Members of the LitR/CarH family are adenosyl B12-based photosensory transcriptional regulator involved in light-inducible carotenoid production in nonphototrophic bacteria. Our study provides the first evidence of the involvement of a class III LitR, which lacks an adenosyl B12-binding domain in the light response of Burkholderia multivorans belonging to betaproteobacteria. Our biochemical analysis suggests that class III LitR protein exhibits features as a photosensor including absorption of light at the UV-A region (λmax = ca. 340 nm), photocyclic response, and light-dependent dissociation. This suggests that class III LitR associates with a UV-A-absorbing molecule, and it has a photosensing mechanism distinguishable from that of the B12-based type.
KEYWORDS: CarH/LitR family, MerR family, light-inducible gene, Burkholderia
ABSTRACT
The LitR/CarH protein family is an adenosyl B12 (AdoB12)-dependent photoreceptor family with DNA-binding activity, and its homologs are widely distributed in the genomes of diverse bacterial genera. In this investigation, we studied the role and functions of a LitR homolog from a Gram-negative soil bacterium, Burkholderia multivorans, which does not possess an AdoB12-binding domain. Transcriptome analysis indicated the existence of 19 light-induced genes, including folE2, cfaB, litS, photolyase gene phrB2, and cryB, located in the region flanking litR. Disruption of litR caused constitutive expression of all the light-inducible genes, while mutation in the light-induced sigma factor gene, litS, abolished the transcription of the phrB2 operon and the cfa operon, indicating that LitR and LitS play a central role in light-inducible transcription. A gel shift assay showed that recombinant protein LitR specifically binds to the promoter regions of litR and the folE2 operon, and its binding was weakened by UV-A illumination. LitR absorbs light at maximally near 340 nm and exhibited a photocyclic response and light-dependent dissociation of multimer into tetramer. The litR mutant produced a 20-fold-higher intracellular level of folate than that of the wild-type strain. Thus, the evidence suggests that LitR light-dependently regulates the transcription of litR itself and the folE2 operon, resulting in the production of folate, and then the expressed RNA polymerase complex containing σLitS directs the transcription of the phrB2 operon and the cfa operon. These light-dependent characteristics suggest that class III LitR, in complex with a UV-A-absorbing molecule, follows a novel light-sensing mechanism.
IMPORTANCE Members of the LitR/CarH family are adenosyl B12-based photosensory transcriptional regulator involved in light-inducible carotenoid production in nonphototrophic bacteria. Our study provides the first evidence of the involvement of a class III LitR, which lacks an adenosyl B12-binding domain in the light response of Burkholderia multivorans belonging to betaproteobacteria. Our biochemical analysis suggests that class III LitR protein exhibits features as a photosensor including absorption of light at the UV-A region (λmax = ca. 340 nm), photocyclic response, and light-dependent dissociation. This suggests that class III LitR associates with a UV-A-absorbing molecule, and it has a photosensing mechanism distinguishable from that of the B12-based type.
INTRODUCTION
The light-induced transcription regulator LitR/CarH protein family comprises MerR-type transcriptional regulators, which retain a DNA-binding domain at the N terminus (encompassing a helix-turn-helix [HTH] motif) and a B12-binding domain at the C terminus (1, 2). The mechanisms underlying light-inducible transcription have been studied for several model bacteria, including Streptomyces coelicolor A3 (2–6), Thermus thermophilus HB27 (7–9), Bacillus megaterium QM B1551 (10), and Myxococcus xanthus (11). A major role of the LitR/CarH family in these bacteria is to control carotenoid production in a light-dependent manner. Light-dependent carotenoid production is important for two reasons (12, 13): (i) carotenoids protect the cells from photooxidative damage by quenching harmful agents, such as reactive oxygen species generated upon illumination, and (ii) carotenoid biosynthesis in response to illumination saves energy under dark conditions. The LitR gene is also frequently flanked by phr, encoding DNA photolyase, the DNA repair enzyme (1). Probably, this is based on the same reason as that deduced for the carotenoid biosynthesis genes.
Biochemical and structural studies have shown that the LitR/CarH family members are novel coenzyme B12 (adenosyl B12 [AdoB12]-based photosensors (9–11). LitR in complex with AdoB12 senses blue and green light, which results in the photolysis of AdoB12 to hydroxocobalamin (OHB12). The photolysis of AdoB12 leads to a large conformational change and subunit dissociation of the LitR-AdoB12 complex, which decreases the DNA-binding affinity for the target promoter and allows RNA polymerase holocomplex to direct the mRNA synthesis of light-inducible genes.
In some bacteria, a complex mechanism underlies light-dependent gene expression. For example, the involvement of not only LitR/CarH proteins but also specific sigma factors σLitS and σCarQ is known for S. coelicolor A3 (2, 3) and M. xanthus, respectively (14). On the other hand, the regulatory mechanism residing in B. megaterium QM B1551 is simple: light-inducible transcription is mediated by only two proteins, the LitR-AdoB12 complex and the housekeeping RNA polymerase σA, allowing a quick response to light stress (10). It is known that the ability to biosynthesize AdoB12 is confined to bacteria and archaea (15, 16), suggesting that the light-sensing mechanism of LitR/CarH family is unique to members of these domains.
Members of the genus Burkholderia are Gram-negative soil-dwelling bacteria, belonging to the β subgroup of proteobacteria. The Burkholderia cepacia complex (BCC) comprises several closely related species, including B. cepacia, B. multivorans, B. cenocepacia, B. vietnamiensis, and other species (17). BCC is isolated from different environments, including animals, plants, streams, soil, rhizospheres, and chemically polluted soil, which suggests that BCC has extraordinary metabolic versatility to adapt to various niches. B. multivorans LMG17588 (ATCC 17616) is isolated from soil enriched with anthranilate (18). The genomic sequence of this bacterium was independently determined by two groups; the bacterium possesses three circular chromosomes of 3.4 Mb, 2.4 Mb, and 0.9 Mb and one 1.6-Mb plasmid, pTGL1 (19, 20). The genomic information of B. multivorans suggests that many genes of this bacterium are involved in carbon metabolism and assimilation of aromatic compounds, including salicylate, benzoate, phthalate, 4-hydroxybenzoate, and 4-hydroxyphenylpyruvate, thus indicating extraordinary catabolic ability/versatility (21).
An intriguing feature of the LitR/CarH family is its wide distribution in phylogenetically diverse bacterial genera (1, 2). In MerR family regulators, the C-terminal domain serves as an effector-binding domain (22). Many of the LitR/CarH homolog proteins have a B12-binding domain at the C terminus, while LitR/CarH homologs without the B12-binding domain are also found, suggesting the existence of other light-sensing mechanisms. This also suggests that nonphototrophic bacteria have a general ability to sense and respond to light and are equipped with diverse light-sensing mechanisms. In the present study, we classified the LitR/CarH family based on the amino acid sequence of the C termini. Furthermore, we performed a genetic and biochemical analysis of a LitR homolog (designated class III LitR) from B. multivorans since it does not possess a B12-binding domain and its function and roles are not known. Our genetic study indicated the existence of the class III LitR-dependent light-inducible gene cluster, and our biochemical analysis suggests that class III LitR possesses a novel light-sensing mechanism and is involved in folate production.
RESULTS
Phylogenetic analysis of LitR/CarH family proteins.
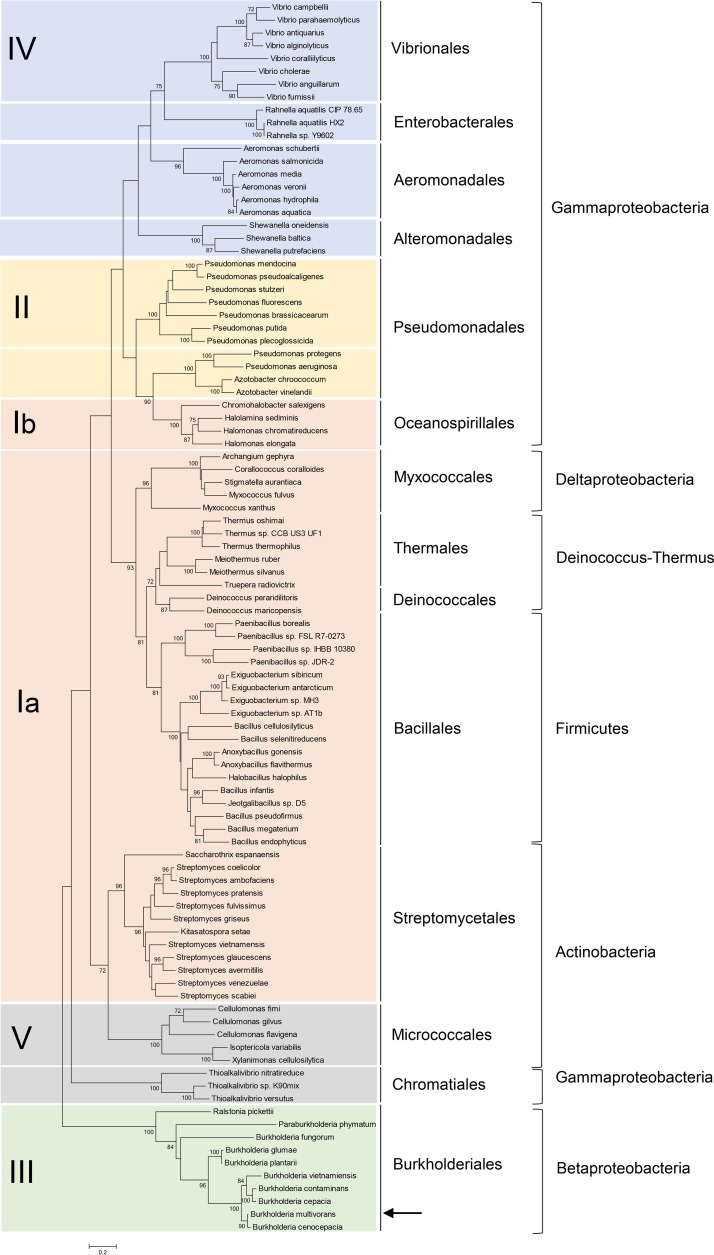
A number of proteins homologous to the LitR/CarH family members are found in the genomes of phylogenetically diverged bacterial genera, including both Gram-positive and Gram-negative bacteria (1, 2). To classify the members of the LitR/CarH family proteins, we performed a phylogenetic analysis using the amino acid sequences of their light-sensing C-terminal domains, because in contrast to the N-terminal HTH DNA-binding domains, they have diverged. The neighbor-joining phylogenetic tree shown in Fig. 1 indicates that the C termini are mainly classified into five classes (designated classes I to V). LitR, which belongs to class I, is an AdoB12-based photosensory regulator studied in several bacteria by us and other groups. Class I LitR proteins retain a complete B12-binding domain composed of two domains: (i) the Rossmann fold domain (Pfam definition, B12-binding domain), which binds B12 via the association of the conserved His residue with the cobalt atom of B12, and (ii) 4-helix bundle domain (Pfam definition, B12-binding 2 domain), which is usually found at the N terminus of the Rossmann fold domain forming a cap over the Rossmann fold domain (1, 2). Class I LitR proteins from the order Oceanospirillales have a unique feature in that they do not retain the conserved His residue within the Rossmann fold domain (23). Thus, we have divided class I LitR proteins into class Ia and class Ib based on the presence of the conserved His residue. LitR proteins of the order Oceanospirillales have been classified into class Ib.
