The mammalian immune system produces reactive oxygen and nitrogen species that kill bacterial pathogens by damaging key cellular components, such as lipids, DNA, and proteins. However, bacteria possess detoxifying and repair systems that mitigate these deleterious effects. The Escherichia coli RIC (repair of iron clusters) protein is a di-iron hemerythrin-like protein that repairs stress-damaged iron-sulfur clusters. E. coli Dps is an iron storage protein of the ferritin superfamily with DNA-binding capacity that protects cells from oxidative stress. This work shows that the E. coli RIC and Dps proteins interact in a fashion that counters RIC protein-induced reactive oxygen species (ROS). Altogether, we provide evidence for the formation of a new bacterial protein complex and reveal a novel contribution for Dps in bacterial redox stress protection.
KEYWORDS: E. coli, di-iron RIC protein, YtfE, Dps, oxidative stress, nitrosative stress, di-iron
ABSTRACT
The RIC (repair of iron clusters) protein of Escherichia coli is a di-iron hemerythrin-like protein that has a proposed function in repairing stress-damaged iron-sulfur clusters. In this work, we performed a bacterial two-hybrid screening to search for RIC-protein interaction partners in E. coli. As a result, the DNA-binding protein from starved cells (Dps) was identified, and its potential interaction with RIC was tested by bacterial adenylate cyclase-based two-hybrid (BACTH) system, bimolecular fluorescence complementation, and pulldown assays. Using the activity of two Fe-S-containing enzymes as indicators of cellular Fe-S cluster damage, we observed that strains with single deletions of ric or dps have significantly lower aconitase and fumarase activities. In contrast, the ric dps double mutant strain displayed no loss of aconitase and fumarase activity with respect to that of the wild type. Additionally, while complementation of the ric dps double mutant with ric led to a severe loss of aconitase activity, this effect was no longer observed when a gene encoding a di-iron site variant of the RIC protein was employed. The dps mutant exhibited a large increase in reactive oxygen species (ROS) levels, but this increase was eliminated when ric was also inactivated. Absence of other iron storage proteins, or of peroxidase and catalases, had no impact on RIC-mediated redox stress induction. Hence, we show that RIC interacts with Dps in a manner that serves to protect E. coli from RIC protein-induced ROS.
IMPORTANCE The mammalian immune system produces reactive oxygen and nitrogen species that kill bacterial pathogens by damaging key cellular components, such as lipids, DNA, and proteins. However, bacteria possess detoxifying and repair systems that mitigate these deleterious effects. The Escherichia coli RIC (repair of iron clusters) protein is a di-iron hemerythrin-like protein that repairs stress-damaged iron-sulfur clusters. E. coli Dps is an iron storage protein of the ferritin superfamily with DNA-binding capacity that protects cells from oxidative stress. This work shows that the E. coli RIC and Dps proteins interact in a fashion that counters RIC protein-induced reactive oxygen species (ROS). Altogether, we provide evidence for the formation of a new bacterial protein complex and reveal a novel contribution for Dps in bacterial redox stress protection.
INTRODUCTION
During the infection process, bacterial pathogens are able to survive aggressive environments through the activation of specific stress resistance genes. One such example of a stress-induced gene is ric. This gene encodes the repair of iron clusters (RIC) protein, which contains a di-iron center and contributes to the protection of bacterial pathogens, such as Escherichia coli, Haemophilus influenzae, Salmonella spp., Yersinia spp., and Clostridium spp. during exposure to nitrosative and/or oxidative stress (1). The ric gene is induced upon exposure to either oxidative or nitrosative stress, and in E. coli, Staphylococcus aureus, Neisseria gonorrhoeae, H. influenzae, and Cryptococcus neoformans, the RIC protein confers stress resistance through maintenance of the activity of various Fe-S-containing enzymes (1–3). Such an effect is well demonstrated for E. coli and S. aureus, where RIC proteins restore the activity of oxidatively and nitrosatively damaged Fe-S clusters in the tricarboxylic acid (TCA) cycle enzymes aconitase and fumarase (1, 4, 5) In E. coli, the RIC protein also acts under nonstress conditions to maintain aconitase and fumarase activities (6). Furthermore, E. coli RIC delivers iron (most likely in the ferrous state) for the assembly of Fe-S clusters in spinach apo-ferredoxin and in the E. coli Fe-S cluster-assembly scaffold protein IscU (7). The RIC protein also contributes to the survival of S. aureus and H. influenzae in activated macrophages, and it is required for full virulence in S. aureus when infecting the wax moth (Galleria mellonella) larva infection model (3, 8). Thus, RIC has an apparent role in bacterial pathogenicity through mediation of Fe-S cluster stability during exposure to redox and/or nitrosative stress.
The RIC proteins of E. coli and S. aureus contain di-iron centers of the histidine/carboxylate type within a four-helix bundle fold (9). The UV-visible spectrum of oxidized RIC exhibits a broad band at ca. 350 nm, and electron paramagnetic resonance (EPR) spectroscopy indicates that the principal g values are below 2 (g = 1.96, 1.92, and 1.88), which is indicative of an S = 1/2 spin state in a mixed-valence and antiferromagnetically coupled Fe(III)-Fe(II) binuclear iron center. Mössbauer spectroscopy showed that the mixed-valence Fe(III)-Fe(II) di-iron center of RIC is more labile than that of the μ-(oxo)-diferric form (7).
RIC proteins possess several highly conserved amino acid residues, of which some have been shown to influence the properties of the di-iron center and/or function of the protein. In particular, substitution of residues His129, Glu133, or Glu208 of the E. coli RIC abrogated its ability to protect the activity of aconitase. Moreover, two μ-carboxylate bridges contributed by Glu133 and Glu208, linking the two di-iron site atoms, were shown to be required for the assembly of a stable di-iron center (10). These studies also demonstrated the important contribution of the conserved His84, His129, His160, His204, Glu133, and Glu208 residues in ligating the di-iron center within the four-helix bundle fold, and these di-iron coordination roles were recently confirmed by X-ray crystallographic structural studies (11).
In the work reported here, we sought to identify proteins that interact with and support the function of RIC of E. coli. For this purpose, an E. coli library was screened for RIC protein interaction partners using the bacterial adenylate cyclase-based two-hybrid (BACTH) system. Potential interacting gene products were further tested by BACTH, bimolecular fluorescence complementation (BiFC), and pulldown assays. Our protein-protein interaction studies revealed that RIC interacts with the DNA-binding protein from starved cells (Dps). Dps is a symmetrical, dodecameric iron storage protein of the ferritin superfamily that contains a di-iron ferroxidation center located at the interface between subunits (12–14). Dps sequesters ferrous iron, which is oxidized preferentially by hydrogen peroxide at its di-iron center and then deposited for storage as Fe(III) oxyhydroxide as an iron core in the central cavity; the sequestered iron can subsequently be released by reduction (14, 15). The ferroxidase activity, DNA-binding, and iron sequestration properties of Dps confer on cells protection from oxidative stress and nutrient deprivation, as judged by the reduced survival of dps mutants under stress conditions, including starvation, oxidative stress, metal toxicity, and thermal stress (16). The physiological relevance of the interaction between the RIC protein and Dps was examined, and the results revealed that Dps modulates the function of RIC.
RESULTS
Identification of novel potential RIC-protein interaction partners by screening a bacterial two-hybrid E. coli library.
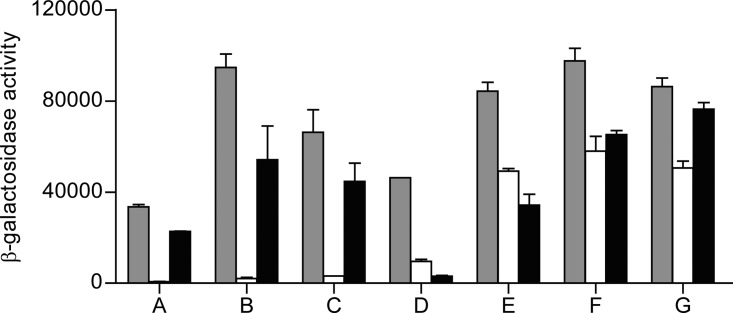
We used a genetic approach to further assess the physiological role of RIC in E. coli by employing the bacterial adenylate cyclase-based two-hybrid (BACTH) system (17) to screen for gene products that could interact with RIC. For this purpose, RIC was fused to the C terminal of the Bordetella pertussis adenylate cyclase T25 fragment and used as “bait” to screen previously constructed partially Sau3A-digested E. coli DNA random libraries that express fusions to the N terminal of the B. pertussis adenylate cyclase T18 fragment (18). We isolated 22 positive recombinant Lac+ colonies, from which plasmids were purified and then transformed into E. coli DHM1 harboring pKT25-RIC, the empty vector pKT25 (negative control), or pKT25-TorD (false-positive control), followed by determination of the β-galactosidase specific activities (Fig. 1). Seven pKT25-RIC transformants, harboring plasmids A to G, exhibited significant β-galactosidase activity indicative of a specific interaction (Fig. 1). Nucleotide sequencing followed by BLAST analysis was used to identify the genes within the inserts of these plasmids. Sequencing data revealed that plasmids A to C contain an ∼2-kb E. coli DNA fragment upstream of the T18 Cya domain and that all included the complete efp and ecnA genes and part of the ecnB gene. The efp gene encodes the elongation factor EF-P, a translation factor that facilitates the in vitro formation of the first peptide bond during translation (19, 20). The gene cluster ecnAB expresses two small cell membrane-associated entericidin lipoproteins, forming EcnAB, a toxin-antitoxin module that regulates programmed bacterial cell death under high osmolarity conditions, with EcnA acting as the antidote for the bacteriolytic entericidin, EcnB (21).
