Abstract
Both baicalin (BA) and jasminoidin (JA) are active ingredients in Chinese herb medicine Scutellaria baicalensis and Fructus gardeniae, respectively. They have been shown to exert additive neuroprotective action in ischemic stroke models. In this study we used transcriptome analysis to explore the pure therapeutic mechanisms of BA, JA and their combination (BJ) contributing to phenotype variation and reversal of pathological processes. Mice with middle cerebral artery obstruction were treated with BA, JA, their combination (BJ), or concha margaritifera (CM). Cerebral infarct volume was examined to determine the effect of these compounds on phenotype. Using the hippocampus microarray and ingenuity pathway analysis (IPA) software, we exacted the differentially expressed genes, networks, pathways, and functions in positive-phenotype groups (BA, JA and BJ) by comparing with the negative-phenotype group (CM). In the BA, JA, and BJ groups, a total of 7, 4, and 11 specific target molecules, 1, 1, and 4 networks, 51, 59, and 18 canonical pathways and 70, 53, and 64 biological functions, respectively, were identified. Pure therapeutic mechanisms of BA and JA were mainly overlapped in specific target molecules, functions and pathways, which were related to the nervous system, inflammation and immune response. The specific mechanisms of BA and JA were associated with apoptosis and cancer-related signaling and endocrine and hormone regulation, respectively. In the BJ group, novel target profiles distinct from mono-therapies were revealed, including 11 specific target molecules, 10 functions, and 10 pathways, the majority of which were related to a virus-mediated immune response. The pure additive effects between BA and JA were based on enhanced action in virus-mediated immune response. This pure mechanistic analysis may provide a clearer outline of the target profiles of multi-target compounds and combination therapies.
Keywords: ischemic stroke, traditional Chinese medicine, baicalin, jasminoidin, combination therapy, additive effects, hippocampus microarray, transcriptome, network pharmacology, Fangjiomics
Introduction
Ischemic stroke (IS) is a common disease with high mortality and disability1,2. Intravenous thrombolysis is the only approved and effective treatment recommended by the FDA3. However, thrombolysis can only be performed within a strictly limited time window, and may also lead to fatal complications3. Thus, it is of great significance to develop a complementary or alternative therapeutic strategy for the disease, beyond extension of the therapeutic window and indications of thrombolysis. As a complex disease, the pathogenesis of IS features multiple common genetic variations and dysfunctions4. The complexity of this disease resists traditional efforts that attempts to seek a “magic bullet” to achieve optimal therapeutic efficacy5,6,7. Therefore, increasing attention has been paid to utilizing one or more multi-target drugs to systematically reverse the disease molecular network to achieve novel homeostasis8,9. It is suggested that multi-target perturbations to the pathological network, especially designed drug arrays (Fangji), may lead to improved effectiveness, reduced adverse effects, and decreased drug resistance9,10,11,12. Herbal medicine, particularly effective ingredients extracted from Chinese medicinal materials, is characterized by low activity and multi-target action13. A combination of several ingredients in the holistic treatment of ischemic stroke, also known as “magic shotguns”, is expected to shed light on a novel therapeutic solution for this complex disease5. Consequently, a series of attempts to identify medicinal herbal ingredients and their underlying pharmacological mechanisms have been made, following conventional methods for drug research and development. However, it remains challenging to clearly elucidate the pharmacological mechanisms of these ingredients due to the complexity of their multiple targets and the potential interrelations between these targets. Polypharmacology, a promising avenue to systemically decipher drug mechanisms of modulating disease molecular networks14, provides an opportunity to fully understand the activities and pharmacological mechanisms of herbal ingredients15,16,17. Therefore, it is appealing to utilize polypharmacology methods to elucidate the pharmacological mechanisms of herbal medicine on multiple targets.
Qingkailing injection, a complementary and alternative therapy for IS that is widely prescribed across China, has been demonstrated to be effective in reducing ischemia-reperfusion injury18. However, its pharmacological mechanism is still far from clear. Baicalin (BA) and jasminoidin (JA), extracted from Scutellaria baicalensis and Fructus gardeniae, respectively, are the major bioactive ingredients in Qingkailing injection according to fingerprint analysis based on high-performance liquid chromatography (HPLC)19. Thus, it is of utmost importance to investigate the biological actions of BA and JA on IS. BA has long been suggested to be a potential therapeutic agent for stroke and a strong candidate for drug discovery20. BA has been reported to reduce neuronal damage, brain edema, and blood-brain barrier permeability and to alleviate memory impairment following ischemia by inhibiting MMP-9 expression, MMP-9-mediated occludin degradation21, and phosphorylation of CaMKII in the hippocampus22. BA is also demonstrated to possess notable neuroprotective effects23, which are mainly attributed to its anti-inflammatory and anti-apoptotic activities, as well as its modulatory effects on multiple pathways such as the TLR2/4 signaling pathway24,25 and NF-κB pathway26,27,28. JA has also been shown to exert neuroprotective effects by enhancing growth factor signaling, reducing apoptosis29, inhibiting oxidative stress, regulating mitochondrial dysfunction30, and inhibiting inflammatory response31,32. Furthermore, the combination of BA and JA has been suggested to significantly improve their effectiveness in treating cerebral ischemia by promoting neuroprotection and neurogenesis, improving anti-oxidation33,34, and regulating apoptosis-related cascades35. In our previous studies, the additive effect between JA and BA was demonstrated, and the additive mechanism included pathway cross-talk in both horizontal and vertical patterns36,37,38. The pathways and processes specific to the combination group were mainly related to apoptosis and survival, gamma-secretase activity, neurophysiological processes, development, reproduction, and regulation of lipid metabolism37.
These prior studies provide systematic “targeting profiles” of BA, JA and their combination on the cerebral ischemia biological network by using polypharmacological methods. Nevertheless, these “targeting profiles” contain all the information about network rewiring responses to perturbations, including not only specific mechanisms contributing to phenotype alterations but also invalid system fluctuations that do not contribute to phenotype variations. What attracted our attention is how to precisely extract the specific pharmacological mechanism that reverses the pathological process. This means that the “most effective shotgun” must be identified from a series of “magic shotguns”, a task that has remained challenging until now. In this paper, using a microarray containing 374 stroke-related genes and ingenuity pathway analysis (IPA) software, we compared the networks perturbed by positive-phenotype ingredients (BA, JA and additive combination BJ) to those perturbed by negative-phenotype ingredients (concha margaritifera, CM) to investigate the pure pharmacological mechanism contributing to phenotype variation by eliminating non-specific roles modulating network rewiring39.
Materials and methods
Animal models
All animals used in our experiments were approved by the Ethics Review Committee for Animal Experimentation, China Academy of Chinese Medical Sciences. The experimental processes and protocols were in compliance with the Prevention of Cruelty to Animals Act 1986 and NIH Guidelines for the Care and Use of Laboratory Animals for Experimental Procedures40.
As described in previous studies41,42,43,44,45, all mice except those in the sham group were subjected to middle cerebral artery obstruction (MCAO) to induce ischemic stroke. Briefly, after anesthesia, the left middle cerebral artery was occluded with an intraluminal filament for 1.5 h and then reperfused to established cerebral ischemia models. Mice in the sham group were subjected to the same surgical procedures, but no filament was inserted. Then, the mice were treated with various compounds, and after the 24-h treatment, the mice were sacrificed for further experiments.
Drug administration and group
All compounds used in this study, including BA, JA, and CM, were prepared according to the standards issued by the National Institutes for Food and Drug Control. Quality control was performed using fingerprint chromatographic methodologies. The three compounds were dissolved in 0.9% NaCl to reach the required concentrations: BA (5 mg/mL), JA (25 mg/mL), and CM (50 mg/mL).
A total of 84 male mice (Kunming strain, 12 weeks old, 38–48 g) were randomly divided into 6 groups: sham, vehicle, BA-treated, JA-treated, CM-treated, and BJ-treated (combination of BA and JA) groups. Mice in the drug-treated groups were injected with corresponding drug solutions (2 mL/kg body weight) via the tail vein 1.5 h after MCAO modeling, and mice in the BJ-treated group were injected with a combination of BA and JA at a 1:1 volume ratio. Mice in the sham and vehicle groups were injected with 0.9% NaCl (2 mL/kg body weight). To determine infarct volume, 9 mice in each group were employed to explore the effect of these compounds on the cerebral ischemia-related phenotype. The remaining 5 mice in each group were used for subsequent microarray analysis to investigate the underlying pharmacological mechanisms.