FIG 1.
Molecular phylogenetic analysis of presumptive LitR/CarH family proteins from bacterial genomic information. A neighbor-joining tree based on the amino acid sequence of C-terminal region of the LitR homolog is shown. Class III LitR of Burkholderia multivorans is shown by an arrow. Numbers at nodes are bootstrap values (expressed as percentages of 100 from resampled data sets; only values >70% are shown). The scale bar shows 0.2 amino acid substitution per amino acid site.
Class II LitR proteins retain only the 4-helix bundle domain. This implies that this class of LitR differs from class I in terms of the mode of association with B12. Class II LitR proteins are mainly distributed within the genus Pseudomonas. The LitR proteins belonging to class III, IV, and V are mainly distributed in Burkholderia, Vibrio, and the genera belonging to Micrococcales, respectively. Class III to V LitR proteins share no similarity in B12-binding domain at their C termini, indicating that their light-sensing mechanisms remain unrevealed. The phylogenetic analysis indicates the diversity of the bacterial light-sensing mechanism; however, current reports are confined to class I LitR proteins. Therefore, we studied a role and function of a class III LitR from Burkholderia multivorans LMG17588 as a representative of this genus.
Identification of light-inducible genes in B. multivorans.
We first examined the light responsiveness of B. multivorans LMG17588, but illumination did not affect cell morphology or pigment production. Thus, we carried out a DNA microarray analysis to primitively screen the light-inducible genes in B. multivorans. Total RNA extracted and purified from the cells cultured under dark and light conditions was used for this analysis (see Materials and Methods). Of the 6,193 genes analyzed, a total of 19 genes were identified as light-inducible genes (Table 1) showing a 2.9- to 26.1-fold-higher transcriptional level under the light condition than under the dark condition; all of these genes are located in the flanking region of litR (Fig. 2). These results suggest that a light-responsive regulatory mechanism also exists in this bacterium and that class III LitR plays a central role in light response.
TABLE 1.
Light-induced genes of B. multivorans identified by DNA microarray analysis
| Gene or operon | Gene name | Fold expression, light/darka | Direction | Annotation for product |
|---|---|---|---|---|
| BMULJ_05674 | 5.8 | + | Hypothetical protein | |
| phrB2 | BMULJ_05675 | 12.0 | − | Deoxyribodipyrimidine photolyase |
| BMULJ_05676 | 6.7 | − | Hypothetical protein | |
| BMULJ_05677 | 5.0 | + | Putative esterase/lipase | |
| litR | BMULJ_05678 | 2.9 | − | MerR family transcriptional regulator |
| cryB | BMULJ_05679 | 7.5 | + | Deoxyribodipyrimidine photolyase-related protein |
| BMULJ_05681 | 4.3 | + | Putative short-chain alcohol dehydrogenase | |
| BMULJ_05682 | 3.4 | + | Hypothetical protein | |
| litS | BMULJ_05687 | 12.6 | − | ECF subfamily RNA polymerase sigma-70 factor |
| folE2 | BMULJ_05688 | 19.1 | − | GTP cyclohydrolase I |
| BMULJ_05689 | 10.2 | − | Hypothetical protein | |
| BMULJ_05690 | 3.2 | − | Hypothetical protein | |
| BMULJ_05691 | 18.5 | − | Hypothetical protein | |
| cfaB1 | BMULJ_05692 | 23.0 | − | Cyclopropane-fatty-acyl-phospholipid synthase |
| BMULJ_05693 | 22.4 | − | Hypothetical protein | |
| BMULJ_05694 | 21.9 | − | Predicted NAD/FAD-binding protein | |
| BMULJ_05695 | 26.0 | − | Outer membrane lipocalin-like protein | |
| cfaB2 | BMULJ_05696 | 26.1 | − | Cyclopropane-fatty-acyl-phospholipid synthase |
| BMULJ_05697 | 12.1 | − | Predicted membrane protein |
Intensity of the WT strain cultured under the light condition normalized to that of the wild-type strain cultured under the dark condition.
FIG 2.
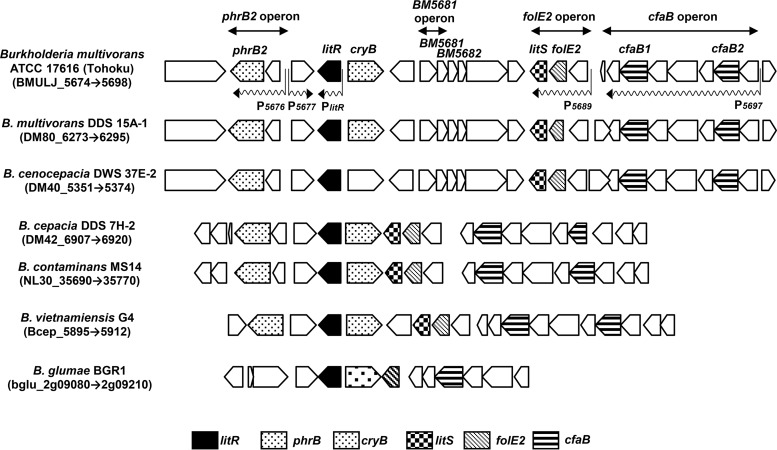
Gene organization and synteny of the light-inducible genes in Burkholderia spp. The light-dependent promoters and putative operon structures (transcriptional units) are shown by wavy arrows. The light-inducible genes include four monocistronic genes (BM5674, BM5677, litR, and cryB) and three operons (phrB2 operon, folE2 operon, and cfaB operon). The annotation for each gene product is shown in Table 1.
The light-inducible genes comprise four monocistronic genes—BM5674, BM5677, litR, and cryB—and four putative operons (Fig. 2). BM5674, BM5677, litR, and cryB encode a hypothetical protein, a putative esterase/lipase, a MerR family transcriptional regulator (class III LitR), and a DNA repair enzyme photolyase-like protein, respectively. The phrB2 operon (BM5676-BM5675) includes two genes encoding a hypothetical protein and a photolyase (phrB2). The BM5681 operon contains genes encoding a putative short-chain alcohol dehydrogenase (BM5681) and hypothetical protein (BM5682). The folE2 operon (BM5689 to BM5687) includes genes encoding a hypothetical protein, GTP cyclohydrolase I (folE2), and an ECF (extracytoplasmic function) subfamily RNA polymerase sigma-70 factor (designated litS). The cfaB operon (BM5697 to 5690) consists of genes encoding a predicted membrane protein, a putative cyclopropane-fatty-acyl-phospholipid synthase (cfaB2), an outer membrane lipocalin-like lipoprotein, a predicted NAD/FAD-binding protein, a hypothetical protein, a putative cyclopropane-fatty-acyl-phospholipid synthase (cfaB1), and a hypothetical protein.
Our comparative genomic analysis showed that most of the identified light-inducible genes are conserved in the genomes of Burkholderia spp. (Fig. 2). The proximity of litR to the three operons phrB2, folE2, and cfaB is conserved in this genus, which indicates a strong functional relationship. This finding indicates that the ability of light sensing is widespread within this genus. Among these light-inducible genes, two transcriptional proteins, LitR and LitS, are conserved, suggesting that they control the expression of the light-inducible genes.
Gene expression analysis by qRT-PCR.
To verify the results obtained with the primitive DNA microarray experiment, we measured the transcription level with regard to some of the light-induced genes by quantitative reverse transcription-PCR (qRT-PCR) analysis. As summarized in Table 2 (see also Fig. S1 in the supplemental material), transcription level of the following genes was elevated 2.0- to 26.7-fold due to illumination in the B. multivorans wild type (WT): BM5674, phrB2, BM5677, litR, cryB, BM5681, BM5682, litS, folE2, BM5689, BM5694, and cfaB2. An essential sigma factor gene, rpoD, showed constitutive expression irrespective of illumination. These results are consistent with those of the DNA microarray analysis. We also examined the type of light affecting gene expression by semi-qRT-PCR. Irradiation with blue light caused remarkable induction of light-inducible genes, while the effect of green and red light was low (Fig. S2). These results indicate that the light-inducible genes are induced by blue, or near to blue, light. We also examined whether treatment with a photooxidative stresser (methylene blue) and a singlet oxygen scavenger (1,4-diazabicyclooctane) affect light-inducible transcription (Fig. S2) (24, 25). The analyses demonstrated that methylene blue treatment with red-light illumination did not induce the light-dependent gene expression and that the addition of 1,4-diazabicyclooctane did not inhibit the induction by blue light. These results indicate that blue light itself generated the transcription.
TABLE 2.
