Abstract
The present study examined the mRNA expression levels of different isoforms of the plasma membrane calcium ATPase 2 (PMCA2) gene generated by alternative splicing at the first intracellular loop (site A) and C-terminal region (site C) in 85 human breast cancer tumor and 69 adjacent non-tumor tissues. Associations were identified between the expression of PMCA2 splice isoforms and the following clinical variables: Estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) status, tumor size, staging and histological classification, and lymph node status. Transcripts including splice site A or splice site C were amplified by reverse transcription-quantitative polymerase chain reaction using PMCA2 isoform-specific primers. Tumor and adjacent tissues were determined to express the different PMCA2 splice isoforms 2w, 2× and 2z (site A), and 2b (site C). The mRNA levels for these variants indicated high biological variability, but increased expression was observed in breast tumor tissues, compared with in adjacent tissues. Significantly increased PMCA2×/b expression levels were detected in breast tumor tissues histologically classified as lobulillar, compared with in ductal-types breast tumor tissues (P<0.028). Furthermore, PMCA2z expression was significantly associated with PR status (P<0.024, compared with in PR-negative tumor tissues), and PMCA2w expression was significantly associated with ER status (P<0.048, increased in ER-positive tumor tissues, compared with ER-negative tumor tissues). Finally, PMCA2b was overexpressed in HER2-positive tumor tissues, compared with in HER2-negative tumor tissues (P<0.014). The data demonstrated the differential mRNA expression of a number of splice site A and C variants of PMCA2 in breast tumor and adjacent tissues, depending on tumor hormone receptor status and histological classification. In agreement with previous data, PMCA2b was overexpressed in HER2-positive tumor tissues, indicating that high mRNA levels of this variant could be a marker of poor prognosis.
Keywords: PMCA2, alternative splicing, breast cancer
Introduction
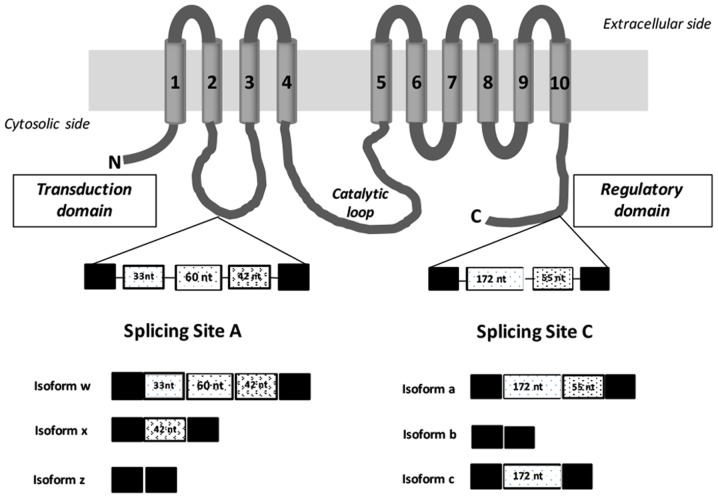
The human plasma membrane calcium ATPase (PMCA2) cooperates with other calcium transport systems and with soluble Ca2+ binding proteins to control calcium homeostasis in eukaryotic cells (1–3). The precise control of intracellular calcium levels is required due to the critical role of Ca2+ in the modulation of vital physiological processes, including lactation, proliferation and apoptosis (3–5). In humans, PMCA2 is expressed in a limited range of tissues, with high expression levels observed in the nervous system, including cerebellar Purkinje cells and cochlear hair cells, and the lactating epithelial mammary gland, and reduced expression levels of the enzyme are observed in pancreatic beta, corneal epithelium and muscle cells (6–10). Recent studies have identified elevated PMCA2 mRNA levels in human breast cancer (11–15). Furthermore, it has been established that two alternative splicing domains at the N-terminal first intracellular loop (site A; 2w, 2×, 2z) and at the C-terminal region (site C; 2a, 2b, 2c) produce >9 different variants through mechanisms in which exons are included or excluded (Fig. 1). Site A includes exons (33, 60 and 42 bp) that encode residues in the vicinity of a phospholipid-responsive domain located in the cytoplasmic loop between the second and third transmembrane domain, which may be incorporated into the mature mRNA in diverse combinations (16–19), affecting protein targeting. Site C comprises of exons (172 and 55 bp) that may be included or excluded in the mature mRNA, inducing changes in the length of the C-terminal tail, which provide different regulatory and trafficking characteristics to the splice isoforms (17–20). All of these alternative-splicing variants have been detected in a tissue-specific gene expression manner, indicating that diversity in PMCA2 expression variants serve an important role in Ca2+ signaling in the physiological and disease conditions; however, splice sites have only been studied independently and for a number of isoforms, these sites have not yet been confirmed at the protein level (7,20). Although the expression of PMCA2 site A or C splice variants has been described in different tissues (18) and breast cancer cell lines (11), there is limited knowledge regarding their expression in different types of human breast tumor tissues. Previous studies regarding tumor cells have identified 2w/b as the most important isoform expressed in breast cancer; however, no comprehensive investigation regarding PMCA2 splice variants has been conducted in human breast and breast cancer tissues. In the present study, the identification of three PMCA2 splice variants (2w, 2× and 2z) produced by alternative exon choice in the N-terminal region of the protein, and one PMCA2 splice variant (2b) in the C-terminal domain in breast cancer tumor and adjacent non-tumor tissues of Spanish patients was reported. Relative expression levels of the different splice isoforms of PMCA2 were also associated with estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and lymph node status, and tumor stage and histological classification. The data presented in the present study indicate variable expression of the different PMCA2 isoform mRNAs in breast tumor and normal tissues and highlight the relevance of this p-type ATPase in the context of breast cancer and the physiology of the breast.
Figure 1.
Diagram illustrating the structure of human plasma membrane calcium ATPase 2 and its splice isoforms. The protein consists of 10 transmembrane domains numbered and indicated in gray tubes. The two alternative splicing sites are depicted in the figure, the splice sites A (N-terminal domain) and C (C-terminal tail) within the regulatory domain (calmodulin-binding domain). The intracellular loop connecting TM4 and TM5 is termed the catalytic loop, and contains the phosphorylation and ATP-binding sites. Alternative splicing of human PMCA2 pump at site A located between transmembrane domains 2 and 3 generates three isoforms w, z and x. At site C three isoforms a, b or c could be generated by alternative splicing in the C-terminal region. Constitutively spliced exons are depicted as black boxes and alternatively inserted exons are depicted in white with different patterns. nt, number of nucleotides; N, N-terminal domain; C, C-terminal domain.