Cerebral infarct volume examination for phenotype-related effect analysis
Twenty-four hours after MCAO modeling, 9 mice in each group were sacrificed to calculate the infarct ratio. The brain was removed and sectioned into five slices in the coronal plane 1, 3, 5, and 7 mm from the prefrontal cortex. The slices were stained with 1% 2,3,5-triphenyl tetrazolium chloride (TTC) at 37 °C for 30 min. Images of these slices were captured with a digital camera (Color CCD camera TP-6001A, Topica Inc, Tokyo, Japan). The area of the infarct region was analyzed using a Pathology Image Analysis System (Topica Inc). The cerebral infarct ratio was the ratio of infarct volume to the total volume43.
RNA extraction and microarray
The hippocampi of 5 mice per group were separated 24 h after MCAO modeling and used for RNA isolation and microarray analysis. The hippocampus was chosen because it should sustain some cell injury or cell death in most models without including any areas of infarction. After the hippocampi were homogenized in TRIzol Reagent, the total RNA was isolated and purified for further experiments, as described in our previous studies41,42,43,44,45,46,47. A total of 374 ischemia-related genes were selected from the Science STKE database to comprise the cDNA microarray in this study. The details of the gene screening process are reported in our previous studies43,46,47.
All microarray analyses were performed based on the Array-Track system (US Food and Drug Administration, Silver Spring, ML, USA). Microarray analysis was performed in accordance with the Minimum Information in a Microarray Experiment (MIAME) guidelines and the Microarray Quality Control (MAQC) project. We submitted the results to the Array Express database. The robust multiarray analysis (RMA) was employed to preprocess the raw cell intensity files (CEL), which contained the fluorescence intensity values for each probe. The data were analyzed and normalized as described in our previous studies43. To investigate the altered genes related to phenotype, we compared the mean expression of genes in the positive-phenotype compound-treated groups (BA, JA, and BJ) with those in the negative-phenotype compound-treated group (CM) using one-way analysis of variance and significance analysis. Genes with a P value < 0.05 and > 1.5-fold increase or < 0.67-fold decrease in expression level were defined as significantly differentially expressed genes for further analysis.
A list of these differentially expressed genes was uploaded to the IPA (ingenuity pathway analysis) system (http://www.ingenuity.com/) to identify the significantly differentially regulated molecules, which were defined as network-eligible molecules. Based on these network-eligible molecules and their connectivity, the regulated target networks were generated algorithmically. Then, the biological functions of these networks were enriched using a right-tailed Fisher's exact test. The significance of canonical pathways based on these genes was measured in two ways: (1) the ratio of the number of genes mapping to the pathway divided by the total number of genes in this canonical pathway and (2) Fisher's exact test, which was used to calculate a P value to determine the probability that these genes are associated with the canonical pathways outside of chance alone. Finally, canonical pathways with a P value < 0.05 were screened for and analyzed36,45.
Results
Phenotype variation on ischemia-induced infarction
In a cerebral ischemia reperfusion injury experiment, BA and JA significantly reduced the infarct volume compared to the vehicle group, whereas CM showed no significant therapeutic effect on the infarct volume. Details of the infarct volume and its significance in each group were reported in our previous studies38,43. Therefore, the BA, JA, and BJ groups were defined as phenotype-positive groups, and the CM group was defined as phenotype-negative group. BJ reduced the infarct volume to a greater extent than BA or JA monotherapies. Further combination effect analysis revealed additive effects between BA and JA with a combination index value close to 1.036.
Divergence and similarity of pure and non-pure pharmacological analysis outcomes
In our previous studies, by comparing the perturbed networks in the compound-treated groups to those in the vehicle group, we investigated the pharmacological mechanisms of BA, JA and BJ, which were named non-pure mechanisms35,36,37,38. Nevertheless, these non-pure mechanisms contain all the information about network rewiring response to perturbations, including not only on-target actions contributing to phenotype variation but also invalid system fluctuations not contributing to phenotype variations. What attracts our attention is how to precisely extract the specific pharmacological mechanism that reverses the pathological process. For the phenotype-negative group (CM group), although the biological networks were perturbed by CM, no significant phenotype variation was found between CM and the vehicle group. Therefore, we defined the perturbation of CM as invalid effects. In this study, we compared the networks perturbed by positive-phenotype compounds to those perturbed by phenotype-negative compound (CM) to eliminate invalid effects (effects of CM) from the holistic actions of positive-phenotype compounds, in order to screen out the on-target actions and corresponding targets, which were defined as pure pharmacological mechanisms, providing more precise effects that contribute to phenotype variation.
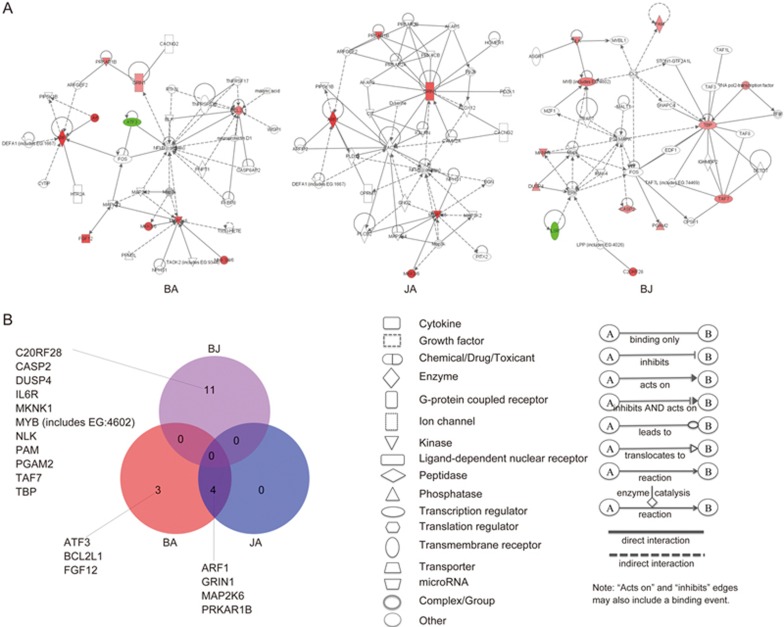
As a result, pure and non-pure pharmacological analysis showed definite distinctions in specific target molecules (genes with EXP fold-change values above 1.0 or below -1.0 were defined as specific target molecules of corresponding compounds). As shown in Figure 1, BA targeted 7 molecules in both pure and non-pure analysis, but only 2 target molecules, ATF3 and BCL2L1, overlapped between pure and non-pure analysis. This indicated that although the other 5 molecules (CAMK2B, CSF1, F5, HTR3A, and JUND) were identified as BA targets in previous non-pure analysis, they did not contribute to phenotype reversal; equally, the other 5 molecules (ARF1, FGF12, GRIN1, MAP2K6, and PRKAR1E) identified only in pure analysis might be considered potential targets of BA, although they were not discovered in non-pure analysis. In the BJ group, 4 overlapping target molecules, ie, IKBKG, IL6R, PGAM2, and TCF12, were detected between the 11 pure and 13 non-pure target molecules. There were 7 and 9 specific target molecules of pure and non-pure analysis in the BJ group, respectively (details shown in Figure 1). In the JA group, none of the target molecules overlapped between the 4 pure and 4 non-pure target molecules.
Figure 1.
Specific target molecules in each group identified by pure and non-pure analyses. Red and green represent up- and down-regulated genes, respectively. Gray represents genes that were not differentially expressed.
Pure network and pathway alterations were analogous in BA and JA groups
In the pure mechanistic analysis, BA and JA showed similar regulating patterns in specific target molecules, canonical pathways, and biological functions. According to pure mechanistic analysis, 41 and 22 differentially expressed genes were identified in the BA- and JA-treated groups, respectively. The results revealed that BA and JA targeted 7 and 4 specific molecules, respectively (Figure 2A, Supplementary Table S1), and all 4 specific target molecules in JA group overlapped with those in the BA group (Figure 2B). Based on these target genes, statistically significant networks were constructed in the BA and JA groups as target networks (Figure 2A). The functions of the target network in the BA group included cell death, genetic disorder, and immunological disease, while the functions of the target network in the JA group included carbohydrate metabolism, lipid metabolism, and small molecule biochemistry.
Figure 2.
Representative significant networks and overlapping patterns of specific target molecules from these networks in each group. (A) Representative significant network in the BA, JA, and BJ groups. Red and green nodes represent up- and down-regulated genes, respectively. Color intensity indicates the fold-change of up- or down-regulation. Uncolored nodes denote genes that were not differentially expressed in our experiment but were integrated into networks based on IPA knowledge. (B) Overlapping patterns of specific target molecules (genes with an EXP fold-change more than 1.0 or less than -1.0) in each group.
