Abstract
ANaerobic AMMonium OXidation (anammox) is an established process for efficient nitrogen removal from wastewater, relying on anammox bacteria to form stable biofilms or granules. To understand the formation, structure, and stability of anammox granules, it is important to determine the composition of the extracellular polymeric substances (EPS). The aim of this research was to elucidate the nature of the proteins, which are the major fraction of the EPS and were suspected to be glycosylated. EPS were extracted from full-scale anammox granular sludge, dominated by “Candidatus Brocadia”, and subjected to denaturing polyacrylamide gel electrophoresis. By further analysis with mass spectrometry, a high abundant glycoprotein, carrying a heterogeneous O-glycan structure, was identified. The potential glycosylation sequence motif was identical to that proposed for the surface layer protein of “Candidatus Kuenenia stuttgartiensis”. The heavily glycosylated protein forms a large fraction of the EPS and was also located by lectin staining. Therefore, we hypothesize an important role of glycoproteins in the structuring of anammox granules, comparable to the importance of glycans in the extracellular matrix of multicellular organisms. Furthermore, different glycoconjugates may have distinct roles in the matrix of granular sludge, which requires more in-depth characterization of different glycoconjugates in future EPS studies.
1. Introduction
Anammox (anaerobic ammonium oxidation) bacteria are a distinct phylogenetic group within the phylum of Planctomycetes, that can convert nitrite and ammonium into nitrogen gas.1 They are widely occurring in nature and applied in wastewater treatment plants (WWTPs) to remove nitrogen from wastewater. This process has the advantage over the conventional nitrogen removal process that it requires no oxygen and no organic carbon source. The anammox process is applied in biofilm or granular sludge systems. Because of the high settling velocity of granules, these systems allow for sufficient biomass retention and require less space compared to reactors with flocculent sludge.2 For the stability and efficiency of WWTP processes, the stability of the granular sludge is of high importance. Although the mechanism of granule formation is not well understood, it is generally accepted that extracellular polymeric substances (EPS) are a key factor.3,4
Similar as in biofilms, in granular sludge, EPS are the components that form the matrix wherein the microorganisms are embedded. EPS are reported as proteins (structural proteins and enzymes), polysaccharides, nucleic acids, and lipids.5 Many functions are assigned to EPS, including functions related to structure and stability, like promoting aggregation, maintaining the physical structure of granules, retaining water, and serving as protective barrier for the cells.6,7 However, in most systems, knowledge on the exact composition of EPS and a link to the function of the individual components has not been established to date. To gain a better understanding of the structure and the stability of the granular sludge EPS matrix, establishment of methods and protocols for both extraction and proper characterization is required.8−10 Especially the subset of EPS components that provide the physical structure (structural EPS) needs to be determined and characterized. Importantly, many of the traditional EPS extractions that are focused on avoiding cell lysis do not solubilize the structural polymers. Therefore, they do not allow to analyze the structural polymers from biofilms and granular sludge.8 In order to find new targets in the EPS matrix to study, it is required to solubilize the matrix. Once new targets are analyzed, their extracellular origin should be verified. With this approach the release of intracellular components during extraction is not a problem.
Characterization of proteins and polysaccharides in EPS has been mostly limited to the use of colorimetric assays. Colorimetric assays have a relatively low specificity and may cause a significant over- or underestimation of individual components.9,10 Moreover, the use of those methods only allows for characterization of the separate classes of molecules but provides no insight into the macromolecular structure of the components. Some studies speculate that the proteins and polysaccharides in EPS are not solely present as separate components but also in various forms of glycoconjugates.11,12 While historically glycosylation of proteins used to be considered to occur exclusively in eukaryotes, today it is accepted that also prokaryotes can perform (complex) protein glycosylation.13 In eukaryotes, glycoproteins fulfill important roles in the extracellular matrix, e.g. in cell–cell interactions, protecting the cells and providing a hydrated gel matrix.14 Prokaryotic protein glycosylation, however, is far from well-studied due to its complexity and enormous diversity. Furthermore, it has been mainly studied in relation with pathogenic traits.15,16 Therefore, studying protein glycosylation in a purely environmental sample such as the extracellular polymers of granular sludge is of special interest. Bourven et al.17 found glycoproteins in anaerobic granular sludge, and very recently, glycosylated amyloid-like proteins in the structural EPS of aerobic granular sludge were reported by Lin et al.18 However, the presence of glycoproteins in EPS was only proposed by colocalization and orthogonal staining experiments but never followed up by in-depth molecular characterization. To find direct proof for glycosylated proteins in EPS, more dedicated methodologies are required.
Here, we are presenting the identification and in-depth characterization of glycoproteins from anammox granular sludge which will aid in providing a better understanding of structural EPS. For this, granular sludge from a full-scale anammox WWTP was used to extract EPS by using an alkaline extraction. Proteins were analyzed using SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) in combination with carbohydrate specific stains, followed by high-resolution mass spectrometry for protein identification and determination of the glycan attachment sites as well as the glycan composition. In addition, lectin staining was applied to localize glycoconjugates in the granules.
2. Materials and Methods
2.1. Anammox Granular Sludge
Anammox granular sludge was collected from the full-scale anammox reactor in Dokhaven (Sluisjesdijk), Rotterdam.19 The VSS (volatile suspended solids) content of the granules was 0.71 g/g granules (determined in accordance with APHA, 2005).20 To identify the dominant anammox species in the granules, a clone library analysis was performed. This was complemented by Fluorescent In Situ Hybridization (FISH), which was performed as described by Johnson et al.21 More detailed information on dominant species analysis is provided in the Supporting Information.
2.2. Extraction of EPS
Freeze-dried anammox granules were incubated in 0.1 M NaOH (50 mg/mL) for 5 h, while being stirred with a magnetic stirrer at 400 rpm. After centrifugation at 4000 rpm for 20 min at 4 °C, the pellet was discarded. Polymers in the supernatant were precipitated out by decreasing the pH to 5 using 1 M HCl. The precipitated polymers were collected by centrifugation at 4000 rpm for 20 min at 4 °C and lyophilized.
2.3. Composition Analysis of Extracted EPS
The elemental composition of lyophilized EPS was analyzed in terms of C, H, N, O, and S. Carbon (C), hydrogen (H), and nitrogen (N) were determined by purge-and-trap chromatography (VARIO Elementar EL, Elementar), and sulfur (S) was deteremined by ion chromatography (IC 883 Plus, Methrom). Oxygen was taken as the residual organic fraction. The proteins in the EPS were measured as BSA (Bovine Serum Albumin) equivalents using the bicinchoninic acid (BCA) method (Interchim Uptima BC assay quantitation kit). The carbohydrates were measured as glucose equivalents using the phenol-sulfuric acid assay.22
2.4. SDS-PAGE Analysis and Staining Experiments
SDS-PAGE was performed by using NuPage Novex 4–12% Bis-Tris gels (Invitrogen). EPS samples were prepared in NuPAGE LDS-buffer, and DTT (dithiothreitol) was added to a final concentration of 10 mM. The proteins were denatured by incubation at 70 °C for 10 min. Subsequently, a 10 μL (16 μg of EPS) sample was loaded per well. The Thermo Scientific Spectra Multicolor Broad Range Protein Ladder was used as molecular weight marker. The gel electrophoresis was performed at 200 V for 35 min. The gels were stained by three different stains afterward.