Light-induced fold change of transcription measured by qRT-PCRa
| Gene | Fold change of transcription at indicated time |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT |
ΔlitR mutant |
ΔlitR/plitR mutant |
ΔlitS mutant |
ΔlitS/plitS mutant |
||||||
| 14 h | 21 h | 14 h | 21 h | 14 h | 21 h | 14 h | 21 h | 14 h | 21 h | |
| BM5674 | 2.0 ± 0.4 | 2.7 ± 1.1 | 0.8 ± 0.0 | 1.3 ± 0.0 | 3.0 ± 0.1 | 0.8 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.0 | 1.7 ± 0.3 | 1.1 ± 0.0 |
| phrB2 | 12.5 ± 1.6 | 8.2 ± 0.6 | 1.2 ± 0.3 | 1.2 ± 0.2 | 8.7 ± 0.7 | 3.5 ± 1.1 | 1.7 ± 0.0 | 0.7 ± 0.0 | 2.6 ± 0.1 | 3.3 ± 0.8 |
| BM5677 | 6.0 ± 0.1 | 6.6 ± 1.4 | 1.2 ± 0.3 | 1.1 ± 0.3 | 6.3 ± 0.1 | 3.0 ± 0.9 | 5.3 ± 0.7 | 0.9 ± 0.1 | 2.3 ± 0.0 | 3.0 ± 0.6 |
| litR | 2.7 ± 0.3 | 1.5 ± 0.1 | 1.1 ± 0.4 | 1.2 ± 0.2 | 1.2 ± 0.2 | 0.9 ± 0.2 | 4.3 ± 0.3 | 3.4 ± 0.0 | 1.1 ± 0.0 | 2.5 ± 0.5 |
| cryB | 6.3 ± 1.8 | 6.7 ± 2.5 | 1.0 ± 0.0 | 1.4 ± 0.2 | 0.3 ± 0.0 | 0.6 ± 0.0 | 2.8 ± 0.5 | 1.6 ± 0.1 | 2.3 ± 0.1 | 2.2 ± 0.0 |
| BM5681 | 11.9 ± 2.9 | 5.2 ± 2.1 | 0.9 ± 0.1 | 1.3 ± 0.2 | 4.2 ± 0.3 | 5.3 ± 0.1 | 14.9 ± 2.7 | 16.1 ± 0.3 | 3.6 ± 0.2 | 14.6 ± 0.2 |
| BM5682 | 10.9 ± 5.8 | 7.3 ± 5.1 | 1.6 ± 0.0 | 1.0 ± 0.1 | 2.1 ± 0.4 | 6.8 ± 0.6 | 16.1 ± 0.1 | 23.8 ± 0.2 | 2.6 ± 0.6 | 19.2 ± 2.4 |
| litS | 18.5 ± 8.3 | 13.8 ± 9.4 | 1.4 ± 0.1 | 1.4 ± 0.2 | 11.0 ± 1.5 | 28.9 ± 2.0 | 13.8 ± 3.6 | 11.7 ± 0.6 | 2.1 ± 0.1 | 5.7 ± 0.1 |
| folE2 | 26.7 ± 11.9 | 19.5 ± 15.0 | 1.5 ± 0.1 | 1.3 ± 0.1 | 22.5 ± 1.3 | 43.1 ± 0.6 | 86.3 ± 23.6 | 84.3 ± 8.4 | 8.0 ± 0.3 | 23.6 ± 0.7 |
| BM5689 | 22.8 ± 3.3 | 11.7 ± 1.3 | 1.1 ± 0.5 | 0.9 ± 0.3 | 15.3 ± 1.3 | 36.7 ± 22.5 | 58.3 ± 16.1 | 59.9 ± 1.6 | 7.7 ± 0.5 | 32.2 ± 16.1 |
| BM5694 | 25.3 ± 2.7 | 23.3 ± 2.3 | 1.1 ± 0.5 | 1.1 ± 0.4 | 26.1 ± 0.0 | 64.2 ± 40.0 | 4.4 ± 0.2 | 1.6 ± 0.1 | 3.8 ± 0.1 | 4.7 ± 1.5 |
| cfaB2 | 22.7 ± 3.1 | 16.5 ± 0.7 | 1.3 ± 0.8 | 1.4 ± 0.4 | 18.7 ± 1.5 | 73.8 ± 42.3 | 2.8 ± 0.8 | 1.3 ± 0.1 | 2.8 ± 0.2 | 4.2 ± 1.4 |
| rpoD | 0.9 ± 0.1 | 1.0 ± 0.0 | 1.0 ± 0.3 | 0.7 ± 0.1 | 1.0 ± 0.0 | 0.8 ± 0.3 | 1.9 ± 0.1 | 1.0 ± 0.1 | 0.7 ± 0.1 | 1.2 ± 0.2 |
Values are ratios (light/dark) of transcriptional intensity.
To reveal the function and role of a class III corresponding to litR and an ECF-type sigma factor corresponding to litS, knockout mutants for litR and litS were constructed (see Materials and Methods). We first attempted to construct a markerless null mutant to avoid a polar effect, but such mutants were not obtained. Therefore, mutants were constructed by single-crossover homologous recombination, in which the whole nonreplicative vector containing a region homologous with the target gene was integrated to separate the target genes. The obtained litR mutant exhibited the same phenotype as the wild-type strain with regard to the colony morphology, growth rate, and pigment production (data not shown). However, the litR mutation significantly affected the transcriptional level of the light-inducible genes; the litR mutation caused constitutive transcription in all of the light-inducible genes examined by the qRT-PCR analysis (Table 2). In this mutant, transcription of each gene was upregulated to the level far exceeding that in the wild type under both the dark and light conditions (Fig. S1). This indicates that LitR serves as a repressor inhibiting the transcription of the light-inducible genes under the dark condition. The introduction of an intact litR gene into the litR mutant restored the transcription to a level similar to that of the wild type (Table 2).
We also examined the effect of the litS mutation on the expression of light-inducible genes. As shown in Table 2 and Fig. S1, the litS mutation markedly decreased the expression of phrB2, BM5694, and cfaB2, and slightly decreased the expression of BM5677 and cryB. The genetic complementation by introducing an intact litS gene partially restored their light dependency, which suggests that RNA polymerase containing σLitS directs the transcription from the promoters preceding each gene and operon. On the other hand, the litS mutation had no effect on the transcription of litR and BM5689 (Table 2 and Fig. S1), suggesting that the promoters of these genes (PlitR and P5689) are directly controlled by LitR.
Characterization of promoters preceding light-inducible genes.
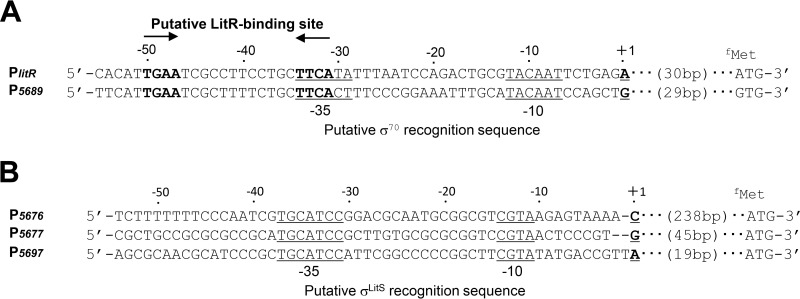
To characterize the promoter structure preceding the light-inducible genes, transcriptional start sites (TSS) were determined by modified 5′ rapid amplification of cDNA ends (RACE) (see Materials and Methods). Each TSS was assigned 31 bp for PlitR, 239 bp for P5676, 46 bp for P5677, 30 bp for P5689, and 20 bp for P5697 upstream from the translational initiation codon (Fig. 3). TSS for PcryB, P5674, and P5681 could not be determined due to their low signal intensities. Among light-inducible promoters, the putative −35 and −10 sequences of PlitR (TTCATAN16TACAAT) and P5689 (TTCACTN16TACAAT) exhibited a high similarity to the sequence recognized by σ70 of Escherichia coli (TGACAN16–18TATAAT) (Fig. 3A) (26). However, the putative −35 and −10 sequences of P5676, P5677, and P5697 were TGCATCCN16CGTA, which did not resemble the consensus sequence of E. coli σ70 (Fig. 3B). The dependence of these promoters on LitS strongly suggests that the promoter sequence is recognized by σLitS. The sequence was conserved in the corresponding region of BM5697 homologs distributed to other Burkholdeari spp. (Fig. S3). These results strongly suggest that σA (rpoD product) directs RNA polymerase to the PlitR and P5689 promoters. Then the expressed σLitS-containing RNA polymerase holoenzyme may recognize the consensus sequence (TGCATCCN16CGTA) and direct transcription from the P5676, P5677, and P5697 promoters.
FIG 3.
Promoters of light-inducible genes. (A) Nucleotide sequence of promoters PlitR and P5689. A putative LitR-binding sequence is shown in bold. A putative σ70 recognition sequence (positions −10 and −35) is underlined, and the transcriptional start site is indicated as +1. (B) Nucleotide sequences of promoters P5676, P5677, and P5697. The putative σLitS recognition sequence (−10 and −35) is underlined.
Light dependence of in vitro DNA-binding activity of LitR.
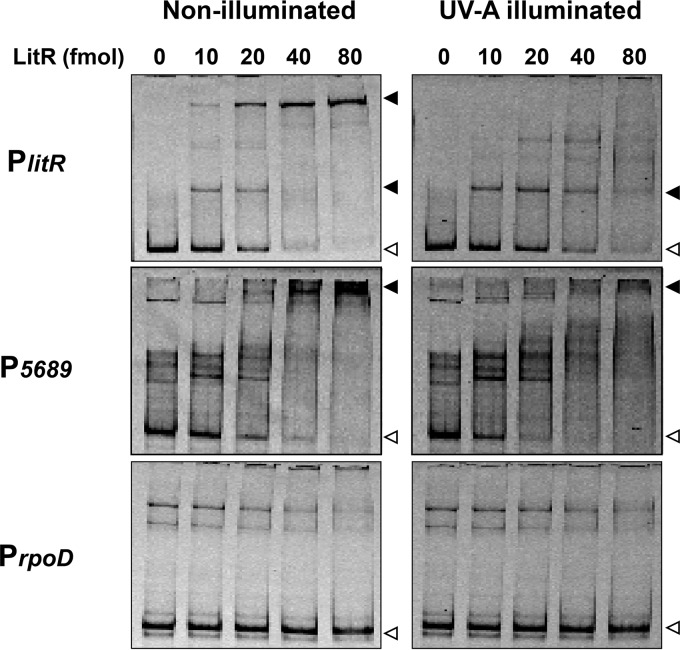
Class III LitR from B. multivorans consists of 323 amino acids (calculated molecular mass: 34,998 Da). The amino acid sequence alignment of class III LitR distributed to the genus of Burkholderia is shown in Fig. S4. A Pfam search showed that its N terminus has similarity with a MerR-type HTH DNA-binding domain, with E values of 1.0e−17, while its C terminus domain shares no similarity with protein domains of known functions (Fig. S4). In order to investigate the function of LitR, a recombinant protein carrying a His tag at its C terminus was overexpressed in E. coli Rosetta 2(DE3)/pLysS and purified to homogeneity by affinity column chromatography (see Materials and Methods and Fig. S5). To analyze the DNA-binding activity of LitR, we performed a gel shift assay. Two promoter regions preceding light-inducible genes (P5689 and PlitR) were used as probes. As a negative control, the promoter region preceding rpoD encoding an essential RNA polymerase sigma factor σ70 (PrpoD) was used. As shown in Fig. 4, LitR bound to P5689 and PlitR in a dose-dependent manner, while no LitR bound to PrpoD. We also carried out a similar analysis with regard to the promoters preceding BM5676 and BM5697 and found that LitR did not bind those regions (data not shown). These results indicate that LitR specifically binds to the promoters P5689 and PlitR. We also examined the effect of UV-A illumination on the DNA-binding activity of LitR, because its recombinant protein exhibited an absorption maximum at 340 nm as described below. The DNA-binding activity was lowered when the LitR protein was illuminated (Fig. 4). These findings imply that LitR has the capacity to function as a photosensitive regulator.
FIG 4.
