Abstract
Cervical cancer (CC) is the second most common type of cancer affecting the female population. The development of CC takes several years, and involves a precancerous stage known as cervical intraepithelial neoplasia (CIN). A key factor in the development of disease is the human papillomavirus (HPV) infection, which initiates carcinogenesis. Furthermore, CC is also impacted by epigenetic changes such as DNA methylation, which causes activation or exclusion of certain genes, and the hypermethylation of cytosines in promoters, thereby switching off previously active genes. The majority of DNA methylation events occur at cytosine-guanine nucleotides, which in the human genome are known as CpG islands. The aim of the present study was to investigate the methylation levels in intronic sequences of the two tumor suppressor genes cell adhesion molecule 1 (CADM1) and T-lymphocyte maturation associated protein (MAL) using cytological samples and to identify potential biomarkers involved in CIN by pyrosequencing. DNA was isolated from cervical smears from patients with CINs, with healthy patients serving as a control group. Samples were converted by treatment with sodium bisulfite and subsequent pyrosequencing to detect the methylation status of the selected genes. The presence of HPV DNA infection analyzed by the polymerase chain reaction, was detected in each sample. Of the total number of samples (n=91), the present study confirmed the presence of one or two high-risk subtypes of HPV in 39 cases (42.85%) and HPV infection was significantly associated with CIN2+ lesions. For the two genes (MAL and CADM1) the present study confirmed that the median methylation was significantly higher in HPV positive patients [P=0.0097, 95% confidence interval (CI): (−0.030, −0.003)/P=0.0024, 95% CI: (−0.06, −0.01)] when compared with patients negative for HPV DNA infection, and the average methylation was demonstrated to be increased with the degree of cervical lesion. The present study used logistical regression to model the dependence between the case/control statuses (control group vs. Dg. 1–4). The area under the curve values for MAL were: 84% for cervical inflammation, 71% for CIN1, 73.4% for CIN2+ and 77% for squamous cell carcinoma (SCC); and for CADM1 were: 88.6% for cervical inflammation, 68% for CIN1, 80% for CIN2+ and 89% for SCC. The present study confirmed that there were statistically significant differences between the methylation levels of individual CpGs and significantly higher median methylation in patients positive for HPV16/18. CADM1 exhibited higher levels of methylation in almost every study group when compared with MAL during the transition of CIN and appeared to be a promising biomarker for future study.
Keywords: cervical cancer, pyrosequencing, bio-markers, DNA methylation, human papillomavirus infection
Introduction
Cervical cancer (CC) is the second most common type of cancer affecting the female population. The World Health Organization (WHO) and GLOBOCAN 2012 have recorded ~528,000 new cases of CC globally (1). The development of CC takes place over several years, and involves a precancerous stage known as cervical intraepithelial neoplasia (CIN), which is divided into three main stages (CIN1, CIN2, CIN3). The primary cause of this disease is infection by human papillomavirus (HPV). At present, over 100 types of HPV have been identified, of which the most prevalent types are HPV16/18, which are responsible for cervical carcinogenesis (2). According to the latest data the most common HPV virus identified in high-grade lesions is HPV16 (47.4%) followed by HPV31 (12.4%), HPV33 (7.1%) and HPV18 (7.1%) (3). The HPV genome is divided into three main regions; the long control region (LCR), which is composed of six open reading frames (ORFs); and the ‘late region’, with two ORFs coding for the viral structural proteins L1 and L2 (4). The major mechanism that engages HPV16/18 in cervical carcinogenesis is the manifestation of two early viral genes: E6 and E7 (known as viral oncogenes). E6 protein binds to the tumor suppressor gene p53 and causes its degradation while E7 inactivates the pRb gene. These mechanisms cause disruptions of cell cycle regulation (5).
Cytological screening has reduced the number of CC-associated mortalities; however, CC is still among the most prevalent oncological diseases, as many women underestimate the importance of regular gynecological examinations.
At present, classical cytology-based screening is being replaced in favor of diagnosing patients with high-risk HPV types, studies are also focused on additional co-factors, such as epigenetics and the search for suitable bio-markers identified via a triage test, PAP smear or HPV test. Epigenetic changes are stable alterations of gene expression without alteration in the DNA sequence itself, and can cause disease even in the absence of a mutation in the gene (6). These changes also include DNA methylation, which is characterized as a covalent chemical modification by the addition of a methylated group to the fifth carbon of the cytosine ring (5mC), thereby preventing access to proteins. DNA methylation is a typical mammalian cellular process and is one of the most well-established markers that defines a molecular landscape that is altered in cancer. Cytosine methylation in vertebrates is found primarily at CG dinucleotides (CpGs). CpGs are usually unmethylated in normal cells, while sporadic CpG sites in the other parts of the genome are methylated. This is associated with gene silencing. Aberrant DNA methylation occurs in the majority of types of cancer, and cause the silencing of some tumor-suppressor genes (TSG) leading to tumor cell growth (7). CpGs hypermethylation is not randomly distributed in carcinogenesis, and therefore may provide a useful signature for tumor diagnosis and prognosis (8).
Following this, several biomarkers (CADM1, DAPK1, CDH1, EPB41L3, FAM19A4, MAL, PAX1, TERT, PRDM14) have been studied which describes the hypermethylation that affect the expression of these genes, thus inducing or accelerating the carcinogenic mechanism, e.g., by deregulation of TSG (9). The present study focused on determining the mean intron region methylation of the two TSGs. They are known as T-lymphocyte maturation associated protein (MAL) and cell adhesion molecule 1 (CADM1).