For visualization of proteins, the Colloidal Blue staining kit (Invitrogen) was used according to the manufacturer’s instructions. For visualization of glycoproteins, the Thermo Scientific Pierce Glycoprotein Staining Kit was used, which is based on the periodic acid-Schiff (PAS) method. This method is specific for glycans bearing vicinal hydroxyl groups. Horseradish Peroxidase was employed as positive control, and Soybean Trypsin Inhibitor was used as negative control. For staining of acidic glycoproteins, Alcian Blue 8GX (Fluka, Sigma-Aldrich) was used. Alcian Blue is a cationic dye. It was employed to stain dissociated (ionic) acidic groups on the carbohydrates. To differentiate between the relatively weaker acidic groups like carboxylate (R-COO–) and the stronger acidic group like sulfate (R-OSO3–), staining with Alcian Blue was performed at different pH values, namely pH 2.5 and pH 1.0. Due to the different dissociation constants (Ka) of the acidic groups, at pH 2.5 carboxylate and sulfate groups are stained by Alcian Blue, while at pH 1.0 only sulfate groups are stained.23 For staining at pH 2.5, an adapted protocol of Møller and Poulsen (2009)24 was used. After electrophoresis, the gels were extensively washed in solution I (25% (v/v) ethanol and 10% (v/v) acetic acid) for 2.5 h while refreshing the solution 4 times. Subsequently, the gel was stained in 0.125% (w/v) Alcian Blue in solution I (the solution was stirred overnight to dissolve the Alcian Blue and centrifuged before use) for 30 min and washed in solution I overnight. For staining of sulfated groups at pH 1.0, the same protocol was performed except that solution I was replaced by solution II (0.1 M HCl and 25% (v/v) ethanol, according to Tobisawa et al.25).
2.5. Enrichment of the 80 kDa Glycoprotein
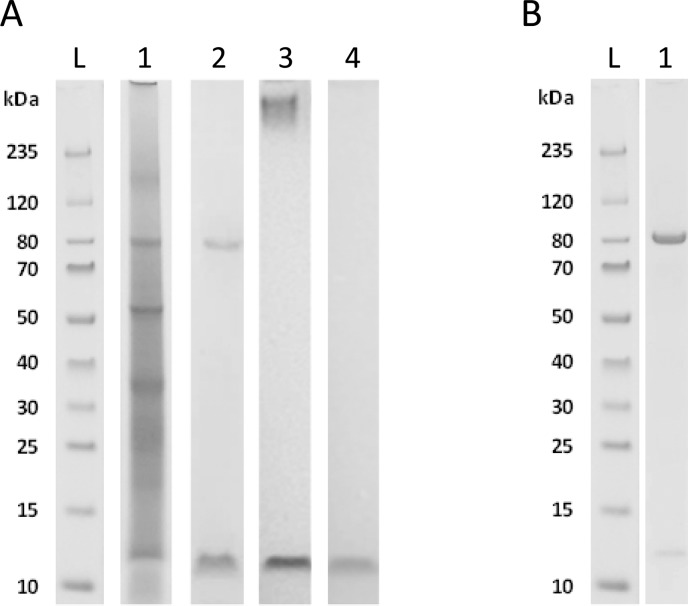
The enrichment of the glycoprotein stained at 80 kDa was achieved following a protocol for the extraction of S-layer proteins with LiCl.26 0.5 g of the extracted EPS was added to 50 mL of 5 M LiCl and stirred on ice for 30 min at 400 rmp. The mixture was centrifuged for 15 min at 13000 rpm. The supernatant was dialyzed at 4 °C against milli-Q water and subsequently lyophilized.
2.6. In-Gel Proteolytic Digestion and Protein Identification
Following SDS-PAGE analysis and Coomassie staining, the 80 kDa gel band originating from approximately 1–2 μg of protein was cut from the gel. After destaining, an in-gel trypsin digestion was performed overnight at 37 °C. Peptides were extracted and subjected to nanoLC-MS/MS analysis on an Ultimate 3000 HPLC (Dionex, San Donato Milanese, Milano, Italy) coupled to a LTQ-Orbitrap mass spectrometer (Thermo Fisher, Bremen, Germany). For each solution a volume of 5 μL was directly injected on a self-made nanocolumn packed with an Aeris Peptide XB-C18 phase (75 μm i.d. × 15 cm, 3.6 μm, 100 Å, Phenomenex, Torrance, CA, USA) and eluted at 300 nL/min flow rate. Solvent A consisted of 3% acetonitrile in H2O containing 0.1% formic acid, and solvent B consisted of 80% acetonitrile in H2O containing 0.1% formic acid. The elution gradient program was as follows: 0 min, 2% B; 40 min, 2% B; 68 min, 15% B; 168 min, 25% B; 228 min, 35% B; 273 min, 50% B; 274 min, 90% B; 288 min, 90% B; 289 min, 2% B; and 309 min, 2% B. Mass spectra were acquired in positive ion mode, setting the spray voltage at 1.8 kV. Data were acquired in data dependent mode with dynamic exclusion enabled; survey MS scans were recorded in the Orbitrap analyzer in the mass range of 300–2000 m/z; then up to five of the most intense ions in each full MS scan were fragmented. Data was analyzed against a bacteria-wide proteome database (SwissProt, 24/11/17) or a “Candidatus (Ca.) Brocadia” focused proteome database (TrEMBL, 24/11/17), using X!tandem (release ALANINE, The GPM) and PEAKS Studio 8.5 (for parameters see the Supporting Information). The best match protein sequence was further analyzed with the InterPro protein analysis tool.27
For further analysis of the glycopeptides, the extracted peptides were injected to a LC-MS/MS system using reverse phase chromatography (ACQUITY M class ultraperformance liquid chromatography (UPLC) connected to an ESI-Q-TOF Premier mass spectrometer, Waters). Solvent A consisted of 3% acetonitrile in H2O containing 0.1% formic acid, and solvent B consisted of 90% acetonitrile in H2O containing 0.1% formic acid. A linear gradient was performed from 5 to 75% solvent B over 30 min at a constant flow rate of 5 μL/min. Data dependent analysis was performed selecting the 2–3 most intense peaks from each scan for collision-induced dissociation (CID).28 Data were analyzed manually using MassLynx 4.1. Peak lists were exported using the msconvertGUI (ProteoWizard) and analyzed with X!tandem (The GPM, release ALANINE) using a “Ca. Brocadia” protein specific database.