Materials and methods
Tissue collection and tumor specimens
In total, 85 tumor and 69 adjacent non-tumor tissue specimens, obtained from 85 Spanish patients with primary breast cancer, were examined in the present study. Paired tumor and non-tumor tissues were available for 69 of the patients with breast cancer. RNA samples were obtained from Biobanc de l'Hospital Clínic de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer (Barcelona, Spain); however, they were unable to provide information regarding age and sex distribution, and the data range of recruitment/sample collection. Informed consent was obtained from all patients who donated samples to this tumor bank. The study protocol was approved by the National Research Ethics Committee of the Hospital Clinic and the Regional Research Ethics committee of the Madrid Community. All tumor tissues were histologically classified as ductal or lobular carcinomas by the Biobanc de l'Hospital Clínic de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer (Table I). Data on ER, PR and HER2 receptor status, determined through immunohistochemistry conducted by the Biobanc de l'Hospital Clínic de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer, were available for 78, 79 and 54 tumor tissues, respectively, and were positive in 55, 52 and 36 cases, respectively (Table I). For 7, 6 and 31 tumor tissues, no data were available regarding ER, PR and HER2 status, respectively. A total of 38 primary breast tumor tissues were lymph-node positive and 42 were lymph-node negative, with no information available for the 5 remaining tumor tissues. A total of 24 of the tumor tissues were <2 cm, 45 were 2–5 cm and 13 were >5 cm; the size data were not available for 3 tumors (Table I). Disease was staged as recommended by the American Joint Committee on Cancer based on tumor extension (T), spread to the lymph nodes (N) and the presence of metastasis (M) [TNM system, (21)]. A total of 18 tumor tissues were classified as stage I, 35 as stage II and 25 as stage III, and 7 were not staged.
Table I.
Relative mRNA levels (mean and standard deviation) for the 2w, 2z, 2× and 2b splice isoforms of PMCA2 in breast tumor tissue vs. adjacent non-tumor tissue by histological and clinical features.
| PMCA2 splice isoform | ||||
|---|---|---|---|---|
| Clinicopathological data | 2w | 2z | 2× | 2b |
| ER statusa | ||||
| Positive (n=55), mean ± SD | 4.14±10.96 | 2.60±5.01 | 2.22±3.09 | 3.25±5.52 |
| Negative (n=23), mean ± SD | 3.64±8.08 | 3.36±4.70 | 2.42±3.69 | 2.11±2.50 |
| P-value | 0.048 | 0.588 | 0.917 | 0.977 |
| PR statusa | ||||
| Positive (n=52), mean ± SD | 2.69±4.27 | 1.77±2.64 | 1.95±2.98 | 3.45±5.68 |
| Negative (n=27), mean ± SD | 6.38±16.12 | 4.77±7.19 | 2.84±3.68 | 1.81±2.19 |
| P-value | 0.352 | 0.024 | 0.215 | 0.717 |
| HER2 statusa | ||||
| Positive (n=36), mean ± SD | 2.62±3.47 | 1.98±3.0 | 2.47±3.44 | 3.90±4.60 |
| Negative (n=18), mean ± SD | 3.11±5.45 | 2.67±3.52 | 1.88±2.52 | 0.175±2.66 |
| P-value | 0.248 | 0.811 | 0.378 | 0.014 |
| Nodal statusa | ||||
| Positive (n=38), mean ± SD | 4.18±13.65 | 2.27±3.82 | 1.65±2.70 | 1.75±1.51 |
| Negative (n=42), mean ± SD | 3.47±4.94 | 3.18±5.65 | 2.74±3.57 | 3.74±6.29 |
| P-value | 0.165 | 0.112 | 0.054 | 0.881 |
| Tumor size (cm)b | ||||
| ≤2 (n=24), mean ± SD | 3.16±5.39 | 3.28±6.97 | 1.97±2.20 | 4.36±7.78 |
| 2–5 (n=45), mean ± SD | 5.09 ± 12.69 | 2.99 ± 3.96 | 2.76±3.91 | 2.46±2.71 |
| ≥5 (n=13), mean ± SD | 0.78±0.66 | 0.89±0.54 | 0.89±0.64 | 1.40±1.50 |
| P-value | 0.100 | 0.07 | 0.168 | 0.476 |
| Histological classificationa | ||||
| Ductal/CDI (n=59), mean ± SD | 4.26±11.48 | 2.58±3.93 | 1.93±2.89 | 2.63±5.27 |
| Lobulillar/CLI (n=26), mean ± SD | 2.59±3.35 | 4.33±9.26 | 3.44±4.70 | 4.27±5.76 |
| P-value | 0.285 | 0.689 | 0.028 | 0.006 |
| Tumor stageb | ||||
| I (n=18), mean ± SD | 3.40±5.81 | 3.32±7.81 | 1.79±2.10 | 4.09±7.88 |
| II (n=35), mean ± SD | 6.34±14.85 | 3.14±4.22 | 3.30±4.47 | 2.61±3.03 |
| III (n=5), mean ± SD | 1.19±1.35 | 1.72±2.61 | 1.13±0.83 | 1.74±1.48 |
| P-value | 0.515 | 0.097 | 0.250 | 0.796 |
To compare variables across different categorical clinicopathological characteristics including ER, PR, and HER2 status, lymph node involvement, and explanatory variables, tumor size, histological classification and stage, the non-parametric Kruskal Wallis test was used, and the Mann Whitney U test was used for pairwise comparison. The pathological staging was conducted as recommended by the American Joint Committee on cancer [Tumor-Node-Metastasis system (21)].
Mann Whitney U test
Kruskal Wallis test. P<0.05 was considered to indicate a statistically significant difference. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor; PMCA2, plasma membrane calcium ATPase 2; SD, standard deviation; CDI, carcinoma ductal invasive; CLI, carcinoma lobulillar invasive.
RNA isolation and cDNA synthesis
RNA samples were directly obtained from Biobanc de l'Hospital Clínic de Barcelona, Institut d'Investigacions Biomèdiques August Pi i Sunyer. RNA was extracted using an RNeasy plus Universal Mini kit (Qiagen, Inc., Valencia, CA, USA), according to the manufacturer's protocols. RNA integrity and concentration was determined using an Agilent Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). cDNA was synthesized at the Universidad Europea de Madrid using Superscript III First-Strand Synthesis SuperMix (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocols. For cDNA synthesis, 250 ng RNA from each sample was used as the starting material. Negative RT was performed according to the manufacturer's protocols without adding SuperScript™ III enzyme for a set of representative samples (3 tumor and 3 adjacent non-tumor tissues).
Design of isoform specific primers
The primers used for reverse transcription-polymerase chain reaction (RT-PCR) and RT-quantitative PCR (RT-qPCR) were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Primer sets for RT-PCR were based on published studies of PMCA2 mRNA expression in different human tissues and breast cancer cell lines (6,16,18), and are included in Table II. In the present study, pair 5AF-5AR was used for splicing site A and pair 8BF-8BR was used for splicing site C (Table II). These primers were gene-specific and selected to flank the alternative splicing sites A (2w, 363 bp; 2×, 270 bp; 2y, 320 bp; and 2z, 228 bp) and C (2a, 584 bp; 2b, 357 bp; and 2c, 529 bp). Primers for RT-qPCR quantification of human PMCA2 mRNA were designated using Primer3 software v3.0.1 (Applied Biosystems; Thermo Fisher Scientific, Inc.). Amplification primer sets for qPCR were based on published human PMCA2 sequences available at www.ensembl.org, and amplicons were intron-spanning and 50–152 bp in length. These primers were isoform-specific and selected to flank alternative splice sites A and/or C (Table II). Designs were optimized to ensure high efficiencies, and verified by generating standard curves using serial dilutions of either cDNA or a purified polymerase PCR template.
Table II.