CADM1 was first observed in patients with non-small cell lung carcinoma (NSCLC) and mapped on chromosome 11q23 (10). This gene was proven to suppress tumor growth through anti-proliferative and pro-apoptotic activity, and the loss of CADM1 expression leads to tumor formation and metastasis (11,12). Hypermethylation of CADM1 is one of the principal causes of gene silencing (13) and was also detected in 40% cases of lung cancer, 32% of prostate cancer cases, 27% of pancreatic adenocarcinoma cases and 83% of cervical carcinoma cases (14,15). The MAL gene is located in the 2q11.1 region and the hypermethylation of its promoter region diminishes its tumor suppressor activity. In 90% of samples from spinocellular carcinoma patients and 93% of samples from patients with diagnosed adenocarcinoma, hypermethylation of certain areas of this TSG has been demonstrated (16). In several studies, DNA methylation of the CADM1/MAL gene promoter has been shown to increase with the severity of cervical disorder (17–19) and that these epigenetic changes point to the presence of more severe high-grade dysplasias (CIN2+). The levels of average methylation in TSG is extremely high in cervical carcinomas and significantly increases in CIN3 lesions in women with high risk (HR)-HPV infection (20).
The aim of the present study was to investigate the methylation levels of CADM1 and MAL using cytological smears by quantitative pyrosequencing analysis. The focus was on specific intronic regions have not been described in much detail previously, in order to examine the significance of the methylation levels of individual CpGs in CADM1 and MAL in HPV16/18 positive patients and also in samples of patients with cervical inflammation.
Materials and methods
Specimens and study population
Cervical specimens were obtained from 91 female patients in collaboration with the Department of Obstetrics and Gynecology at Jesenius Faculty of Medicine in Martin (Martin, Slovakia) and the Department of Molecular Biology and Division of Oncology (Biomedical Center Martin JFM CU, Slovakia). Cytological smears from the uterine cervix were collected and stored in a LBC/APTIMA transport fluid medium. The present study was approved by the Ethical Committee of Jessenius Faculty of Medicine in Martin and all patients provided written informed consent. The patient's clinical protocols were reviewed for clinical data, diagnosis and age of patients. The smears were divided into two groups (1st group: divided into 4 subgroups according to dysplasia, 2nd group: controls) according to the diagnosis of the patients. 1st group included patients who had been operated or diagnosed with cervical carcinoma or had visited an outpatient clinic for due to the presence or deterioration of a cervical lesion. Based on the condition and progression of cervical neoplasia, the following four subgroups were established diagnosis group (Dg).1: cervical inflammation without dysplasia, 20 samples, Dg.2: CIN1, 14 samples, Dg.3: 19 samples of CIN2+ [CIN2, CIN3, carcinoma in situ (CIS) included] and Dg.4, 7 patients with squamous cell carcinoma (SCC). Clinical diagnosis of individual samples were confirmed by histological examination. As the 2nd group: controls (Dg. 0), 31 samples were collected from patients without uterine cervical lesions, with normal onco-cytology outcome and who had not previously received cervical surgery. A total of 20 inflammation samples, 40 CIN1-CIN3/CIS or carcinomas and 31 control samples were collected. The highest median age (48.7 years) was in the SCC group (range, 32–68 years) and 41.5 years in patients with cervical inflammation (range, 22–64 years). The lowest median age was in the CIN2+ group (32 years) with range 21–65 years, followed by CIN1 (33 years) with a range of 19–67 years. The control group only has a slightly higher median age (38 years) than the CINs (range 23–75 years (Table I). In this case we did not confirm any statistical significance and therefore the differences in the median age between groups did not affect the results.
Table I.
Median age of the patients and positivity rates for HPV16/18 in the cervical scrapes in regard to their most severe underlying histological diagnosis.
| Dg. | Histology | Total n | Median age (range), years | HPV 16/18 positive, n (%) |
|---|---|---|---|---|
| 0 | Control | 31 | 38 (23–75) | 6 (19) |
| 1 | Inflammation | 20 | 41.5 (22–64) | 7 (35) |
| 2 | CIN 1 | 14 | 33 (19–67) | 6 (42.8) |
| 3 | CIN 2+ | 19 | 32 (21–65) | 13 (68.4) |
| 4 | SCC | 7 | 48.7 (32–68) | 7 (100) |
Dg, diagnosis group; Dg. 0, controls; Dg.1, inflammation samples; Dg.2, CIN1; Dg.3, CIN2+; Dg.4, SCC; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1; HPV, human papillomavirus.
DNA extraction and bisulfite conversion
Cervical cells (from all groups) yielded from swab smears were stored in LBC/APTIMA vials, and nucleic acids were extracted using a kit, subject to the type of medium in which the samples were stored. The first type contained dissolved cervical cells in the LBC medium, and cells were isolated by the DNase Blood and Tissue kit (Qiagen GmbH, Hilden, Germany). For the second type of DNA extraction a commercially available MasterPure™ Complete DNA and RNA Purification kit (Epicentre; Illumina, Inc., San Diego, CA, USA) was used, for the cells contained in 500 µl of APTIMA transport medium. The concentration and quality of the sample was determined by measuring the sample purity in a UV spectrophotometer, and loaded onto 1.5% agarose gel. Subsequently, 1–2 µg of DNA was bisulfite-treated using an EpiTect Bisulfite kit (Qiagen, Inc., Valencia, CA, USA). Up to 2 µg of DNA were used in a total reaction volume of 20 µl; the total amount of bisulfite reaction was 140 µl (also containing 85 µl bisulfite mix and 35 µl of DNA Protect Buffer). The exact protocol for this reaction was described previously (21). Following bisulfite treatment genomic DNA was stored at −20°C until polymerase chain reaction (PCR) analysis.
HPV DNA and detection
A PCR reaction was used to diagnose the most common HPV genotypes (HPV16 and 18) by using primers that have been described and published in previous literature (22). The primers were designed according to the sequences from the PGMY09/11 primer set (23). The resulting sequence and the optimization of the PCR conditions were described previously (23).