2.7. In-Gel Glycan Release and Composition Analysis
As described in section 2.6, following SDS-PAGE analysis and Coomassie staining, the 80 kDa gel band, originating from approximately 1–2 μg of protein, was cut from the gel. The O-glycans were further released from the protein backbone by in-gel reductive beta-elimination using 1 M NaBH in 0.5 M NaOH at 50 °C and overnight incubation.29 Prior to MS analysis, released O-glycans were purified using a Hypercarb SPE cartridge (Thermo Fisher Scientific).30 The purified fraction was further analyzed using LC-ESI-MS/MS with a Hypercarb Porous Graphitic Carbon (PGC) stationary phase (0.32 × 150 mm, 5 μm).29,31 Solvent A consisted of 3% acetonitrile in H2O containing 0.1% formic acid, and solvent B consisted of 90% acetonitrile in H2O containing 0.1% formic acid. A linear gradient was performed from 5 to 70% solvent B within 15 min maintaining a constant flow rate of 9 μL/min using a UPLC pump system (ACQUITY M class, Waters). The UPLC system was coupled to an ESI-Q-TOF mass spectrometer (Waters Premier), which was operated in positive ion mode (ES+). Fragmentation experiments were performed on the identified glycan peaks using CID in separate analysis runs. Data were analyzed using MassLynx 4.1, and annotation of fragment ion peaks was done by using Glycoworkbench 2.1.32
2.8. Imaging of Glycoconjugates
The granules were stained and mounted in coverwell chambers with a 1 mm spacer in order to avoid squeezing of the samples. Glycoconjugates of the anammox granules were examined by means of fluorescence lectin bar-coding.33 Thus, all commercially available lectins (FITC or Alexa488) were applied as an individual probe to one granule. For 3d imaging a TCS SP5X confocal laser scanning microscope (Leica, Germany) was employed. The upright microscope was equipped with a super continuum light source and controlled by the software LAS AF 2.4.1. The confocal data sets were recorded by using 25× NA 0.95 and 63× NA 1.2 water immersion lenses. Excitation was at 490 nm (laser power 70% at laser, 50% in software), and emission signals were detected simultaneously with two photomultipliers from 485 to 495 nm (reflection) and 505–600 nm (fluorescence). Image data sets were deconvolved with Huygens version 16.05 using the CMLE algorithm (SVI, The Netherlands) and projected with Imaris version 9.1.2 (Bitplane, Switzerland).
3. Results
3.1. Extraction and Composition of EPS from Anammox Granular Sludge
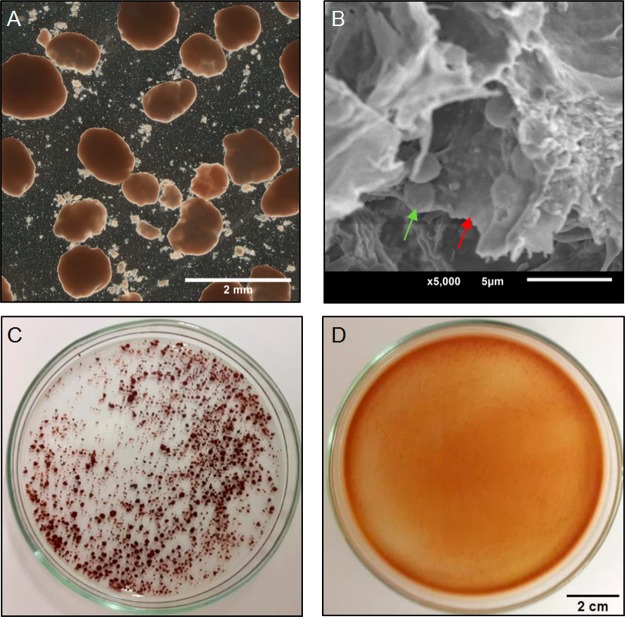
The granular sludge collected from the wastewater treatment plant Dokhaven was enriched with a species that was very close to “Ca. Brocadia sp. 40”, which is renamed to “Ca. Brocadia sapporoensis”.34 (See supplemental Figure S1.) For solubilization of the granular sludge, the granules were incubated in a stirred 0.1 M NaOH solution for 5 h at room temperature. This procedure fully disintegrated the granular structure (Figure 1). A mixture of sol-like liquid and mineral particles was formed, indicating the organic matrix of the granules was solubilized. After precipitating the polymers with HCl, 0.20 ± 0.04 g/g organic dry weight was recovered from the granular sludge. In Table 1 the basic characterization of the composition of the recovered EPS is given. The main part of the recovered EPS consists of proteins, which is in agreement with the high content of nitrogen from the elemental composition analysis. Interestingly also the sulfur content was relatively high (in comparison with the average sulfur content in protein, which is ∼0.3%).
Figure 1.
A) Optical microscope image of anammox granular sludge from the WWTP. B) Scanning electron microscope image of the inside of a broken granule where bacteria (green arrow) can be seen, embedded in the EPS matrix (red arrow). C) Anammox granules before and D) after incubation for 5 h in 0.1 M NaOH.
Table 1. Protein and Carbohydrate Contents and the Elemental Composition of the Extracted EPS from Anammox Granules.
| elemental
composition (weight-%) |
||||||
|---|---|---|---|---|---|---|
| proteins mg/g EPS | carbohydrates mg/g EPS | C | H | N | O | S |
| 599 ± 4 | 49.0 ± 2 | 40.5 | 6.6 | 9.0 | 35.7 | 1.4 |
3.2. SDS-PAGE Analysis
The proteins in the extracted EPS were further characterized using SDS-PAGE in combination with different stains. Following Coomassie Blue staining, bands were observed at molecular weights of ca. 80, 55, 35, 20, and 12 kDa (Figure 2A, lane 1). Although proteins were the major component of the extracted EPS material, only a few predominant bands were observed. This is different from intracellular protein profiles, which typically show bands over the whole molecular weight range. PAS stained 2 bands, at approximately 80 kDa and 12 kDa, confirming the presence of carbohydrates in the protein extract (Figure 2A, lane 2). In addition, Alcian Blue staining was applied with pH 2.5 (Figure 2A, lane 3) and pH 1.0 (Figure 2A, lane 4). With pH 2.5, carboxylate and sulfate groups are stained, while with pH 1.0 only the sulfate groups are stained. It was observed that at pH 1.0, only the band at 12 kDa was stained by Alcian Blue, indicating the presence of sulfate groups. This is in correspondence with the high sulfur content that was measured by the elemental composition analysis. It was also observed that a smear appeared at the high molecular weight range (above 235 kDa), when the gel was stained with Alcian Blue at pH 2.5. However, no band was observed at this position when using Coomassie Blue or PAS, indicating either a heavily negatively charged carbohydrate polymer or aggregate-like structure. Carbohydrate branches may shield the protein backbone from staining by Coomassie Blue. Furthermore, the large amount of acidic carbohydrate residues may lead to an unevenly distributed charge on the molecules, inhibiting the formation of a dense band on the gel.24 This smeared band in the high molecular weight range is similar to what is observed for mucin-like proteins, when stained with Alcian Blue at pH 2.5.35,36
Figure 2.