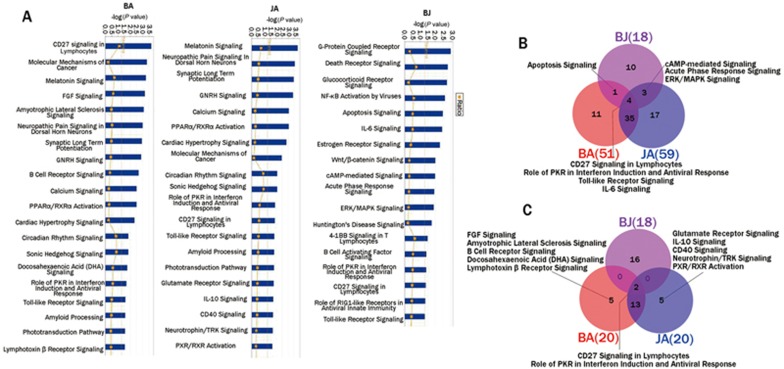
According to pure mechanistic analysis, a total of 51 and 59 statistically significant pathways involving differentially expressed genes were identified as canonical pathways in the BA and JA groups, respectively (Supplementary Table S2). The P-values of the top 20 canonical pathways in the BA and JA groups were compared and listed in Figure 3A. As shown in Figure 3B, 39 canonical pathways overlapped between the BA and JA groups, accounting for 76.47% and 66.10% of all canonical pathways in the BA and JA groups, respectively. We roughly classified these pathways based on prior knowledge, published literature and relevant databases. As a result, the overlapping canonical pathways between BA and JA were closely related to the nervous system, the immune system and inflammation. The non-overlapping canonical pathways in the BA group tended to contribute to inflammation and apoptosis, while those in JA group mainly involved hormone and endocrine regulation (Table 1). Among the top 20 canonical pathways (canonical pathways with minimum P-values), such an overlapping phenomenon was also observed; up to 75% of the top 20 canonical pathways in both BA and JA groups overlapped between the two groups (Figure 3C).
Figure 3.
Top 20 canonical pathways and overlapping patterns of canonical pathways in each group. (A) Top 20 canonical pathways in each group; (B) overlapping patterns for all canonical pathways in each group; (C) overlapping patterns for top 20 canonical pathways in each group.
Table 1. Pathways and their categories in each group and overlapping patterns of these pathways.
| BA (11) | Pathway category |
|---|---|
| Docosahexaenoic Acid (DHA) Signaling | nervous system |
| Lymphotoxin β Receptor Signaling | inflammation and immune |
| Induction of Apoptosis by HIV1 | inflammation and immune viral infection immunity |
| IL-15 Signaling | inflammation and immune |
| GM-CSF Signaling | inflammation and immune |
| Small Cell Lung Cancer Signaling | apoptosis and cancer |
| VEGF Signaling | others |
| Bladder Cancer Signaling | apoptosis and cancer |
| p53 Signaling | apoptosis and cancer |
| PTEN Signaling | apoptosis and cancer |
| Chronic Myeloid Leukemia Signaling | apoptosis and cancer |
| JA (17) | Pathway category |
| Type I Diabetes Mellitus Signaling | endocrine and hormone |
| Renin-Angiotensin Signaling | endocrine and hormone |
| Corticotropin Releasing Hormone Signaling | endocrine and hormone |
| Androgen Signaling | endocrine and hormone |
| Cardiac β-adrenergic Signaling | endocrine and hormone |
| Relaxin Signaling | endocrine and hormone |
| Insulin Receptor Signaling | endocrine and hormone |
| Cellular Effects of Sildenafil (Viagra) | endocrine and hormone |
| Inositol Phosphate Metabolism | others |
| Hepatic Cholestasis | others |
| Germ Cell-Sertoli Cell Junction Signaling | endocrine and hormone |
| NF-κB Signaling | inflammation and immune |
| Tight Junction Signaling | others |
| RAR Activation | others |
| CREB Signaling in Neurons | nervous system |
| Ephrin Receptor Signaling | nervous system |
| NRF2-mediated Oxidative Stress Response | others |
| BJ (10) | Pathway category |
| G-Protein Coupled Receptor Signaling | inflammation and immune viral infection immunity |
| Wnt/β-catenin Signaling | others |
| Death Receptor Signaling | apoptosis and cancer |
| Glucocorticoid Receptor Signaling | endocrine and hormone |
| NF-κB Activation by Viruses | inflammation and immune viral infection immunity |
| Estrogen Receptor Signaling | endocrine and hormone |
| Huntington's Disease Signaling | nervous system |
| 4-1BB Signaling in T Lymphocytes | inflammation and immune viral infection immunity |
| B Cell Activating Factor Signaling | inflammation and immune viral infection immunity |
| Role of RIG1-like Receptors in | inflammation and immune |
| Antiviral Innate Immunity | viral infection immunity |
| BA and JA (35) | Pathway category |
| Molecular Mechanisms of Cancer | apoptosis and cancer |
| Melatonin Signaling | nervous system |
| FGF Signaling | others |
| Amyotrophic Lateral Sclerosis Signaling | nervous system |
| Neuropathic Pain Signaling in Dorsal Horn Neurons | nervous system |
| Synaptic Long Term Potentiation | nervous system |
| GNRH Signaling | nervous system |
| B Cell Receptor Signaling | inflammation and immune |
| Calcium Signaling | others |
| PPARα/RXRα Activation | others |
| Cardiac Hypertrophy Signaling | others |
| Circadian Rhythm Signaling | nervous system |
| Sonic Hedgehog Signaling | nervous system |
| Amyloid Processing | nervous system |
| Phototransduction Pathway | nervous system |
| Glutamate Receptor Signaling | nervous system |
| IL-10 Signaling | inflammation and immune |
| CD40 Signaling | inflammation and immune |
| Neurotrophin/TRK Signaling | nervous system |
| PXR/RXR Activation | others |
| IL-17 Signaling | inflammation and immune |
| Acute Myeloid Leukemia Signaling | nervous system |
| LPS-stimulated MAPK Signaling | inflammation and immune |
| Nitric Oxide Signaling in the Cardiovascular System | others |
| Dopamine Receptor Signaling | nervous system |
| BMP signaling pathway | others |
| Melanocyte Development and Pigmentation Signaling | nervous system |
| CDK5 Signaling | nervous system |
| IGF-1 Signaling | others |
| α-Adrenergic Signaling | nervous system |
| HMGB1 Signaling | inflammation and immune |
| p38 MAPK Signaling | inflammation and immune |
| Nicotinate and Nicotinamide Metabolism | others |
| Fc Epsilon RI Signaling | inflammation and immune |
| Cholecystokinin/Gastrin-mediated Signaling | others |
| BA and BJ (1) | Pathway category |
| Apoptosis Signaling | apoptosis and cancer |
| JA and BJ (3) | Pathway category |
| cAMP-mediated Signaling | inflammation and immune endocrine and hormone |
| Acute Phase Response Signaling | others |
| ERK/MAPK Signaling | inflammation and immune |
| BA, JA and BJ (4) | Pathway category |
| CD27 Signaling in Lymphocytes | inflammation and immune |
| Role of PKR in Interferon Induction and Antiviral Response | inflammation and immune viral infection immunity |
| Toll-like Receptor Signaling | inflammation and immune |
| IL-6 Signaling | inflammation and immune |
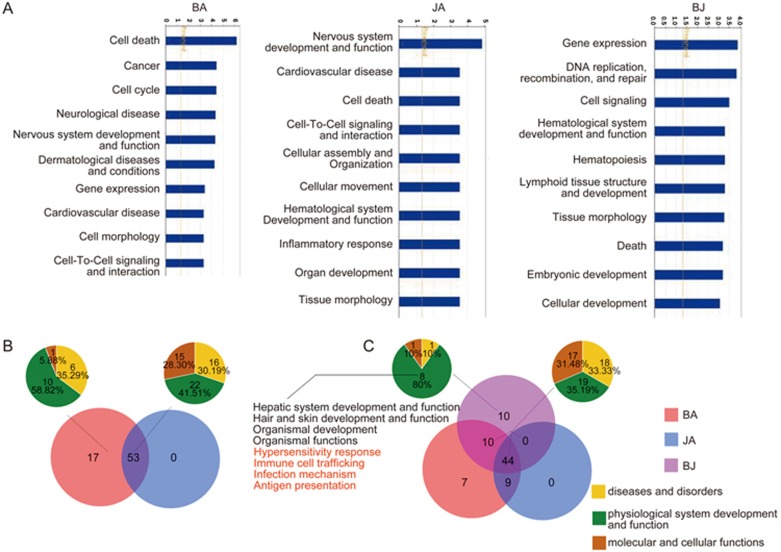
Based on the differentially expressed genes in the BA and JA groups compared to the CM group, IPA also identified 70 and 53 significant biological functions in the BA and JA groups (Supplementary Table S3), respectively. The P-values of the top 10 functions in the BA and JA groups were compared and are listed in Figure 4A. As shown in Figure 4B, all 53 functions in JA group were included in the BA group, accounting for 75.71% of BA functions. These 53 overlapping functions were divided into three categories: 16 (30.19%) were classified as diseases and disorders, 22 (41.51%) as physiological system development and function, and the other 15 (28.30%) as molecular and cellular functions. Additionally, among the 17 non-overlapping functions of the BA group, 10 functions (58.82%) were associated with physiological system development and function, 6 (35.29%) belonged to diseases and disorders, and the remaining 1 (5.88%) was related to molecular and cellular functions (Figure 4B). No unique functions were identified in the JA group.