CpG assays and analysis of selected regions of CADM1 and MAL
For the amplification of bisulfite-converted DNA a PyroMark PCR kit (Qiagen, Inc.) was used. The total PCR reaction volume was 25 µl. For analysis of selected regions of CADM1 (3 CpG) and MAL (4 CpG) genes, commercially available CpG assays [PyroMark CpG Assay (200) Hs_CADM1_01_PM (978746, PM00049686), PyroMark CpG Assay (200) Hs_MAL_01_PM (978746, PM00011935, Qiagen GmbH)] (21) were used and with the exact sequence region for analysis (Figs. 1 and 2). Briefly, the obtained PCR products were stored overnight in the refrigerator (+4°C), and 5 µl of the product was removed for electrophoretic analysis (1.75% agarose gel with ethidium bromide staining). Subsequently, 10–20 µl of PCR products were subjected to pyrosequencing by PyroMark Q96 ID (Qiagen, Inc). Completely methylated and unmethylated DNA were used as control samples (EpiTect Control DNA, methylated/EpiTect Control DNA, unmethylated; Qiagen GmbH). For the immobilization of the PCR product to the beads, a mixture of 10 µl optimized biotinylated PCR product, 1.5 µl streptavidin-coated sepharose beads, 40 µl Binding buffer and 28.5 µl deionized water was prepared. Bisulfite modified DNA was placed into a thermal cycler with the following program: Denaturation (95°C, 5 min), incubation (60°C, 25 min), denaturation (95°C, 5 min), incubation (60°C, 85 min), denaturation (95°C, 175 min), incubation (60°C, 25 min) and incubation (20°C). The total volume of a capture was pipetted into each well of the Pyromark plate low. Analysis was conducted according to the manufacturer's protocol, which was described previously (21).
Figure 1.
Map of CpG methylation sites in the first intron of T-lymphocyte maturation associated protein. Chromosomal location: Chr2: 95691554-95691815. Individual CpGs are underlined in red. CpG, CG dinucleotides.
Figure 2.
Map of CpG methylation sites in the first intron of cell adhesion molecule 1. Chromosomal location: Chr11: 115374815-115374991. Individual CpGs are underlined in red. CpG, CG dinucleotides.
Statistical data analysis
The two-sample test for equality of proportions with a continuity correction was used to examine the hypothesis of the equality of proportions of HPV+ and HPV- in patients and controls. Robust one-way analysis of variance was used to test the hypothesis of equality of medians in the patients groups for each CpG island. The test was followed by the Tukey honest significant difference post-hoc test. The effect size was quantified by the 95% CI. Univariate and multivariate logistic regression models were used to analyze the dependence between a response and predictor(s). The predictive ability was visualized by the receiver operating curve (ROC) and quantified by the AUC, the area under ROC. The cut-off on the class probability was determined by the Youden method, and translated into a cutoff on the median methylation. All analyses were performed using R version 3.2.3 (24). P<0.05 was considered to indicate a statistically significant difference.
Results
HPV 16/18 detection and genotyping
The present study confirmed the presence of one or two high-risk genotypes of HPV in 39 cases (42.85%); however, the number of positive subtypes of HPV DNA per patient were not discerned. HPV DNA was present in 19% (6/31) of controls, in 35% (7/20) of patients with inflammation, in 42.8% (6/14) of CIN1 cases, in 68.4% (13/19) of CIN2+ cases and in 100% (7/7) of SCC (Table I). Logistic regression was used to analyze the dependence between the status and age in the patient and control groups. However, the association was not statistically significant (P=0.636). The two-sample test for equality of proportions with a continuity correction was used to examine the hypothesis of the equality of proportions of HPV+ and HPV- in patients and controls. HPV infection has been significantly present in CIN2+ cases (P=0.001528), with a 95% CI of −0.78 and −0.19, and in SCC cases (P=0.0002933), 95% CI: (−1, −0.58).
Pyrosequencing
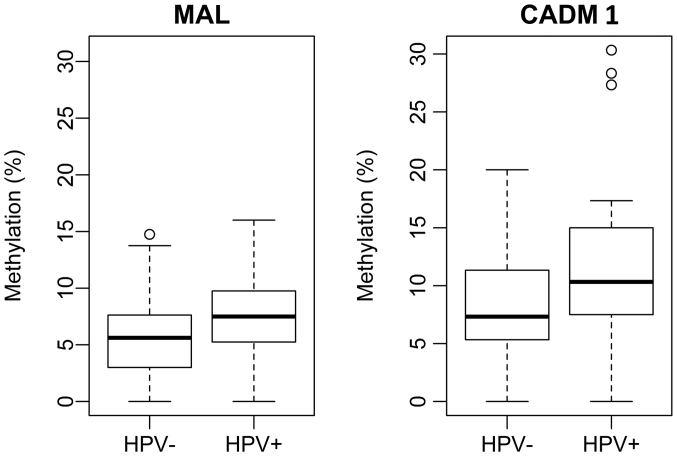
Samples modified with sodium bisulfite were subsequently used in the PCR reaction. Using methylate-specific primers, we converted cytosine to uracil, respectively thymine in the PCR product while methylated cytosine remained unchanged. The present study focused on the detection of methylated regions in the sequence of two TSGs (MAL and CADM1) by pyrosequencing. By logistic regression it was identified that the MAL/CADM1 methylation status in the control group was not associated with age of patients (P-values are not presented). By the Welch two-sample test with the one-side alternative the median methylation in HPV+/− DNA in all patient groups were compared. For MAL and CADM1 the median methylation was identified as significantly higher in HPV positive patients [P=0.0097, 95% CI: (−0.030, −0.003) for MAL/P=0.0024, 95% CI: (−0.06, −0.01) for CADM1] than in the patients negative for HPV DNA (Fig. 3).
Figure 3.
Welch's two-sample test with one-side alternative comparisons of the median methylation of MAL and CADM1 in HPV+/− DNA. The boxplot represents the median and the interquartile range. MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1; HPV, human papillomavirus.