A) Proteins of EPS from anammox granules were analyzed by SDS-PAGE and stained with Coomassie Blue for proteins (lane 1), with PAS for (neutral) carbohydrates (lane 2), with Alcian Blue at pH 2.5 for acidic carbohydrates (lane 3), and with Alcian Blue at pH 1.0 for the presence of sulfate groups (lane 4). Besides bands only stained by Coomassie Blue, there was one major band at 80 kDa which also showed a strong PAS staining. A smeared band above 235 kDa marker was only visible with Alcian Blue staining at pH 2.5. A low molecular weight band at 12 kDa was stained with Alcian Blue also at pH 1.0 indicating the presence of sulfate groups. B) Coomassie Blue staining shows the enrichment of the glycoprotein at approximately 80 kDa from the bulk EPS following a protocol of Lortal et al.26 established for LiCl based S-layer protein extraction (lane 1). Lane L is the molecular weight ladder.
In summary, besides a few major bands stained with Coomassie Blue, carbohydrate and acid specific staining revealed three bands in the solubilized EPS extract: (i) a high molecular weight smeared band containing acidic groups, (ii) a band at 80 kDa with both protein and carbohydrate positive staining, and (iii) at approximately 12 kDa a band stained positive for sulfate groups. Based on these results, our further analysis was focused on the protein band with an apparent molecular weight of 80 kDa because it showed an abundant band with a clear staining with both Coomassie and PAS. The protein appeared at a molecular weight which fits in the range of surface layer (S-layer) proteins, making it an interesting target. The high molecular weight smear may be associated with a large aggregate or a mucin-like structure. The relatively low molecular weight 12 kDa band, on the other hand, may originate from a carbohydrate/peptide-like structure, or another highly sulfated polymer, rather than from a glycoprotein.
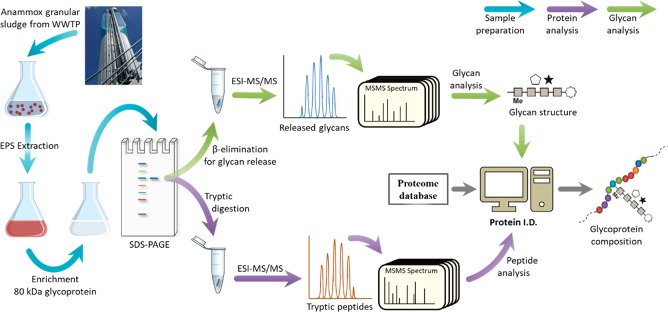
3.3. Enrichment and MS Analysis of the 80 kDa Glycoprotein
The strategy to characterize the target glycoprotein that appeared at 80 kDa is illustrated in Figure 3. First, the glycoprotein was enriched using a LiCl based surface-layer protein extraction protocol (Figure 2B). The Coomassie stained band was excised from the gel and subjected to proteolytic digestion using trypsin. The extracted peptides were analyzed by tandem mass spectrometry using a LTQ-Orbitrap mass spectrometer. There was no match found with any known S-layer protein from the database. Instead, the database search, using a bacteria-wide and further a “Ca. Brocadia” focused proteome database, uncovered the protein sequence of a hypothetical protein of “Ca. Brocadia sapporoensis” (WP_070066018.1) as strongest match, as well as a sequence identical with a small C-terminal fragment of WP_070066018.1 (OQD46794.1). (For the summary of the database search results see supplemental Tables S1–S4.) Unfortunately, it was noticed that the sequence for WP_070066018.1 was retracted from the protein database recently. On the other hand, the same protein shows high sequence homology to another protein from “Ca. Brocadia sapporoensis” (WP_070066019.1). (For results for sequence alignment see Figure S2.)
Figure 3.
Workflow for the identification of glycoproteins from granular sludge sample, following recently established protocols.29,37
3.4. Analysis of Glycopeptides and Glycan Structure Following in-Gel Glycan Release
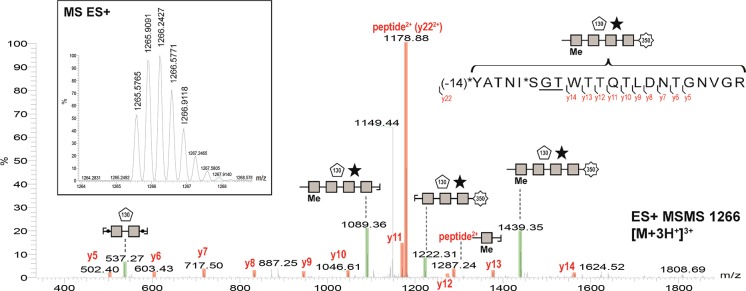
Following a closer investigation of the tandem-MS fragmentation spectra of the 80 kDa gel band, an approximately 1.5 kDa glycan was found attached to at least four different peptides, all matching the sequence of WP_070066018.1 very closely. The fragmentation spectrum of one of the glycopeptides is shown in Figure 4. The peak at 1287.24 represents a fragment that contains both the protein and the glycan and therefore confirms the identification of a glycoprotein. In addition, following further manual investigation of the spectra, indications for additional glycosylation sites were found, which could not be clearly assigned to any annotated protein sequence due to a limited number of fragments. In all four glycopeptides identified, the sequence motif GTX (Glycine-Threonine-any amino acid) was present. Overall, the GTX motif was found 15 times within the sequence of WP_070066018.1 which would indicate the potential for a very high degree of glycosylation (see supplemental Figure S5). The molecular weight of the best matched protein is 53 kDa, while the apparent molecular weight of the glycoprotein on the SDS-PAGE was approximately 80 kDa. A substantially higher apparent molecular weight on the SDS-PAGE compared to the theoretical molecular weight has been frequently observed for glycosylated proteins.
Figure 4.
Fragmentation spectrum of the glycopeptide matching the sequence WP_070066018.1 of “Ca. Brocadia sapporoensis”. The peak at 1287.24 represents a fragment that contains a part of both the protein and the glycan.
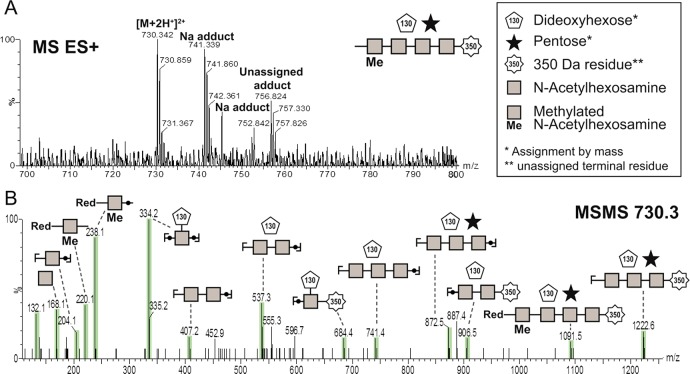
To determine the composition of the discovered glycan, in-gel β-elimination was applied to release the presumed O-linked glycan from the protein backbone. Subsequent PGC-MS/MS analysis of the released structure confirmed the presence of a glycan with a monoisotopic mass of 1456.6 Da. Fragmentation experiments further showed that the glycan is a heptameric structure with 4 different types of residues, as indicated in Figure 5 (and supplemental Figure S7). A methylated N-acetyl hexosamine (HexNAc) backbone, substituted with a pentose and a dideoxyhexose residue, was annotated by mass. In addition, the glycan carried a 350 Da large terminal residue. The 350 Da terminal residue is unique and, to the best of the knowledge of the authors, has not been reported before and therefore could not be further specified by mass.