FIG 1.
E. coli RIC interactions investigated by BACTH. β-Galactosidase activities of E. coli DHM1 cell lysates separately expressing plasmids A to G, which were extracted from the BamHI library and cotransformed with the pKTN25-RIC (gray bars), pKTN25 empty plasmid (white bars), and pKTN25 fused to TorD (black bars). Each bar represents the mean value ± standard error from results of at least three independent cultures.
The other four plasmids, D to G, also contained an ∼2-kb insert located upstream of the T18 Cya domain, but in these cases the inserts carried the entire rhtA gene, encoding an inner membrane transporter involved in resistance to homoserine/threonine (22), and the dps gene, encoding the DNA-binding and iron storage protein from starved cells (12). Like RIC, E. coli Dps has been implicated in oxidative stress protection, which raised the possibility of a functional association between these two proteins that might be dependent on their direct interaction. For this reason, the potential interaction between the two proteins was investigated further in order to establish its validity and determine its physiological purpose.
E. coli RIC protein interacts with Dps.
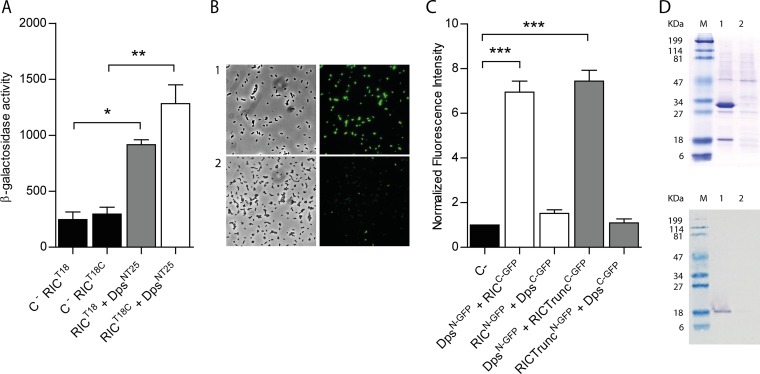
To determine whether the interaction between RIC and Dps, as identified through the screening of the pUT18 library, is indeed genuine, further BACTH experiments were performed. To enable such experiments, the gene encoding the RIC protein was cloned into pUT18C and pUT18 vectors (to create T18-RIC and RIC-T18 fusions), and the dps-coding region was introduced into the pKNT25 vector (to give Dps-T25 fusions), following which the β-galactosidase activities of the corresponding cotransformants were measured. High β-galactosidase activities were recorded for both sets of the RIC-Dps BACTH combinations tested, with activities 4 to 6 times greater than those of the controls (Fig. 2A), indicative of interaction between the RIC protein and Dps within the cytosol of E. coli.
FIG 2.
E. coli RIC interacts with Dps. (A) Bacterial two-hybrid assay. The interaction of the RIC protein, linked to the C terminal (white bar) or N terminal (gray bar) of the T18-Cya domain and expressed from pUT18 or pUT18C, respectively, was evaluated in E. coli DHM1 cotransformed with pKTN25 containing a Dps linked to the N terminal of the T25-Cya domain. E. coli cells harboring simultaneously empty pKTN25 and pUT18/pUT18C vectors expressing ric fusions served as negative controls (black bars). (B and C) BiFC assays. Cells were cotransformed with vectors expressing either RICC-GFP (pMRBAD-link-C-GFP-RIC) or RIC-TruncatedC-GFP (pMRBAD-link-C-GFP-RICTrunc) with DpsN-GFP (pET11a-link-N-GFP-Dps). The inverse configurations were also included. (B) Cells expressing RICC-GFP and DpsN-GFP were analyzed by light microscopy (bright field, left upper panel) and fluorescence microscopy (right upper panel). Lower panels depict images of cotransformed cells with empty vectors. Images were acquired using a 100× objective, and a fluorescein isothiocyanate (FITC) filter was used for the acquisition of the fluorescence images. (C) Fluorescence quantification was performed using MetaMorph microscopy automation and image analysis software. Fluorescence values for the negative control (empty plasmid vectors) were normalized to 1. (D) Pulldown assays. Lane 1, cells expressing pET-28a-RIC(HisTag) (RIC protein linked to a N-terminal His-tagged RIC [∼30 kDa]) and pACYCDuet-1-Dps (nonlabeled Dps [∼18 kDa]). Lane 2, cells expressing pET-28a (empty vector) and pACYCDuet-1-Dps (nonlabeled Dps [∼18 kDa]). Protein fractions were eluted from the Ni-chelating column at 100 mM imidazole and analyzed by SDS-PAGE (upper panel) and Western blotting using the anti-E. coli Dps antibody (bottom panel). Values are means ± standard errors from at least three independent cultures analyzed in duplicate. ***, P < 0.0005 (one-way analysis of variance [ANOVA] multiple-comparison test).
A second approach was used to test the proposed RIC-Dps interaction, which involved a bimolecular fluorescence complementation (BiFC) assay. In this method, one of the two proteins of interest is fused to the N-terminal half of the green fluorescent protein (GFP), and the other protein of interest is fused to the C-terminal half; the assay depends upon an interaction between the two proteins that promotes the reassembly of the two halves of GFP, such that emission of fluorescence is restored (23). Thus, GFP fusions (both the N- and C-terminal domains) were generated for both the RIC protein and Dps, and the fluorescence intensity of the corresponding E. coli cells containing plasmids coexpressing the RIC and Dps fusions was measured (Fig. 2B and C). The data showed that cells expressing RICC-GFP and DpsN-GFP exhibit an approximately 6-fold higher fluorescence relative to that of the control, while transformants expressing RICN-GFP and DpsC-GFP presented fluorescence levels similar to that of the control samples.
RIC consists of two domains, namely, a short N-terminal so-called ScdA_N domain of ∼60 residues of unclear function with a highly conserved pair of Cys residues (11) and a larger C-terminal “hemerythrin” domain of ∼140 residues that forms a di-iron center. We tested the BiFC interaction between Dps and a truncated form of RIC that lacks the Scd_N domain to determine which of the two RIC protein domains is responsible for the observed interaction with Dps. The results showed that the degree of interaction between the truncated RIC and Dps is similar to that observed when using the full-length protein (Fig. 2C). Thus, the interaction between RIC protein and Dps appears to occur through the C-terminal hemerythrin domain of RIC.
The interaction between RIC and Dps was also investigated by a pulldown assay. To this end, cells containing plasmids that express nonlabeled Dps and N-terminally His-tagged RIC were treated with formaldehyde, as described in Materials and Methods, to promote in vivo cross-linking. The cell extract was loaded into a Ni-chelating column, and the His-tagged RIC was eluted at 100 mM imidazole buffer. The fraction was analyzed by denaturing SDS-PAGE and by Western blotting in which the E. coli Dps antibody was used. Also, cells expressing only the nonlabeled Dps were treated and analyzed similarly to serve as a control. The results depicted in Fig. 2D show that elution of His-tagged RIC occurred together with a band that has a molecular mass corresponding to that of Dps. This band was proved by Western blotting to be the E. coli Dps (Fig. 2D). Therefore, the pulldown assays support the interaction between RIC and Dps.
Dps modulates the function of the RIC protein in maintaining Fe-S cluster status.