Gel shift assay using LitR recombinant protein. LitR (0 to 80 fmol) was incubated with the probes for PlitR (306 bp), P5689 (378 bp), and PrpoD (436 bp) and loaded into a nondenaturing polyacrylamide gel. LitR was illuminated with UV-A at 0.06 μmol · s−1 · m−2 for 180 s prior to the addition of Cy5-labeled probe. Open and closed arrowheads indicate the probe alone and protein-probe complex, respectively.
We propose the LitR-binding site based on the nucleotide sequence alignment of P5689 and PlitR. These regions contained an identical inverted repeat sequence, TGAAN12TTCA, between the −30 and −50 regions (Fig. 3). The inverted repeat sequence was found in the corresponding region of other Burkholderia spp. (Fig. S3).
Light-dependent change in absorbance of LitR recombinant protein.
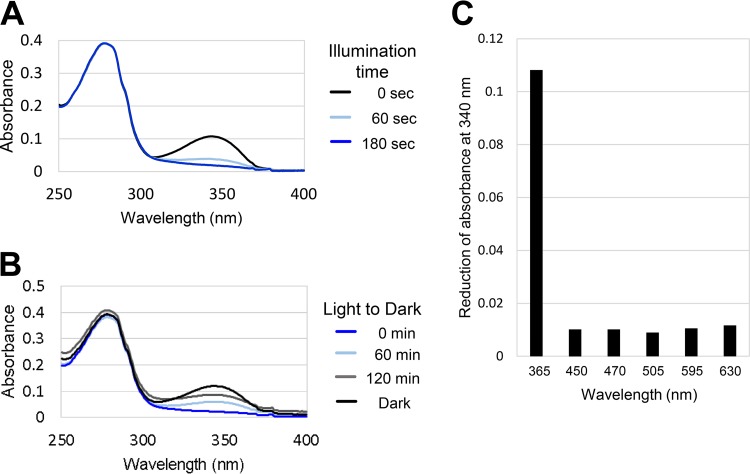
The LitR recombinant protein was colorless and exhibited an absorption maximum at 340 nm in the UV-A region (Fig. 5A). This implies that LitR associates with a compound that absorbs the range of light and that the compound is also synthesized in E. coli. We also examined the effect of illumination on the absorbance spectrum of LitR. UV-A illumination for 180 s decreased the absorbance at a maximum of 340 nm (Fig. 5A). Furthermore, the absorption peak disappeared due to UV-A illumination was restored by subsequent incubation under the dark condition (Fig. 5B). This mode of light/dark-dependent spectral change is called photocyclic response, known as a typical characteristic of photosensors such as LOV (light-oxygen-voltage) and BLUF (blue-light sensing using flavin) (27–29). The photocycle was also observed with regard to the recombinant protein LitRbvi of B. vietnamiensis G4, which shares 88% identity with LitR of B. multivorans (Fig. S4, S5, and S6A). Dark incubation of the illuminated LitR proteins restored their absorbance (Fig. S6B). We also examined which type of light affects the absorption at 340 nm. As shown in Fig. 5C and Fig. S6C, illumination of light at λmax of 365 nm reduced the peak absorption, whereas illumination at λmax of 450, 530, and 630 nm did not. This result was consistent with above-mentioned result of in vivo transcriptional analysis (Fig. S2).
FIG 5.
Light-dependent change of absorbance spectrum of LitR recombinant protein. (A) Absorbance spectrum of the recombinant protein LitR. The absorbance spectrum of LitR was recorded after the sample was illuminated for 60 s and 180 s using LED lamps, UV-A (λmax = 365 nm) at 0.06 μmol · s−1 · m−2, on the sample. (B) Dark-state recovery measured at 60 and 120 min. (C) Effects of illumination of various wavelengths of light. The type of light reducing the absorbance spectrum at 340 nm of LitR was examined. Illumination time was 180 s.
To identify the domain responsible for the UV-A absorption, N-terminally truncated LitR proteins were generated as a glutathione S-transferase (GST)-fused recombinant proteins (GST-LitR81–323, GST-LitR100–323, and GST-LitR200–323) (Fig. S5). All three truncated proteins exhibited the same profile as the wild-type LitR in terms of absorption spectrum and insensitivity to UV-A illumination (Fig. S7). This indicates that the C-terminal domain encompassing the region from amino acids 200 to 323 is responsible for the UV-A absorption. These results suggest that the UV-A-absorbing chromophore is associated with the C-terminal domain and the LitR-chromophore complex serves as a photosensor.
Light-dependent change in conformation of LitR.
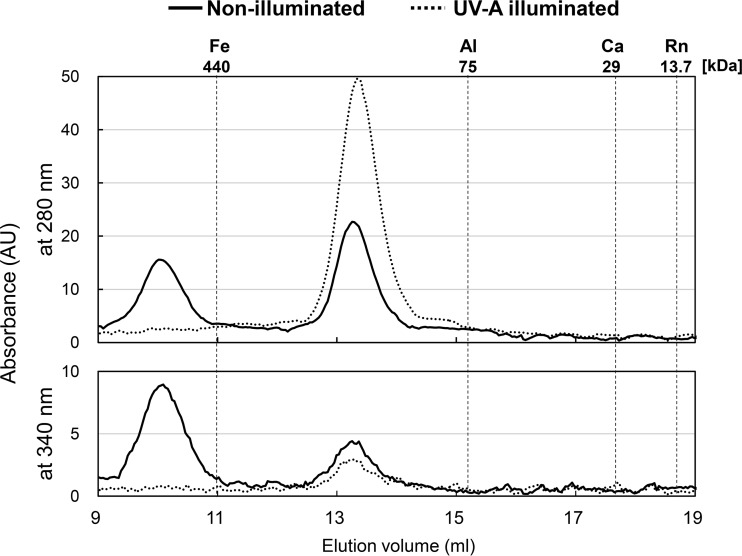
It is known that class I LitR dissociates from a tetrameric complex into its monomer form in a light-dependent manner (9). To examine the conformational change in class III LitR with a C-terminal His tag (calculated molecular mass: 36,232 Da), we performed gel filtration chromatography to estimate relative molecular weight (Mr) (see Materials and Methods). The absorbance spectra were monitored at 280 and 340 nm. Mr was estimated using a calibration curve (see Materials and Methods). As shown in Fig. 6, the dark-incubated LitR protein exhibited two major peaks, while the UV-A-illuminated LitR protein exhibited a single peak. The Mr corresponding to high molecular weight of the nonilluminated protein was not estimated since it was outside the calibration curve. However, the large size (>440 kDa) indicates that the dark-state LitR forms an oligomeric structure consisting of 16 or more LitR molecules. The later peak of the dark-incubated protein was estimated at a relative Mr of 155,600 (average of three replicates: 153,900), suggestive of a tetrameric structure. In contrast, the UV-A-illuminated LitR was eluted only at a single position Mr of 150,600 (average of three replicates: 154,000), corresponding to the tetrameric structure. SDS-PAGE and silver staining analyses verified that each peak contained only LitR. These results indicate that illumination causes the dissociation of LitR from oligomer to tetramer. The same dissociation was observed with regard to N-terminally truncated LitR81–323 protein carrying a C-terminal His Tag (calculated molecular mass: 26,987 Da) (Fig. S5 and S8). The dark-incubated LitR81–323 was eluted at two positions corresponding to Mrs of approximately 440,000 and 107,200, while UV-A illuminated LitR81–323 protein was eluted at a single position corresponding to an Mr of approximately 102,400. These results indicate that the region from amino acids 81 to 323 contains the domain involved in the formation of multimer conformation as well as that for photoresponse.
FIG 6.
Gel filtration chromatography for the analysis of light-induced dissociation of LitR protein. To estimate the relative molecular masses of the nonilluminated (solid line) and UV-A-illuminated (dashed line) LitR, the recombinant protein LitR was loaded onto a Superdex 200 10/300 GL column and the absorbance spectra were monitored at 280 and 340 nm. The molecular mass standards used were ferritin (Fe), conalbumin (Al), carbonic anhydrase (Ca), and RNase (Rn), indicating 440, 75, 29, and 13.7 kDa, respectively. AU, arbitrary units.
Role of the conserved cysteine residues.
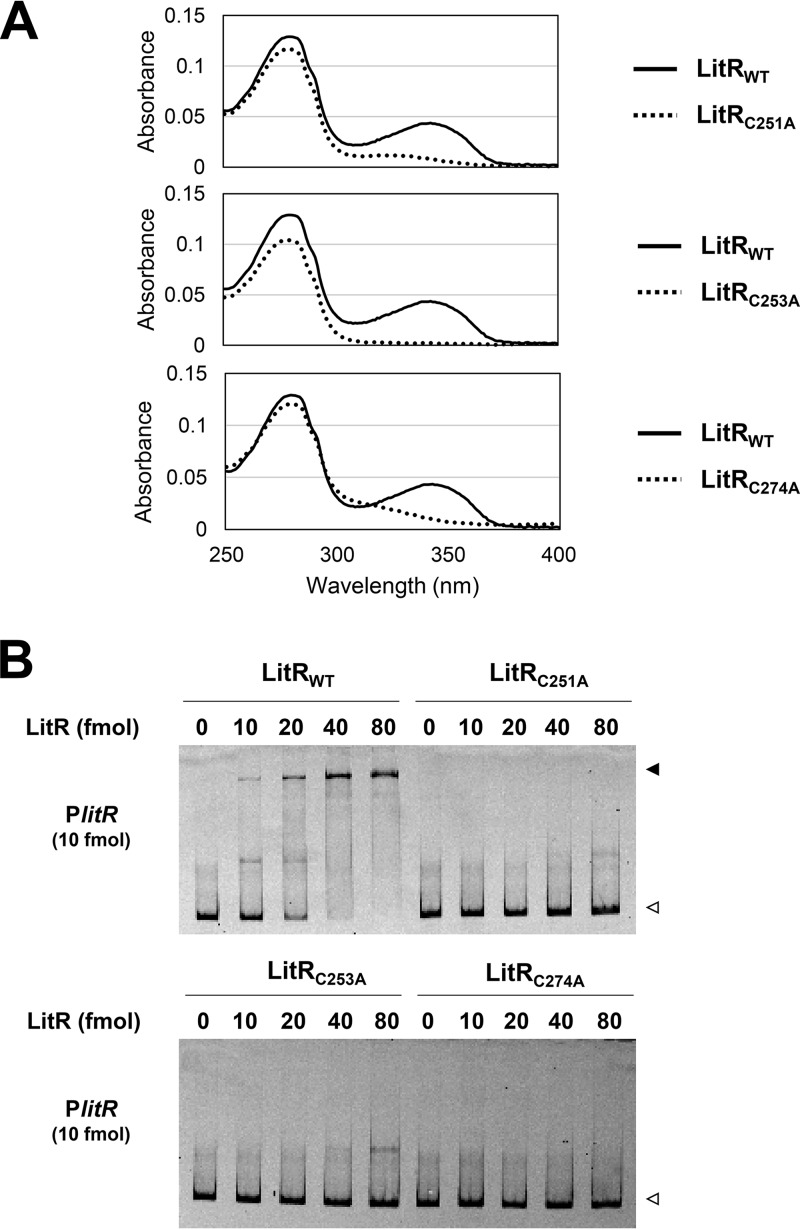
The result of a multiple amino acid alignment analysis showed the presence of three conserved cysteine (Cys) residues at the C-terminal positions 251, 253, and 274 (designated Cys251, Cys253, and Cys274, respectively) (Fig. S4 and S5). Generally, Cys residues play an important role in protein structure and function, including the formation of disulfide bonds serving as redox sensors and metal binders to modulate protein function (30). Therefore, we constructed Ala substitution mutants with regard to the three Cys residues (designated LitRC251A, LitRC253A, and LitRC274A). The three mutant proteins did not show UV-A absorbance (Fig. 7A) and lost their PlitR-binding activity (Fig. 7B). The mutant LitR proteins also lost their ability to form multimeric structures consisting of 16 or more LitR molecules observed in the dark-state LitRWT, though they retained their ability to form a tetrameric structure (Fig. S9). These results indicate that the Cys residues in the C terminus play a fundamental role in the function of LitR with respect to DNA binding and light sensing.