Primers for PMCA isoforms expression analysis using quantitative polymerase chain reaction analysis and for normal reverse transcription-polymerase chain reaction (RT-PCR) (5AF-5AR; 8BF-8BR).
| Splicing isoform site | Strand | Sequence (5′→3′) | Amplicon size (bp) | Isoform |
|---|---|---|---|---|
| Oligo name for PMCA-2 Site A | ||||
| 1AF | Forward | GTGTGAAGAAGGGGGATGGC | 104 | 2w |
| 1AR | Reverse | CTGCATTTTACCATTGACTAGGCTGG | ||
| 2AF | Forward | TCAGACTGGCATCATCTTTACCCTCC | 80 | 2× |
| 2AR | Reverse | CCTGCATTTTACCTTTTTTGTCTTTCTTCTC | ||
| 3AF | Forward | GTGTGAAGAAGGGGGATGGC | 109 | 2y |
| 3AR | Reverse | CCGTCCTGTTGTTTGGCATTGAC | ||
| 4AF | Forward | GACAAAAAAGCCAAACAACAGGACGG | 73 | 2z |
| 4AR | Reverse | CTCGGCACTCTTGAGGGGCTG | ||
| 5AF | Forward | TGACTGCTGTGGGTGTG | 363 | 2w |
| 5AR | Reverse | CAGCTTGGTGAGCTTGC | 270 | 2× |
| 320 | 2y | |||
| 228 | 2z | |||
| Oligo name for PMCA-2 Site C | ||||
| 6BF | Forward | GCCCTAGTCGCGTGTCGTTGTCC | 77 | 2a |
| 6BR | Reverse | CTAGCCCTGCCCGGCTGACG | ||
| 7BF | Forward | GGCAGGCAGGCTCACACAGAAG | 52 | 2b |
| 7BR | Reverse | CACGACGCGGATCTGTGTCTGG | ||
| 8BF | Forward | CACCATCCCTACCAGCAGGCT | 584 | 2a |
| 357 | 2b | |||
| 529 | 2c | |||
| 8BR | Reverse | GCTCGAGTTCTGCTTGAGCGC | ||
| Housekeeping oligo name | ||||
| 9HF | Forward | TACTTGGATAACTGTGGTAATTCTAGAG | 110 | 18S |
| 9HR | Reverse | AGGGGCTGACCGGGTTGG | rRNA |
PMCA2, plasma membrane calcium ATPase 2; bp, base pair.
Nomenclature
A naming system was used for full-length PMCA molecules described by Chicka and Strehler (19) and also was used to label splice variants with the isoform number and letter. Accordingly, 2w refers to the ‘w’ splice variant of PMCA2, and PMCA2w/b refers to PMCA2 with variant 2w at splice-site A and variant 2b at splice-site C. Furthermore, PMCA2w refers to PMCA2 with 2w at splice-site A and either variant at splice-site C.
RT-PCR analysis for alternative splicing of PMCA2
RNA aliquots of 250 ng were subjected to RT as aforementioned. The obtained cDNA (25 µl) was used to assess the PMCA2 alternative splicing at sites A and C. PCR was performed using recombinant Taq DNA polymerase (Invitrogen; Thermo Fisher Scientific, Inc.) and 2 mM primers, as previously described (6,16,18). For amplification of isoforms PMCA2 cDNA of site A, following an initial 5 min at 95°C, amplification of 38 cycles consisted of 95°C for 1 min, 52°C for 1 min and 72°C for 1 min, and then a final step of 72°C for 10 min. For amplification of isoforms PMCA2 cDNA of site C, following an initial 5 min at 95°C, amplification of 38 cycles consisted of 95°C for 1 min, 59°C for 1 min and 72°C for 1 min, and then a final step of 72°C for 10 min. The PCR product bands were visualized by electrophoresis on 2% agarose gels for bands of site A, and 1.5% agarose for bands of site C using 0.5 mg/ml ethidium bromide for UV transillumination. Analysis of bands was performed with Image Lab 6.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). In order to confirm the specificity of the PCR products obtained for sites A and C, nested PCR was performed. For each PMCA2 isoform, an aliquot of the product of the first round of PCR was used as template. In the second round of PCR, specific oligos for each PMCA2 isoform (Table II) and identical conditions to RT-qPCR quantification were used. The nested PCR product bands were visualized by electrophoresis on 3% agarose gels using RedSafe for UV transillumination. Analysis of bands was performed with Image Lab 6.0 software.
RT-qPCR for alternative splicing of PMCA2
RT-qPCR was performed in triplicate using a Bio-Rad CFX96 Real-Time PCR Thermocycler (Bio-Rad Laboratories, Inc.). Using Bio-Rad CFX Manager 3.0 software (Bio-Rad Laboratories, Inc.), cycle quantifications (Cq) were calculated for each reaction. Cycle-fluorescence growth curves for tumor and non-tumor samples were measured using Bio-Rad CFX Manager 3.0 software (Bio-Rad Laboratories, Inc.). By plotting fluorescence against the cycle number, the real-time PCR instrument generates an amplification plot that represents the accumulation of product over the duration of the entire PCR reaction. All samples were assayed in 96-well plates. The results provided are based on the means of three experiments. All expression data were normalized to 18S ribosomal RNA (rRNA). To determine mRNA expression for PMCA2 and 18S rRNA, SYBR® Green assays in a final volume of 10 µl/well were conducted. The thermocycling conditions for each isoform were 95°C for 30 sec, followed by 38 cycles at 95°C for 5 sec and 65°C (isoforms: 2b and 2w) or 63°C (isoforms: 2y, 2× and 2z) for 10 sec for all assays. For all reactions quantifying 18S rRNA, primers were included at 50 nM and were cycled in separate plates at a 1:1,000 dilution of total cDNA using the same method for each specific target. For melting curve analysis, a dissociation step (50°C for 5 sec, and then increased by 0.5°C every 5 sec until 95°C) was added. The melting peak curve was obtained using Bio-Rad CFX Manager 3.0 software. The change in fluorescence/change in temperature (−ΔF/ΔT) is plotted against temperature to obtain a clear view of the melting dynamics. Agarose gel electrophoresis of the amplified products for all primer sets revealed single bands of the expected size (Table II). The bands were extracted using the MinElute Gel Extraction kit (Qiagen GmbH, Hilden, Germany; 10 µl final volume). An aliquot (3 µl) of purified product was sequenced (NZYTech, Lda., Lisbon, Portugal) using the RT-qPCR specific primers for each isoform (Table II). Sequences were analyzed using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) and human PMCA2 gene sequence information from Ensembl [ATP2B2 ENSG00000157087; www.ensembl.org, Ensembl Release 92 (22)].
Data and statistical analysis
PMCA2 mRNA RT-qPCR data were analyzed using the comparative Cq method. Normalized mRNA expression was calculated using the ΔCq method (23) by subtracting the mean Cq-value of triplicates of selected isoforms from the mean Cq-value of triplicates of the housekeeping gene. Additionally, the expression levels of each isoform of PMCA2 mRNA relative to their expression levels in adjacent non-tumor tissues was estimated using the following steps and equations: i) Normalized level of mRNA expression of each PMCA isoform in all tumor and adjacent non-tumor samples: ΔCq=Cq of PMCA2 isoform-Cq housekeeping gene (18S rRNA). ii) Relative mRNA expression of each isoform in tumor tissue: ΔΔCq=ΔCq tumor sample-ΔCq adjacent non-tumor tissue. When no corresponding non-tumor tissue was available, relative mRNA expression was calculated vs. a non-tumor tissue sample from another patient with the identical tumor type. The relative expression (E) of each isoform (mean ± standard deviation) in a tumor sample=2(−ΔΔCq), where E>1 indicates over-expression and E<1 indicates under-expression.