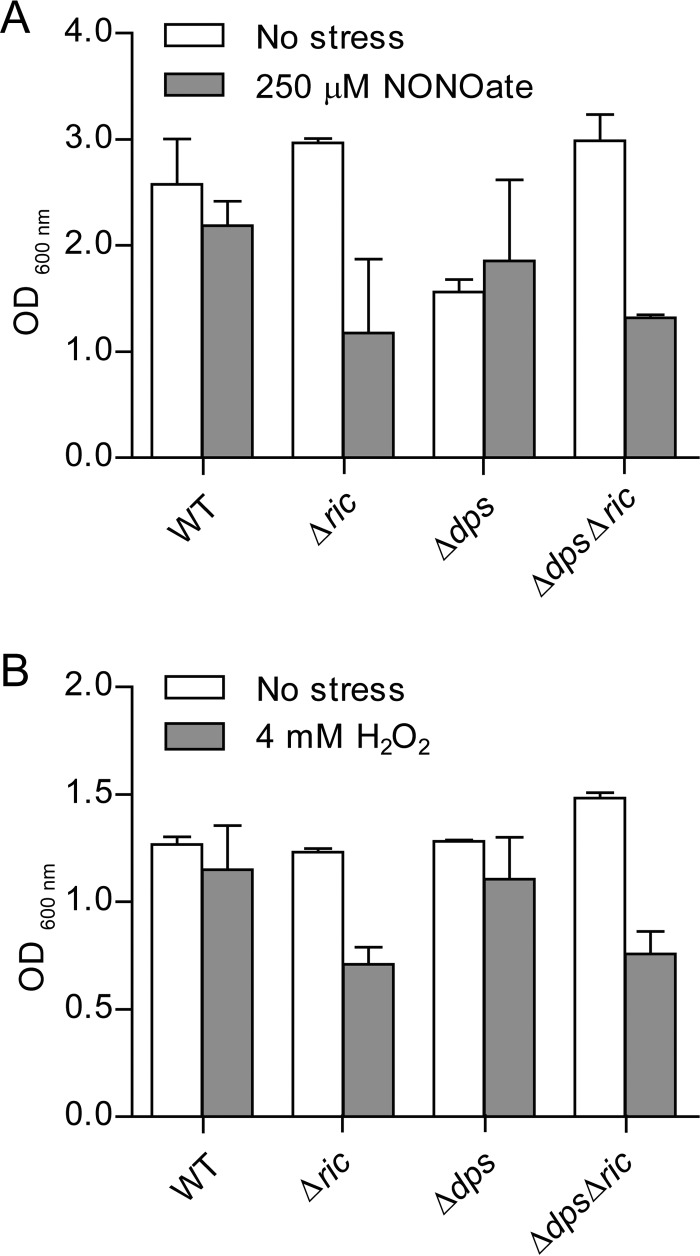
The RIC protein has been linked to the resistance of E. coli to oxidative and nitrosative stresses, as its inactivation decreases the survival of E. coli upon exposure to hydrogen peroxide or nitric oxide donors (4). Due to the interaction of the RIC and Dps proteins shown above, we questioned whether Dps could contribute to the stress protection afforded by RIC. To consider this possibility, a Δdps Δric double mutant was constructed, and the growth of E. coli wild type and Δric, Δdps, and Δdps Δric mutants under oxidative and nitrosative stress conditions was tested (Fig. 3). The experiments showed that inactivation of ric resulted in impaired growth under stress conditions imposed by 4 mM H2O2 or 250 μM spermine NONOate (Fig. 3), which is consistent with previous reports (4). However, under these conditions, the dps mutation had little impact on growth. Combining the Δdps mutation with the Δric mutation did not result in any further growth reduction, i.e., the Δdps Δric strain grew similarly to the Δric strain under the oxidative and nitrosative stress conditions employed (Fig. 3). Thus, Dps does not notably compensate for the lack of RIC under peroxide- or NO-induced stress.
FIG 3.
Growth of E. coli Δric, Δdps, and Δdps Δric strains under nitrosative and oxidative stress. E. coli wild-type, Δric, Δdps, and Δdps Δric strains were grown under anaerobic (A) and aerobic (B) conditions. At an OD600 of 0.3, cells were left untreated (No stress), treated with 250 μM spermine NONOate for 7 h (A), or treated with 4 mM H2O2 for 5 h (B). Error bars are ±standard deviations (SD) for experiments carried out at least three times.
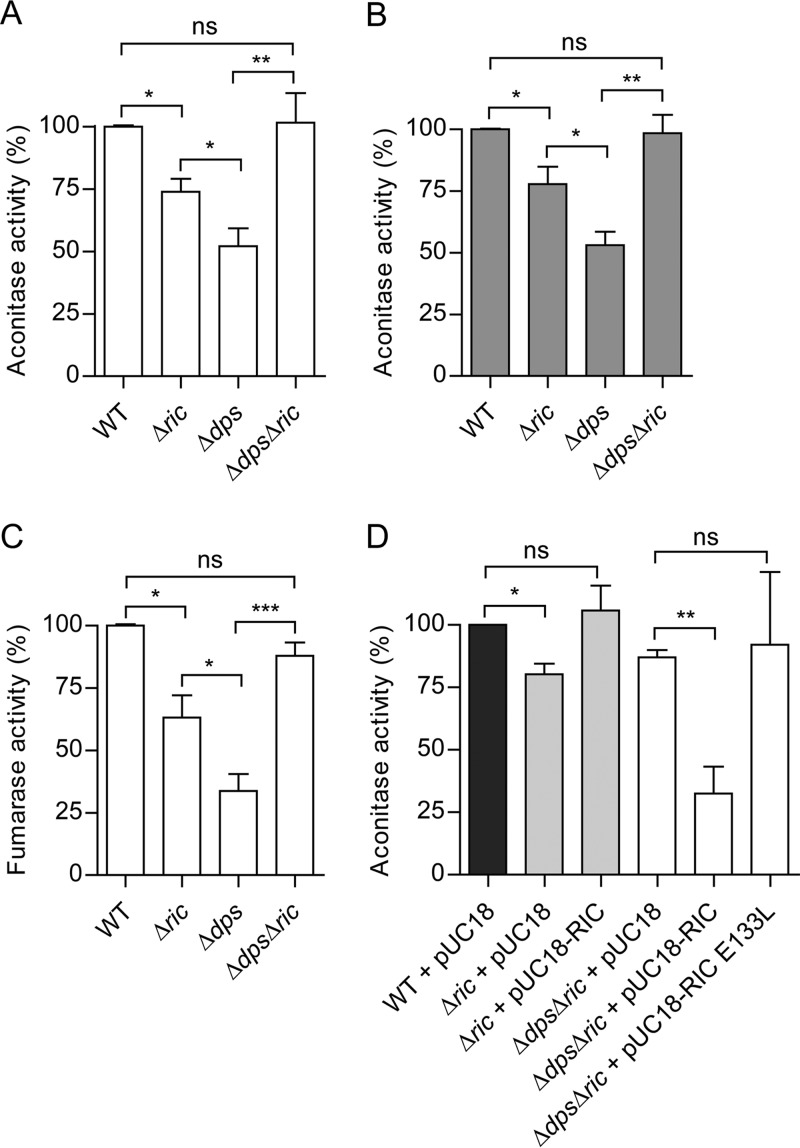
Another characteristic of the E. coli ric mutant is the reduced endogenous activity of Fe-S cluster-containing proteins, such as aconitase and fumarase, that contain solvent-exposed Fe-S clusters with a marked sensitivity to redox and nitrosative stress (4). Therefore, the possible contribution of Dps to this phenotype was explored by comparing the aconitase activity of the Δdps and Δdps Δric strains to that of the wild type and Δric mutant. The results showed that the Δdps mutation caused a 50% reduction in aconitase activity in log phase (Fig. 4A), consistent with a role for Dps in maintaining Fe-S cluster status. As expected, a similar effect was observed for the Δric mutant, although the reduction in activity (30%) was only approximately half as great as that observed for the Δdps mutant (Fig. 4A). Surprisingly, the Δdps Δric mutant exhibited aconitase activity that was higher than that of the corresponding single mutants and similar to that of the wild type (Fig. 4A). These aconitase activity effects were apparent in both the early log and the post-exponential phases (optical density at 600 nm [OD600] values of 0.6 and 2, respectively) (Fig. 4A and B), suggesting that the phenotype is independent of growth stage (note that dps is stationary-phase induced).
FIG 4.
Aconitase and fumarase activity of E. coli Δric and Δdps mutant strains. Aconitase activity of E. coli wild-type, Δric, Δdps, and Δdps Δric strains grown aerobically to OD600 of 0.6 (A and D) or 2.0 (B). Fumarase activity of E. coli wild-type, Δric, Δdps, and Δdps Δric strains grown aerobically to an OD600 of 0.6 (C). Complementation experiments of aconitase activity (D) were performed using Δric and Δdps Δric strains transformed with pUC18, and with pUC18 encoding the RIC protein and the mutated Glu133Leu-RIC (a RIC protein where glutamate 133 underwent site-directed mutation by leucine [10]). Values are represented as normalized to the aconitase or fumarase activity of wild-type cells. Values are means (±standard errors) from at least two independent cultures analyzed in duplicate. *, P < 0.05; **, P < 0.005; ns, not significant (unpaired Student's t test).