FIG 7.
Features of LitR mutant proteins (A) Absorbance spectra. The absorbance spectra of the dark-incubated LitRC251A (top graph), LitRC253A (middle graph), and LitRC274A (bottom graph) (5.6 μM) are shown by dashed lines. The absorbance spectrum of the wild-type LitR (LitRWT) is shown by solid lines. (B) Gel shift assay. LitR mutant proteins (0 to 80 fmol) were incubated with the Cy5-labeled probes (10 fmol) for PlitR (306 bp) and loaded onto a nondenaturing polyacrylamide gel. Open and closed arrowheads indicate the probe alone and probe-protein complex, respectively.
Folate production.
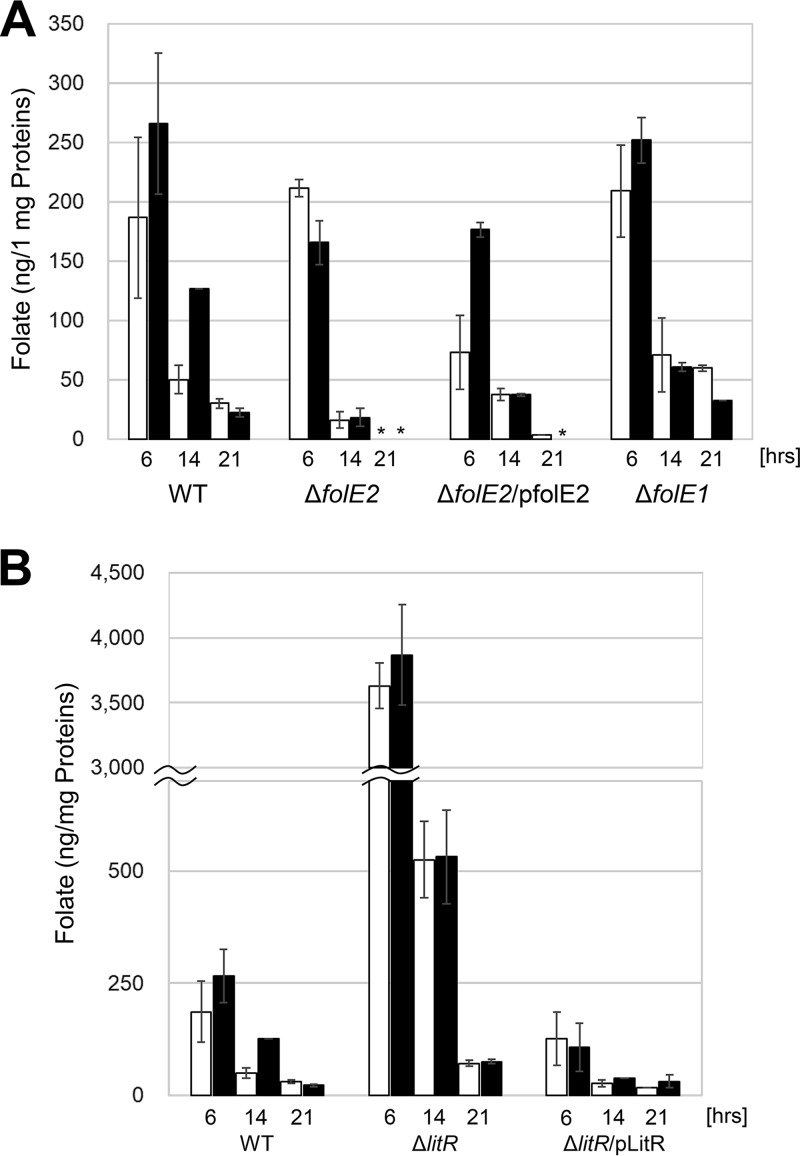
The aforementioned result of transcriptional analysis (Table 2) indicated that the expression of folE2 is under the regulation of LitR. Hence, we examined whether illumination actually affects folate production by a bioassay using a folate-requiring bacterium, Enterococcus hirae NBRC 3181 (see Materials and Methods). In wild-type B. multivorans, folate production increased in response to illumination at 6 h and 14 h (Fig. 8A), and blue light illumination was most effective in folate production (Fig. S10). Folate biosynthesis is based on the supply of pterin, the intermediate generated from GTP due to the activity of GTP cyclohydrolase I encoded by folE2. To verify the role of folE2 in the folate biosynthesis, a knockout mutant was generated. The constructed folE2 mutant showed a lower level of folate production than did the wild type (Fig. 8A). Genetic complementation by introducing an intact folE2 gene with P5689 into the mutant partially restored folate production. B. multivorans retains folE paralogs (folE1 and folE2). We also generated a mutant for folE1, a paralogous gene. As shown in Fig. 8A, folE1 mutation did not affect folate production. This indicates that FolE2 plays the major role in folate production. However, folE2 disruption did not affect the growth in LB medium (data not shown). These results imply the existence of a complex system maintaining the intracellular folate level based on the complementation between FolE proteins as well as the incorporation of folate from the culture medium.
FIG 8.
Intracellular folate produced by B. multivorans. (A) Intracellular folate produced by the wild type (WT), a folE2 mutant (ΔfolE2), a genetically complemented folE2 mutant strain (ΔfolE2/pfolE2), and a folE1 mutant (ΔfolE1) was quantified. Strains were grown for 6, 14, and 21 h under dark (open bars) and blue-light (solid bars) (17.3 μmol · s−1 · m−2) conditions. (B) Intracellular folates produced by the wild type, a litR mutant (ΔlitR), and a genetically complemented strain of litR mutant (ΔlitR/plitR) were quantified. Illumination conditions were the same as for panel A. Data presented are the means of duplicated measurements. The error bars indicate standard errors.
We also examined the effect of litR mutation on folate production. As shown in Fig. 8B, the litR mutant produced about 20-fold-larger amounts of folate than did the wild-type strain, as expected from the upregulation of folE2 in the litR mutation background (Table 2). The litR mutant introduced with an intact litR exhibited a folate production level similar to that of the wild type (Fig. 8B). These results indicate that LitR negatively controls folate production by repressing the transcription of folE2.
DISCUSSION
Accumulating genome sequence information has reinforced the view that even nonphototrophic bacteria are equipped with a variety of photosensing apparatuses. The well-characterized systems are blue-light-sensing domains called LOV (light-oxygen-voltage) and BLUF (blue-light sensing using flavin). Both LOV and BLUF domains bind flavin as a photoreceptor and linked to diverse effector/regulator functions of nonphototrophic bacteria (27–29). Some bacteria retaining these systems are affected in their dynamic behavior, such as motility and biofilm formation, by illumination (27–29).
In this study, we assessed the diversity content of the LitR/CarH family transcriptional regulators and discovered that the protein family can be classified into several subgroups (Fig. 1). The B12-dependent class I is widely distributed in both Gram-negative and -positive bacteria, indicating that this type of LitR is a major form of this kind of transcriptional regulator. In contrast, distribution of other subgroups of LitR (classes II to V) appears to be limited to a certain genus or order, suggesting that those regulators have evolved based on their adaptation to specific lineages or environments.
The possible effector-binding domains in the C-terminal region of the class II to V LitR proteins shared no distinct similarities with domains of known function. This suggests that those types of LitR sense light by a mechanism different from that of the known systems. The diversity of light-sensing mechanism and its correlation with the phylogenetic position of each organism is an interesting issue in terms of the genetic evolution of a specific function. Probably, the evolutionary traits reflect the illumination condition of the niche of corresponding bacteria. The availability of ligand molecule may also be a critical factor that defines the preference.
We discovered that class III LitR specifically distributed to Burkholderia actually exhibits features as a photosensor. The recombinant LitR proteins derived from B. multivorans and B. vietnamiensis absorbed light in the UV-A region (λmax = ca. 340 nm), suggesting that a photosensing molecule associating with this type of LitR absorbs UV-A light to induce its light-dependent conformational change. Since the recombinant protein was prepared in E. coli, the possible light-absorbing molecule was likely to be a relatively general molecule synthesized in not only Burkholderia but also E. coli. Based on these observations, we assessed for the possibility of NADH (λmax = 340 nm) or tetrahydropterin (λmax = 360 nm) to be the chromophore but have not yet obtained any positive result. We also tried to extract the ligand molecule from LitR by denaturing treatment but were not successful in detecting the corresponding molecule.
Our gel filtration analysis demonstrated that UV-A irradiation caused dissociation of class III LitR from oligomeric to tetrameric state (Fig. 6). The same mode of dissociation was observed with regard to the N-terminally truncated form of the protein (see Fig. S8 in the supplemental material), indicating that the corresponding C-terminal region contains the domain for protein-protein interaction as well as that for photosensing. Similar light-dependent dissociation is known for class I, the B12-dependent type. In the case of this class, tetrameric form binds DNA under the dark condition, and its transformation into dimeric structures is induced by illumination. The dimer form exhibits a low DNA-binding activity and hence dissociates from DNA. In analogy with this, the class III LitR may form an oligomer to strongly bind DNA, and its dissociation into tetramers due to illumination releases the protein from DNA. Currently, we estimate the oligomeric structure to consist of 16 LitR molecules (tetramer × 4) based on its apparent molecular weight in the gel filtration analysis. We should carefully assess whether the oligomeric structure is actually the form essential for the repression in the dark state, since the recombinant protein was vulnerable to aggregation. It is possible that the DNA-binding form is also tetrameric but with a conformation different from that under the light condition.