The Kolmogorov-Smirnov test was used to confirm the normality of the data. In order to compare variables across different categorical clinicopathological characteristics including ER, PR and HER2 status, lymph node involvement and explanatory variables, including tumor size, histological classification and stage, the non-parametric Kruskal-Wallis test was used and the Mann-Whitney U test was used for pairwise comparison. All statistical tests were performed using the software package SPSS (version 21; IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Characterizing alternative splice variant PMCA2 mRNA expression by qualitative RT-PCR
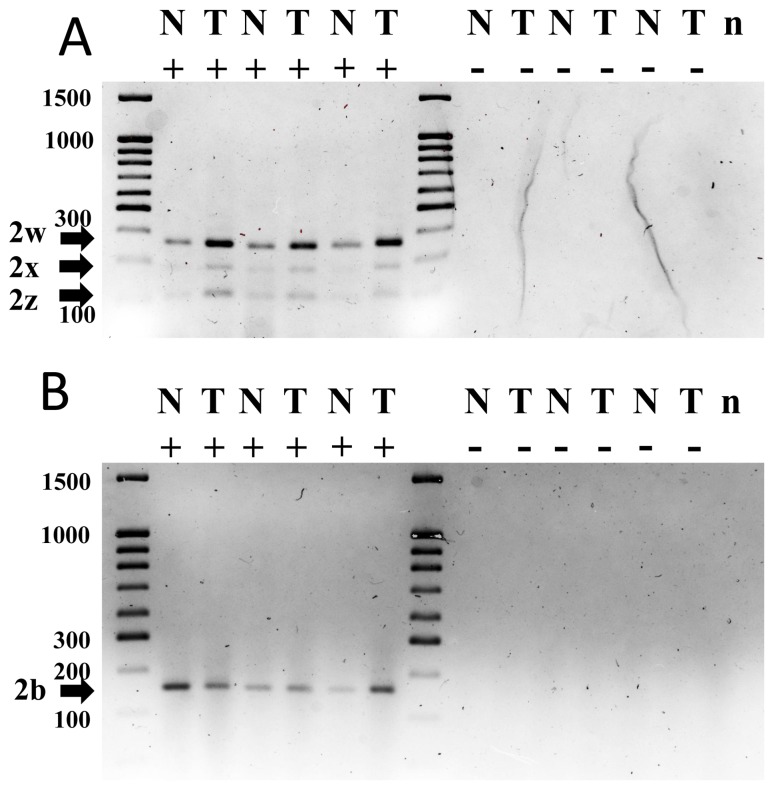
RT-PCR for PMCA2 using splice variant-sensitive primers for sites A and C determined PMCA2 products in tumor and adjacent non-tumor samples. A total of 20 samples were analyzed by RT-PCR and representative examples of samples for sites A and C are depicted in Fig. 2A and B. According to the oligos used for amplification, for site A, four RT-PCR product bands (363, 320, 270 and 228 bp) should be observed; however, the present tumor and non-tumor samples were determined to express three mRNA isoforms [PMCA2w (363 bp), PMCA2× (270 bp) and PMCA2z (228 bp)], and it was not possible to detect PMCA2y (320 bp) using this technique (Fig. 2A). For site C, three RT-PCR product bands (584, 357 and 529 bp) should have been detected; however, tumor and non-tumor samples primarily expressed PMCA2b (357 bp) mRNA, with PMCA2a (584 bp) and PMCA2c (520 bp) not detected (Fig. 2B). Bands corresponding to particular PMCA2 splice variants were then identified by comparing bp sizes of the RT-PCR products to those of published expected bp sizes (6,16,18).
Figure 2.
RT-PCR analysis of PMCA2 performed with oligos, as previously described, in order to amplify sites A and C isoforms. Total RNA was isolated from representative human breast tumor samples and adjacent tissue samples. The RT-PCR products for sites A and C were run on 2 and 1.6% electrophoresis agarose gels, respectively. RT-PCR amplifications were performed without or with reverse transcriptase. (A) For site A, three PCR amplification products were detected in the tumor and non-tumor tissue samples: PMCA2w (363 bp), PMCA2× (270 bp) and PMCA2z (228 bp). (B) For site C, one band corresponding to PMCA2b mRNA (357 bp) was observed in all the breast tissue samples and no other products were detected. The sizes of a number of DNA molecular weight markers are depicted. bp, base pairs; n, negative PCR control without complementary DNA; N, adjacent tissue samples; T, human breast tumor samples; -, without reverse transcriptase; +, with reverse transcriptase; RT-PCR, reverse transcription, polymerase chain reaction; PMCA2, plasma membrane calcium ATPase 2.
