW, Stanton C (2017). Feeding the microbiota: transducer of nutrient signals for the host. Gut 66: 1709–1717. [DOI] [PubMed] [Google Scholar]
- Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J et al (2018). Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross‐sectional study. Schizophr Res. 10.1016/j.schres.%202018.01.002 [DOI] [PubMed] [Google Scholar]
- Söderholm JD, Yates DA, Gareau MG, Yang P‐C, MacQueen G, Perdue MH (2002). Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. Am J Physiol Gastrointest Liver Physiol 283: G1257–G1263. [DOI] [PubMed] [Google Scholar]
- Takasuna K, Hagiwara T, Hirohashi M, Kato M, Nomura M, Nagai E , et al (1998). Inhibition of intestinal microflora β‐glucuronidase modifies the distribution of the active metabolite of the antitumor agent, irinotecan hydrochloride (CPT‐11) in rats. Cancer Chemother Pharmacol 42: 280–286. [DOI] [PubMed] [Google Scholar]
- Takeno S, Nakagawa M, Sakai T (1990). Teratogenic effects of nitrazepam in rats. Res Commun Chem Pathol Pharmacol 69: 59–70. [PubMed] [Google Scholar]
- Takeno S, Sakai T (1991). Involvement of the intestinal microflora in nitrazepam‐induced teratogenicity in rats and its relationship to nitroreduction. Teratology 44: 209–214. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Bowman AA, Young VB, Ellermeier CD (2016). Antibiotic‐induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSPHERE 1: e00045–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursby E, Juge N (2017). Introduction to the human gut microbiota. Biochem J 474: 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticinesi A, Milani C, Lauretani F, Nouvenne A, Mancabelli L, Lugli GA et al (2017). Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep 7: 11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K et al (2015). Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav 138: 179–187. [DOI] [PubMed] [Google Scholar]
- Turnheim K (2003). When drug therapy gets old: pharmacokinetics and pharmacodynamics in the elderly. Exp Gerontol 38: 843–853. [DOI] [PubMed] [Google Scholar]
- Ulmer JE, Vilén EM, Namburi RB, Benjdia A, Beneteau J, Malleron A et al (2014). Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont bacteroides thetaiotaomicron reveals the first GAG‐specific bacterial endosulfatase. J Biol Chem 289: 24289–24303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hogezand RA, Kennis HM, van Schaik A, Koopman JP, van Hees PA, van Tongeren JH (1992). Bacterial acetylation of 5‐aminosalicylic acid in faecal suspensions cultured under aerobic and anaerobic conditions. Eur J Clin Pharmacol 43: 189–192. [DOI] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ (2005). pH and peptide supply can radically alter bacterial populations and short‐chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71: 3692–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tong Q, Shou J‐W, Zhao Z‐X, Li X‐Y, Zhang X‐F et al (2017). Gut microbiota‐mediated personalized treatment of hyperlipidemia using berberine. Theranostics 7: 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weich S, Nazareth I, Morgan L, King M (2007). Treatment of depression in primary care. Br J Psychiatry 191: 164–169. [DOI] [PubMed] [Google Scholar]
- Wilson ID, Nicholson JK (2017). Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res 179 (Supplement C): 204–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Inserra A, Lewis MD, Mastronardi CA, Leong L, Choo J et al (2016). Inflammasome signaling affects anxiety‐ and depressive‐like behavior and gut microbiome composition. Mol Psychiatry 21: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kurita A, Asahara T, Takakura A, Katono K, Iwasaki M et al (2008). Metabolism of irinotecan and its active metabolite SN‐38 by intestinal microflora in rats. Oncol Rep 20: 727–730. [PubMed] [Google Scholar]
- Yoo DH, Kim IS, Van Le TK, Jung IH, Yoo HH, Kim DH (2014). Gut microbiota‐mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos 42: 1508–1513. [DOI] [PubMed] [Google Scholar]
- Yoo HH, Kim IS, Yoo DH, Kim DH (2016). Effects of orally administered antibiotics on the bioavailability of amlodipine: gut microbiota‐mediated drug interaction. J Hypertens 34: 156–162. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T et al (2016). Population‐based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]