Figure 5.
Analysis of the glycan structure composition following in-gel glycan release via β-elimination. A) shows the precursor ion and B) shows the spectrum after collision-induced dissociation (CID).
3.5. Lectin Staining
To demonstrate that glycoproteins are located outside the cell or within the EPS matrix the granules were subjected to fluorescence lectin staining. After screening all commercially available lectins, Vicia graminea (VGA) labeled with fluoresceine isothiocyanate (FITC) was selected as it showed a strong signal of the granule substructure. VGA is mostly known from medical applications for detecting O-linked Galactose/GalNAc epitopes. Interestingly, VGA is reported to only recognize peptide linked carbohydrate conjugates, while it does not react with free carbohydrates.38,39 In Figure 6 a maximum intensity projection of 53 images recorded by confocal laser microscopy is presented indicating the high abundance of glycoproteins throughout the granule. It is reasonable to assume VGA binding to the HexNAc constituents of the 80 kDa protein identified here, as it is a highly abundant O-glycosylated protein.
Figure 6.
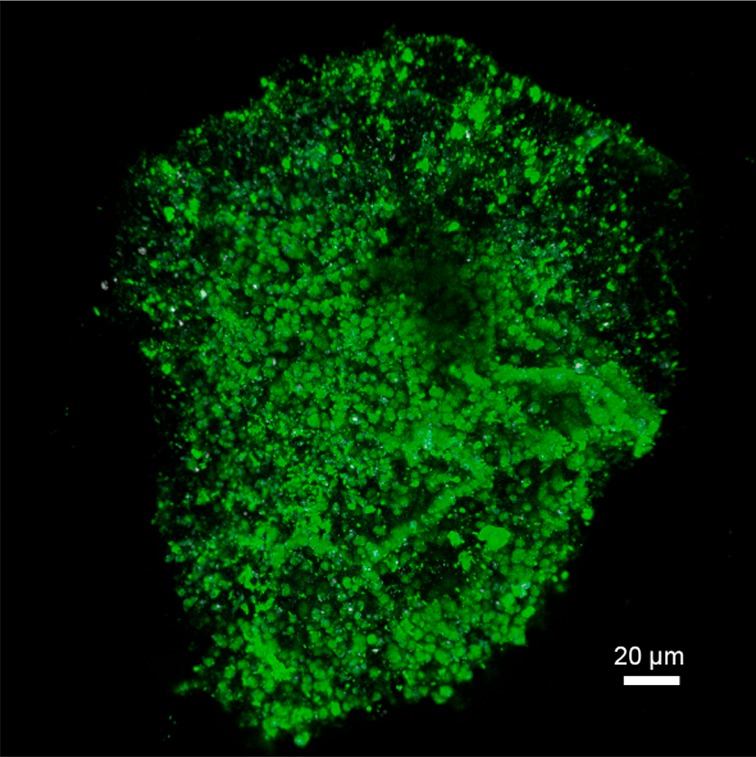
Confocal laser scanning microscopy showing a maximum intensity projection of 53 optical sections 1 μm apart. The anammox granule was stained with the lectin from Vicia graminea (VGA-FITC). VGA is reported to be specific for peptide linked glycan conjugates, while it does not bind to the free carbohydrates.38,39
4. Discussion
EPS are the key factor when it comes to understanding the structure and the stability of anammox granular sludge. In the present study, EPS were extracted from granular sludge from a full-scale anammox WWTP. An alkaline extraction was used, because it solubilized the granular shape, indicating that a significant part of the structural polymers was solubilized. Because the EPS are a complex network, there is no single method to extract all the EPS components. To study other possible EPS components, different extraction methods are required. Alkaline treatment can extract negatively charged components and can break disulfide bonds in proteins, making it easier to extract them.40 Due to the acidic nature of the EPS, solubilizing them can cause a decrease in pH. A high concentration of NaOH was used to maintain a high pH during the whole extraction. The alkaline extraction resulted in new targets to study. Glycoproteins and acidic (including sulfated) glycoconjugates were found with the appropriate staining experiments. A relatively high abundant glycoprotein with an apparent molecular weight of 80 kDa was further characterized. It was found to be heavily glycosylated with a heterogeneous O-glycan. To our knowledge, little or no literature provides direct evidence of glycoproteins in granular sludge or natural biofilms. In previous EPS studies, glycoproteins would have been overlooked due to the fact that proteins and polysaccharides are often studied separately, and unspecific colorimetric assays are commonly used.
The apparent molecular weight of the 80 kDa glycoprotein fitted in the molecular weight range for S-layer proteins and the glycoprotein was successfully enriched with a protocol for S-layer extraction. This suggested the targeted glycoprotein to be an S-layer protein. S-layer proteins are often glycosylated and are one of the most commonly observed cell surface structures of prokaryotes.41 Since they form the outermost layer of the cells, they are directly involved in the interactions between the cell and its environment, which makes it an interesting target in biofilm research. Mass spectrometric analysis of the 80 kDa glycoprotein showed that the sequence of the target protein was not identical to any known anammox S-layer protein annotated in the database. This could be due to the fact that there is in general low homology among S-layer proteins.42 The identified glycoprotein might be a not yet annotated protein from a “Ca. Brocadia” species, since there were strong indications for single amino acid polymorphism within the matched sequence regions and some other segments of the sequence were not matched at all. Mass spectrometric analysis revealed a previously retracted protein sequence from “Ca. Brocadia sapporoensis” (WP_070066018.1) and a C-terminal fragment of the same protein (OQD46794.1) as the closest match.