Average methylation and determining the degree of methylation of individual CpG islands
By the Robust One-way ANOVA the hypothesis of equality of medians in the patients groups for each CpG island were examined. The ANOVA P-values were P=0.0002 for MAL (1st CpG), P=0.0066 for MAL (2nd CpG), P=0.1099 for MAL (3rd CpG), P=0.0207 for MAL (4th CpG)/P=0.0074 for CADM1 (1st CpG), P=0.0063 for CADM1 (2nd CpG), P=0.0007 for CADM1 (3rd CpG). Tukey's post-hoc HSD test was used to perform the multiple comparisons testing. Pairs (CpG islands) with significantly different methylations are presented in Table II.
Table II.
Multiple comparisons testing of each CpG island in the two tumor-suppressor genes.
| Dg. | MAL 1. | MAL 2. | MAL 3. | MAL 4. | CADM1 1. | CADM1 2. | CADM1 3. |
|---|---|---|---|---|---|---|---|
| 0 vs. 1 | 0.00000c | 0.00000c | 0.01914a | 0.00031c | 0.00010c | 0.00006c | 0.00002c |
| 0 vs. 2 | 0.00632b | 0.01790a | 0.11492 | 0.46309 | 0.57563 | 0.46030 | 0.03467a |
| 0 vs. 3 | 0.00692b | 0.00179b | 0.02417a | 0.01624a | 0.00147b | 0.02081a | 0.00170b |
| 0 vs. 4 | 0.01825a | 0.06606 | 0.95381 | 0.12388 | 0.02095a | 0.04557a | 0.01732a |
| 1 vs. 2 | 0.36317 | 0.06729 | 0.60254 | 0.03027a | 0.02851a | 0.05680 | 0.17161 |
| 2 vs. 3 | 0.57294 | 0.83631 | 0.88263 | 0.24036 | 0.02276a | 0.21970 | 0.80962 |
| 2 vs. 4 | 0.41027 | 0.59283 | 0.18941 | 0.25177 | 0.03047a | 0.06590 | 0.30761 |
P<0.05
P<0.01
P<0.001. Dg, diagnosis group; Dg.0, controls; Dg.1, inflammation samples; Dg.2, CIN1; Dg.3, CIN2+; Dg.4, SCC; CIN, cervical intraepithelial neoplasia; SCC, squamous cell carcinoma; MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1.
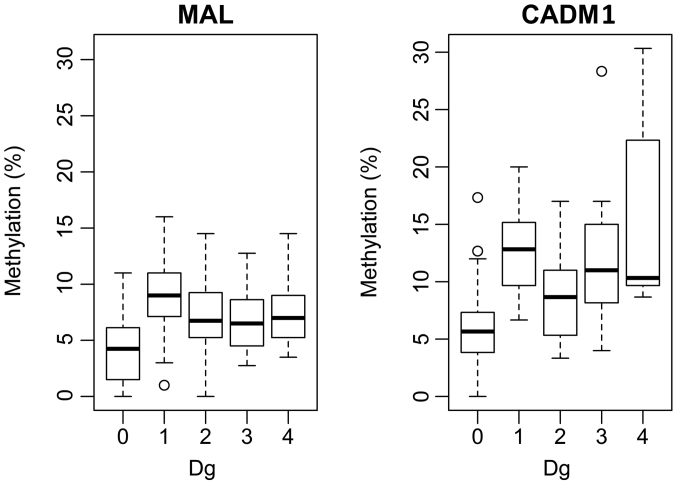
An ANOVA with Tukey's post hoc test, was used to test the hypothesis of equality of medians in the patient groups, for averaged 4 resp. 3 CpG islands. The ANOVA P-values were: P=0.0026 for MAL and P=0.0001 for CADM1. Fig. 4 illustrated the pairs with significantly different methylations. MAL was demonstrated to have a significantly lower median methylation compared with CADM1 for each group (Dg.0: P=0.010145, 95% CI: −0.036, −0.003; Dg.1: P=0.0001447, 95% CI: −0.050, −0.0180; Dg.2: P=0.001147, 95% CI: −0.0378, −0.003; Dg.3: P=0.0003808, 95% CI: −0.0745, −0.0235; Dg.4: P=0.04694).
Figure 4.
Boxplot of the methylation of the controls and the four levels of lesion (MAL/CADM1). MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1; Dg, diagnosis group.
Logistic regression models using the most promising markers were investigated to see whether marker combinations could be used to improve the discrimination between cases and the control group. Multinomial logistic regression was used to examine whether HPV status impacted DNA methylation at specific CpG islands or if there is any dependence between age of patient and increasing risk of neoplasia or HPV. For CADM1 the present study confirmed that the last CpG (3rd) was significantly more methylated in samples with inflammation (P=0.001), CIN1 (P=0.086), CIN2+ (P=0.038) compared with the other examined CpGs (1st, 2nd). In MAL it was also discovered that the first CpG was more significantly methylated than other CpGs (P=0.043 for inflammation, P=0.017 for CIN1, P=0.155 for CIN2+). For SCC (Dg.4) the second and third CpGs for the gene MAL were significantly more methylated (P=0 for 2nd CpG, P=0.012 for 3rd CpG) than in other groups. For CADM1 no statistically significant differences were identified in the SCC (Dg.4) group (Table III).
Table III.
Multinomial logit model of diagnosis as a function of age, HPV, MAL 1–4 and CADM1 1–3.
| Dg. no. | Age | HPV | MAL 1 | MAL 2 | MAL 3 | MAL 4 | CADM1 1 | CADM1 2 | CADM1 3 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.054 | 0.622 | 0.043 | 0.697 | 0.111 | 0.315 | 0.989 | 0.678 | 0.001 |
| 2 | 0.321 | 0.093 | 0.017 | 0.195 | 0.875 | 0.043 | 0.091 | 0.584 | 0.086 |
| 3 | 0.423 | 0.028 | 0.155 | 0.721 | 0.867 | 0.451 | 0.183 | 0.261 | 0.038 |
| 4 | 0.584 | 0.000 | 0.909 | 0.000 | 0.012 | 0.227 | 0.346 | 0.233 | 0.316 |
Dg, diagnosis group; Dg.1, inflammation samples; Dg.2, CIN1; Dg.3, CIN2+; Dg.4, SCC; CIN, cervical intraepithelial neoplasia; MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1; HPV, human papillomavirus.