Similar effects were observed when testing the activity of another Fe-S enzyme, namely, fumarase. The data showed a reduction of 70% in fumarase activity in the Δdps mutant compared to that in the wild type during the early log phase (OD600 = 0.6). Also, in the Δric mutant there was a reduction in fumarase activity of about 40%, while the Δdps Δric double mutant displayed a fumarase activity similar to that of the wild type (Fig. 4C).
The restoration of aconitase and fumarase activity to wild-type levels in the Δdps Δric double mutant (with respect to the corresponding single mutants) suggests that the negative impact of the lack of the RIC protein on such activity is dependent on the presence of Dps (and vice versa), and this in turn indicates a hitherto unrecognized functional interdependence for these two proteins.
The association of the above aconitase activity effects with the RIC protein was confirmed by complementation using a multicopy plasmid bearing the wild-type ric gene under the control of its natural promoter. Complementation of the single Δric mutant led to the recovery of aconitase activity to levels similar to those of the wild type (Fig. 4D). More importantly, provision of a wild-type version of ric (in a multicopy plasmid) caused a large (60%) and significant reduction in the aconitase activity of the Δdps Δric double mutant (Fig. 4D). Thus, as anticipated, the ric-complemented double mutant exhibited the same phenotype as the dps mutant. This confirms that RIC is responsible for the decreasing aconitase activity in the dps-negative background.
To investigate whether the role of RIC in lowering the aconitase activity in the dps mutant is dependent on a biochemically functional version of the RIC protein, the ability of a RIC variant (lacking a complete di-iron site due to an E133L substitution; see reference 10) was tested in complementation experiments (Fig. 4D). Data clearly showed that the nonfunctional E133L-RIC variant does not enable a notable decrease in aconitase activity when expressed in the Δdps Δric strain (Fig. 4D).
In summary, the results suggest that, in the absence of Dps, the RIC protein has a deleterious effect on aconitase and fumarase activities, but that such an effect is not exhibited when Dps is present. This would imply that the interaction between Dps and RIC ensures that neither of these two proteins can participate in processes that negatively impact the activity of these Fe-S enzymes.
RIC does not interact with other E. coli iron storage proteins.
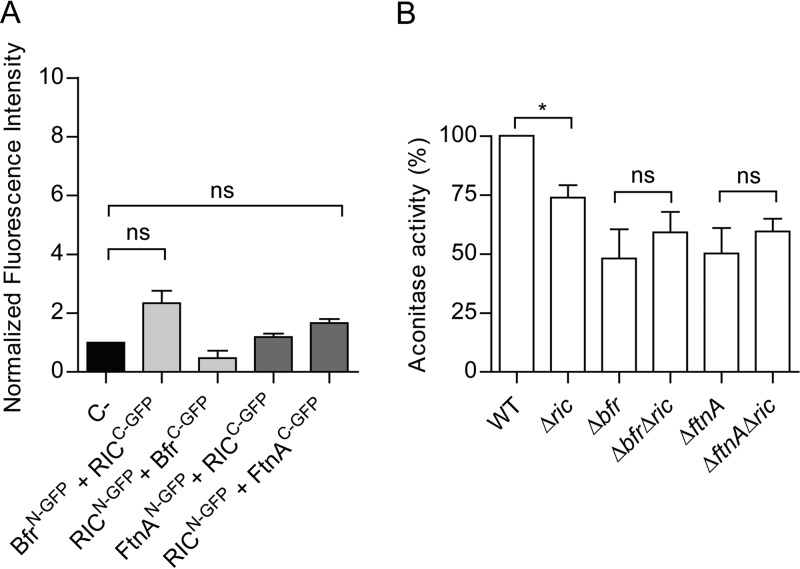
Escherichia coli Dps is an iron-sequestering protein composed of 12 identical subunits forming a shell surrounding a central cavity where up to ∼500 ferric iron atoms can be sequestered. As E. coli encodes two other iron storage proteins, namely, bacterioferritin (Bfr) and ferritin (FtnA), the possibility that RIC interacts with these other iron storage proteins was also investigated. Thus, corresponding BiFC experiments were performed in cells carrying recombinant plasmids that express RIC with either Bfr or FtnA as N- or C-terminal fusions to GFP domains. The resulting fluorescence intensity data failed to support any protein-protein interaction between RIC and Bfr or FtnA (Fig. 5A).
FIG 5.
Iron storage proteins Bfr and FtnA do not interact with RIC. (A) The possible interaction between RIC and Bfr or FtnA was analyzed by fluorescence microscopy and quantified by MetaMorph microscopy automation and image analysis software. Cells were cotransformed with vectors expressing RICC-GFP (pMRBAD-C-GFP-RIC) and BfrN-GFP (pET11a-N-GFP-Bfr) or FtnAN-GFP (pET11a-N-GFP-FtnA). The inverse conformations were also examined. Fluorescence values for negative control (empty plasmid vectors) were normalized to 1. Values are means ± standard errors from at least three independent cultures analyzed in duplicate. ns, not significant (one-way ANOVA multiple-comparison test). (B) E. coli cells of single (Δric, Δbfr, and ΔftnA) and double mutant strains (Δbfr Δric and ΔftnA Δric) were grown aerobically, collected at an OD of 0.6, and the aconitase activity was determined. Values are represented as normalized to the aconitase activity of wild-type cells. Values are means ± standard errors from at least two independent cultures analyzed in duplicate. *, P < 0.05; ns, not significant (unpaired Student's t test).
In a second set of experiments, the aconitase activities of wild-type, Δric, Δbfr, ΔftnA, Δbfr Δric, and ΔftnA Δric strains grown to the exponential phase (OD600 = 0.6), were determined. Similarly to the Δdps strain, the Δbfr and ΔftnA strains both displayed ∼50% lower aconitase activity levels (Fig. 5B). But, contrary to the effect of combining the Δdps and Δric mutations, the combined absence of RIC and Bfr or FtnA resulted in cellular aconitase activities similar to those present in the corresponding single-mutant strains (Fig. 5B). Thus, the lower aconitase activity caused by the Δric mutation is not additive with respect to the lower activity resulting from the Δbfr or ΔftnA mutations. Furthermore, it can be concluded that (unlike Dps) Bfr and FtnA do not interact with RIC, and that their absence does not result in a RIC dependent decrease in aconitase activity.
RIC increases intracellular ROS levels when Dps is absent.
Dps protects cells from oxidative stress due to its ability to couple the reduction of hydrogen peroxide to water with the oxidation of free ferrous iron to sequestered ferric iron. In addition, its association with DNA helps to prevent ROS-induced DNA damage (24). This suggests that the role of Dps in preventing RIC protein-induced inhibition of aconitase activity may arise from the ability of Dps to detoxify ROS that might be produced by the di-iron center of the RIC protein (e.g., through binding and reduction of oxygen). Therefore, the ROS contents of Δric, Δdps, and Δdps Δric strains were compared with those found in the wild type to determine whether the presence of RIC in the absence of Dps, results in raised levels of ROS (Fig. 6A). Data show that the wild-type and Δric mutant strains contain similar amounts of ROS, while the Δdps strain had significantly higher levels (∼2-fold) (Fig. 6A). This is as expected, given the known role of Dps in redox stress resistance (24). However, introduction of the ric mutation into the dps mutant eliminated the increased intracellular ROS levels of the Δdps single mutant (Fig. 6A). This suggests that the raised ROS levels of the dps single mutant are a consequence of an increase in RIC dependent ROS production, which thus supports a role for Dps in interacting with RIC to restrict its release of ROS.
FIG 6.