Figure 4.
Top 10 biological functions and overlapping patterns of biological functions in each group. (A) Top 10 biological functions in each group; (B) overlapping patterns of all biological functions between the BA and JA groups; and (C) overlapping patterns of all biological functions among the BA, JA, and BJ groups. The small pie charts represent the constitutive categories of corresponding sections of biological functions. Yellow, green, and brown represent diseases and disorders, physiological systems development and function, and molecular and cellular functions, respectively. The biological functions in red are related to immunity.
Combination group produced novel variations based on pure mechanism analysis
To investigate the phenotype-related additive effects between BA and JA on improving cerebral ischemia, we also compared the BJ group to the phenotype-negative CM group. According to pure mechanistic analysis, the combination group had novel targets entirely different from those of BA or JA. A total of 11 genes were detected as specific target molecules of BJ (Figure 2A, Supplementary Table S1). Notably, none of these 11 target molecules overlapped with those of the BA or JA groups (Figure 2B). Four networks including these target genes were constructed for the BJ group. The functions of the 4 target networks included gene expression, DNA replication, recombination, and repair, cellular development; gene expression, cell cycle, tissue morphology; cell death, skeletal and muscular disorders, neurological disease; cancer, gastrointestinal disease; and hepatic system disease.
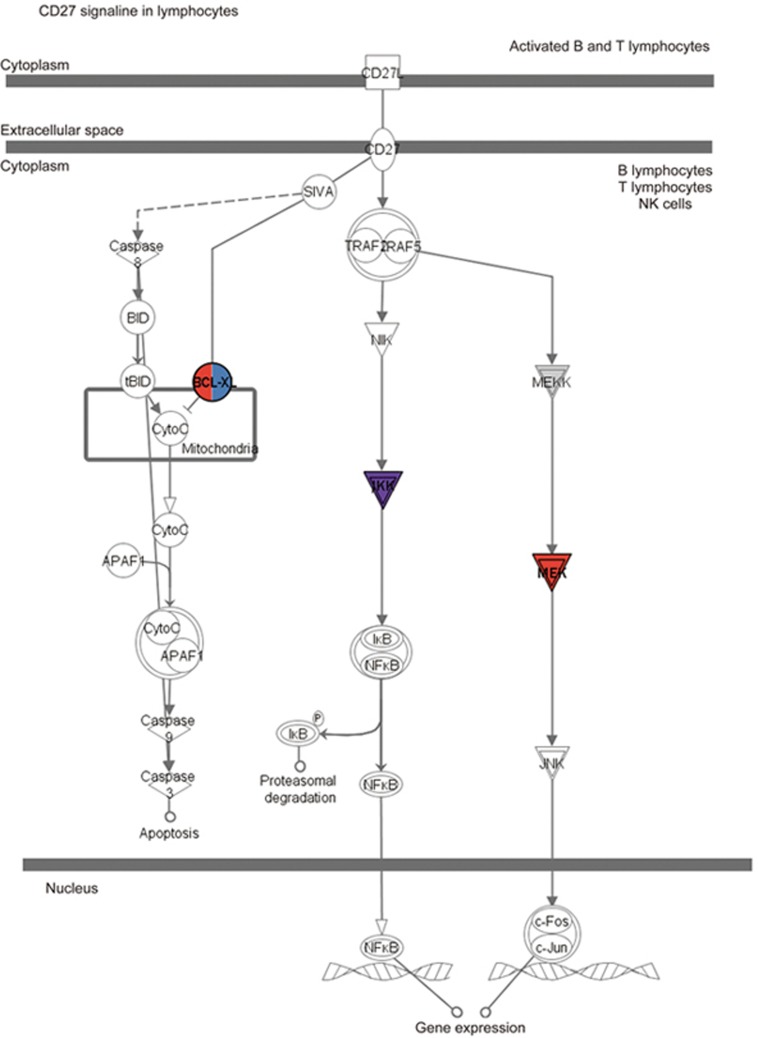
Compared with the CM group, 18 significant pathways were identified in the BJ group (Supplementary Table S2). Among these pathways, more than half (10 pathways, accounting for 55.56%) were included exclusively in the BJ group. Four pathways overlapped with both the BA and JA groups, including CD27 signaling in lymphocytes, the role of PKR in interferon induction and antiviral response, Toll-like receptor signaling, and IL-6 signaling. The target molecules of BA, JA, and BJ in these pathways are shown in Supplementary Table 4, and the CD27 signaling pathway was set as an example to show the target molecules in each group (Figure 5). One overlapping pathway, apoptosis signaling, was found between BA and BJ but not JA, and three overlapping pathways, ie, cAMP-mediated signaling, acute phase response signaling, and ERK/MAPK signaling, were detected between JA and BJ but not BA (Figure 3B). Additionally, we compared the top 20 canonical pathways in the BJ group with those in the BA and JA groups. Only 2 overlapping pathways were detected among the three groups, and the remaining 16 pathways in the BJ group did not overlap with those in either the BA or JA groups (Figure 3C).
Figure 5.
Targets of BA, JA, and BJ in the CD27 signaling pathway in lymphocytes. The target molecules of BA, JA, and BJ are red, blue and purple, respectively.
Based on the differentially expressed genes, IPA also identified 64 significant functions in the BJ group, most of which (54 functions, accounting for 84.38%) overlapped with those in the BA and/or JA groups. Among these 54 overlapping functions, 18 (33.33%) were classified as diseases and disorders, 19 (35.19%) were classified as physiological system development and function, and the other 17 (31.48%) were classified as molecular and cellular functions. Among the 10 non-overlapping functions in the BJ group, the majority (8/10, accounting for 80%) were related to physiological system development and function (Figure 4C). Notably, 4 of these 8 functions were related to immune reaction, including hypersensitivity response, immune cell trafficking, infection mechanism, and antigen presentation.
Discussion
Herbal medicines, especially ingredients extracted from traditional Chinese medicine, feature multiple targets, which constitute target networks (on-modules) of herbal medicines in cellular network rewiring48. In our previous studies, the pharmacological mechanisms of BA, JA, and their combination BJ were explored, and the target pathways of these compounds were partially investigated. However, these target pathways included not only specific mechanisms contributing to phenotype alteration but also invalid system fluctuations that do not contribute to phenotype variations. What deserves more investigation is the specific pharmacological mechanism that actually reverses the pathological process. In our previous studies we proposed that the target pathways of BA, JA and BJ could be classified into three categories, ie, immune-related, neuron-related, and ERK/MAPK signaling cascade-related pathways36. Further investigation by GeneGo MetaCore analyses showed that the specific pathways of BJ mainly involved apoptosis and survival, gamma-secretase, neurophysiological processes, development, reproduction, and regulation of lipid metabolism37. In this study, we compared the networks perturbed by positive-phenotype ingredients to those perturbed by negative-phenotype ingredients to precisely extract the pure pharmacological mechanism of phenotype variation39.