The draft genome of “Ca. Brocadia sp. 40” was published in 2016 by Ali et al.43 In 2017 the bacteria were analyzed in more detail and were renamed “Ca. Brocadia sapporoensis”.34 Although the closest match sequence (WP_070066018.1) is retracted, it is very similar to WP_070066019.1 of “Ca. Brocadia sapporoensis”, and also other “Ca. Brocadia species” have a similar protein (see supplemental Figure S8). Therefore, the sequence WP_070066018.1 was considered representative and was used to predict structural aspects of the identified glycoprotein. These aspects were found to be comparable to the 250 kDa S-layer glycoprotein Kustd1514. This glycoprotein was recently identified in another anammox species, “Ca. Kuenenia stuttgartiensis”, by van Teeseling et al.44,45 (The similarities are also valid for WP_070066019.1.) Interestingly, using the InterPro protein analysis tool, the 80 kDa glycoprotein was predicted to contain an Immunoglobulin-like (Ig-like) domain. This is also found for Kustd1514. (See supplemental Figures S3 and S4 for sequence analysis.) Ig-like domains are observed in cell surface proteins and have different functions, including cell–cell recognition and cell surface receptor functions.46 The highly abundant amino acids, 16% threonine, 9% serine, and 9% glycine, suggest a structural role, similar as for Kustd1514 (14% threonine, 12% serine, and 10% glycine). Also in terms of glycan attachment site (GTX) and glycan structure the 80 kDa glycoprotein was comparable with Kustd1514.45 In this study at least 4 O-glycan attachment sites could be identified. Considering the number of GTX motifs within the proposed protein sequence, an even higher degree of glycosylation could be assumed. The HexNAc backbone, substituted with a pentose and a dideoxyhexose residue, was also comparable to Kustd1514.45
Based on the findings of this study, we hypothesize that the found glycoprotein is an S-layer protein. Although S-layer proteins are one of the most commonly observed cell surface structures of prokaryotes,42 there is no general function assigned. One of the functions that are proposed in literature is a role in biofilms. For example, S-layer proteins of Tannerella forsythia have been found to be up-regulated when grown as biofilms.29 It would be of interest to study the role of S-layers in the structure of the matrix of granules. The best way to identify S-layers is by visualizing their specific pattern on the cell surface with microscopy. However, because of the compact EPS around the cells it is more difficult to visualize the S-layer proteins in mature granules than in suspended biomass from lab-scale reactors. In addition, S-layers can be shed of the cells47 and therefore can be potentially integrated in the matrix. To elucidate the potential role of the identified glycoprotein in granules, more specific studies on the localization of the protein are required to study whether it is attached to the outside of the cell wall or integrated within the matrix. Apart from a potential structural role in supporting the formation of a gel matrix, the heterologous glycan structure which is linked to the protein via a methylated HexNAc may provide a very efficient protective layer against degradation.48 This could be of high importance for slow growing micro-organisms.
In addition to the O-glycosylated 80 kDa protein, the staining experiments showed the presence of other glycoconjugates: a large conjugate (>235 kDa) or polymer-like structure with carboxylate residues and a sulfate containing structure of approximately 12 kDa. Currently the extracellular matrix of biofilms is recognized to be a highly complex and organized structure and sometimes even considered comparable to the extracellular matrix of multicellular organisms.49,50 Glycoproteins and (sulfated) proteoglycans are major components of the extracellular matrix of mammalian cells.14 In the current research, glycoconjugates with both carboxylate and sulfate groups are present in the recovered EPS, and glycoproteins were detected in situ throughout the whole granule. In addition, the high molecular weight smear that appeared on the SDS-PAGE resembled properties of mucin-like compounds. Mucins are glycoproteins which form mucus gels as a protective barrier around the epithelial cells and aid against infection and dehydration.51 Likewise, the EPS matrix of granules is considered as a hydrogel-like matrix which protects the cells and allows nutrients to diffuse to the cells.52 The finding of the various glycoconjugates in the anammox granules reflects the idea of similarity between biofilms and multicellular organisms, regarding the extracellular matrix.
Remarkably, most of the prokaryotic protein glycosylation with higher complexity have been associated with pathogenic traits of bacteria but were rarely described in natural biofilm communities. The results presented in this study demonstrate that it is necessary to include glycoproteins as a major target in the EPS research field. The proteins and glycans should be studied in the context of an integrated structure, since the physical properties and the biological function of glycoproteins are determined by the combination of both parts.53 To achieve this, in-depth molecular analyses that will allow a deeper understanding of the matrix structure in biofilms and granular sludge are significantly required.
Acknowledgments
This research was funded by the SIAM Gravitation Grant 024.002.002, The Netherlands Organization for Scientific Research. We thank Tommaso Lotti (University of Florence, Italy) and Francesca Boscaro (CISM, University of Florence, Italy) for performing the LTQ-orbitrap MS measurements. Yingyu Law and Thi Quynh Ngoc Nguyen (SCELSE, Singapore) are acknowledged for performing the clone library analysis. We also thank Laura van Niftrik and Huub Op den Camp (Radboud University, The Netherlands) for helpful discussions about S-layer proteins and comments on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b03180.
The authors declare no competing financial interest.
Supplementary Material
References
- Neumann S.; van Teeseling M. C. F.; van Niftrik L.. Cell Biology of Anaerobic Ammonium-Oxidizing Bacteria: Unique Prokaryotes with an Energy-Conserving Intracellular Compartment. In Planctomycetes: Cell Structure, Origins and Biology; Humana Press: 2013; pp 89–123, 10.1007/978-1-62703-502-6_4. [DOI] [Google Scholar]
- Nicolella C.; van Loosdrecht M. C. M.; Heijnen J. J. Wastewater Treatment with Particulate Biofilm Reactors. J. Biotechnol. 2000, 80 (1), 1–33. 10.1016/S0168-1656(00)00229-7. [DOI] [PubMed] [Google Scholar]