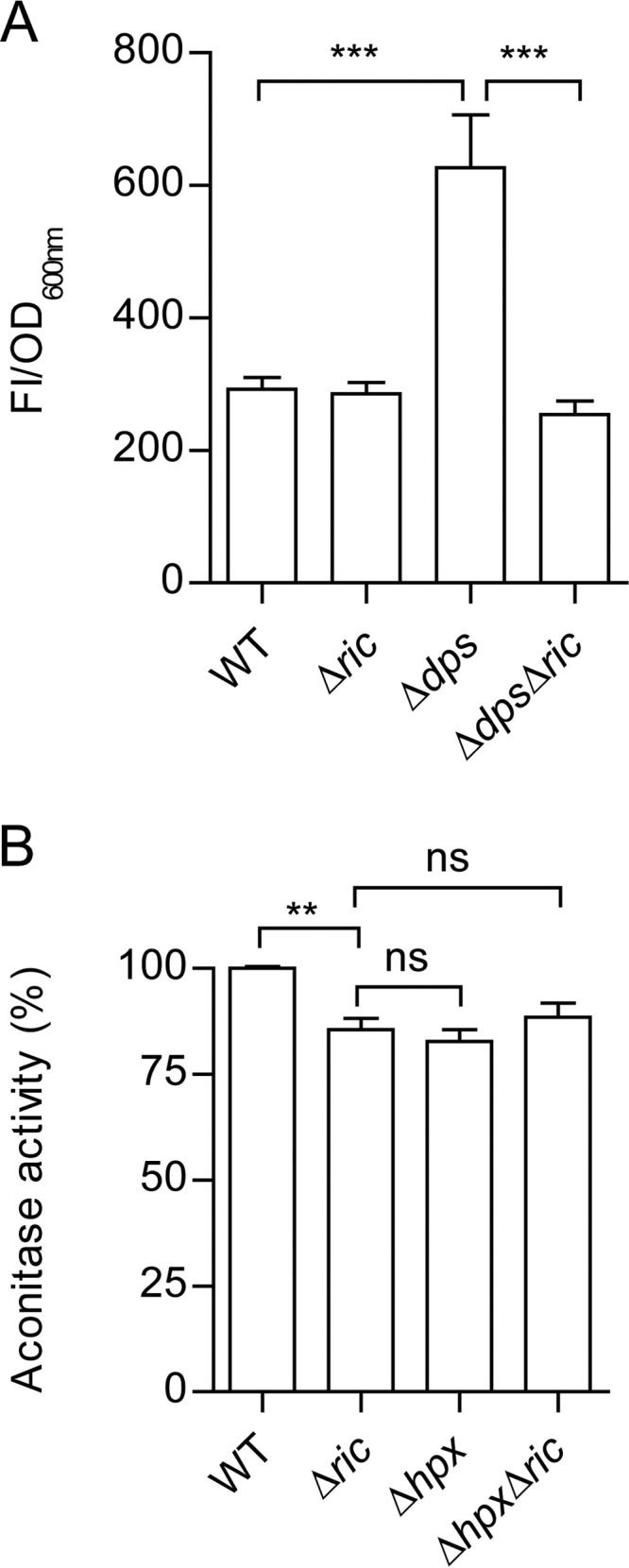
Intracellular ROS content of Δdps and Δric mutant strains and the effect of combining the Δhpx and Δric mutations on aconitase activity. (A) The E. coli wild-type and isogenic Δric, Δdps, and Δdps Δric mutant strains were grown to OD600 of 0.6, and the intracellular ROS levels were measured by incubation of cell suspensions with 10 μM dichloro-dihydro-fluorescein diacetate (DCFH-DA) for 2 h. Fluorescence intensity was normalized according to the final OD (FI/OD). (B) Aconitase activity of the E. coli wild-type, Δric, Δhpx (ΔahpCF ΔkatE ΔkatG), and Δhpx Δric strains grown aerobically to the log phase (OD600 = 0.6). Values are represented as normalized to the aconitase activity of wild-type cells. Values are means (±standard errors) from at least two independent cultures analyzed in duplicate. ***, P < 0.0005; ns, not significant (unpaired Student's t test).
To discover whether other elements of the redox stress resistance response might also act to lessen RIC induced ROS production, the Δric mutation was introduced into a strain (Δhpx) that lackes the capacity to degrade hydrogen peroxide due to inactivation of both catalase genes, as well as of the alkyl-hydroperoxide reductase genes (Table 1) (25, 26). Aconitase assays showed that the Δhpx Δric quadruple mutant has activity levels similar to those determined for the Δric and Δhpx mutants (Fig. 6B). Therefore, we concluded that the three major peroxidases (KatE, KatG, and AhpCF) of E. coli are not involved in countering any RIC protein-mediated ROS production, at least under conditions where Dps is active.
TABLE 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| DHM1 | F′ cya-854 recA1 endA1 gyrA96 (Nalr) thi1 hsdR17 spoT1 rfbD1 glnV44(AS) | Euromedex |
| Wild type | K-12 ATCC 23716 | ATCC |
| Δric strain | K-12 Δric::cat | 36 |
| Δdps strain | JS091 Δdps::kan | 37 |
| Δbfr strain | JW3298 Δbfr::kan | 37 |
| ΔftnA strain | MC4100 ΔftnA::spc | 28 |
| Δdps Δric strain | K-12 Δdps::kan Δric::cat | This study |
| Δbfr Δric strain | K-12 Δbfr::kan Δric::cat | This study |
| ΔftnA Δric strain | K-12 ΔftnA::spc Δric::cat | This study |
| MG1655 | F− WT | 38 |
| SJ90 | BW25113 Δric::cat | 39 |
| LC106 (hpx) | ΔahpCF ′kan::′ahpF Δ(katG17::Tn10)1 Δ(katE12::Tn10)1 | 26 |
| Δhpx Δric strain | LC106 Δric1::cat | This study |
| XL2-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB+ lacIqZΔM15 Tn10 (Tetr)] | Agilent |
| BL21Gold(DE3) | E. coli B F− ompT hsdS(rB− mB−) dcm+ Tetr gal λ(DE3) endA Hte | Agilent |
| Plasmids | ||
| pUT18/pUT18C | Vector that allows construction of in-frame fusions at the N terminal/C terminal of T18 fragment (amino acids 225–399 of CyaA) | 17 |
| pKT25/pKNT25 | Vector that allows construction of in-frame fusions at the N terminal/C terminal of T25 fragment (amino acids 1–224 of CyaA) | 17 |
| pUT18/pUT18C-RIC | RIC fused to T18 fragment in N/C terminal | This study |
| pKT25/pKNT25-RIC | RIC fused to T25 fragment in N/C terminal | This study |
| pUT18/pUT18C-Dps | Dps fused to T18 fragment in N/C terminal | This study |
| pKT25/pKNT25-Dps | Dps fused to T25 fragment in N/C terminal | This study |
| pUT18-Zip | Leucine zipper fused to T18 fragment in the N terminal | 17 |
| pKT25-Zip | Leucine zipper fused to T25 fragment in the C terminal | 17 |
| pUT18-TorD | TorD fused to T18 fragment in N terminal | 18 |
| pKT25-TorD | TorD fused to T25 fragment in C terminal | 18 |
| BamHI | pUT18 plasmid that contains chromosomal fragments obtained by partial digestion of the MC4100 chromosomal DNA with Sau3A1 and cloned into the BamHI site | 18 |
| pUC18 | Expression vector | ATCC |
| pUC18-RIC | Vector for expression of RIC | 4 |
| pUC18-RIC-Glu133Leu | Vector for expression of RIC-Glu133Leu | 10 |
| pET11a-link-GFP | Vector for expression of fusions with N-terminal fragment of GFP | 33 |
| pMRBAD-link-GFP | Vector for expression of fusions with C-terminal fragment of GFP | 33 |
| pET11a-RIC-GFP | RIC fused to N-terminal GFP fragment | This study |
| pMRBAD-RIC-GFP | RIC fused to C-terminal GFP fragment | This study |
| pET11a-Dps-GFP | Dps fused to N-terminal GFP fragment | This study |
| pMRBAD-Dps-GFP | Dps fused to C-terminal GFP fragment | This study |
| pET11a-Bfr-GFP | Bfr fused to N-terminal GFP fragment | This study |
| pMRBAD-Bfr-GFP | Bfr fused to C-terminal GFP fragment | This study |
| pET11a-FtnA-GFP | FtnA fused to N-terminal GFP fragment | This study |
| pMRBAD-FtnA-GFP | FtnA fused to C-terminal GFP fragment | This study |
| pET11a-RICTrunc-GFP | Truncated RIC fused to N-terminal GFP fragment | This study |
| pMRBAD-RICTrunc-GFP | Truncated RIC fused to C-terminal GFP fragment | This study |
| pET-28a | Expression vector | Novagen |
| pET-28a-RIC(HisTag) | Vector for expression of N-terminal poly-His-tagged RIC | This study |
| pACYCDuet-1-Dps | Vector for expression of Dps | This study |
WT, wild type; GFP, green fluorescent protein.
DISCUSSION
Aconitase and fumarase are enzymes of the TCA cycle that are prone to oxidative stress damage. We previously showed that the di-iron RIC repairs these enzymes and is able to transfer iron to Fe-S containing proteins (4–7). In the work described here, we screened an E. coli BACTH library in order to identify proteins that interact with RIC and thus might be required to assist its function. As a consequence of our screening, Dps emerged as a RIC protein interaction candidate. This interaction was supported by generation and analysis of additional Dps and RIC protein BACTH constructs and by GFP complementation and pulldown assays. Dps belongs to the ferritin superfamily, which led us to investigate the possible interaction of RIC with the two other ferritins present in E. coli, namely, ferritin and bacterioferritin. However, none of these proteins were found to interact with RIC or to influence its activity in vivo.