Pure pharmacological analysis provides a clearer outline of target profiles than non-pure analysis
First, the pure target analysis provides us an opportunity to distinguish ineffective targets from targets contributing to phenotype alterations, which may help to narrow the scope of potential action targets. By comparing the target molecules found by pure and non-pure analyses, three molecules, F5, HTR3A, and HTR2C, were identified in the BA, JA or BJ groups in non-pure analysis but not in pure analysis. Thus, we speculate that regulation of the three molecules is not specific to phenotype variation. Noticeably, all three molecules were exactly the target molecules of CM. This also supported the hypothesis that regulation of the 3 molecules might be insufficient to reverse the pathological phenotype. Similar findings were also observed in canonical pathway analysis. For example, in the JA group, the G-protein coupled receptor (GPCR) signaling pathway was identified in non-pure analysis35 but not in pure analysis. The GPCR signaling pathway was also detected in the CM group39. This indicates that the GPCR signaling pathway is not the key pathway leading to phenotype alteration; in other words, only regulation of GPCR signaling is insufficient to alter the pathological phenotype. Therefore, peeling off ineffective target molecules or pathways from “target profiles” may facilitate more precise on-target action of multi-target compounds.
Second, pure target analysis provides an opportunity to discover new therapeutic targets. In this study, pure analysis revealed a set of potential targets that were not identified in non-pure analysis. For example, a total of 5 molecules, namely, ARF1, FGF12, GRIN1, MAP2K6 and PRKAR1B, were detected as specific targets of BA in pure analysis but not in non-pure analysis. More remarkably, all 4 specific target molecules of JA identified by pure analysis were entirely different from those identified by non-pure analysis, and 7 specific target molecules of BJ were identified by pure analysis but not by non-pure analysis. All specific targets can be regarded as potential targets of these compounds for further pharmacological analysis. Additionally, multiple pathways were also identified as potential targets of these compounds. For example, the CD40 signaling and IL-17 signaling pathways, which were not discovered in non-pure analysis35, were identified as potential target pathways of JA in pure pharmacological analysis. These pathways may add to the “target profiles” of these compounds.
Third, the pure target analysis validates target molecules and pathways detected by non-pure analysis. Several molecules overlapped between pure and non-pure analyses, indicating that these molecules contributed not only to differences between perturbed and unperturbed biological systems but also to differences between phenotype-positive and phenotype-negative perturbed systems. As a result, we infer that these molecules definitely account for some phenotypic variations. For instance, both pure and non-pure analyses identified BCL2L1 and IKBKG as the target molecule in BA and BJ groups, respectively. We propose that the two molecules might be part of the primary causes of phenotype alteration. In addition, several pathways, such as ERK/MAPK signaling and GNRH signaling, were identified as specific target pathways in the JA group by both pure and non-pure analyses35. Therefore, we hypothesize that phenotype alteration can be partially attributed to these pathways. In fact, as shown in the diagram of target pathways (Figure 6), the two pathways were in the core position among all pathway cross-talk, which also supported the hypothesis above.
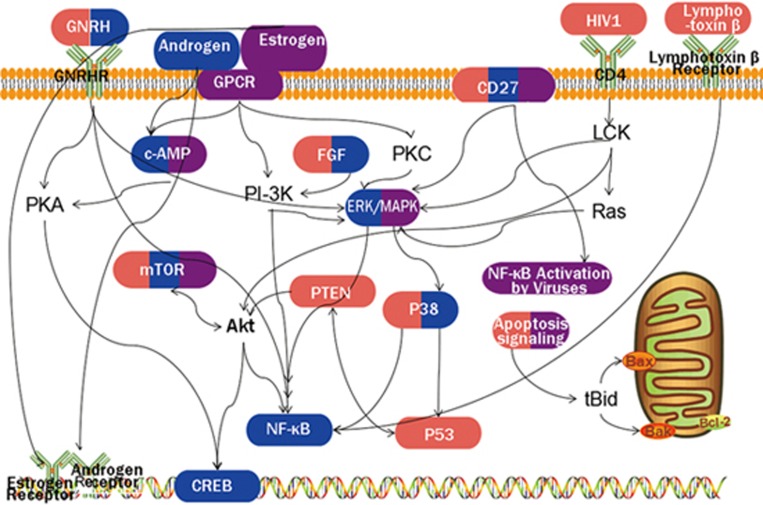
Figure 6.
A diagram of target pathways of BA, JA, and BJ. Pathways activated by BA, JA, and BJ are labeled red, blue and purple, respectively. The solid line represents the association between pathways. It should be noted that the diagram only displays some of the classic pathways as an example to illustrate the pure additive mechanism between BA and JA and does not include all target pathways for these compounds.
Pure pharmacological mechanisms of analogous ingredients: similarities and diversities
It was demonstrated in our previous study37 that BA and JA have many similar molecular mechanisms. The similarities of regulatory mechanisms between BA and JA were more obvious in pure mechanistic analysis. All 4 specific target molecules of JA, ie, ARF1, GRIN1, MAP2K6, and PRKAR1B, overlapped with those of BA (Figure 2). MAP2K6 is a component in the MAPK pathway, which is a stress and inflammation-related pathway49,50, and can be activated by ARF151. GRIN1 is believed to play a key role in neuron function51,52,53. Mutations of GRIN1 or PRKAR1B lead to nervous system disease or neurodegenerative disorder54,55. These overlapping target molecules, which are associated with the nervous system and inflammation, might be the cellular basis of similar pharmacological actions between BA and JA.
Notably, among the 39 overlapping pathways between BA and JA, 16 pathways were associated with development and signal propagation of the nervous system, and 12 pathways were related to inflammation and immune response. For example, suppression of synaptic long-term potentiation can be induced by cerebral ischemia56,57, and up-regulation of sonic hedgehog signaling underlies neurogenesis after cerebral ischemia58. For inflammatory and immune pathways, the Toll-like receptor signaling pathway mediates the inflammatory reaction in cerebral ischemia and is involved in the extension of cerebral infarction and the aggravation of ischemic brain damage59,60, which is also a critical component of the innate immune system that has been shown recently to mediate ischemic injury61. The pathway of CD27 signaling in lymphocytes has an impact on the balance between adaptive responses and immunopathology62, and the role of PKR in interferon induction and antiviral response and Toll-like receptors also affects the innate immune response63,64. Therefore, we infer that the similar molecular mechanisms between BA and JA in treating cerebral ischemia may be attributed to their effects on nervous system as well as inflammation and immune response. Furthermore, all of the biological functions in JA group overlapped with those in BA group, which also demonstrated the similarities between BA and JA.
However, diverse regulatory mechanisms between BA and JA were also observed. Among the 12 non-overlapping canonical pathways of BA, in addition to those related to the nervous system and inflammation response, the remaining 7 pathways were closely related to cancer, including small cell lung cancer signaling, VEGF signaling, bladder cancer signaling, p53 signaling, PTEN signaling, chronic myeloid leukemia signaling, and apoptosis signaling. Based on published reports, the VEGF signaling pathway is suggested to mediate central nervous system involvement in acute lymphoblastic leukemia65, and regulation of VEGF may improve neurobehavioral recovery after cerebral ischemic stroke66. The PTEN signaling pathway is involved in osteosarcoma67, T-cell acute lymphoblastic leukemia68, and glioblastomas69,70; PTEN is also reported to be related to nervous regeneration and repair71,72,73, and promotion of PTEN degradation may prevent hippocampal neuronal loss and memory impairment74. Thus, we speculate that apoptosis and cancer-related signaling pathways are unique targets of BA in treating cerebral ischemia, which are quite different from those of JA.
Among the 20 non-overlapping canonical pathways of JA, 9 pathways, ie, type I diabetes mellitus signaling, renin-angiotensin signaling, corticotropin-releasing hormone signaling, androgen signaling, cardiac β-adrenergic signaling, relaxin signaling, insulin receptor signaling, and cellular effects of sildenafil (Viagra), and germ cell-sertoli cell junction signaling, were associated with endocrine regulation, such as adrenocortical hormone, insulin, and gonadal hormone. Activation of the renin-angiotensin system exaggerates ischemic brain damage75. Androgens are suggested to be an important factor in cerebrovascular disease76,77. Cerebral ischemia induces insulin synthesis and secretion in the pancreas78. Therefore, we infer that endocrine and hormone regulation may be a mechanism of JA in improving cerebral ischemia.