- Seviour T.; Yuan Z.; van Loosdrecht M. C. M.; Lin Y. Aerobic Sludge Granulation: A Tale of Two Polysaccharides?. Water Res. 2012, 46 (15), 4803–4813. 10.1016/j.watres.2012.06.018. [DOI] [PubMed] [Google Scholar]
- Grotenhuis J. T. C.; Smit M.; van Lammeren A. A. M.; Stams A. J. M.; Zehnder A. J. B. Localization and Quantification of Extracellular Polymers in Methanogenic Granular Sludge. Appl. Microbiol. Biotechnol. 1991, 36 (1), 115–119. 10.1007/BF00164710. [DOI] [Google Scholar]
- Flemming H.-C.; Wingender J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8 (9), 623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Wingender J.; Neu T. R.; Flemming H.-C.. What Are Bacterial Extracellular Polymeric Substances? In Microbial Extracellular Polymeric Substances: Characterization, Structure and Function; Wingender J., Neu T. R., Flemming H.-C., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 1999; pp 1–19. [Google Scholar]
- Neu T. R.; Lawrence J. R.. The Extracellular Matrix–an Intractable Part of Biofilm Systems. Perfect Slime—Microbial Extracell. Subst.; Flemming H.-C., Wingender J., Neu T. R., Eds.; 2016; pp 25–60. [Google Scholar]
- Felz S.; Al-Zuhairy S.; Aarstad O. A.; van Loosdrecht M. C. M.; Lin Y. M. Extraction of Structural Extracellular Polymeric Substances from Aerobic Granular Sludge. J. Vis. Exp. 2016, 10.3791/54534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le C.; Stuckey D. C. Colorimetric Measurement of Carbohydrates in Biological Wastewater Treatment Systems: A Critical Evaluation. Water Res. 2016, 94, 280–287. 10.1016/j.watres.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Le C.; Kunacheva C.; Stuckey D. C. Protein” Measurement in Biological Wastewater Treatment Systems: A Critical Evaluation. Environ. Sci. Technol. 2016, 50 (6), 3074–3081. 10.1021/acs.est.5b05261. [DOI] [PubMed] [Google Scholar]
- Görner T.; de Donato P.; Ameil M.-H.; Montarges-Pelletier E.; Lartiges B. S. Activated Sludge Exopolymers: Separation and Identification Using Size Exclusion Chromatography and Infrared Micro-Spectroscopy. Water Res. 2003, 37 (10), 2388–2393. 10.1016/S0043-1354(02)00553-5. [DOI] [PubMed] [Google Scholar]
- Jorand F.; Boué-Bigne F.; Block J. C.; Urbain V. Hydrophobic/hydrophilic Properties of Activated Sludge Exopolymeric Substances. Water Sci. Technol. 1998, 37 (4–5), 307–315. 10.2166/wst.1998.0652. [DOI] [Google Scholar]
- Messner P. Prokaryotic Protein Glycosylation Is Rapidly Expanding from “curiosity” to “ubiquity.. ChemBioChem 2009, 10 (13), 2151–2154. 10.1002/cbic.200900388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. L.; Lehninger A. L.; Cox M. M.. Lehninger Principles of Biochemistry; Macmillan: 2008; 10.1007/978-3-662-08289-8. [DOI] [Google Scholar]
- Tan F. Y. Y.; Tang C. M.; Exley R. M. Sugar Coating: Bacterial Protein Glycosylation and Host-Microbe Interactions. Trends Biochem. Sci. 2015, 40 (7), 342–350. 10.1016/j.tibs.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Nothaft H.; Szymanski C. M. Bacterial Protein N-Glycosylation: New Perspectives and Applications. J. Biol. Chem. 2013, 288 (10), 6912–6920. 10.1074/jbc.R112.417857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourven I.; Bachellerie G.; Costa G.; Guibaud G. Evidence of Glycoproteins and Sulphated Proteoglycan-like Presence in Extracellular Polymeric Substance from Anaerobic Granular Sludge. Environ. Technol. 2015, 36 (19), 2428–2435. 10.1080/09593330.2015.1034186. [DOI] [PubMed] [Google Scholar]
- Lin Y.; Reino C.; Carrera J.; Pérez J.; van Loosdrecht M. C. M. Glycosylated Amyloid-like Proteins in the Structural Extracellular Polymers of Aerobic Granular Sludge Enriched with Ammonium-Oxidizing Bacteria. MicrobiologyOpen 2018, e00616. 10.1002/mbo3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Star W. R. L.; Abma W. R.; Blommers D.; Mulder J.-W.; Tokutomi T.; Strous M.; Picioreanu C.; van Loosdrecht M. C. M. Startup of Reactors for Anoxic Ammonium Oxidation: Experiences from the First Full-Scale Anammox Reactor in Rotterdam. Water Res. 2007, 41 (18), 4149–4163. 10.1016/j.watres.2007.03.044. [DOI] [PubMed] [Google Scholar]
- American Public Health Association (APHA), Standard Methods for the Examination of Water and Wastewater, 21st ed.; 2005; Washington, DC. [Google Scholar]
- Johnson K.; Jiang Y.; Kleerebezem R.; Muyzer G.; van Loosdrecht M. C. M. Enrichment of a Mixed Bacterial Culture with a High Polyhydroxyalkanoate Storage Capacity. Biomacromolecules 2009, 10 (4), 670–676. 10.1021/bm8013796. [DOI] [PubMed] [Google Scholar]
- Dubois M.; Gilles K. A.; Hamilton J. K.; Rebers Pa.; Smith F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28 (3), 350–356. 10.1021/ac60111a017. [DOI] [Google Scholar]
- Shori D. K.; Kariyawasam H. H.; Knight R. A.; Hodson M. E.; Genter T.; Hansen J.; Koch C.; Kalogeridis A. Sulphation of the Salivary Mucin MG1 (MUC-5B) Is Not Correlated to the Degree of Its Sialylation and Is Unaffected by Cystic Fibrosis. Pfluegers Arch. 2001, 443 (1), S50–S54. 10.1007/s004240100644. [DOI] [PubMed] [Google Scholar]
- Møller H. J.; Poulsen J. H.. Staining of Glycoproteins/proteoglycans on SDS-Gels. In The Protein Protocols Handbook; Springer: 2009; pp 773–777, 10.1385/1-59259-169-8:773. [DOI] [Google Scholar]
- Tobisawa Y.; Imai Y.; Fukuda M.; Kawashima H. Sulfation of Colonic Mucins by N-Acetylglucosamine 6-O-Sulfotransferase-2 and Its Protective Function in Experimental Colitis in Mice. J. Biol. Chem. 2010, 285 (9), 6750–6760. 10.1074/jbc.M109.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortal S.; van Heijenoort J.; Gruber K.; Sleytr U. B. S-Layer of Lactobacillus Helveticus ATCC 12046: Isolation, Chemical Characterization and Re-Formation after Extraction with Lithium Chloride. J. Gen. Microbiol. 1992, 138 (3), 611–618. 10.1099/00221287-138-3-611. [DOI] [Google Scholar]
- Finn R. D.; Attwood T. K.; Babbitt P. C.; Bateman A.; Bork P.; Bridge A. J.; Chang H.-Y.; Dosztányi Z.; El-Gebali S.; Fraser M.; Gough J.; Haft D.; Holliday G. L.; Huang H.; Huang X.; Letunic I.; Lopez R.; Lu S.; Marchler-Bauer A.; Mi H.; Mistry J.; Natale D. A.; Necci M.; Nuka G.; Orengo C. A.; Park Y.; Pesseat S.; Piovesan D.; Potter S. C.; Rawlings N. D.; Redaschi N.; Richardson L.; Rivoire C.; Sangrador-Vegas A.; Sigrist C.; Sillitoe I.; Smithers B.; Squizzato S.; Sutton G.; Thanki N.; Thomas P. D.; Tosatto S. C. E.; Wu C. H.; Xenarios I.; Yeh L.-S.; Young S.-Y.; Mitchell A. L. InterPro in 2017-beyond Protein Family and Domain Annotations. Nucleic Acids Res. 2017, 45 (D1), D190–D199. 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M.; Catalina M. I.; Deelder A. M.; Hokke C. H. Glycoproteomics Based on Tandem Mass Spectrometry of Glycopeptides. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2007, 849 (1–2), 115–128. 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Posch G.; Pabst M.; Brecker L.; Altmann F.; Messner P.; Schäffer C. Characterization and Scope of S-Layer Protein O-Glycosylation in Tannerella Forsythia. J. Biol. Chem. 2011, 286 (44), 38714–38724. 10.1074/jbc.M111.284893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer N. H.; Lawson M. A.; Jardine D. R.; Redmond J. W. A General Approach to Desalting Oligosaccharides Released from Glycoproteins. Glycoconjugate J. 1998, 15 (8), 737–747. 10.1023/A:1006983125913. [DOI] [PubMed] [Google Scholar]