We also observed that inactivation of RIC resulted in lower aconitase and fumarase activity, which is consistent with previous findings indicating that this protein contributes to the protection of solvent accessible Fe-S clusters from ROS damage observed under aerobic growth conditions (6). Similar results were obtained here for the dps, ftnA, and bfr single-mutant strains, indicating that lack of any of these gene products results in lower endogenous aconitase activity. The role of FtnA and Bfr in aconitase protection was previously demonstrated, as the two ferritins promote the reactivation of aconitase activity following stress damage in Salmonella enterica serovar Typhimurium (27). In contrast with our findings in E. coli, no loss of aconitase activity was observed for S. enterica ftnA or bfr single mutants in the absence of stress; this discrepancy may be related to different physiological roles and regulation of ferritins in Salmonella species and E. coli (27, 28).
A surprising finding was that the defective aconitase activity of the Δdps and Δric single mutant strains was reversed when these two mutations were combined in the Δdps Δric double mutant, such that the activity was restored to that measured in the wild type. This result, together with the smaller amounts of ROS observed in the Δdps Δric mutant compared to that in the Δdps mutant, suggests that RIC is responsible for the generation of ROS but only in the absence of Dps and, thus, that the interaction of Dps and RIC serves to enable Dps to restrict ROS release (which is presumed to damage the Fe-S cluster of aconitase and fumarase and hence lower the observed activity of these enzymes in a dps mutant) by the RIC protein. Interestingly, other redox stress resistance components (KatE, KatG, and AhpCF) failed to impact the RIC mediated inhibition of aconitase activity (at least in the presence of Dps). These results suggest that the protective effect of Dps on the ROS generation by RIC is one that is highly specific and not replicated by the other peroxide-consuming cytosolic factors. Indeed, our findings indicate that a direct interaction is required to enable Dps to quench the ROS-generating activity of RIC. The exact mechanism involved in the quenching is unclear, and a better understanding will require in vitro reaction studies combining the Dps and RIC proteins. However, two processes by which Dps could exert a ROS-quenching action upon RIC can be considered, as follows: (i) Dps might sequester iron released from the di-iron site of RIC and thus restrict Fe-driven Fenton chemistry, or (ii) Dps could consume hydrogen peroxide (or hydroxyl radicals [see references 15 and 29]) generated by RIC due to the reaction of its di-iron site with molecular oxygen.
Although the absence of RIC in the presence of Dps resulted in reduced aconitase and fumarase activity, lack of RIC had no impact on ROS levels when Dps was present. The reason for this effect is unclear, but it may indicate a role for the RIC in supplying iron from Dps for Fe-S cluster repair and/or synthesis.
The proposed role of RIC (4) is to repair damaged Fe-S clusters of proteins, such as aconitase and fumarase, by donating iron from its di-iron center. This leads to the formation of an intermediate mononuclear iron center that is prone to react with oxygen to generate ROS, such as hydrogen peroxide. In this process, the interaction with Dps would fulfill two roles, namely, trapping ROS released by RIC and providing a sink for iron liberated from the di-iron center of RIC.
In conclusion, we report an interaction between the Dps and RIC proteins of E. coli which represents the first example of a protein that interacts with the ferritin-like Dps. In addition, our results indicate that the Dps-RIC protein interaction contributes to the function of RIC, which is one of the few known bacterial proteins involved in repair.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli strains used in this work are listed in Table 1 and were grown at 37°C. E. coli XL2-Blue and E. coli reporter strain DHM1 carrying a nonreverting adenylate cyclase deficiency (cya) gene were used as host strain and for detection of protein-protein interactions, respectively.
Construction of the E. coli double mutant strains was performed by bacteriophage P1-mediated transduction (30), and the corrected mutations were confirmed by PCR using the primers listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primer purpose and name | Sequence |
|---|---|
| Construction of plasmids used in BACTH | |
| ric_Fw | GAGGTGTCGACTATGGCTTATC |
| ric_Rv | CTTTTAGGATCCTCACCCGCC |
| dps_Fw | GTTAATTACTGGGATCCAACATCAAGAGG |
| dps_Rv | TCCTGTCAGGTACCCGCTTTTATC |
| T18_Fw | CATTAGGCACCCCAGGCTTTAC |
| T18_Rv | GAGCGATTTTCCACAACAAGTC |
| T18C_Fw | CATACGGCGTGGCGGGGAAAAG |
| T18C_Rv | AGCGGGTGTTGGCGGGTGTCG |
| T25_Fw | ATGCCGCCGGTATTCCACTG |
| T25_Rv | CGGGCCTCTTCGCTATTACG |
| NT25_Fw | CACCCCAGGCTTTACACTTTATGC |
| NT25_Rv | CAATGTGGCGTTTTTTTCCTTCG |
| Construction of plasmids used in BiFC | |
| ric_xhoFw | GAATGAGGTCTCGAGTATGGCTTATC |
| ric_bamRv | GCGCAATGGGATCCAGCTTTTAGA |
| ric_ncoFw | GAGGTATCAGCCATGGCTTATCG |
| ric_aatRv | CCAGCTTTTAGACGTCTCACCC |
| dps_xhoFw | CGTTAATTACTCGAGCATAACATCAAG |
| dps_bamRv | GTACTAAGGATCCGCACCATCAGC |
| dps_sphFw | CAAGAGGATATGCATGCATGAGTACCGCTA |
| dps_aatRv | CATCAGCGATGGGACGTCTCGATGTTAG |
| bfr_xhoFw | GAGTGGAAGCGCTCGAGTCAAAAAATG |
| bfr_bamRv | GGAGGGTTCTGGATCCCGACACG |
| bfr_ncoFw | GAAGGAGTCAAACCATGGAAGGTGATAC |
| bfr_aatRv | CGGACGTCCCTTCTTCGCGGATC |
| truncric_xhoFw | CTTTAAGAAGGCTCGAGACATATGGCTG |
| truncric_ncoFw | GGAGATATACCCATGGCTGAACAAC |
| ftna_xhoFw | CAAATATAACCTTTCTCGAGCACTATC |
| ftna_bamRv | TGAAACGGATCCAGTAAACCTGC |
| ftna_ncoFw | GAGCACTACCATGGTGAAACCAGAAAT |
| ftna_aatRv | CGGAGAGGACGTCTTTTGTGTGTC |
| Construction of plasmids used for protein expression | |
| pric_ndeFw | AAGAATGAGGTATCACATATGGCTTATCGC |
| pric_ecoriRv | GGCTGTTTATTGGTAAGAATTCGGCTGCTG |
| pdps_ndeFw | GAGGATATGAACATATGAGTACCGC |
| pdps_kpnRV | GTACTAAAGTTCGGTACCATCAGCG |
| Double mutant construction confirmation | |
| Conf_dps_Fw | CAGAATAGCGGAACACATAGC |
| Conf_dps_Rv | GATGCACTAAATAAGTGCGTTG |
| Conf_bfr_Fw | CTCTTCAAAGAGTGGAAGCG |
| Conf_bfr_Rv | GATCTCTTATTAACCGGGAGG |
| Conf_ftnA_Fw | CAAATTATAGTGACGCCACAG |
| Conf_ftnA_Rv | ACCGATCAGAGTAAGATTTGC |
| Conf_ric_Fw | AAGAATGAGGTATCACATATGGCTTATCGC |
| Conf_ric_Rv | GGCTGTTTATTGGTAAGAATTCGGCTGCTG |
E. coli cells were grown in LB medium under aerobic conditions in flasks containing a 1/5 volume of culture or under anaerobic conditions in rubber seal-capped flasks filled with medium and extensively bubbled with nitrogen prior to growth. For the stress assays, cells were grown, at 37°C and 150 rpm in M9B minimal medium [60 mM K2HPO4, 33 mM KH2PO4, 7.6 mM (NH4)2SO4, 1.7 mM sodium citrate, 1 mM MgSO4, and 10 μM MnCl2 (pH 7)] supplemented with 10 μg/ml thiamine and 40 μg/ml l-arginine, l-leucine, l-proline, l-threonine, and 40 mM glucose. Cultures at an OD600 of 0.3 were either left untreated or exposed to 4 mM H2O2 for 6 h or 250 μM spermine NONOate for 9 h.
BACTH experiments.
The bacterial adenylate cyclase-based two-hybrid (BACTH) system assay (17) was used to identify RIC-interacting proteins. E. coli RIC protein was fused to the C terminal of the Bordetella pertussis Cya T25 domain (pKT25-RIC) and used to screen an E. coli MC4100 gene library containing chromosomal fragments fused to the N terminal of the B. pertussis Cya T18 domain. The DNA fragments were obtained by partial digestion with Sau3AI and cloning into the BamHI site of pUT18 plasmids (18). About 1 μg of pUT18BamHI DNA library was transformed, together with pKT25-RIC, into E. coli DHM1 cells by electroporation. Blue colonies present in Ampr Cmr selective plates (l-agar with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal]) were identified after incubation at 30°C for 36 h, and cells with the highest β-galactosidase activity were considered to contain recombinant plasmids harboring genes encoding polypeptides that interact with the E. coli RIC protein. A total of 22 colonies were obtained, and the corresponding plasmids were isolated, cotransformed with pKT25-RIC plasmid in E. coli DHM1, and the strength of the protein-protein interactions was again estimated by quantification of the β-galactosidase activity. Seven isolates considered positive were named A through G (Fig. 1) and sequenced using primer T18_Fw (Table 2). To identify the encoded proteins, the sequences were screened against the E. coli K-12 MG1655 genome using BLAST.