Determination of the methylation cut-off using logistic regression (MAL, CADM1)
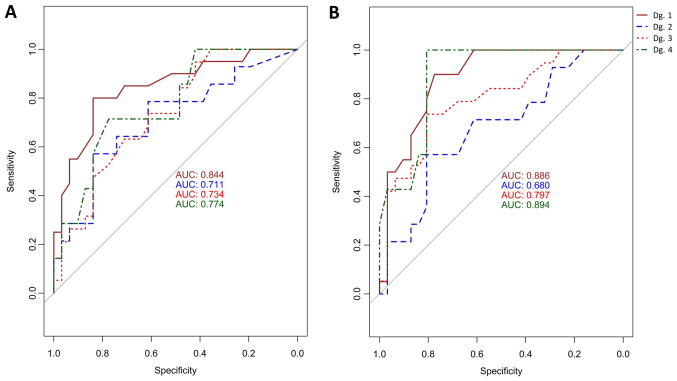
Logistic regression was used to examine to the dependence between the case/control statuses (control group vs. Dg. 1–4). The AUC values for MAL were: 84% for cervical inflammation, 71% for CIN1, 73.4% for CIN2+, 77% for SCC; and for CADM1 were: 88.6% for cervical inflammation, 68% for CIN1, 80% for CIN2+ and 89% for SCC. The ROC graph is shown in Fig. 5 (left) for MAL and Fig. 5 (right) for CADM1. The cut-off on the class probability was determined by the Youden method. The cutoff for MAL and CADM1 on the median methylation corresponding to the cutoff on the class probabilities was 0.09. Using this cutoff value, MAL had the highest sensitivity and specificity in inflammation samples (80 and 83.9% respectively) followed by SCC samples (71.4 and 77.4% respectively). CIN1 was less sensitive (57.1%) but still had high specificity (83.9%). However, for the MAL gene CIN2+ group had the highest sensitivity (94.7%) but the lowest specificity (41.9%). CADM1 had the highest sensitivity (100%) and also specificity (80.6%) in SCC and also in CIN2+ where the sensitivity was 73.7% with an even higher specificity (80.6%). In patients with inflammation a high sensitivity (90%) and specificity (77.4%) were observed. The lowest sensitivity (57.1%) but still high specificity (80.6%) for the median methylation was found in CIN1. The dependence between women with inflammation and CIN1 was also calculated (AUC=68% for MAL, AUC=74.4% for CADM1), CIN1 and CIN2+ (AUC=50.5% for MAL, AUC=66.5% for CADM1), CIN1 and SCC (AUC=56% for MAL, AUC=78.5% for CADM1).
Figure 5.
(A) ROC curve with AUC values (logistic regression) for samples from the control group and Dg. 1 (brown), 2 (blue), 3 (red) and 4 (green) depending on the median methylation of the MAL. (B) ROC curve with AUC values (logistic regression) for samples from the control group and Dg. 1 (brown), 2 (blue), 3 (red) and 4 (green) depending on the median methylation of the CADM1. ROC, receiver operating curve; AUC, area under the curve; MAL, T-lymphocyte maturation associated protein; CADM1, cell adhesion molecule 1; Dg, diagnosis group.
Discussion
Epidemiological and molecular studies have demonstrated that persistent infection of HR-HPV is one of the causes of cervical carcinogenesis (25,26) and the major mechanism that engages HPV16/18 in cervical carcinogenesis is the manifestation of two early viral genes E6 and E7 (4). Only a minority of all persistent HR-HPV infections alters the expression of viral genes E6 and E7, consequently resulting in an increased expression of oncoproteins (26). This process is histologically known as CIN2/3 and, without treatment can ultimately lead to CC. Recent studies have demonstrated that the inclusion of testing to detect the presence of HR-HPV DNA in cytology after 6 months of patient treatment has significantly increased the sensitivity of CIN2+ detection (27,28). HR-HPV testing is an attractive modality for primary screening due to its extremely high sensitivity for CIN3+ lesions. However, the specificity of HR-HPV testing is lower compared with cytology (approximately 4–6%) (29). In order to manage the increased number of colposcopy referrals, HR-HPV positive women should be further stratified by secondary triage tests such as p16/Ki67 immunostaining and methylation markers (29). In the present study, HPV DNA was detected in 42.8% CIN1, 68.4% CIN2+ and 100% of SCC samples. From these results, an increase in the incidence of HPV DNA positive samples was observed as the severity of the lesion increased.
Recent results suggest that other factors, such as genetic and epigenetic changes (CpG DNA methylation), are required for the onset of carcinogenesis (30). Approximately ten human genes have consistently elevated methylation in cervical pre-cancers including CADM1, EPB41L3, FAM19A4, MAL, miR-124, PAX1 and SOX1. Methylation testing is still in the early stages of development; however, it is exhibiting promise as an accurate molecular classifier (31).
Methylation of CpG regions of multiple TSGs occurs at different stages of development of CIN and in the process of transition to invasive cervical carcinoma, of which CADM1 and MAL are the most commonly methylated genes (17,32,33). The principal effect of DNA methylation in promotor/intronic regions of CADM1 and MAL is their downregulation. MAL was identified to be an essential component of the glycolipid-enriched membrane (GEM) rafts, which have been implicated in the polarized sorting of apical proteins (34,35). This down-regulation may disturb regular apical sorting, thereby disrupting cellular polarity, which is a phenomenon observed in epithelial cells infected with HPV. The down-regulation of CADM1 may result in the loss of the Rb tumor suppressor pathway signaling, which represents a relatively common event in cervical carcinogenesis (36). However, the molecular mechanism of CADM1/MAL involvement in cervical carcinogenesis has not yet been thoroughly investigated and the definitive confirmation that the down-regulation of these genes in primary CCs will require further study.