Product specificity of RT-qPCR analysis
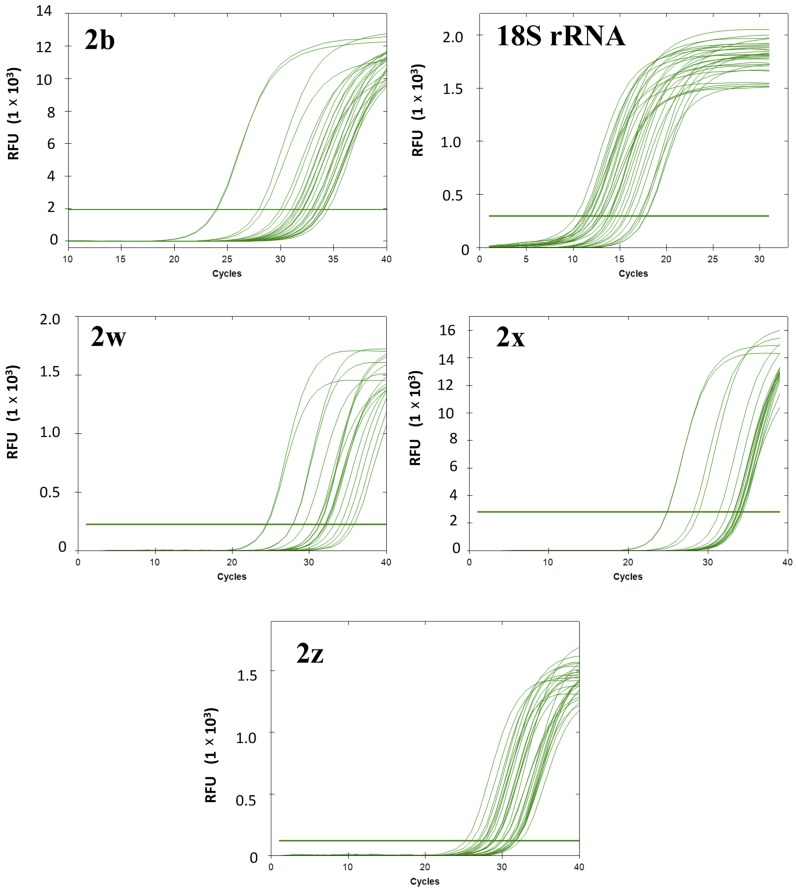
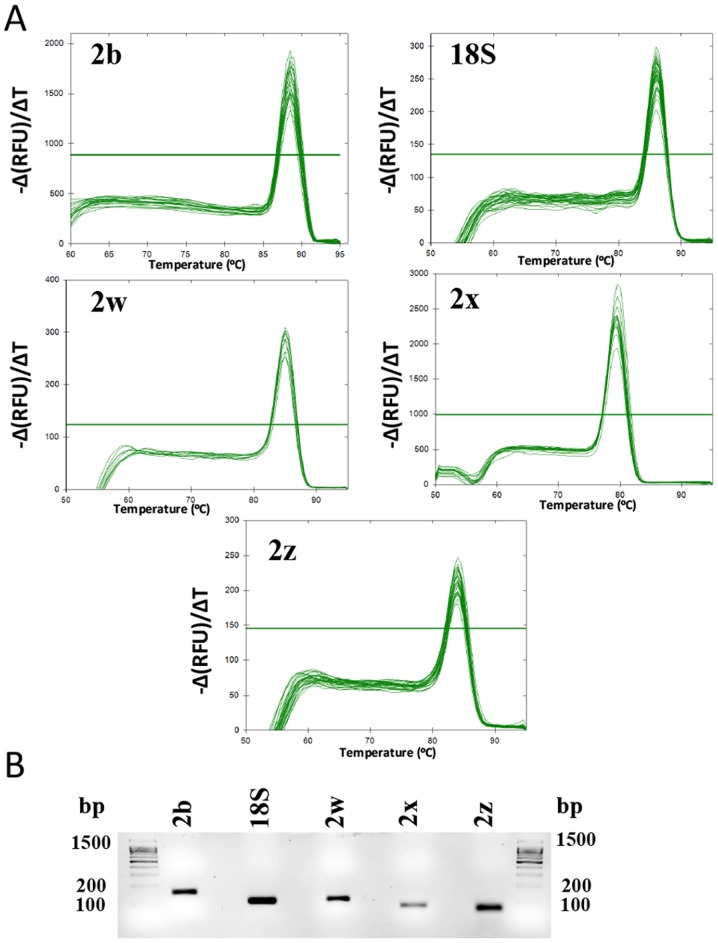
The expression of PMCA2 splice isoforms and of the reference gene (18S rRNA) was determined in tumor and non-tumor samples. As an example of the RT-qPCR results, cycle-fluorescence growth curves for each of the splicing isoforms (PMCA2b, PMCA2w, PMCA2z and PMCA2×) and 18S rRNA in a number of representative samples are depicted in Fig. 3. The PMCA2 splicing isoforms exhibited Cq values ranging from 11.05–17.61 for the reference gene (18S rRNA), 24.28–34.63 for PMCA2b, 25.36–33.34 for PMCA2z, 25.58–34.63 for PMCA2× and 23.87–34.72 for PMCA2w. In the melt peak analysis, a single homogeneous peak for all primer sets was detected (Fig. 4A). Agarose gel electrophoresis analysis of the amplified products for all primer sets revealed single bands of the expected size (Table II; Fig. 4B). Furthermore, the specificity of the PCR products was confirmed by a nested PCR for each PMCA2 isoform, where single bands of the expected size were observed for each of the PMCA2 isoforms (Fig. 5). Sequence data analysis of the qPCR products (Fig. 4B) using BLAST verified the identity of each of the amplified products as Human ATP2B2 using [Ensemble version: ENSG00000157087.18; Ensembl Release 92 (22)]. PMCA2b: ATP2B2-202 ENST00000360273.6, ATP2B2-205 ENST00000452124.2 and ATP2B2-204 ENST00000397077.6; PMCA2w: ATP2B2-202 ENST00000360273.6 and ATP2B2-214 ENST00000645850.1; PMCA2×: ATP2B2-205 ENST00000452124.2 and ATP2B2-215 ENST00000643662.1; PMCA2z: ATP2B2-204 ENST00000397077.6; and Human 18S rRNA: GenBank: KY962518.1 and Chromosome 21: 8,437,037-8,437,147 [www.ensembl.org, Ensembl Release 92 (22)].
Figure 3.
Representative reverse transcription-quantitative polymerase chain reaction. The figure depicts cycle-fluorescence growth curves for the 18S rRNA reference gene and for the four plasma membrane calcium ATPase 2 gene isoforms for a portion (20%) of the total tumor and adjacent tissue samples analyzed in the present study. Cycle quantification was measured at the quasilinear region of the curve. RFU, relative fluorescence units.
Figure 4.
Product specificity of reverse transcription-quantitation polymerase chain reaction analysis through melting curve analysis. (A) Melting curves for the 18S rRNA reference gene and the four PMCA2 gene isoforms depicting single peaks. (B) Subsequently, 3% agarose gel electrophoresis of one representative tumoral sample resulted in the amplification of a single product of the expected size for the four PMCA2 isoforms and the 18S rRNA reference gene. PMCA2, plasma membrane calcium ATPase 2; bp, base pairs; RFU, relative fluorescence units.
Figure 5.
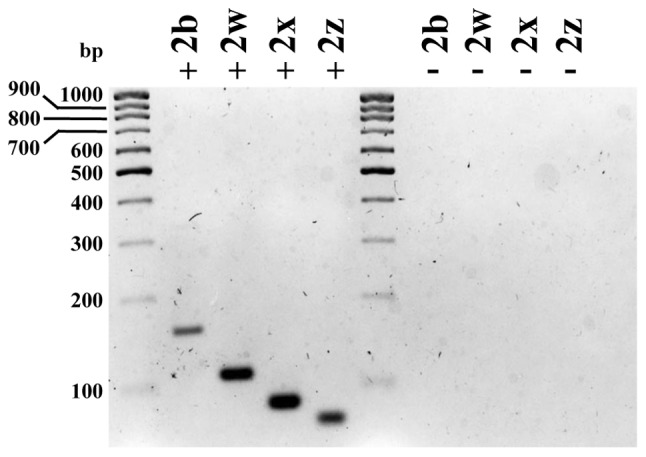
Nested PCR for plasma membrane calcium ATPase 2 using specific RT-qPCR oligos revealed single bands of the expected size, confirming the specificity of the PCR products: 2w, 152 bp; 2w, 104 bp; 2×, 80 bp; 2z, 73 bp. RT-PCR was performed using total RNA isolated from tissue adjacent to the tumor (N). For electrophoresis, PCR products were run on 3% agarose gel. The sizes of a number of DNA molecular weight markers are depicted. bp, base pairs; +, with cDNA; -, without cDNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Differential expression of alternative splicing PMCA2 mRNA isoforms in breast tumor tissues, compared with in adjacent tissues, by qPCR
mRNA expression levels of PMCA2 splice isoforms in the 85 mammary carcinoma tissues and 69 adjacent non-tumor tissues were determined by qPCR. These experiments demonstrated that, for site A, the isoforms expressed in tumor and adjacent tissues were PMCA2w, PMCA2× and PMCA2z. It was not possible to quantify the expression of isoform PMCA2y. Table I lists the relative mRNA expression levels of the different splice isoforms (PMCA2w, PMCA2z, PMCA2× and PMCA2b) recorded in tumor tissues, compared with in non-tumor tissues. These experiments confirmed that, for site C, the isoform PMCA2b was expressed in tumor and adjacent tissues, and it was not possible to quantify the expression of the PMCA2a isoform. mRNA levels for the different isoforms demonstrated high biological variability and increased expression was observed in the breast tumor tissues, compared with in adjacent tissues (Table I).