Pure additive mechanisms: combination of paralleled compounds enhances the effect on viral infection immunity
The molecular mechanisms underlying the BJ combination in treating cerebral ischemia were remarkably distinct from those of BA or JA monotherapies. We found that all specific target molecules of BJ were completely different from those of BA or JA. Moreover, a total of 10 canonical pathways were identified exclusively in the BJ group. We noted that 5 of the 10 non-overlapping pathways were related to immune response and signaling, including the role of RIG1-like receptors in antiviral innate immunity, 4-1BB signaling in T lymphocytes, B cell activating factor signaling, NF-κB activation by viruses, and G-protein coupled receptor signaling. It is reported that cerebral ischemia induced interference in the function of the innate and adaptive immune cells, resulting in systemic immunosuppression. This post-stroke immunodeficiency could potentially protect the brain by reducing autoimmune reactions to brain antigens. However, any potential brain protective effect of stroke-induced immunosuppression might be confounded by an increased incidence of infection79,80,81. Regulation of the immune response will contribute to a long-term prognosis of stroke. In our studies, the above mentioned 5 immune response related non-overlapping pathways were related to either innate or adaptive immunity. Notably, these 5 pathways were also suggested to be related to the viral infection-mediated immune response and signaling. For instance, RIG1-like receptors play a critical role in antiviral immunity82, both 4-1BB signaling and B cell activating factor signaling are involved in viral infection processes83,84, and a constitutively active viral G protein-coupled receptor (vGPCR) contributes to the pathogenesis of viral oncogenesis85. This may play a regulatory role in the post-stroke infection pathological process. Thus, we hypothesize that immune response, especially viral infection immunity, may be a specific target mechanism of BJ, which is fairly different from BA or JA. This hypothesis is also supported by the biological functions of BJ: in the 10 total non-overlapping biological functions of BJ, 4 functions (immune cell trafficking, infection mechanism, antigen presentation, hypersensitivity response) were associated with immunity.
The other 5 non-overlapping canonical pathways of BJ were related to apoptosis and cancer, endocrine and hormone systems, and the nervous system (Table 1), similar to those of BA or JA. Therefore, we proposed that the additive effects between BA and JA were generated not only by inheriting the targets of each monotherapy but also by integrating the mechanisms of single ingredients to produce novel pharmacological actions, which are indicated as enhanced effects on viral infection immunity.
Based on pure mechanistic analysis, we concluded that the underlying pharmacological mechanisms of BA and JA in treating cerebral ischemia were similar, and mainly involved the nervous system, inflammation and immune response. In addition, the unique targets of BA were related to apoptosis and cancer-related signaling, while the unique mechanisms of JA were associated with endocrine and hormone regulation. When BA was combined with JA, their impacts on infection immunity were enhanced and additive effects were produced.
Conclusion
In this study, the pure therapeutic mechanisms of BA, JA and BJ in treating cerebral ischemia were explored by comparing differentially expressed microarrays to those with a negative phenotype. Our methodologies for the pure mechanistic analysis of multi-target compounds may contribute substantially to the interpretation of the precise pharmacological actions of multi-target compounds. Further investigation with a larger number of genes and higher precision will be attempted in future studies.
Abbreviation
BA, Baicalin; JA, Jasminoidin; CM, Concha margaritifera; BJ, Baicalin combined with jasminoidin; MCAO, Middle cerebral artery obstruction; IPA, Ingenuity pathway analysis; IS, Ischemic stroke; FDA, U S Food and Drug Administration; HPLC, High-performance liquid chromatography; MMP, Matrix metalloproteinase; CaMKII, Calmodulin-dependent protein kinase II; TLR, Toll-like receptors; NF-κB, Nuclear factor-κB; STKE, Science signal transduction knowledge environment; PKR, Protein kinase R; IL, Interleukin; cAMP, Cyclic adenosine monophosphate; ERK, Extracellular signal-regulated kinase; MAPK, Mitogen-activated protein kinases; GPCR, G-protein coupled receptor; GnRH, Gonadotropin releasing; VEGF, Vascular endothelial growth factor; PTEN, Phosphatase and tensin homolog deleted on chromosome ten; HIV, Human Immunodeficiency Virus; mTOR, Mammalian target of rapamycin; FGF, Fibroblast growth factor; CREB, cAMP-response element binding protein.
Author contribution
Zhong WANG designed and directed the research; Peng-qian WANG performed the research and drafted the manuscript; Jun LIU, Qiong LIU, Ying-ying ZHANG, Wen-juan XU, and Bing LI performed the histological experiment and revised the manuscript; Ya-nan YU analyzed the experimental data.
Acknowledgments
This study was supported by the 9th autonomously selected subject projects of the China Academy of Chinese Medical Sciences (No Z0405).
Footnotes
Supplementary files are available at the website of Acta Pharmacologica Sinica.
Supplementary Information
Specific target molecules in BA, JA and BJ groups.
Canonical pathways and corresponding P-values in each group.
Biological functions and corresponding P-values in each group.
Target molecules of BA, JA, BJ in the 4 overlapped pathways.
References
- Feigin VL, Roth GA, Naghavi M, Parmar P, Krishnamurthi R, Chugh S, et al. Global burden of stroke and risk factors in 188 countries, during 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet Neurol 2016; 15: 913–24. [DOI] [PubMed] [Google Scholar]
- Krishnamurthi RV, Moran AE, Feigin VL, Barkercollo S, Norrving B, Mensah GA, et al. Stroke prevalence, mortality and disability-adjusted life years in adults aged 20-64 years in 1990-2013: data from the global burden of disease 2013 study. Neuroepidemiology 2015; 45: 190–202. [DOI] [PubMed] [Google Scholar]
- Jr AH, Del ZG, Alberts MJ, Bhatt DL, Brass L, Furlan A, et al. Guidelines for the early management of adults with ischemic stroke: a guideline from the american heart association/american stroke association stroke council, clinical cardiology council, cardiovascular radiology and intervention council, and the atherosc. Stroke 2007; 115: 1056–83. [Google Scholar]
- Matthew T, Martin F, Holliday EG, Cathie S, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 2012; 11: 951–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov 2004; 3: 353–9. [DOI] [PubMed] [Google Scholar]
- Frantz S. Drug discovery: playing dirty. Nature 2005; 437: 942–3. [DOI] [PubMed] [Google Scholar]
- Hopkins AL. Drug discovery: Predicting promiscuity. Nature 2009; 462: 167–8. [DOI] [PubMed] [Google Scholar]
- Gu J, Gui Y, Chen L, Yuan G, Lu HZ, Xu X. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS One 2013; 8: e62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu J, Cheng Y, Wang Y. Fangjiomics: in search of effective and safe combination therapies. J Clin Pharmacol 2011; 51: 1132–51. [DOI] [PubMed] [Google Scholar]
- Duan DD, Wang Z, Zhang BL, Wang YY. Fangjiomics: revealing adaptive omics pharmacological mechanisms of the myriad combination therapies to achieve personalized medicine. Acta Pharmacol Sin 2015; 36: 651–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Z. Diverse array-designed modes of combination therapies in Fangjiomics. Acta Pharmacol Sin 2015; 36: 680–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Duan DD, Wang YY. Editorial: Combination therapy of vascular diseases and Fangjiomics: when west meets east in the era of phenomics. Curr Vasc Pharmacol 2015; 13: 420–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Zhang Q, Ung CO, Wang Y, Han Y, Hu Y, et al. An analysis of chemical ingredients network of chinese herbal formulae for the treatment of coronary heart disease. PLoS One 2015; 10: e0116441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhu F, Ma X, Cao Z, Li Y, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov 2009; 8: 111–28. [DOI] [PubMed] [Google Scholar]
- Li P, Chen J, Zhang W, Fu B, Wang W. Transcriptome inference and systems approaches to polypharmacology and drug discovery in herbal medicine. J Ethnopharmacol 2017; 195: 127. [DOI] [PubMed] [Google Scholar]
- Gu P, Chen H. Modern bioinforma meets traditional chinese medicine. Brief Bioinform 2014; 15: 984–1003. [DOI] [PubMed] [Google Scholar]
- Huang C, Zheng C, Li Y, Wang Y, Lu A, Yang L. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Briefings Bioinformatics 2014; 15: 710. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang X, Zhang B. Qingkailing injection for the treatment of acute stroke: a systematic review and meta-analysis. J Tradit Chin Med 2014; 34: 131–9 [DOI] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of People's Republic of China. Beijing: Chinese Medical Science and Technology Press; 2010. p 1529–1531.