A previous study reported that there was a correlation between methylation and the progression of neoplasia to a higher stage. There were no difference in the median methylation between the control group and CIN1 or between CIN1 and CIN2+. Based on these results they considered that the methylation degree of individual CpG islands in the promoter region of the CADM1 gene was lower in normal squamous epithelium than in CINs (37). However, when CIN passed to the invasive cervical cancer (ICC), the average methylation of specific CpG regions increased significantly, which could lead to increased or complete inhibition of gene expression and result in a loss of cell adhesion and tumor-suppressor function of the gene and thereby inducing malignant transformation cells (37). The authors in the above-mentioned study also confronted the results of the median methylation in TSG and progression of disease with results reported by other study (17). These publications demonstrated that the frequency of average methylation in the promoter region of the gene increases with the severity of disease and that the presence of methylation in the ICC was significantly different from the degree of methylation occurring in other types of lesions.
Based on these results the present study hypothesized that progression of disease was dependent on the degree of the median methylation or HPV infection and not on the age of the patient. The same analysis was also applied in an additional study (38) and confirmed statistical significance (P=0.05). A previous study monitored the degree of methylation of the CADM1/MAL genes from the uterine cervix specimens using the quantitative methylation-specific PCR method and demonstrated that the median methylation of these genes is a significant predictor of the CIN1 status: positive patients (35%), CIN2: positive patients (32.8%), CIN3: positive patients (59%). They also identified a statistically significant association between the HR-HPV infection positivity and the transition of the disease to a more advanced stage (39).
The MassARRAY technique EpiTYPER DNA was used for analysis of the 15 methylated regions of the CADM1 in ICC, CIN1, CIN2/3 tissue samples and a control group. In 9 CpGs they exhibited an increased methylation status in ICC compared with the control group, and the average degree of methylation (2, 3, 4. and 5. CpG) in the selected promoter region was higher than in the remaining CpG islands. However, CpGs 9 and 10 were less methylated. This provides evidence for varying degrees of methylation in the individual CpG regions in the CADM1 gene (37). In our selected area the final CpG located behind the first exon was significantly more methylated in each group than the first two CpGs (P-values: 0.001 for inflammation samples, 0.086 for CIN1, 0.038 for CIN2+). The present study also identified that one CpG (1st CpG) was significantly more methylated in gene MAL than other 3 CpGs (P-values: 0.043 for inflammation samples, 0.017 for CIN1, 0.155 for CIN2+).
In several studies, DNA methylation of the CADM1/MAL gene promoter region has been demonstrated to increase with the severity of cervical disorders (17–19) and that these epigenetic changes suggest the presence of CINs. These findings are also supported by the results of an additional study, which demonstrated that levels of the median methylation in tumor suppressor genes are extremely high in cervical carcinomas and significantly increase in CIN3 lesions in women with HR-HPV infections that persists over a period of time longer than 5 years (20). The present study confirmed in both genes (CADM1/MAL) that the median methylation is significantly higher in HPV positive patients than in HPV negative patients. A higher median methylation and more HPV positive patients were identified in SCC than in CIN1 cases.
In a previous study, by analysis (pyrosequencing) of the promoter regions of the genes on tumor cell lines (C33A HPV negative, HeLa HPV18 positive, SiHa and CaSki HPV16 positive) the hypermethylation of the CADM1 gene in the CaSki, SiHa and HeLa cell lines were detected; however, not in the C33A cell line. The methylation status of the MAL was positive for the CaSki and SiHa cell lines but not for the HeLa and C33A cell lines. Based on these results, hypermethylation of the CADM1 gene was demonstrated to be associated with HR-HPV infection. Hypermethylation of the MAL gene was also associated with infection with high-risk HPV types. Although the methylation stage indicates that the HeLa and C33A cell lines are not statistically significant (P=0.148) (40). In an additional study the methylation state of the promoter region of the CADM1 gene (nucleotides −444 to −305) were analyzed by pyrosequencing. They demonstrated that methylation was significantly increased in HPV positive HeLa cell lines (71.7%), SiHa (84.8%), CaSki (95.2%) and significantly decreased in HPV negative cell lines C33A (2.6%) (41). For further analysis cell lines were not used; however, tissue taken from patients with L-SIL and H-SIL as well as from patients with cervical carcinoma were analyzed. The degree of average methylation for the CADM1 was 3.5 (95% CI, 3.0–4.0) in the L-SIL group, 5.6 (95% CI, 4.0–4.7) in the H-SIL group and 17.7 (95% CI, 10.8–29.1) in carcinomas. For the MAL gene, the average methylation rate was 2.7 (95% CI, 2.5–3.0) in the L-SIL group, 3.7 (95% CI, 3.0–4.6) in the H-SIL and 13 (95% CI, 7.6–22) in carcinomas. For both genes P<0.001 (40). The degree of average methylation increased significantly in carcinoma samples as well as in pre-invasive cervical lesion samples (39). The present study also confirmed that the median methylation significantly increased from CIN1 cases trough CIN2+ cases to carcinomas in both genes, while the median methylation of CADM1 was higher trough each group than the median methylation of the gene MAL.
In the present study, statistically significant differences were identified in the inflammation samples when compared with the control or CIN1 group, which may suggest that hypermethylation of the TSGs in inflammatory samples is not random and that may be indicative of the early stages of carcinogenesis. Previous studies generally did not contain an inflammatory group, which could lead to an underestimation of at-risk patients. It is well-established that inflammation is a possible precursor of CIN1, and may culminate in CC (38), therefore it would be appropriate to include a cervical inflammation group in further methylation studies.