Associating PMCA2 mRNA levels with the clinical and histological characteristics of human breast tumor tissues
The expression of PMCA2 splice isoform mRNA was also assessed in association with the clinical characteristics of the patients, including tumor histological classification and size, disease stage, and lymph node involvement (Table I). Significant associations were determined between PMCA2 expression and tumor histology, with PMCA2b expression levels being significantly reduced in ductal-type tumor tissues, compared with in lobulillar-type tumor tissues (P<0.006; Table I), and PMCA2× expression was significantly increased in lobulillar-type tumor tissues, compared with in ductal-type tumor tissues (P<0.028). Additionally, non-significant associations were demonstrated between PMCA2z expression and tumor size (P<0.07). Furthermore, PMCA2× mRNA expression was increased in lymph node-negative tumor tissues, compared with lymph node-positive tumor tissues, but this difference was not significant (P=0.054; Table I).
Differential expression of alternative splicing PMCA2 mRNA isoforms, according to the hormone receptor status
The splice PMCA2 isoform mRNA expression was investigated in association with the molecular markers of breast cancer ER, PR and HER2 status (Table I). Significant associations were determined between PMCA2z expression and PR status (P<0.024), with significantly increased expression in PR-negative tumor tissues, compared with in PR-positive tumor tissues. Furthermore, when comparing expression levels of mRNA of the PMCA2 isoforms according to the ER status, significantly increased PMCA2w expression determined in the ER-positive tumor tissues, compared with in the ER-negative tumor tissues (P<0.048; Table I) and PMCA2b overexpression observed in the HER2-positive tumor tissues, compared with in HER2-negative tumor tissues.
Discussion
In humans, the expression of PMCA2 mRNA is limited to a reduced number of tissues, including the brain and mammary gland (20). Besides a role in mammary gland physiology, PMCA2 mRNA levels are elevated in breast cancer cell lines (11,12,24) and a number of studies have detected a strong association between high PMCA2 mRNA expression and reduced survival time (3,14). The present data indicated that PMCA2 mRNA is expressed in different types of human breast tumor and adjacent non-tumor tissues. Despite high variability in expression levels, these were increased in the tumor specimens, compared with in adjacent healthy tissue (Table I). The differential expression of PMCA2 could induce changes in Ca2+ efflux and this capacity has been associated with the diminished sensitivity of cells to apoptosis and an enhanced response of cancer cells to proliferative stimuli (25–28). Following thorough consideration, a cohort of Spanish patients with breast tumor types were selected to compare the expression levels of PMCA2 splice variants in breast tumor and adjacent non-tumor tissues. These were then used to examine the relevance of PMCA2 in a context of human breast cancer and the physiology of the human breast. Currently, the expression of PMCA2 splice isoforms has been described in specialized tissues (20,29), including inner ear hair cells, the nervous system and the lactating gland. PMCA2 exhibits the highest binding affinity for calmodulin, but simultaneously, in the absence of calmodulin, it exhibits high basal activity indicating its importance in specialized cells, where there is a requirement to pump Ca2+ at a high rate (30,31). Splicing at Site A inserts up to 3 exons and at site C one or two exons (Fig. 1), indicating that up to nine different variants may be produced by alternative splicing. The present data indicated that at least three different PMCA2 mRNA splice variants (PMCA2w/b PMCA2z/b and PMCA2×/b) are expressed at different levels in a number of subtypes of breast tumor and normal breast tissue. This observation is consistent with data from rodent and pre-lactation human breast models, indicating that PMCA2 is a key pump responsible for Ca2+ efflux from the maternal compartment into milk (32–34), but prompts further questions due to >1 splice isoform were detected. Previous studies regarding the mammary gland in pregnant and lactating rats demonstrated that following parturition and during lactation, total PMCA2 protein expression is upregulated in the rat mammary gland due to a predominant increase in the PMCA2w/b splice variant (32,34–37), in order to match the demands of Ca2+ homeostasis in the mammary gland. This data is consistent with functional studies indicating that the 2b splice variant has an increased affinity for calmodulin, compared with the splice variant 2a (31,32), and the 2w splice option at splice site A results in the localization of PMCA2 at apical membranes (10,19,38–40). Consistently, high expression levels of the PMCA2w/b isoform have been observed on the epithelial cell luminal membrane in tissue specimens from a patient with breast cancer in the third trimester of pregnancy (41); however, we are unaware of the significance and localization of the other two isoforms 2z/b and 2×/b detected in this study in the normal physiology of the human mammary gland and in breast cancer. Previous studies regarding recombinant protein expression have indicated that alternative splicing at site A influences the apical or basolateral localization of PMCA2, including PMCA2w, which is located at the apical membrane, and PMCA2× and PMCA2z, which are located at the basolateral membrane, and this occurs regardless of whether the COOH terminal splices correspond to the 2b or 2a isoforms (19); therefore, it would be notable to examine whether the isoforms 2z/b and 2×/b are translated into functional proteins. Furthermore, it would also be notable to examine the 5′untranslated region (UTR) of PMCA2 in humans, due to the four different mice transcriptional start regions being described in this UTR region (36), but only two are specifically used to control PMCA2 expression in the mammary gland during lactation. PMCA2w/a expression was not detected in the present human mammary tissue samples. This variant results in a truncated form of the pump due to the frame shift, which has been detected exclusively in the outer hair cells of the inner ear (29).
In the present study, there was a significantly increased expression of PMCA2×/b in lobulillar-type tumor tissues, compared with in ductal-type tumor tissues (P<0.028). This data is similar to recent evidence indicating that lobulillar and ductal are two separate subtypes of breast cancer at the clinical and molecular level, as demonstrated by different gene profiles (42). The detection of high levels of PMCA2 mRNA is consistent with the data of previous studies demonstrating that PMCA2 overexpression results in the reduced transcription activity of nuclear factor of activated T cells (NFAT) in breast cancer (2,43). Furthermore, previous studies conducted in PC12 cells indicated that NFAT inhibition is involved in the regulation of the PMCA2× splice variant (44). These results highlighted the complex association that exists between PMCA2 and NFAT signaling in breast cancer.
The present results indicated high PMCA2z expression in PR-negative tumor tissues, while PMCA2w was significantly overexpressed in ER-positive tumor tissues, indicating differences in tumor characteristics that may be associated with hormone levels, impacting the mRNA levels of these two variants. In agreement with previous data, the present data also demonstrated that PMCA2b was significantly overexpressed in HER2-positive tumor tissues (15); therefore, PMCA2b could be used as a marker for HER2-positive tumor tissues, which have been associated with a poor prognosis (45). A limitation of the present study is the lack of availability of clinical data on patients with breast cancer. Furthermore, PMCA2× mRNA expression was increased in lymph node-negative tumor tissues, compared with in lymph node-positive tumor tissues, but this difference was not significant (P=0.054); therefore, increased PMCA2× expression may be associated with less aggressive tumor behavior.