Although we have not yet identified the chemical structure responsible for light absorption in class III LitR, useful information was obtained from the mutational analysis regarding the conserved three Cys residues. Substitution of each Cys with Ala abolished the ability of the protein to absorb UV-A light and to bind DNA. This indicates that the three Cys residues play an essential role for the light response of class III LitR. This makes us think of the possibility that the Cys residues constitute the ligand binding site. Alternatively, an architecture based on the formation of disulfide bridge between the Cys residues serves as the photosensing apparatus, like the redox-sensing structure of OxyR and OhrR (30). The information regarding the role of Cys residues may provide us a clue to clarify the photoreceiving molecular structure of the novel type of transcriptional regulator.
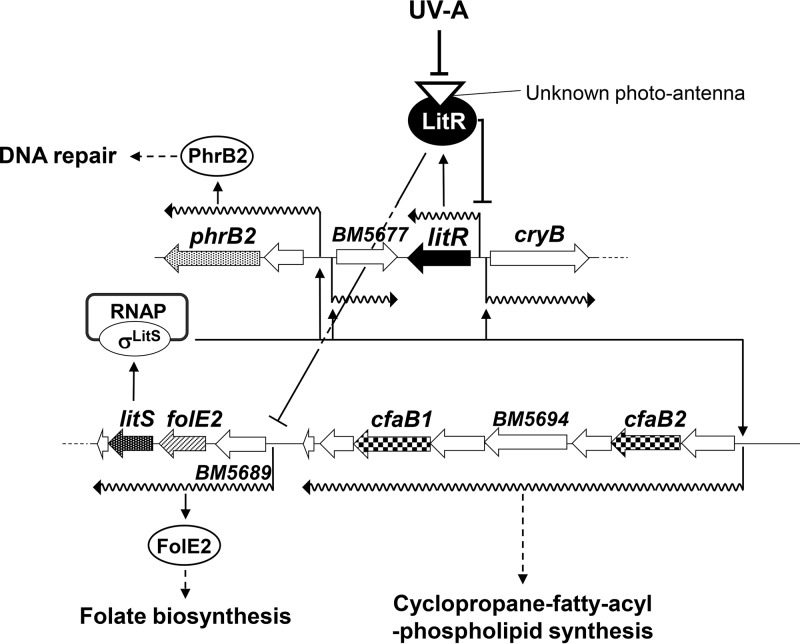
The results of our genetic analysis suggest that the class III LitR plays a central role in light-dependent gene expression in B. multivorans. Figure 9 shows the current working hypothesis for the regulatory network. The evidence obtained in this study indicates that LitR and LitS act as a photosensitive transcriptional repressor and a photoinducible RNA polymerase sigma factor, respectively. LitR, in complex with the UV-A-absorbing molecule, inhibits the expression of light-inducible genes under the dark condition. Illumination of UV-A-containing light inactivates the repression, possibly by dissociating the oligomeric structure to a tetrameric one. This allows the expression of litS and hence the production of σLitS. The σLitS-containing RNA polymerase specifies the transcription from the σLitS-dependent promoters preceding the BM5677, phrB2, and cfaB operons. The expression of these genes results in the production of BM5677, PhrB2, CfaB1, CfaB2, and related proteins, which protect cells from damage caused by illumination.
FIG 9.
Working hypothesis for the light response mechanism of B. multivorans. LitR protein associating with a compound absorbing near the UV-A region represses the transcription at the litR promoter and folE2 operon under the dark condition. By sensing UV-A light, LitR dissociates from an oligomer to a tetramer. The dissociation lowers the DNA-binding ability of LitR in the target promoters, which, in turn, allows the expression of the RNA polymerase ECF-type sigma factor LitS and FolE2. Then, folate production is enhanced, and σLitS-containing RNA polymerase (RNAP) directs the transcription of other light-inducible genes. Thus, expressed PhrB2 functions as a DNA repair enzyme, and CfaB1, CfaB2, and related proteins synthesize cyclopropane-fatty-acyl-phospholipid to protect the cell membrane from photooxidative stress generated by illumination.
LitR also directed the light-dependent transcription of folE2 encoding GTP-cyclohydrolase I. The reaction catalyzed by this enzyme is fundamental to the synthesis of pterin and folate derivatives. Actually, knockout of litR caused remarkable elevation of intracellular folate content (Fig. 8B). Recently, Romine et al. reported that PhrR, a class I LitR, controls folate and ubiquinone biosynthesis in Halomonas (23). The researchers also revealed that the mutation in phrR caused the uncontrolled production of tetrahydrofolate. Since folE and litR are found in adjacent regions in many bacterial genomes, we speculate that similar light-dependent control of folE expression occurs in those bacteria. The light-induced activation of FolE may have a defensive role. In a marine planktonic cyanobacterium, Oscillatoria, biopterin glucoside protects cells from UV-A damage (31). Alternatively, the overproduction of folate may fulfill the requirement of DNA photolyase (Phr), whose production is also induced in a light-dependent manner. The DNA repair enzyme binds flavin adenine dinucleotide and 5,10-methenyltetrahydrofolate, which serve as a chromophore and a photoantenna molecule, respectively (32).
An intriguing fact is that B. multivorans retains multiple copies of homologous genes for phrB2, folE2, and cfaB in different loci from the litR cluster. Such homologs, including phrB1, cfaA, and folE1, were transcribed both under light and dark conditions (our unpublished data). This implies that the constitutive expression of those genes is fundamental to the growth of the organism. The basal level of expression, however, may not be sufficient if cells were illuminated, hence the existence of a set of light-induced genes. The system, enabling both basal expression under the dark condition and upregulation under the light condition, may benefit the cell by being energy saving and adaptive to hazardous environment. The accumulation of the related knowledge will deepen our insight into the ecology of nonphototrophic bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
The wild-type strain of Burkholderia multivorans LMG17588 was obtained from the BCCM/LMG Bacteria Collection (Ghent, Belgium). DNA of Burkholderia vietnamiensis G4 (DSM11737) was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ), a German collection of microorganisms and cell cultures. E. coli HST08 (TaKaRa Bio Inc., Shiga, Japan) and E. coli Rosetta 2(DE3)/pLysS (Merck KGaA, Darmstadt, Germany) were used for general cloning and as an expression host for recombinant proteins, respectively. E. coli S17-1 λpir, BW25141, and pK18mobsacB were obtained from National BioResource Project (NIG, Japan). Enterococcus hirae NBRC 3181 was obtained from Biological Resource Center, NITE (NBRC). E. coli and B. multivorans were grown on LB medium supplemented with the following antibiotics: 20 μg/ml of kanamycin, 20 μg/ml of chloramphenicol, and 50 μg/ml of ampicillin. All reagents were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan) unless otherwise indicated.
Molecular phylogenetic analysis.
A neighbor-joining phylogenetic tree based on the amino acid sequence of the C-terminal domain of LitR homologs was constructed as follows: amino acid sequences of the C-terminal domain were obtained from the database resource KEGG (http://www.genome.jp/kegg/), which are regions (except for the HTH domain) predicted by Pfam. The amino acid sequences were analyzed using CLUSTAL W (33) and MEGA 5 (34). Molecular phylogenetic trees were reconstructed by the neighbor-joining method (35). The distance matrix was calculated using Kimura's two-parameter model (36).
Light irradiation of cells and proteins.
Light irradiation of cells was performed using an illuminating incubator (BR-180LF; Taitech, Saitama, Japan) equipped with white, blue, green, and red fluorescent lamps (20 W; Toshiba, Tokyo, Japan). Light-emitting diode (LED) lamps, UV-A (λmax = 365 nm), blue light (λmax = 450 nm), green light (λmax = 530 nm), and red light (λmax = 630 nm) (Optocode Corp., Tokyo, Japan), were used for the irradiation of protein samples. Light intensity was measured using a model Li-250A light meter (LI-COR Inc., Lincoln, NE).
Total RNA preparation.
A B. multivorans wild-type strain and mutants were cultured in LB liquid medium under dark and light conditions at 28°C by using an illuminating shaker. One milliliter of culture was sampled at 14 h and 21 h. After centrifugation at 10,000 × g, the pellets were fixed with RNAprotect bacterial reagent (Qiagen GmbH, Hilden, Germany) to avoid degradation of RNA during the RNA purification step. Total RNA was purified using an RNeasy minikit (Qiagen). To remove any genomic DNA contaminating the partially purified total RNA, DNase I (TaKaRa Bio) treatment was performed twice. The concentration of total RNA was measured with NanoDrop Lite (Thermo Fisher Scientific, Rockford, IL).
DNA microarray.
The B. multivorans wild-type strain was cultured at 28°C for 14 and 21 h in LB liquid medium under dark and light conditions. Total RNA was purified as described above. A single DNA microarray experiment and data analysis were carried out by the sequencing service of Roche NimbleGen (Tokyo, Japan). The 385K 4-plex custom array contained 5,981 genes from B. multivorans ATCC 17616 fixed on a glass slide obtained by fixing two sets of six unique probes composed of 60-mer synthetic oligonucleotides for each gene.
DNA sequencing of cloned genes.
The accuracy of the nucleotide sequences of the DNA fragments cloned in this study was verified by the sequencing service of Eurofins Genomics K.K. (Tokyo, Japan) or using an ABI 3100 genetic analyzer (Thermo Fisher Scientific).
Gene disruption.
To disrupt the litR (BMULJ_05678) gene, pK18mslitR was constructed as follows. The internal region of litR was amplified by PCR with a high-fidelity PrimeSTAR HS DNA polymerase (TaKaRa Bio) and primer set DislitRbmjF/DislitRbmjR (see Table S1). The PCR amplicon was cloned into pUC118 by blunt-end cloning. The sequence-verified clone was digested by EcoRI and HindIII and inserted into the same site of pK18mobsacB to yield pK18mslitR. The constructed pK18mslitR was introduced into B. multivorans by conjugative transfer using E. coli S17-1 λpir as described previously (20, 21). Among the obtained kanamycin-resistant colonies, the expected single-crossover mutant generated by chromosomal integration of pK18mslitR into the target region was checked for correct recombination by PCR with an appropriate primer pair.
The disruptants for litS, folE1, and folE2 were constructed similarly to that for litR. Each disruption plasmid (pK18mslitS for litS, pK18msfolE1 for folE1, and pK18msfolE2 for folE2) retained an internal region of each gene. Internal regions were amplified with the primer sets DislitSbmjF/DislitSbmjR for litS, DisfolE1F/DisfolE1R for folE1, and DisfolE2F/DisfolE2R for folE2. These plasmids were introduced into B. multivorans by conjugative transfer, and the resultant single-crossover mutants were confirmed for correct recombination by PCR with appropriate primer pairs.
Complementation vectors.