Genes coding for RIC and Dps were PCR amplified from E. coli K-12 genomic DNA using the oligonucleotides described in Table 2 and cloned into pKT25 (fused to Cya C-terminal T25 domain), pKNT25 (fused to Cya N-terminal T25 domain), pUT18 (fused to Cya N-terminal T18 domain), and pUT18C (fused to Cya C-terminal T18 domain) plasmids, and the enzyme Pfu DNA polymerase (Thermo Scientific). The resulting recombinant plasmids encoded Dps or RIC with either a C- or N-terminally linked T25 or T18 domain from the B. pertussis Cya protein. Two complementary plasmids, one carrying a T25 fragment and the other a T18 fragment, were cotransformed into the E. coli DHM1 strain (lacking cya). E. coli DHM1 cells containing the ric-encoding pUT18 or pUT18C plasmids were cotransformed with complementary pKTN25 empty plasmids that served as negative controls.
In all cases, false positives were tested by cotransformation of E. coli DHM1 with plasmids containing each gene and pKT25-TorD, which expresses E. coli TorD, which binds nonspecifically to a wide variety of polypeptides (31).
For β-galactosidase activity determination (32), at least 3 representative colonies of each transformation plate were inoculated, in duplicate, in LB medium, and following an overnight growth at 37°C, transformant cultures were reinoculated (at a 0.01 dilution) into LB with ampicillin (100 μg/ml), kanamycin (50 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 0.5 mM). When cultures reached an OD600 of 0.5 (after approximately 16 h of growth, at 30°C), 1 ml of each culture was collected by centrifugation (5,000 × g, 5 min at 4°C). The pellets were lysed by incubation with 100 μl BugBuster HT 1× (Novagen) at 37°C, for 30 min. Cellular debris were then removed by centrifugation, and the β-galactosidase activities were assayed in 20-μl suspensions in a microplate reader. The assays were initiated by addition of a reaction mixture comprising 0.27% β-mercaptoethanol (vol/vol) and 0.9 mg/ml ONPG (o-nitrophenyl-β-d-galactopyranoside) in buffer A (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 1 mM MgSO4 · 7H2O, and 10 mM KCl). Reaction mixtures were incubated at 28°C, and the absorbance was recorded at 420 nm at 2-min intervals for 90 min. The β-galactosidase specific activity was defined as ONPG/min/milligram of protein. Interactions were considered positive for those reactions, whereas β-galactosidase activity was at least four times higher than that of the negative control.
Bimolecular fluorescence complementation (BiFC) assays.
BiFC assay was performed essentially as described previously (33). For this purpose, the genes encoding RIC a truncated version of the RIC (lacking the first 57 amino acid residues in the N terminal [10]), Dps, Bfr, and FtnA were PCR amplified from genomic DNA of E. coli K-12 using the oligonucleotides described in Table 2. The DNA fragments were cloned into vectors (pET11a-link-N-GFP and pMRBAD-link-C-GFP [33]) that express the green fluorescent protein (GFP) to allow formation of corresponding N- or C-terminal GFP fusions, respectively. Cloning was achieved using XhoI and BamHI sites (for cloning into pET11a-link-N-GFP) or NcoI and AatII sites (for cloning into pMRBAD-link-C-GFP) sites, except for Dps, for which SphI replaced NcoI. All recombinant plasmids were sequenced, confirming the integrity of the genes and the absence of undesired mismatches. Cells harboring pET11a-link-N-GFP and pMRBAD-link-C-GFP served as negative controls.
E. coli BL21(DE3)Gold (Agilent) was cotransformed with the resulting recombinant pET11a-link-N-GFP and pMRBAD-link-C-GFP vectors in various combinations (RIC/Dps, truncated RIC/Dps, RIC/Bfr, and RIC/FtnA), and plated on selective LB agar. Colonies were inoculated in LB medium, grown overnight at 37°C and 150 rpm, and plated onto inducing LB agar medium containing 20 μM IPTG and 0.2% of arabinose. After an overnight incubation at 30°C followed by 2 days of incubation at room temperature, colonies were suspended in phosphate-buffered saline (PBS) and spread onto 1.7% agarose slides. Cells were examined for green fluorescence in a Leica DM6000 B upright microscope coupled to an Andor iXon+ camera, using 1000× amplification and a fluorescein isothiocyanate (FITC) filter. The images were analyzed using the MetaMorph microscopy automation and image analysis software.
Pulldown and Western blot assays.
The genes encoding RIC and Dps were amplified from E. coli K-12 genomic DNA by PCR using the oligonucleotides listed in Table 2, cloned into pET-28a and pACYCDuet-1 vectors, respectively, and sequenced, which confirmed their integrity and the absence of undesired mutations. E. coli BL21(DE3)Gold was transformed with the following pair of plasmids: (i) pET-28a-RIC(HisTag) (expressing RIC fused to an N-terminal His tag) and pACYCDuet-1-Dps (expressing a nonlabeled Dps); and (ii) pET-28a (empty vector) together with pACYCDuet-1-Dps. Cells harboring the latter pair of recombinant plasmids served as control samples. Cells were grown in LB medium supplemented with 10 μM Fe and the appropriate antibiotics at 30°C to an OD600 of 0.3. At this time, 0.3 mM IPTG was added to induce the expression of the His-tagged RIC and Dps proteins, and after 4 h, the cross-linking agent formaldehyde (1% final concentration) was added to the cells. The cross-linking reaction (34) was carried out at 37°C for 20 min, and the reaction was stopped by incubation with glycine (final concentration of 0.5 M) at room temperature for 5 min. Bacterial cells were harvested by centrifugation, washed twice with PBS, and resuspended in PBS. Cells were disrupted in a French press (Thermo), and cell debris were removed by centrifugation. The total protein concentration of the supernatants was determined by the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). For the pulldown experiments, these supernatants were loaded into Ni-chelating Sepharose Fast Flow columns (GE Healthcare), which were first washed with 10 mM Tris-HCl (pH 7.5), and the proteins were eluted with imidazole-containing buffers. The protein fractions were analyzed by denaturing 12.5% SDS-PAGE, in which the cross-linking promoted by formaldehyde was reversed by heating and analyzed by Western blotting.
For Western blot analysis, samples that were first resolved by SDS-PAGE were transferred to a nitrocellulose blotting membrane (GE Healthcare) in a Trans-Blot Semi-Dry cell apparatus (Bio-Rad). The membrane was blocked by addition of Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.5] and 500 mM NaCl) containing 5% of dried skimmed milk and incubation at room temperature for 1 h. Then, the membrane was incubated with the primary antibody against E. coli K-12 Dps (1:1,000 dilution in TBS-T [TBS plus 0.05% Tween 20] plus 5% of dried skimmed milk). Following an overnight incubation at 4°C, the membrane was washed with TBS-T and incubated with the secondary antibody (anti-rabbit IgG–alkaline phosphatase) (Sigma) diluted 1:10,000 in TBS-T plus 5% dried skimmed milk). The reaction proceeded for 1 h at room temperature, and the color was developed by addition of 10 μl of NBT-BCIP (nitroblue tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate; Sigma) in 10 ml buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, and 5 mM MgCl2).
Enzyme activity assays and determination of endogenous ROS.
E. coli wild-type, Δric, Δdps and Δdps Δric strains that were transformed with either pUC18, pUC18-RIC, or pUC18-RIC-E133L (prepared as described in reference 10) were tested for endogenous aconitase and fumarase activities. To this end, the E. coli cells strains were grown in LB medium at 37°C, under aerobic conditions, to OD600 values of 0.6 and 2, as indicated in each case.
For the aconitase assays, cells grown to the desired cell density were centrifuged, washed in reaction buffer (50 mM Tris-HCl and 0.6 mM MnCl2 [pH 8]), and the pellets were frozen in liquid nitrogen. The following experiments were performed under anaerobic conditions. Prior to the activity assay, the cell pellets were resuspended in reaction buffer containing 0.5 mg/ml lysozyme and 0.2 mg/ml DNase, incubated on ice for 10 min, and then centrifuged at 9,600 × g for 10 min at 4°C. The aconitase activity was determined in these supernatants (50 μl) in reaction mixtures that also contained 200 μM NADP+, 1 U isocitrate dehydrogenase, and 30 mM sodium citrate (10), and by recording the formation of NADPH at 340 nm.