In conclusion, the present data determined the expression of a number of splice variants of PMCA2 in breast tumor and adjacent tissues, including PMCA2w, PMCA2z and PMCA2× for site A, and PMCA2b for site C. Notably, the expression in of the PMCA2z and PMCA2× variants, which are primarily expressed in brain tissue, was also determined in the breast tumor types. The differential distribution and expression of PMCA2 splice variants was dependent on hormone receptor status and histological classification of the tumor. PMCA2b was significantly overexpressed in HER2-positive tumor tissues, indicating that high mRNA levels of PMCA2b could be a marker of poor prognosis. The present data indicated PMCA2 isoform-specific differences in human breast tissues and provides direction for future studies designed to produce further insight into the role of the alternative splicing isoforms PMCA2w/b, PMCA2z/b and PMCA2×/b in breast cancer.
Acknowledgements
The authors would like to thank the Biobanc Core Facility of the Institut d'Investigacions Biomèdiques August Pi i Sunyer (Barcelona, Spain) for technical help. The authors would also like to thank Professor Margarita Rubio (Department of Basic Biomedical Sciences, Faculty of Biomedical and Health Sciences, Universidad Europea de Madrid, Madrid, Spain) for help with the statistical analysis, Mrs Maria Gregoria Montalvo-Lominchar (Doctoral Studies and Research School, Universidad Europea de Madrid, Villaviciosa de Odón, Madrid, Spain) and Mrs Carolina Sánchez (Doctoral Studies and Research School, Universidad Europea de Madrid) for technical help with the RT-qPCR assays, and Professor Mr Pablo Gómez del Arco, (Molecular Biology Department, Universidad Autónoma de Madrid, Madrid, Spain) for technical help with the sequence analysis of splicing isoforms of PMCA2 and 18S rRNA reference gene.
Glossary
Abbreviations
- bp
base pairs
- Cq
cycle quantification
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- PMCA2
plasma membrane calcium ATPase 2
- PR
progesterone receptor
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- site A
first intracellular loop of ATPase 2
- site C
C-terminal region of ATPase 2
- TNM system
Tumor-Node-Metastasis system
- 18S rRNA
18S ribosomal RNA
- nt
nucleotides
- UTR
untranslated region
Funding
The present study was funded by the Universidad Europea de Madrid (project 2014/UEM005).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ARL performed the primer design, cDNA synthesis and qPCR assays, contributed to the study design and contributed to the writing of the paper. MG performed qualitative PCR assays and contributed to the writing of the paper. ALA contributed to the study design and provided input on drafting the work and revising it critically for intellectual content. AFS contributed to the study design, statistical analysis and contributed to the writing of the paper. AN performed qPCR assays, contributed to the study design, statistical analysis and drafted the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Informed consent was obtained from all patients who donated samples to this tumor bank. The study protocol was approved by the National Research Ethics Committee of the Hospital Clinic and the Regional Research Ethics committee of the Madrid Community.
Patient consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Strehler EE, Filoteo AG, Penniston JT, Caride AJ. Plasma-membrane Ca(2+) pumps: Structural diversity as the basis for functional versatility. Biochem Soc Trans. 2007;35:919–922. doi: 10.1042/BST0350919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padányi R, Pászty K, Hegedűs L, Varga K, Papp B, Penniston JT, Enyedi Á. Multifaceted plasma membrane Ca(2+) pumps: From structure to intracellular Ca(2+) handling and cancer. Biochim Biophys Acta. 2016;1863:1351–1363. doi: 10.1016/j.bbamcr.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Stafford N, Wilson C, Oceandy D, Neyses L, Cartwright EJ. The plasma membrane calcium ATPases and their role as Major new players in human disease. Physiol Rev. 2017;97:1089–1125. doi: 10.1152/physrev.00028.2016. [DOI] [PubMed] [Google Scholar]
- 4.Brini M, Calì T, Ottolini D, Carafoli E. The plasma membrane calcium pump in health and disease. FEBS J. 2013;280:5385–5397. doi: 10.1111/febs.12193. [DOI] [PubMed] [Google Scholar]
- 5.Bruce JIE. Metabolic regulation of the PMCA: Role in cell death and survival. Cell Calcium. 2018;69:28–36. doi: 10.1016/j.ceca.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Váradi A, Molnár E, Ashcroft SJ. A unique combination of plasma membrane Ca2+-ATPase isoforms is expressed in islets of Langerhans and pancreatic beta-cell lines. Biochem J. 1996;314:663–669. doi: 10.1042/bj3140663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strehler EE. Plasma membrane calcium ATPases: From generic Ca(2+) sump pumps to versatile systems for fine-tuning cellular Ca(2.) Biochem Biophys Res Commun. 2015;460:26–33. doi: 10.1016/j.bbrc.2015.01.121. [DOI] [PubMed] [Google Scholar]
- 8.Talarico EF, Jr, Kennedy BG, Marfurt CF, Loeffler KU, Mangini NJ. Expression and immunolocalization of plasma membrane calcium ATPase isoforms in human corneal epithelium. Mol Vis. 2005;11:169–178. [PubMed] [Google Scholar]
- 9.Talarico EF, Jr, Mangini NJ. Alternative splice variants of plasma membrane calcium-ATPases in human corneal epithelium. Exp Eye Res. 2007;85:869–879. doi: 10.1016/j.exer.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calì T, Brini M, Carafoli E. Regulation of cell calcium and role of plasma membrane calcium ATPases. Int Rev Cell Mol Biol. 2017;332:259–296. doi: 10.1016/bs.ircmb.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Lee WJ, Roberts-Thomson SJ, Holman NA, May FJ, Lehrbach GM, Monteith GR. Expression of plasma membrane calcium pump isoform mRNAs in breast cancer cell lines. Cell Signal. 2002;14:1015–1022. doi: 10.1016/S0898-6568(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 12.Lee WJ, Roberts-Thomson SJ, Monteith GR. Plasma membrane calcium-ATPase 2 and 4 in human breast cancer cell lines. Biochem Biophys Res Commun. 2005;337:779–783. doi: 10.1016/j.bbrc.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 13.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 14.VanHouten J, Sullivan C, Bazinet C, Ryoo T, Camp R, Rimm DL, Chung G, Wysolmerski J. PMCA2 regulates apoptosis during mammary gland involution and predicts outcome in breast cancer. Proc Natl Acad Sci USA. 2010;107:11405–11410. doi: 10.1073/pnas.0911186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong J, VanHouten JN, Dann P, Kim W, Sullivan C, Yu H, Liotta L, Espina V, Stern DF, Friedman PA, Wysolmerski JJ. PMCA2 regulates HER2 protein kinase localization and signaling and promotes HER2-mediated breast cancer. Proc Natl Acad Sci USA. 2016;113:E282–E290. doi: 10.1073/pnas.1516138113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heim R, Hug M, Iwata T, Strehler EE, Carafoli E. Microdiversity of human-plasma-membrane calcium-pump isoform 2 generated by alternative RNA splicing in the N-terminal coding region. Eur J Biochem. 1992;205:333–340. doi: 10.1111/j.1432-1033.1992.tb16784.x. [DOI] [PubMed] [Google Scholar]