- Zhang Z, Wu R, Li P, Liu F, Zhang W, Zhang P, et al. Baicalin administration is effective in positive regulation of twenty-four ischemia/reperfusion-related proteins identified by a proteomic study. Neurochem Int 2009; 54: 488–96. [DOI] [PubMed] [Google Scholar]
- Tu XK, Yang WZ, Liang RS, Shi SS, Chen JP, Chen CM, et al. Effect of baicalin on matrix metalloproteinase-9 expression and blood-brain barrier permeability following focal cerebral ischemia in rats. Neurochem Res 2011; 36: 2022–8. [DOI] [PubMed] [Google Scholar]
- Wang P, Cao Y, Yu J, Liu R, Bai B, Qi H, et al. Baicalin alleviates ischemia-induced memory impairment by inhibiting the phosphorylation of CaMKII in hippocampus. Brain Res 2016; 1642: 95–103. [DOI] [PubMed] [Google Scholar]
- Gaire BP, Moon SK, Kim H. Scutellaria baicalensis in stroke management: nature's blessing in traditional Eastern medicine. Chin J Integr Med 2014; 20: 712–20. [DOI] [PubMed] [Google Scholar]
- Tu XK, Yang WZ, Shi SS, Chen Y, Wang CH, Chen CM, et al. Baicalin inhibits TLR2/4 signaling pathway in rat brain following permanent cerebral ischemia. Inflammation 2011; 34: 463–70. [DOI] [PubMed] [Google Scholar]
- Li HY, Yuan ZY, Wang YG, Wan HJ, Hu J, Chai YS, et al. Role of baicalin in regulating Toll-like receptor 2/4 after ischemic neuronal injury. Chin Med J (Engl) 2012; 125: 1586–93. [PubMed] [Google Scholar]
- Tu XK, Yang WZ, Shi SS, Wang CH, Chen CM. Neuroprotective effect of baicalin in a rat model of permanent focal cerebral ischemia. Neurochem Res 2009; 34: 1626–34. [DOI] [PubMed] [Google Scholar]
- Xu M, Chen X, Gu Y, Peng T, Yang D, Chang RC, et al. Baicalin can scavenge peroxynitrite and ameliorate endogenous peroxynitrite-mediated neurotoxicity in cerebral ischemia-reperfusion injury. J Ethnopharmacol 2013; 150: 116–24. [DOI] [PubMed] [Google Scholar]
- Xue X, Qu XJ, Yang Y, Sheng XH, Cheng F, Jiang EN, et al. Baicalin attenuates focal cerebral ischemic reperfusion injury through inhibition of nuclear factor κB p65 activation. Biochem Biophys Res Commun 2010; 403: 398–404. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Li L, Hölscher C. Neuroprotective effects of geniposide in the MPTP mouse model of Parkinson's disease. Eur J Pharmacol 2015; 768: 21–7. [DOI] [PubMed] [Google Scholar]
- Zhao C, Lv C, Li H, Du S, Liu X, Li Z, et al. Geniposide protects primary cortical neurons against oligomeric Aβ1-42-induced neurotoxicity through a mitochondrial pathway. PLoS One 2016; 11: e0152551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li G, Hölscher C, Li L. Neuroprotective effects of geniposide on Alzheimer's disease pathology. Rev Neurosci 2015; 26: 371–83. [DOI] [PubMed] [Google Scholar]
- Lv C, Wang L, Liu X, Yan S, Yan SS, Wang Y, et al. Multi-faced neuroprotective effects of geniposide depending on the RAGE-mediated signaling in an Alzheimer mouse model. Neuropharmacology 2015; 89: 175–84. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li P, Wang Y, Liu J, Zhang Z, Cheng W, et al. Ameliorative effects of a combination of baicalin,jasminoidin and cholic acid on ibotenic acid-induced dementia model in rats. PLoS One 2013; 8: e56658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li P, Wang Y, Liu J, Zhang Z, Cheng W, et al. The components of Huang-Lian-Jie-Du-Decoction act synergistically to exert protective effects in a rat ischemic stroke model. PLoS One 2013; 8: e56658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Li P, Wang Z, Li PT, Zhang WS, Sun ZH, et al. A comparative study on the individual and combined effects of baicalin and jasminoidin on focal cerebral ischemia-reperfusion injury. Brain Res 1123: 188–95. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Li HX, Chen YY, Fang H, Yu YN, Liu J, et al. Convergent and divergent pathways decoding hierarchical additive mechanisms in treating cerebral ischemia–reperfusion injury. CNS Neurosci Ther 2014; 20: 253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yu Y, Zhang Y, Liu J, Li H, Dang H, et al. Vertical and horizontal convergences of targeting pathways in combination therapy with baicalin and jasminoidin for cerebral ischemia. CNS Neurol Disord Drug Targets 2016; 15: 740–50. [DOI] [PubMed] [Google Scholar]
- Guo L, Meng FY, Zhang GD, Zhao J, Zhang ZJ, Zhou CX, et al. Baicalin and jasminoidin effects on gene expression and compatibility in the hippocampus following focal cerebral ischemia. Neural Regen Res 2011; 6: 165–170. [Google Scholar]
- Wang PQ, Li B, Liu J, Zhang YY, Yu YN, Zhang XX, et al. Phenotype-dependent alteration of pathways and networks reveals a pure synergistic mechanism for compounds treating mouse cerebral ischemia. Acta Pharmacol Sin 2015; 36: 734–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (US) Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. Guide for the Care & Use of Laboratory Animals 1996, 103, 1072–3. [Google Scholar]
- Liu J, Zhou CX, Zhang ZJ, Wang LY, Jing ZW, Wang Z. Synergistic mechanism of gene expression and pathways between jasminoidin and ursodeoxycholic acid in treating focal cerebral ischemia-reperfusion injury. CNS Neurosci Ther 2012; 18: 674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jing ZW, Zhou CX, Zhang L, Cheng J, Zhang ZJ, et al. Fusion of core pathways reveals a horizontal synergistic mechanism underlying combination therapy. Eur J Pharmacol 2011; 667: 278–86. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang ZJ, Zhou CX, Wang Y, Cheng YY, Duan DY, et al. Outcome-dependent global similarity analysis of imbalanced core signaling pathways in ischemic mouse hippocampus. CNS Neurol Disord Drug Targets 2012; 11: 1070–82. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhou C, Yu Y, Liu J, Jing Z, Lv A, et al. Variations in target gene expression and pathway profiles in the mouse hippocampus following treatment with different effective compounds for ischemia-reperfusion injury. Naunyn Schmiedebergs Arch Pharmacol 201; 385: 797–806. [DOI] [PubMed] [Google Scholar]
- Chen Y, Meng F, Fang H, Yu Y, Liu J, Jing Z, et al. Hierarchical profiles of signaling pathways and networks reveal two complementary pharmacological mechanisms. CNS Neurol Disord Drug Targets 2013; 12: 882–93. [DOI] [PubMed] [Google Scholar]
- Wang Z, Du Q, Wang F, Liu Z, Li B, Wang A, et al. Microarray analysis of gene expression on herbal glycoside recipes improving deficient ability of spatial learning memory in ischemic mice. J Neurochem 2004; 88: 1406–15. [DOI] [PubMed] [Google Scholar]
- Zhang ZJ, Wang Z, Zhang XY, Ying K, Liu JX, Wang YY. Gene expression profile induced by oral administration of baicalin and gardenin after focal brain ischemia in rats. Acta Pharmacol Sin 2005; 26: 307–14. [DOI] [PubMed] [Google Scholar]
- Li B, Liu J, Zhang YY, Wang PQ, Yu YN, Kang RX, et al. Quantitative identification of compounds-dependent on-modules and differential allosteric modules from homologous ischemic networks. CPT Pharmacometrics Syst Pharmacol 2016; 5: 575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liang W, Li J, Long J. Knockdown of FSTL1 inhibits oxLDL-induced inflammation responses through the TLR4/MyD88/NF-κB and MAPK pathway. Biochem Biophys Res Commun 2016; 478: 1528–33. [DOI] [PubMed] [Google Scholar]
- Ma JQ, Li Z, Xie WR, Liu CM, Liu SS. Quercetin protects mouse liver against CCl4-induced inflammation by the TLR2/4 and MAPK/NF-κB pathway. Int Immunopharmacol 2015; 28: 531–9. [DOI] [PubMed] [Google Scholar]
- Zhou F, Dong C, Davis JE, Wu WH, Surrao K. Wu G.The mechanism and function of mitogen-activated protein kinase activation by ARF1. Cell Signal 2015; 27: 2035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafford G, Jasnow AM, Ressler KJ. Grin1 receptor deletion within CRF neurons enhances fear memory. PLoS One 2014; 9: e111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemori J, Takao K, Koshimizu H, Hattori S, Furuse T, Wakana S, et al. ENU-mutagenesis mice with a non-synonymous mutation in Grin1 exhibit abnormal anxiety-like behaviors, impaired fear memory, and decreased acoustic startle response. BMC Res Notes 2013; 6: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Shiina M, Tohyama J, Haginoya K, Lerman-Sagie T, Okamoto N, et al. GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia 2015; 56: 841–8. [DOI] [PubMed] [Google Scholar]