In conclusion, the present study focused on the detection of methylation in the region of two TSGs (MAL and CADM1). Several major studies have confirmed a significant increase of the median methylation in the specific promoter region of these genes tested on cell lines, while the percentage of methylation increased with the progression of neoplasia to cervical carcinoma (39,40). However, in the majority of studies, pyrosequencing was not used as a sensitive tool for quantitative analysis of CpG methylation. Since many patients underestimate the importance of regular medical check-ups, it is important during examinations to provide them with the necessary comfort, not to exacerbate their stress and try to capture the dysplasia at an early stage. The specimens used in the present study therefore constituted of cytological smears from the cervix. CADM1 was confirmed to be more methylated in almost every study group compared with MAL, and that third CpG in CADM1 exhibited higher methylation levels during the transition of CIN and has the potential to serve as a promising biomarker for future study. There was also a significantly higher median methylation in HPV+ patients than in HPV-patients which supports the suitability of this combination (methylation levels/HPV positivity) as an early detection tool for this severe oncological disease.
Acknowledgements
Not applicable.
Funding
The present study was funded by the project implementation Biomedical Center Martin (grant no. 26220220187), and was supported by the Operational Program Research and Innovation funded by the ERDF, the project ‘Molecular Diagnostics of Cervical Cancer’ (grant no. 26220220113) and by VEGA (grant no. 1/0380/18).
Availability of data and materials
The datasets generated/analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SM and ZL designed the study. SM and VH performed the experiments. MG conducted the statistical analysis. JV, MN, EK and TB obtained and handled cervical specimens and clinical data. SM analyzed the data and was a major contributor in writing the manuscript. MK performed the histological examination of the samples and clinical data. ZL, PZ and JD supervised the entire study, and participated in study design and coordination. All authors read and revised the manuscript, and approved the final version to be published.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Jessenius Faculty of Medicine in Martin. All patients provided written informed consent prior to their inclusion in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Resende LS, Rabelo-Santos SH, Sarian LO, Alves Figueiredo RR, Ribeiro AA, Zeferino LC, Derchain S. A porsingle and multiple HPV type infections in Brazilian women of differentrait of t age strata with squamous or glandular cervical lesions. BMC Infect Dis. 2014;22:214. doi: 10.1186/1471-2334-14-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruni 1, Barrionuevo-Rosas L, Albero G, et al. ICO information centre on HPV and cancer (HPV Information Centre) Human Papillomavirus Relat Dis Eur. 2017 [Google Scholar]
- 4.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piyathilake CJ, Mascaluso M, Alvares RD, Chen M, Badiga S, Edberg JC, Partridge EE, Johanning GL. A higher degree of methylation of the HPV 16 E6 gene is associated with a lower likelihood of being diagnosed with cervical intraepithelial neoplasia. Cancer. 2011;117:957–963. doi: 10.1002/cncr.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balch C, Fang F, Matei DE, Huang TH, Nephew KP. Minireview: Epigenetic changes in ovarian cancer. Endocrinology. 2009;150:4003–4011. doi: 10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 8.Qureshi SA, Bashir MU, Yaqinuddin A. Utility of DNA methylation markers for diagnosing cancer. Int J Surg. 2010;8:194–198. doi: 10.1016/j.ijsu.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Mersakova S, Nachajova M, Szepe P, Kasajova PS, Halasova E. DNA methylation and detection of cervical cancer and precancerous lesions using molecular methods. Tumour Biol. 2016;37:23–27. doi: 10.1007/s13277-015-4197-1. [DOI] [PubMed] [Google Scholar]
- 10.Murakami Y, Nobukuni T, Tamura K, Maruyama T, Sekiya T, Arai Y, Gomyou H, Tanigami A, Ohki M, Cabin D, et al. Localization of tumor suppressor activity important in nonsmall cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci USA. 1998;95:8153–8158. doi: 10.1073/pnas.95.14.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ando K, Ohira M, Ozaki T, Nakagawa A, Akazawa K, Suenaga Y, Nakamura Y, Koda T, Kamijo T, Murakami Y, Nakagawara A. Expression of TSLC1, a candidate tumor suppressor gene mapped to chromosome 11q23, is downregulated in unfavorable neuroblastoma without promoter hypermethylation. Int J Cancer. 2008;123:2087–2094. doi: 10.1002/ijc.23776. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Shimada Y, Hashimoto Y, Kaganoi J, Kan T, Watanabe G, Murakami Y, Imamura M. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer Res. 2003;63:6320–6326. [PubMed] [Google Scholar]
- 13.Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004;23:5632–5642. doi: 10.1038/sj.onc.1207756. [DOI] [PubMed] [Google Scholar]
- 14.Steenbergen RD, Kramer D, Braakhuis BJ, Stern PL, Verheijen RH, Meijer CJ, Snijders PJ. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 15.Overmeer RM, Henken FE, Snijders PJ, Claassen-Kramer D, Berkhof J, Helmerhorst TJ, Heideman DA, Wilting SM, Murakami Y, Ito A, et al. Association between dense CADM1 promoter methylation and reduced protein expression in high-grade CIN and cervical SCC. J Pathol. 2008;215:388–397. doi: 10.1002/path.2367. [DOI] [PubMed] [Google Scholar]
- 16.Momparler RL, Bovenzi V. DNA methylation and cancer. J Cell Physiol. 2000;183:145–154. doi: 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 17.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CH. Clinical implications of (epi) genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 18.Overmeer RM, Louwers JA, Meijer CJ, van Kemenade FJ, Hesselink AT, Daalmeijer NF, Wilting SM, Heideman DA, Verheijen RH, Zaal A, et al. Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int J Cancer. 2011;129:2218–2225. doi: 10.1002/ijc.25890. [DOI] [PubMed] [Google Scholar]