- 17.Hilfiker H, Guerini D, Carafoli E. Cloning and expression of isoform 2 of the human plasma membrane Ca2+ ATPase. Functional properties of the enzyme and its splicing products. J Biol Chem. 1994;269:26178–26183. [PubMed] [Google Scholar]
- 18.Stauffer TP, Hilfiker H, Carafoli E, Strehler EE. Quantitative analysis of alternative splicing options of human plasma membrane calcium pump genes. J Biol Chem. 1993;268:25993–26003. [PubMed] [Google Scholar]
- 19.Chicka MC, Strehler EE. Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem. 2003;278:18464–18470. doi: 10.1074/jbc.M301482200. [DOI] [PubMed] [Google Scholar]
- 20.Strehler EE, Zacharias DA. Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev. 2001;81:21–50. doi: 10.1152/physrev.2001.81.1.21. [DOI] [PubMed] [Google Scholar]
- 21.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. 7th. Springer (eds); New York: 2010. AJCC (American Joint Committee on Cancer) Cancer Staging Manual. [Google Scholar]
- 22.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Monteith GR, Davis FM, Roberts-Thomson SJ. Calcium channels and pumps in cancer: Changes and consequences. J Biol Chem. 2012;287:31666–31673. doi: 10.1074/jbc.R112.343061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts-Thomson SJ, Curry MC, Monteith GR. Plasma membrane calcium pumps and their emerging roles in cancer. World J Biol Chem. 2010;1:248–253. doi: 10.4331/wjbc.v1.i8.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu L, Tuck D, Katsaros D, Lu L, Schulz V, Perincheri S, Menato G, Scarampi L, Harris L, Yu H. Favorable outcome associated with an IGF-1 ligand signature in breast cancer. Breast Cancer Res Treat. 2012;133:321–331. doi: 10.1007/s10549-012-1952-5. [DOI] [PubMed] [Google Scholar]
- 27.Curry MC, Luk NA, Kenny PA, Roberts-Thomson SJ, Monteith GR. Distinct regulation of cytoplasmic calcium signals and cell death pathways by different plasma membrane calcium ATPase isoforms in MDA-MB-231 breast cancer cells. J Biol Chem. 2012;287:28598–28608. doi: 10.1074/jbc.M112.364737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curry M, Roberts-Thomson SJ, Monteith GR. PMCA2 silencing potentiates MDA-MB-231 breast cancer cell death initiated with the Bcl-2 inhibitor ABT-263. Biochem Biophys Res Commun. 2016;478:1792–1797. doi: 10.1016/j.bbrc.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 29.Krebs J. The plethora of PMCA isoforms: Alternative splicing and differential expression. Biochim Biophys Acta. 2015;1853:2018–2024. doi: 10.1016/j.bbamcr.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Lopreiato R, Giacomello M, Carafoli E. The plasma membrane calcium pump: New ways to look at an old enzyme. J Biol Chem. 2014;289:10261–10268. doi: 10.1074/jbc.O114.555565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brini M, Di Leva F, Ortega CK, Domi T, Ottolini D, Leonardi E, Tosatto SC, Carafoli E. Deletions and mutations in the acidic lipid-binding region of the plasma membrane Ca2+ pump: A study on different splicing variants of isoform 2. J Biol Chem. 2010;285:30779–30791. doi: 10.1074/jbc.M110.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhardt TA, Horst RL. Ca2+-ATPases and their expression in the mammary gland of pregnant and lactating rats. Am J Physiol. 1999;276:C796–C802. doi: 10.1152/ajpcell.1999.276.4.C796. [DOI] [PubMed] [Google Scholar]
- 33.Brini M, Carafoli E. Calcium pumps in Health and disease. Physiol Rev. 2009;89:1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- 34.Reinhardt TA, Lippolis JD, Shull GE, Horst RL. Null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2 impairs calcium transport into milk. J Biol Chem. 2004;279:42369–42373. doi: 10.1074/jbc.M407788200. [DOI] [PubMed] [Google Scholar]
- 35.Reinhardt TA, Lippolis JD. Mammary gland involution is associated with rapid down regulation of major mammary Ca2+-ATPases. Biochem Biophys Res Commun. 2009;378:99–102. doi: 10.1016/j.bbrc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Silverstein RS, Tempel BL. Atp2b2, encoding plasma membrane Ca2+-ATPase type 2, (PMCA2) exhibits tissue-specific first exon usage in hair cells, neurons, and mammary glands of mice. Neuroscience. 2006;141:245–257. doi: 10.1016/j.neuroscience.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 37.Faddy HM, Smart CE, Xu R, Lee GY, Kenny PA, Feng M, Rao R, Brown MA, Bissell MJ, Roberts-Thomson SJ, Monteith GR. Localization of plasma membrane and secretory calcium pumps in the mammary gland. Biochem Biophys Res Commun. 2008;369:977–981. doi: 10.1016/j.bbrc.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill JK, Williams DE, LeMasurier M, Dumont RA, Strehler EE, Gillespie PG. Splice-site A choice targets plasma-membrane Ca2+-ATPase isoform 2 to hair bundles. J Neurosci. 2006;26:6172–6180. doi: 10.1523/JNEUROSCI.0447-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antalffy G, Caride AJ, Pászty K, Hegedus L, Padanyi R, Strehler EE, Enyedi A. Apical localization of PMCA2w/b is enhanced in terminally polarized MDCK cells. Biochem Biophys Res Commun. 2011;410:322–327. doi: 10.1016/j.bbrc.2011.05.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padányi R, Xiong Y, Antalffy G, Lór K, Pászty K, Strehler EE, Enyedi A. Apical scaffolding protein NHERF2 modulates the localization of alternatively spliced plasma membrane Ca2+ pump 2B variants in polarized epithelial cells. J Biol Chem. 2010;285:31704–31712. doi: 10.1074/jbc.M110.164137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters AA, Milevskiy MJ, Lee WC, Curry MC, Smart CE, Saunus JM, Reid L, da Silva L, Marcial DL, Dray E, et al. The calcium pump plasma membrane Ca(2+)-ATPase 2 (PMCA2) regulates breast cancer cell proliferation and sensitivity to doxorubicin. Sci Rep. 2016;6:25505. doi: 10.1038/srep25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desmedt C, Zoppoli G, Sotiriou C, Salgado R. Transcriptomic and genomic features of invasive lobular breast cancer. Semin Cancer Biol. 2017;44:98–105. doi: 10.1016/j.semcancer.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Holton M, Yang D, Wang W, Mohamed TM, Neyses L, Armesilla AL. The interaction between endogenous calcineurin and the plasma membrane calcium-dependent ATPase is isoform specific in breast cancer cells. FEBS Lett. 2007;581:4115–4119. doi: 10.1016/j.febslet.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 44.Kosiorek M, Podszywalow-Bartnicka P, Zylinska L, Pikula S. NFAT1 and NFAT3 cooperate with HDAC4 during regulation of alternative splicing of PMCA isoforms in PC12 cells. PLoS One. 2014;9:e99118. doi: 10.1371/journal.pone.0099118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.