- Wong TH, Chiu WZ, Breedveld GJ, Li KW, Verkerk AJ, Hondius D, et al. PRKAR1B mutation associated with a new neurodegenerative disorder with unique pathology. Brain 2014; 137: 1361–73. [DOI] [PubMed] [Google Scholar]
- Ding J, Fu G, Zhao Y, Cheng Z, Chen Y, Zhao B, et al. EGCG ameliorates the suppression of long-term potentiation induced by ischemia at the Schaffer collateral-CA1 synapse in the rat. Cell Mol Neurobiol 2012; 32: 267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Chen L, Sokabe M. Neurosteroid estradiol rescues ischemia-induced deficit in the long-term potentiation of rat hippocampal CA1 neurons. Neuropharmacology 2007; 52: 1124–38. [DOI] [PubMed] [Google Scholar]
- Spaccapelo L, Galantucci M, Neri L, Contri M, Pizzala R, D'Amico R, et al. Up-regulation of the canonical Wnt-3A and Sonic hedgehog signaling underlies melanocortin-induced neurogenesis after cerebral ischemia. Eur J Pharmacol 2013; 707: 78–86. [DOI] [PubMed] [Google Scholar]
- Tu XK, Yang WZ, Chen JP, Chen Y, Ouyang LQ, Xu YC, et al. Curcumin inhibits TLR2/4-NF-κB signaling pathway and attenuates brain damage in permanent focal cerebral ischemia in rats. Inflammation 2014; 37: 1544–51. [DOI] [PubMed] [Google Scholar]
- Famakin B, Mou Y, Spatz M, Lawal M, Hallenbeck J. Downstream Toll-like receptor signaling mediates adaptor-specific cytokine expression following focal cerebral ischemia. J Neuroinflammation 2012; 9: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh BJ, Williams-Karnesky RL, Stenzel-Poore MP. Toll-like receptor signaling in endogenous neuroprotection and stroke. Neuroscience 2009; 158: 1007–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte MA, van Olffen RW, van Gisbergen KP, van Lier RA. Timing and tuning of CD27-CD70 interactions: the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol Rev 2009; 229: 216–31. [DOI] [PubMed] [Google Scholar]
- Wei X, Li XZ, Zheng X, Jia P, Wang J, Yang X, et al. Toll-like receptors and interferon associated immune factors responses to spring viraemia of carp virus infection in common carp (Cyprinus carpio). Fish Shellfish Immunol 2016; 55: 568–76. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xiang W, Wang G, Yan Z, Zhu Z, Guo Z, et al. Interferon β (IFN-β) production during the double-stranded RNA (dsRNA) response in hepatocytes involves coordinated and feed forward signaling through Toll-like receptor 3 (TLR3), RNA-dependent protein kinase (PKR), inducible nitric oxide synthase (iNOS), and Src protein. J Biol Chem 2016; 291: 15093–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch V, Trentin L, Herzig J, Demir S, Seyfried F, Kraus JM, et al. Central nervous system involvement in acute lymphoblastic leukemia is mediated by vascular endothelial growth factor. Blood 2017; 03: 769315. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhu YM, Li YN, Li PY, Wang D, Liu Y, et al. The 15-LO-1/15-HETE system promotes angiogenesis by upregulating VEGF in ischemic brains. Neurol Res 2017; 39: 795–802. [DOI] [PubMed] [Google Scholar]
- Zheng D, Wu W, Dong N, Jiang X, Xu J, Zhan X, et al. Mxd1 mediates hypoxia-induced cisplatin resistance in osteosarcoma cells by repression of thePTEN tumor suppressor gene. Mol Carcinog 2017. doi: 10.1002/mc.22676. [DOI] [PubMed]
- Tesio M, Trinquand A, Ballerini P, Hypolite G, Lhermitte L, Petit A, et al. Age-related clinical and biological features of PTEN abnormalities in T-cell acute lymphoblastic leukaemia. Leukemia 2017. doi: 10.1038/leu.2017.157. [DOI] [PubMed]
- Park SH, Won J, Kim SI, Lee Y, Park CK, Kim SK, et al. Molecular Testing of Brain Tumor. J Pathol Transl Med 2017; 51: 205–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Bush EC, Toonen JA, Ma Y, Solga AC, Sims PA, et al. Whole tumor RNA-sequencing and deconvolution reveal a clinically-prognostic PTEN/PI3K-regulated glioma transcriptional signature. Oncotarget 2017. doi:10.18632/oncotarget.17193. [DOI] [PMC free article] [PubMed]
- Huang Z, Hu Z, Xie P, Liu Q. Tyrosine-mutated AAV2-mediated shRNA silencing of PTEN promotes axon regeneration of adult optic nerve. PLoS One 2017; 12: e0174096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino M, Ichimura M, Nakano N, Minami A, Kitagishi Y, Matsuda S. Roles of PTEN with DNA repair in Parkinson's disease. Int J Mol Sci 2016; 17: E954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y, Chen M, Zhou L. MiR-17 targets PTEN and facilitates glial scar formation after spinal cord injuries via the PI3K/Akt/mTOR pathway. Brain Res Bull 2017; 128: 68–75. [DOI] [PubMed] [Google Scholar]
- Chen X, Du YM, Xu F, Liu D, Wang YL. Propofol prevents hippocampal neuronal loss and memory impairment in cerebral ischemia injury through promoting pten degradation. J Mol Neurosci 2016; 60: 63–70. [DOI] [PubMed] [Google Scholar]
- Inaba S, Iwai M, Tomono Y, Senba I, Furuno M, Kanno H, et al. Exaggeration of focal cerebral ischemia in transgenic mice carrying human renin and human angiotensinogen genes. Stroke 2009; 40: 597–603. [DOI] [PubMed] [Google Scholar]
- Uchida M, Palmateer JM, Herson PS, DeVries AC, Cheng J, Hurn PD. Dose-dependent effects of androgens on outcome after focal cerebral ischemia in adult male mice. J Cereb Blood Flow Metab 2009; 29: 1454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillinan N, Deng G, Grewal H, Herson PS. Androgens and stroke: good, bad or indifferent? Exp Neurol 2014; 259: 10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi K, Harada S, Tokuyama S. Pancreatic changes in nerve growth factor/TRKA associated with insulin secretion in cerebral ischemia. Biol Pharm Bull 2015; 38: 1747–51. [DOI] [PubMed] [Google Scholar]
- Miró-Mur F, Urra X, Gallizioli M, Chamorro A, Planas AM. Antigen presentation after stroke. Neurotherapeutics 2016; 13: 719–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelgesang A, Becker KJ, Dressel A. Immunological consequences of ischemic stroke. Acta Neurol Scand 2014; 129: 1–12. [DOI] [PubMed] [Google Scholar]
- Esmaeili A, Dadkhahfar S, Fadakar K, Rezaei N. Post-stroke immunodeficiency: effects of sensitization and tolerization to brain antigens. Int Rev Immunol 2012; 31: 396–409. [DOI] [PubMed] [Google Scholar]
- Tsutsui-Takeuchi M, Ushio H, Fukuda M, Yamada T, Niyonsaba F, Okumura K, et al. Roles of retinoic acid-inducible gene-I-like receptors (RLRs), Toll-like receptor (TLR) 3 and 2'-5' oligoadenylate synthetase as viral recognition receptors on human mast cells in response to viral infection. Immunol Res 2015; 61: 240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezys V, Penaloza-MacMaster P, Barber DL, Ha SJ, Konieczny B, Freeman GJ, et al. 4-1BB signaling synergizes with programmed death ligand 1 blockade to augment CD8 T cell responses during chronic viral infection. J Immunol 2011; 187: 1634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HC, Huang J, Khairnar V, Duhan V, Pandyra AA, Grusdat M, et al. Deficiency of the B cell-activating factor receptor results in limited CD169+ macrophage function during viral infection. J Virol 2015; 89: 4748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Cheng A, Zou Z, Yang YS. Sumpter RM Jr, Huang CL, et al. Endolysosomal trafficking of viral G protein-coupled receptor functions in innate immunity and control of viral oncogenesis. Proc Natl Acad Sci U S A 2016; 113: 2994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specific target molecules in BA, JA and BJ groups.
Canonical pathways and corresponding P-values in each group.
Biological functions and corresponding P-values in each group.
Target molecules of BA, JA, BJ in the 4 overlapped pathways.