For a genetic complementation assay of the constructed litR and litS mutants, a chromosome integration vector based on the transposon pUT-miniTn5 Cm (Funakoshi, Tokyo, Japan) carrying the minitransposon Tn5 and chloramphenicol resistance as a selectable marker was used. To construct plitR harboring an intact litR gene and its promoter region used for the genetic complementation of litR mutant, the litR gene was amplified by PCR with primer set litRcmpF/litRcmpR, and then the PCR amplicon was cloned into the NotI site of pUT-miniTn5 Cm. For litS, a fused DNA fragment containing an intact litS and a BM5689 promoter region was amplified by a two-stage PCR with primer set P5689cmpF/P5689cmpMR(Nd) and litScmpMF/litScmpR and then cloned into the NotI site of pUT-miniTn5 Cm, yielding plitS, used for the genetic complementation of the litS mutant. For folE2, a fused DNA fragment containing an intact folE2 and a BM5689 promoter region was amplified with primer sets P5689cmpF/P5689cmpMR(Bm) and folE2cmpMF/folE2cmpR and cloned into the NotI site of pUT-miniTn5 Cm, yielding pfolE2. All primers used in the aforementioned cloning experiment are listed in Table S1. The constructed vectors were introduced into the mutants by conjugative transfer, as described above, and proper integration was confirmed by PCR with an appropriate primer set.
Determination of TSS.
Transcriptional start sites (TSS) were determined with the service (modified 5′ RACE) of DNAFORM (Yokohama, Japan) using total RNA purified from the light-irradiated cells. In brief, RNA quality was assessed by Bioanalyzer (Agilent Technologies, Santa Clara, CA) to ensure that the RNA integrity number (RIN) was over 7.0 and the A260/280 and A260/230 ratios were over 1.7. The library construction protocol was as follows. The Ribo-Zero rRNA Removal kit was used to deplete rRNA from 1 μg of total RNA. cDNA was synthesized using Superscript III (Thermo Fisher Scientific), according to the manufacturer's protocol. RNA was digested by RNase H to purify single-stranded cDNA. Adapters comprising a barcode sequence as well as priming sites for the Illumina platform were ligated to both ends of the cDNA. Libraries were sequenced using the Illumina NextSeq500 sequencing system. Data Processing Pipeline (Illumina) real-time analysis (RTA) software was used for base calling. Sequenced reads were mapped to the whole genome of B. multivorans ATCC 17616 using a Burrows-Wheeler alignment tool (BWA) without changing any parameters. TSS clustering and tags per million reads (TPM) were calculated using CAGEr (37).
Semiquantitative reverse transcription-PCR (semi-qRT-PCR) analysis.
cDNA was synthesized with SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). Total RNA (2 μg) was used for cDNA synthesis. GoTaq green master mix, 2× (Promega Corp., Madison, WI), was used for PCR. The protocol for PCR was as follows: initial denaturation at 95°C for 180 s and amplification for 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, followed by 72°C for 3 min. The oligonucleotide primers used are listed in Table S1 in the supplemental material. The PCR products were analyzed by agarose gel electrophoresis, and the band intensity was indicative of gene expression level.
qRT-PCR.
To quantify the gene expression levels of the light-inducible genes, qRT-PCR analysis was performed. Total RNA was purified from cells cultured in LB liquid medium for 14 h and 21 h under dark and blue light conditions at an intensity of 18.31 μmol · s−1 · m−2. Total RNA was purified as described above. cDNA was synthesized using 2 μg of total RNA with a SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). The quantification of cDNA was performed by using an Applied Biosystems 7500 real-time PCR system (Thermo Fisher) and PowerUp SYBR green master mix (Thermo Fisher Scientific) according to the manufacturer's instructions. The PCR protocol was as follows: initial denaturation at 50°C for 120 s and 95°C for 120 s and then amplification by 40 cycles of 95°C for 15 s and 60°C for 60 s. The gene-specific oligonucleotide primers designed using Primer 3 (https://primer3plus.com/) are listed in Table S1. For sample normalization, dnaA (a housekeeping gene) was used as an internal standard to quantify the relative expression of the analyzed genes. Relative quantification of gene expression was calculated by the relative quantitative threshold cycle (2−ΔΔCT) method (38) using the dnaA signals as internal references. All reactions were performed in triplicate.
Construction of LitR protein expression vectors.
To construct a LitR expression vector with an N-terminal GST tag and a C-terminal His tag, a double-tag-containing vector, pGEX-GFP-His, was first constructed. DNA fragments containing enhanced green fluorescent protein (EGFP) with His×6 were amplified by PCR using the EGFP-F/EGFP-R primer set and pIJ786 as a template, and the amplicon was cloned between the BamHI and XhoI sites of pGEX-6P-2 to yield pGEX-GFP-His. litR was amplified by PCR with the litRbmj-F/litRbmj-His6-R primer set. The amplicon was cleaved by BamHI-EcoRI and then inserted into the same sites of pGEX-GFP-His to yield pGEXlitR. Mutant alleles of LitR, in which cysteine 251 (C251), cysteine 253 (C253), and cysteine 274 (C274) were replaced with alanine, were constructed by a two-stage PCR procedure. To construct expression vectors for mutant LitR proteins, the following primer sets were used for two-stage PCR: litRbmj-F/litRbmjMR(C251A) and litRbmjMF(C251A)/litRbmj-His6-R for LitRC251A, litRbmj-F/litRbmjMR(C253A) and litRbmjMF(C253A)/litRbmj-His6-R for LitRC253A, and litRbmj-F/litRbmjMR(C274A) and litRbmjMF(C274A)/litRbmj-His6-R for LitRC274A. Each amplicon was cloned into the BamHI site of pGEX-GFP-His. To construct expression vectors for truncated LitR proteins, the following primer sets were used for PCR: litRbmjF(81–323)/litRbmj-His6-R for GST-LitR81–323, litRbmjF(100–323)/litRbmj-His6-R for GST-LitR100–323, and litRbmjF(200–323)/litRbmj-His6-R for GST-LitR200–323. Each amplicon was cloned into the BamHI site of pGEX-GFP-His. For the expression vector of LitRbvi recombinant protein from B. vietnamiensis G4, litRbvi was amplified by PCR with the primer set litRbviF(6P2)/litRbviR(6P2), and the amplicon was cloned into the BamHI-EcoRI sites of pGEX-GFP-His.
Preparation of a LitR recombinant proteins.
E. coli Rosetta2(DE3)/pLysS strains retaining litR expression vectors were precultured with LB liquid medium containing chloramphenicol and ampicillin at 28°C overnight. The seeded culture was inoculated into 100 ml of LB liquid medium in a 500-ml baffled Erlenmeyer flask at a final concentration of 1%. Culture with rotary shaking (135 rpm) was performed for 3 h at 28°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at a final concentration of 0.1 mM, with continued incubation at 28°C for 4 h. The E. coli cells were harvested by centrifugation, and the cell pellets were suspended in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 1.8 mM KH2PO4). The cell wash was performed 2 times with PBS (pH 7.5), and subsequently, the cells were disrupted by ultrasonication using an Astrason Ultrasonic Processor XL (Misonix Inc., Farmingdale, NY) or APV 1000 Gaulin mechanical cell presser (APV Homogenizers, As, Denmark). To obtain cell extract, the supernatant was centrifuged and filtered. The samples were loaded onto a GSTrap HP column equipped with an ÄKTA explorer 10S (GE Healthcare, UK Ltd., England) according to the manufacturer's instructions. The elution peak corresponding to GST-LitR-His confirmed by SDS-PAGE was dialyzed with dialysis buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 200 mM l-(+)-arginine monohydrochloride, and 1 mM EDTA at 4°C overnight. GST-LitR-His (2 mg) was incubated with 1 mg of PreScission protease at 4°C for 4 h to cleave the GST tag. The LitR-His was purified to homogeneity in a cOmplete His tag purification column (5 ml of resin) (Roche, Basel, Switzerland) mainly according to the manufacturer's instructions. The immobilized-metal affinity chromatography (IMAC) binding buffer used contained 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 200 mM l-arginine monohydrochloride. The IMAC elution buffer used contained 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 200 mM l-arginine monohydrochloride, and 500 mM imidazole. The peak corresponding to LitR-His was dialyzed with precooled IMAC binding buffer at 4°C overnight. The sample was centrifuged at 15,000 rpm for 15 min at 4°C, and the supernatant was then used in the biochemical experiments. The protein concentration was measured with a Bio-Rad protein assay kit (Bio-Rad, Laboratories, Hercules, CA), and the absorbance was measured with a Multiskan GO spectrophotometer (Thermo Fisher Scientific).
Gel shift assay.
The DNA-binding activity of LitR protein was evaluated by gel shift assay. DNA fragments containing the promoter regions were amplified by PCR with primer sets PlitRF/PlitRR for PlitR, PBM5689F/PBM5689R for PBM_5689, and PrpoDF/PrpoDR for PrpoD. These DNA fragments were cloned into a TA cloning vector, pMD19 (TaKaRa Bio). To prepare a Cy5-labeled probe, two primer sets, a Cy5-labeled pMD19F(Cy5) and a nonlabeled pMD19R, both of which anneal to the pMD19 vector, were used to amplify the cloned DNA fragments. The purified LitR (0 to 32 fmol) and DNA probe (10 fmol) were coincubated at 28°C for 30 min in 10 mM Tris-HCl (pH 7.2), 50 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.5 μg of poly(dI-dC) and loaded onto a nondenaturing polyacrylamide gel containing 6% polyacrylamide. The light irradiation of the recombinant proteins was carried out by using light-emitting diode lamps with UV-A light (λmax = 365 nm) for 180 s. The resulting polyacrylamide gel was scanned with a Typhoon FAL9500 image analyzer (GE Healthcare).
Spectroscopic analysis.
The absorbance spectra for LitR recombinant proteins were measured with a Cary 60 UV-visible (UV-Vis) spectrophotometer (Agilent Technologies). To examine the light-dependent absorption changes, UV-A (λmax = 365 nm) irradiation of 25.0 μM recombinant protein LitR, purified to homogeneity by affinity chromatography, was carried out. The absorbance spectrum of LitR was recorded after the sample was illuminated for 60 s and 180 s by UV-A (λmax = 365 nm) at 0.06 μmol · s−1 · m−2 onto the sample. Dark-state recovery was measured after proteins were incubated for 60 and 120 min. The type of light reducing the absorbance spectrum of LitR at 365 nm was also examined. Illumination of various wavelength of light was also carried out with the following light intensities for 180 s: 0.06 (λmax = 365 nm), 60.41 (450 nm), 14.02 (470 nm), 22.13 (505 nm), 21.76 (595 nm), and 75.93 (630 nm) μmol · s−1 · m−2.
Gel filtration chromatography.