For the fumarase activity assays (35), once the cells reached the desired cell density, they were centrifuged, washed with 50 mM sodium phosphate (pH 7.3) buffer, and frozen in liquid nitrogen. Cell pellets were resuspended in 2 ml of the same phosphate buffer and lysed by five freeze-thaw cycles that used liquid nitrogen and a water bath at room temperature. The resulting cell extracts were cleared by addition of sodium deoxycholate to a final concentration of 0.5%. Fumarase activity was determined under anaerobic conditions in reaction mixtures that contained the cell lysates (10 μl) and 50 mM l-malate (which is quickly converted to fumarate) and by following the consumption of fumarate at 240 nm.
Endogenous reactive oxygen species content was determined in E. coli wild-type, Δric, Δdps, Δdps Δric, Δbfr, Δbfr Δric, ΔftnA, and ΔftnA Δric strains (Table 1). Cells were grown aerobically to an OD600 of 0.6, collected by centrifugation, resuspended in PBS, and distributed in 96-well microtiter plates. Following the addition of 10 μM dichloro-dihydro-fluorescein diacetate (DCFH-DA), the fluorescence was measured for 2 h in a Cary spectrofluorimeter (Varian, Agilent) at an excitation wavelength (λex) of 485 nm and an emission wavelength (λem) of 538 nm. The fluorescence intensity (FI) was normalized in relation to the optical density of each culture at 600 nm.
ACKNOWLEDGMENTS
We thank Hirotada Mori (Graduate School of Biological Sciences, Nara Institute of Science and Technology, Ikoma, Nara, Japan) for provision of the mutants from the Keio collection, and James Imlay (Department of Microbiology, University of Illinois at Urbana-Champaign, Urbana, IL) for the Hpx mutant. We are also grateful to Tracy Palmer and Frank Sargent (Centre for Bacterial Cell Biology, Newcastle University, Newcastle upon Tyne, UK) for providing the BACTH E. coli library.
This work was financially supported by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular), funded by FEDER funds through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI), and by national funds through FCT (Fundação para a Ciência e a Tecnologia) for grants PTDC/BBB-BQB/5069/2014 and SFRH/BD/118545/2016 (LSOS). This project has also received funding from the European Union's Horizon 2020 research and innovation program under grant agreement number 810856.
REFERENCES
- 1.Overton TW, Justino MC, Li Y, Baptista JM, Melo AMP, Cole JA, Saraiva LM. 2008. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J Bacteriol 190:2004–2013. doi: 10.1128/JB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow ED, Liu OW, O'Brien S, Madhani HD. 2007. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Curr Genet 52:137–148. doi: 10.1007/s00294-007-0147-9. [DOI] [PubMed] [Google Scholar]
- 3.Harrington JC, Wong SMS, Rosadini CV, Garifulin O, Boyartchuk V, Akerley BJ. 2009. Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated ytfE gene. Infect Immun 77:1945–1958. doi: 10.1128/IAI.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Justino MC, Almeida CC, Teixeira M, Saraiva LM. 2007. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J Biol Chem 282:10352–10359. doi: 10.1074/jbc.M610656200. [DOI] [PubMed] [Google Scholar]
- 5.Balasiny B, Rolfe MD, Vine C, Bradley C, Green J, Cole J. 2018. Release of nitric oxide by the Escherichia coli YtfE (RIC) protein and its reduction by the hybrid cluster protein in an integrated pathway to minimize cytoplasmic nitrosative stress. Microbiology 164:563–575. doi: 10.1099/mic.0.000629. [DOI] [PubMed] [Google Scholar]
- 6.Justino MC, Almeida CC, Gonçalves VL, Teixeira M, Saraiva LM. 2006. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol Lett 257:278–284. doi: 10.1111/j.1574-6968.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 7.Nobre LS, Garcia-Serres R, Todorovic S, Hildebrandt P, Teixeira M, Latour J-M, Saraiva LM. 2014. Escherichia coli RIC is able to donate iron to iron-sulfur clusters. PLoS One 9:e95222. doi: 10.1371/journal.pone.0095222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva LO, Nobre LS, Mil-Homens D, Fialho A, Saraiva LM. 2018. Repair of iron centers RIC protein contributes to the virulence of Staphylococcus aureus. Virulence 9:312–317. doi: 10.1080/21505594.2017.1389829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todorovic S, Justino MC, Wellenreuther G, Hildebrandt P, Murgida DH, Meyer-Klaucke W, Saraiva LM. 2008. Iron-sulfur repair YtfE protein from Escherichia coli: structural characterization of the di-iron center. J Biol Inorg Chem 13:765–770. doi: 10.1007/s00775-008-0362-y. [DOI] [PubMed] [Google Scholar]
- 10.Nobre LS, Lousa D, Pacheco I, Soares CM, Teixeira M, Saraiva LM. 2015. Insights into the structure of the diiron site of RIC from Escherichia coli. FEBS Lett 589:426–431. doi: 10.1016/j.febslet.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Lo F-C, Hsieh C-C, Maestre-Reyna M, Chen C-Y, Ko T-P, Horng Y-C, Lai Y-C, Chiang Y-W, Chou C-M, Chiang C-H, Huang W-N, Lin Y-H, Bohle DS, Liaw W-F. 2016. Crystal structure analysis of the repair of iron centers protein YtfE and its interaction with NO. Chemistry 22:9768–9776. doi: 10.1002/chem.201600990. [DOI] [PubMed] [Google Scholar]
- 12.Almiron M, Link A, Furlong JD, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 13.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol 5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 14.Calhoun LN, Kwon YM. 2011. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol 110:375–386. doi: 10.1111/j.1365-2672.2010.04890.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem 277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 16.Karas VO, Westerlaken I, Meyer AS. 2015. The DNA-Binding protein from starved cells (Dps) utilizes dual functions to defend dells against multiple stresses. J Bacteriol 197:3206–3215. doi: 10.1128/JB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J 23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaha G, Stanley RE, Steitz TA. 2009. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science 325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganoza MC, Aoki H. 2000. Peptide bond synthesis: function of the efp gene product. Biol Chem 381:553–559. doi: 10.1515/BC.2000.071. [DOI] [PubMed] [Google Scholar]
- 21.Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol 280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- 22.Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV. 2003. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res Microbiol 154:123–135. doi: 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 23.Magliery TJ, Wilson CGM, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc 127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 24.Chiancone E, Ceci P. 2010. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta 1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Park S, You X, Imlay J a. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc Natl Acad Sci U S A 102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seaver LC, Imlay JA. 2004. Are respiratory enzymes the primary sources of intracellular hydrogen peroxide? J Biol Chem 279:48742–48750. doi: 10.1074/jbc.M408754200. [DOI] [PubMed] [Google Scholar]
- 27.Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol Microbiol 63:1495–1507. doi: 10.1111/j.1365-2958.2007.05600.x. [DOI] [PubMed] [Google Scholar]
- 28.Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol 181:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellapadrona G, Ardini M, Ceci P, Stefanini S, Chiancone E. 2010. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic Biol Med 48:292–297. doi: 10.1016/j.freeradbiomed.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 30.Lennox ES. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 31.Pommier J, Mejean V, Giordano G, Iobbi-Nivol C. 1998. TorD, a cytoplasmic chaperone that interats with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J Biol Chem 273:16615–16620. doi: 10.1074/jbc.273.26.16615. [DOI] [PubMed] [Google Scholar]
- 32.Thibodeau SA, Fang R, Joung JK. 2004. High-throughput β-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36:410–415. doi: 10.2144/04363BM07. [DOI] [PubMed] [Google Scholar]
- 33.Wilson CGM, Magliery TJ, Regan L. 2004. Detecting protein-protein interactions with GFP-fragment reassembly. Nat Methods 1:255–262. doi: 10.1038/nmeth1204-255. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira E, Giménez R, Aguilera L, Guzmán K, Aguilar J, Badia J, Baldomà L. 2013. Protein interaction studies point to new functions for Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Res Microbiol 164:145–154. doi: 10.1016/j.resmic.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Puchegger S, Redl B, Stoffler G. 1990. Purification and properties of a thermostable fumarate hydratase from the archaeobacterium Sulfolobus solfataricus. J Gen Microbiol 136:1537–1541. doi: 10.1099/00221287-136-8-1537. [DOI] [PubMed] [Google Scholar]
- 36.Vine CE, Justino MC, Saraiva LM, Cole J. 2010. Detection by whole genome microarrays of a spontaneous 126-gene deletion during construction of a ytfE mutant: confirmation that a ytfE mutation results in loss of repair of iron-sulfur centres in proteins damaged by oxidative or nitrosative stress. J Microbiol Methods 81:77–79. doi: 10.1016/j.mimet.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 37.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang S, Imlay JA. 2010. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol 78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]