- 19.De Strooper LM, van Zummeren M, Steenbergen RD, Bleeker MC, Hesselink AT, Wisman GB, Snijders PJ, Heideman DA, Meijer CJ. CADM1, MAL and miR124-2methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;67:1067–1071. doi: 10.1136/jclinpath-2014-202616. [DOI] [PubMed] [Google Scholar]
- 20.Bierkens M, Hesselink AT, Meijer CJ, Heideman DA, Wisman GB, van der Zee AG, Snijders PJ, Steenbergen RD. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;133:1293–1299. doi: 10.1002/ijc.28138. [DOI] [PubMed] [Google Scholar]
- 21.Meršaková S, Visnovsky J, Holubekova V, Nachajova M, Kudela E, Danko J, Lasabova Z. Detection of methylation of the promoter region of the MAL and CADM1 genes by pyrosequencing in cervical carcinoma. Neuro Endocrinol Lett. 2014;35:619–623. [PubMed] [Google Scholar]
- 22.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlée F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. Improved amplification of genital human papillomaviruses. J Clin Microiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janusicova V, Mendelova A, Zubor P, Kapustova I, Svecova I, Kudela E, Burjanivova T, Lasabova Z, Danko J. mRNA expression in cervical specimens for determination of severe dysplasia or worse in HPV-16/18-positive squamous lesions. J Low Genit Tract Dis. 2014;18:273–280. doi: 10.1097/LGT.0000000000000000. [DOI] [PubMed] [Google Scholar]
- 24.RC Team: R: A language and environment for statistical computing, corp-author. R foundation for statistical computing. Vienna, Austria: 2015. URL. [Google Scholar]
- 25.Lazo PA. The molecular genetics of cervical carcinoma. Br J Cancer. 1999;80:2008–2018. doi: 10.1038/sj.bjc.6690635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25(Suppl 1):S2–S23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocken M, Helmerhorst TJ, Berkhof J, Louwers JA, Nobbenhuis MA, Bais AG, Hogewoning CJ, Zaal A, Verheijen RH, Snijders PJ, Meijer CJ. Risk of recurrent high-grade cervical intraepithelial neoplasia after successful treatment: A long-term multi-cohort study. Lancet Oncol. 2011;12:441–450. doi: 10.1016/S1470-2045(11)70078-X. [DOI] [PubMed] [Google Scholar]
- 28.Kocken M, Uijterwaal MH, de Vries AL, Berkhof J, Ket JC, Helmerhorst TJ, Meijer CJ. High-risk human papillomavirus testing versus cytology in predicting post-treatment disease inwomen treated for high-grade cervical disease: A systematic review and meta-analysis. Gynecol Oncol. 2012;125:500–507. doi: 10.1016/j.ygyno.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Hesselink AT, Heideman DA, Steenbergen RD, Coupé VM, Overmeer RM, Rijkaart D, Berkhof J, Meijer CJ, Snijders PJ. Combined promoter methylation analysis o CADM1 and MAL: An obkective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011;17:2459–2465. doi: 10.1158/1078-0432.CCR-10-2548. [DOI] [PubMed] [Google Scholar]
- 30.Yang HJ. Aberrant DNA methylation in cervical carcinogenesis. Chin J Cancer. 2013;32:42–48. doi: 10.5732/cjc.012.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cushieri K, Ronco G, Lorincz A, Smith L, Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders P, et al. Eurogin roadmap 2017: Triage strategies for the management of HPV positivie women in cervical screening programs. Int J Cancer. 2018;143:735–745. doi: 10.1002/ijc.31261. [DOI] [PubMed] [Google Scholar]
- 32.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tornesello ML, Buonaguro L, Giorgi-Rossi P, Buonaguro FM. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed Res Int. 2013;2013:519619. doi: 10.1155/2013/519619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Belmonte F, Arvan P, Alonso MA. MAL mediates apical transport of secretory proteins in polarized epithelial Madin-Darby canine kidney cells. J Biol Chem. 2001;276:49337–49342. doi: 10.1074/jbc.M106882200. [DOI] [PubMed] [Google Scholar]
- 35.Hatta M, Nagai H, Okino K, Onda M, Yoneyama K, Ohta Y, Nakayama H, Araki T, Emi M. Down-regulation of members of glycolipid-enriched membrane raft gene family, MAL and BENE, in cervical squamous cell cancers. J Obstet Gynaecol Res. 2004;30:53–58. doi: 10.1111/j.1341-8076.2004.00156.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Xie HY, Ding SM, Xing CY, Chen A, Lai MC, Zhou L, Zheng SS. CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb-E2F pathway in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2016;15:289–296. doi: 10.1016/S1499-3872(16)60099-1. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Tao L, Tan Q, Dong Y, Pan X, Pang L, Qi Y, Zou H, Liang W, Liu W, et al. CpG island methylation of the CADM1 gene correlates with cervical carcinogenesis in the Uighur and Han populations of Xinjiang, China. Int J Clin Exp Pathol. 2016;9:6977–6987. [Google Scholar]
- 38.van Baars R, van der Marel J, Snijders PJ, Rodriquez-Manfredi A, ter Harmsel B, van den Munckhof HA, Ordi J, del Pino M, van de Sandt MM, Wentzensen N, et al. CADM1 and MAL methylation status in cervical scrapes is representative of the most severe underlying lesion in women with multiple cervical biopsies. Int J Cancer. 2016;138:463–471. doi: 10.1002/ijc.29706. [DOI] [PubMed] [Google Scholar]
- 39.Uijterwaal MH, van Zummeren M, Kocken M, Luttmer R, Berkhof J, Witte BI, van Baal WM, Graziosi GCM, Verheijen RHM, Helmerhorst TJM, et al. Performance of CADM1/MAL-methylation analysis for monitoring of women treated for high-grade CIN. Gynecol Oncol. 2016;143:135–142. doi: 10.1016/j.ygyno.2016.07.089. [DOI] [PubMed] [Google Scholar]
- 40.Ki EY, Lee KH, Hur SY, Rhee JE, Kee MK, Kang C, Park JS. Methylation of cervical neoplastic cells infected with human papillomavirus 16. Int J Gynecol Cancer. 2016;26:176–183. doi: 10.1097/IGC.0000000000000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo HJ, Kim SJ, Song KJ, Kim SS, Yoon CH, Choi BS, Rhee JE. Hypermethylation of the tumor-suppressor cell adhesion molecule 1 in human papillomavirus-transformed cervical carcinoma cells. Int J Oncol. 2015;46:2656–2662. doi: 10.3892/ijo.2015.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated/analyzed during the current study are available from the corresponding author on reasonable request.