To estimate the relative molecular weight (Mr) of the LitR protein, 0.1 mg of LitR protein, purified by affinity chromatography, was analyzed using an ÄKTA explorer 10S system equipped with a Superdex 200 10/300 GL column (GE Healthcare). The column was equilibrated with buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 200 mM l-arginine monohydrochloride at a flow rate of 0.2 ml/min. l-Arginine monohydrochloride was added to avoid aggregation of LitR (39). Gel filtration calibration kit LMW (GE Healthcare; comprises RNase A at an Mr of 13,700, carbonic anhydrase at an Mr of 29,000, ovalbumin at an Mr of 43,000, and conalbumin at an Mr of 75,000) and gel filtration calibration kit HMW (GE Healthcare; comprises aldolase at an Mr of 158,000 and ferritin at an Mr of 440,000) were used as standard Mr markers. The sample was illuminated with UV-A light (365 nm) at approximately 0.06 μmol · s−1 · m−2 for 180 s prior to being loaded onto the Superdex 200 10/300 GL column. The relative Mr of LitR was estimated based on the molecular weight markers and results of triplicate individual experiments. Gel filtration chromatography was performed three times using three independent protein preparations. To confirm the eluted protein, proteins were separated by SDS-PAGE and visualized by silver staining.
Extraction and quantification of intracellular folate.
Folate was quantified by a bioassay with Difco folic acid assay medium (Becton, Dickinson and Co., Sparks, MD) according to the manufacturer's instructions and previous studies (40, 41). To extract intracellular folate, cells were cultured with LB medium at 28°C. The cultured cells were harvested by centrifugation, washed twice with PBS, and then suspended in PBS. The cell suspension was disrupted with a sonicator (Astrason Ultrasonic Processor XL), and the cell extract was centrifuged at 12,000 × g for 10 min to remove the undisrupted cell debris and membrane fraction. After boiling at 121°C for 15 min, the supernatant was centrifuged at 12,000 × g for 15 min and filtered. Enterococcus hirae NBRC 3181, used as an indicator strain, was cultured at 37°C for 16 h in Lactobacilli Broth AOAC (Becton, Dickinson and Co.) for seed culture. Folic acid (pteroylglutamic acid) was used as a standard. Protein concentration was measured with a protein assay kit (Bio-Rad Laboratories).
Accession number(s).
The microarray data obtained in this study have been registered in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE112472.
Supplementary Material
ACKNOWLEDGMENTS
We thank Masayuki Shimamura for qRT-PCR analysis. We also thank Yoshiyuki Ohtsubo and Yuji Nagata for technical advice on gene disruption.
This study was supported by a grant-in-aid for scientific research for the support of scientists (no. 16K07676) from the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00285-18.
REFERENCES
- 1.Takano H. 2016. The regulatory mechanism underlying light-inducible production of carotenoids in nonphototrophic bacteria. Biosci Biotechnol Biochem 80:1264–1273. doi: 10.1080/09168451.2016.1156478. [DOI] [PubMed] [Google Scholar]
- 2.Padmanabhan S, Jost M, Drennan CL, Elías-Arnanz M. 2017. A new facet of vitamin B12: gene regulation by cobalamin-based photoreceptors. Annu Rev Biochem 86:485–514. doi: 10.1146/annurev-biochem-061516-044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takano H, Obitsu S, Beppu T, Ueda K. 2005. Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J Bacteriol 187:1825–1832. doi: 10.1128/JB.187.5.1825-1832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takano H, Hagiwara K, Ueda K. 2015. Fundamental role of cobalamin biosynthesis in the developmental growth of Streptomyces coelicolor A3(2). Appl Microbiol Biotechnol 99:2329–2337. doi: 10.1007/s00253-014-6325-z. [DOI] [PubMed] [Google Scholar]
- 5.Takano H, Beppu T, Ueda K. 2006. The CarA/LitR-family transcriptional regulator: its possible role as a photosensor and wide distribution in non-phototrophic bacteria. Biosci Biotechnol Biochem 70:2320–2324. doi: 10.1271/bbb.60230. [DOI] [PubMed] [Google Scholar]
- 6.Takano H, Asker D, Beppu T, Ueda K. 2006. Genetic control for light-induced carotenoid production in non-phototrophic bacteria. J Ind Microbiol Biotechnol 33:88–93. doi: 10.1007/s10295-005-0005-z. [DOI] [PubMed] [Google Scholar]
- 7.Takano H, Agari Y, Hagiwara K, Watanabe R, Yamazaki R, Beppu T, Shinkai A, Ueda K. 2014. LdrP, a cAMP receptor protein/FNR family transcriptional regulator, serves as a positive regulator for the light-inducible gene cluster in the megaplasmid of Thermus thermophilus. Microbiology 160:2650–2660. doi: 10.1099/mic.0.082263-0. [DOI] [PubMed] [Google Scholar]
- 8.Takano H, Kondo M, Usui N, Usui T, Ohzeki H, Yamazaki R, Washioka M, Nakamura A, Hoshino T, Hakamata W, Beppu T, Ueda K. 2011. Involvement of CarA/LitR and CRP/FNR family transcriptional regulators in light-induced carotenoid production in Thermus thermophilus. J Bacteriol 193:2451–2459. doi: 10.1128/JB.01125-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jost M, Fernandez-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY, Kang G, Padmanabhan S, Elias-Arnanz M, Drennan CL. 2015. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526:536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takano H, Mise K, Hagiwara K, Hirata N, Watanabe S, Toriyabe M, Shiratori-Takano H, Ueda K. 2015. Role and function of LitR, an adenosyl B12-bound light-sensitive regulator of Bacillus megaterium QM B1551, in regulation of carotenoid production. J Bacteriol 197:2301–2315. doi: 10.1128/JB.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elías-Arnanz M. 2011. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A 108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edge R, McGarvey DJ, Truscott TG. 1997. The carotenoids as anti-oxidants—a review. J Photochem Photobiol B Biol 41:189–200. doi: 10.1016/S1011-1344(97)00092-4. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong GA. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu Rev Microbiol 51:629–659. doi: 10.1146/annurev.micro.51.1.629. [DOI] [PubMed] [Google Scholar]
- 14.Galbis-Martínez M, Padmanabhan S, Murillo FJ, Elías-Arnanz M. 2012. Carf mediates signaling by singlet oxygen, generated via photoexcited protoporphyrin IX, in Myxococcus xanthus light-induced carotenogenesis. J Bacteriol 194:1427–1436. doi: 10.1128/JB.06662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang H, Kang J, Zhang D. 2017. Microbial production of vitamin B12: a review and future perspectives. Microb Cell Fact 16:15. doi: 10.1186/s12934-017-0631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 17.Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. 2006. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol 14:277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu H, Imura Y, Ohori A, Nagata Y, Tsuda M. 2003. Distribution and organization of auxotrophic genes on the multichromosomal genome of Burkholderia multivorans ATCC 17616. J Bacteriol 185:3333–3343. doi: 10.1128/JB.185.11.3333-3343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuhara S, Komatsu H, Goto H, Ohtsubo Y, Nagata Y, Tsuda M. 2008. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 154:1763–1774. doi: 10.1099/mic.0.2007/015537-0. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama E, Ohtsubo Y, Nagata Y, Tsuda M. 2010. Identification of Burkholderia multivorans ATCC 17616 genes induced in soil environment by in vivo expression technology. Environ Microbiol 12:2539–2558. doi: 10.1111/j.1462-2920.2010.02227.x. [DOI] [PubMed] [Google Scholar]
- 22.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol Rev 27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 23.Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, Carre A, Li X, Xu C, Clauss TRW, Kim Y-M, Metz TO, Wright AT. 2017. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc Natl Acad Sci U S A 114:E1205–E1214. doi: 10.1073/pnas.1612360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaeser J, Klug G. 2005. Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 151:1927–1938. doi: 10.1099/mic.0.27789-0. [DOI] [PubMed] [Google Scholar]
- 25.Singh H, Vadasz JA. 1977. Singlet oxygen quenchers and the photodynamic inactivation of E. coli ribosomes by methylene blue. Biochem Biophys Res Commun 76:391–397. doi: 10.1016/0006-291X(77)90737-9. [DOI] [PubMed] [Google Scholar]
- 26.Shimada T, Yamazaki Y, Tanaka K, Ishihama A. 2014. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One 9:e90447. doi: 10.1371/journal.pone.0090447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losi A, Gärtner W. 2017. Solving blue light riddles: new lessons from flavin-binding LOV photoreceptors. Photochem Photobiol 93:141–158. doi: 10.1111/php.12674. [DOI] [PubMed] [Google Scholar]
- 28.Herrou J, Crosson S. 2011. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol 9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujisawa T, Masuda S. 2018. Light-induced chromophore and protein responses and mechanical signal transduction of BLUF proteins. Biophys Rev 10:327–337. doi: 10.1007/s12551-017-0355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imlay JA. 2015. Transcription factors that defend bacteria against reactive oxygen species. Annu Rev Microbiol 69:93–108. doi: 10.1146/annurev-micro-091014-104322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wachi Y, Grant Burgess J, Iwamoto K, Yamada N, Nakamura N, Matsunaga T. 1995. Effect of ultraviolet-A (UV-A) light on growth, photosynthetic activity and production of biopterin glucoside by the marine UV-A resistant cyanobacterium Oscillatoria sp. Biochim Biophys Acta 1244:165–168. doi: 10.1016/0304-4165(94)00219-N. [DOI] [PubMed] [Google Scholar]
- 32.Sancar A. 2008. Structure and function of photolyase and in vivo enzymology: 50th anniversary. J Biol Chem 283:32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 36.Givens GD, Seidemann MF. 1977. Middle ear measurements in a difficult to test mentally retarded population. Ment Retard 15:40–42. [PubMed] [Google Scholar]
- 37.Haberle V, Forrest ARR, Hayashizaki Y, Carninci P, Lenhard B. 2015. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res 43:e51. doi: 10.1093/nar/gkv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Arakawa T, Ejima D, Tsumoto K, Obeyama N, Tanaka Y, Kita Y, Timasheff SN. 2007. Suppression of protein interactions by arginine: a proposed mechanism of the arginine effects. Biophys Chem 127:1–8. doi: 10.1016/j.bpc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Masuda M, Ide M, Utsumi H, Niiro T, Shimamura Y, Murata M. 2012. Production potency of folate, vitamin B12, and thiamine by lactic acid bacteria isolated from Japanese pickles. Biosci Biotechnol Biochem 76:2061–2067. doi: 10.1271/bbb.120414. [DOI] [PubMed] [Google Scholar]
- 41.Arcot J, Shrestha A. 2005. Folate: methods of analysis. Trends Food Sci Technol 16:253–266. doi: 10.1016/j.tifs.2005.03.013. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.