- Pabst M.; Altmann F. Influence of Electrosorption, Solvent, Temperature, and Ion Polarity on the Performance of LC-ESI-MS Using Graphitic Carbon for Acidic Oligosaccharides. Anal. Chem. 2008, 80 (19), 7534–7542. 10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- Ceroni A.; Maass K.; Geyer H.; Geyer R.; Dell A.; Haslam S. M. GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res. 2008, 7 (4), 1650–1659. 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Neu T. R.; Kuhlicke U. Fluorescence Lectin Bar-Coding of Glycoconjugates in the Extracellular Matrix of Biofilm and Bioaggregate Forming Microorganisms. Microorganisms 2017, 5 (1), 5. 10.3390/microorganisms5010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita Y.; Zhang L.; Kimura Z.; Ali M.; Fujii T.; Okabe S. Enrichment and Physiological Characterization of an Anaerobic Ammonium-Oxidizing Bacterium “Candidatus Brocadia Sapporoensis.. Syst. Appl. Microbiol. 2017, 40 (7), 448–457. 10.1016/j.syapm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Pearson R.; Tellam R.; Xu B.; Zhao Z.; Willcox M.; Kongsuwan K.; Kritaya K. Isolation, Biochemical Characterization and Anti-Adhesion Property of Mucin from the Blue Blubber Jellyfish (Catostylus Mosaicus). Biosci. Methods 2016, 2 (4), 21–30. 10.5376/bm.2011.02.0004. [DOI] [Google Scholar]
- Fallon M. A.; Latchney L. R.; Hand A. R.; Johar A.; Denny P. A.; Georgel P. T.; Denny P. C.; Culp D. J. The Sld Mutation Is Specific for Sublingual Salivary Mucous Cells and Disrupts Apomucin Gene Expression. Physiol. Genomics 2003, 14 (2), 95–106. 10.1152/physiolgenomics.00151.2002. [DOI] [PubMed] [Google Scholar]
- Kolarich D.; Jensen P. H.; Altmann F.; Packer N. H. Determination of Site-Specific Glycan Heterogeneity on Glycoproteins. Nat. Protoc. 2012, 7 (7), 1285–1298. 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
- Prigent M. J.; Verez Bencomo V.; Sinaÿ P.; Cartron J. P. Interaction of Synthetic Glycopeptides Carrying Clusters of O-Glycosidic Disaccharide Chains (β-D-Gal(1–3)-α-D-GalNAc) with β-D-Galactose-Binding Lectins. Glycoconjugate J. 1984, 1 (1), 73–80. 10.1007/BF01875414. [DOI] [Google Scholar]
- Duk M.; Lisowska E.; Kordowicz M.; Wasniowska K. Studies on the Specificity of the Binding Site of Vicia Graminea Anti-N Lectin. Eur. J. Biochem. 1982, 123 (1), 105–112. 10.1111/j.1432-1033.1982.tb06505.x. [DOI] [PubMed] [Google Scholar]
- Nielsen P. H.; Jahn A.. Extraction of EPS. In Microbial Extracellular Polymeric Substances: Characterization, Structure and Function; Wingender J., Neu T. R., Flemming H.-C., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 1999; pp 49–72. [Google Scholar]
- Sleytr U. B.; Messner P.; Pum D.; Sara M. Crystalline Bacterial Cell Surface Layers (S Layers): From Supramolecular Cell Structure to Biomimetics and Nanotechnology. Angew. Chem., Int. Ed. 1999, 38 (8), 1034–1054. . [DOI] [PubMed] [Google Scholar]
- Sleytr U. B.; Schuster B.; Egelseer E.-M.; Pum D. S-Layers: Principles and Applications. FEMS Microbiol. Rev. 2014, 38 (5), 823–864. 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.; Haroon M. F.; Narita Y.; Zhang L.; Rangel Shaw D.; Okabe S.; Saikaly P. E. Draft Genome Sequence of the Anaerobic Ammonium-Oxidizing Bacterium “ Candidatus Brocadia Sp. 40.. Genome Announc. 2016, 4 (6), e01377-16. 10.1128/genomeA.01377-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling M. C. F.; de Almeida N. M.; Klingl A.; Speth D. R.; Op den Camp H. J. M.; Rachel R.; Jetten M. S. M.; van Niftrik L. A New Addition to the Cell Plan of Anammox bacteria: “Candidatus Kuenenia Stuttgartiensis” has a Protein Surface Layer as the Outermost Layer of the Cell. J. Bacteriol. 2014, 196 (1), 80–89. 10.1128/JB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Teeseling M. C. F.; Maresch D.; Rath C. B.; Figl R.; Altmann F.; Jetten M. S. M.; Messner P.; Schäffer C.; van Niftrik L. The S-Layer Protein of the Anammox Bacterium Kuenenia Stuttgartiensis Is Heavily O-Glycosylated. Front. Microbiol. 2016, 7, 1721. 10.3389/fmicb.2016.01721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavkov T.; Egelseer E. M.; Tesarz M.; Svergun D. I.; Sleytr U. B.; Keller W. The Structure and Binding Behavior of the Bacterial Cell Surface Layer Protein SbsC. Structure 2008, 16 (8), 1226–1237. 10.1016/j.str.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Sleytr U. B.; Beveridge T. J. Bacterial S-Layers. Trends Microbiol. 1999, 7 (6), 253–260. 10.1016/S0966-842X(99)01513-9. [DOI] [PubMed] [Google Scholar]
- Van Klinken B. J.; Dekker J.; Büller H. A.; Einerhand A. W. Mucin Gene Structure and Expression: Protection vs. Adhesion. Am. J. Physiol. 1995, 269 (5), G613–G627. 10.1152/ajpgi.1995.269.5.G613. [DOI] [PubMed] [Google Scholar]
- Nikolaev Y. A.; Plakunov V. K. Biofilm—“City of Microbes” or an Analogue of Multicellular Organisms?. Microbiology 2007, 76 (2), 125–138. 10.1134/S0026261707020014. [DOI] [PubMed] [Google Scholar]
- Flemming H. C.; Wingender J. Relevance of Microbial Extracellular Polymeric Substances (EPSs) - Part I: Structural and Ecological Aspects. Water Sci. Technol. 2001, 43 (6), 1–8. 10.2166/wst.2001.0326. [DOI] [PubMed] [Google Scholar]
- Christensen B. E.Physical and Chemical Properties of Extracellular Polysaccharides Associated with Biofilms and Related Systems. In Microbial Extracellular Polymeric Substances: Characterization, Structure and Function; Wingender J., Neu T. R., Flemming H.-C., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 1999; pp 143–154, 10.1007/978-3-642-60147-7_8. [DOI] [Google Scholar]
- Seviour T.; Pijuan M.; Nicholson T.; Keller J.; Yuan Z. Understanding the Properties of Aerobic Sludge Granules as Hydrogels. Biotechnol. Bioeng. 2009, 102 (5), 1483–1493. 10.1002/bit.22164. [DOI] [PubMed] [Google Scholar]
- Krištić J.; Lauc G.. Ubiquitous Importance of Protein Glycosylation. In High-Throughput Glycomics and Glycoproteomics: Methods and Protocols; Lauc G., Wuhrer M., Eds.; Springer New York: New York, NY, 2017; pp 1–12, 10.1007/978-1-4939-6493-2_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.