Abstract
Extended-spectrum β-lactamases (ESBLs) confer resistance to clinically important third-generation cephalosporins, which are often used to treat invasive salmonellosis. In the United States, ESBLs are rarely found in Salmonella. However, in 2014, the US Food and Drug Administration found blaCTX-M-65 ESBL-producing Salmonella enterica serotype Infantis in retail chicken meat. The isolate had a rare pulsed-field gel electrophoresis pattern. To clarify the sources and potential effects on human health, we examined isolates with this pattern obtained from human surveillance and associated metadata. Using broth microdilution for antimicrobial susceptibility testing and whole-genome sequencing, we characterized the isolates. Of 34 isolates, 29 carried the blaCTX-M-65 gene with <9 additional resistance genes on 1 plasmid. Of 19 patients with travel information available, 12 (63%) reported recent travel to South America. Genetically, isolates from travelers, nontravelers, and retail chicken meat were similar. Expanded surveillance is needed to determine domestic sources and potentially prevent spread of this ESBL-containing plasmid.
Keywords: antibacterial agents, antimicrobial resistance, beta-lactamases, drug resistance, bacteria, poultry, Salmonella, extended-spectrum β-lactamase–producing, ESBL, CTX-M-65, serotype Infantis, travel, treatment failure, United States, Salmonella enterica
A leading cause of bacterial foodborne disease in the United States is nontyphoidal Salmonella (1). Salmonella enterica serotype Infantis (hereafter called Salmonella Infantis) is one of the most common Salmonella serotypes in the United States and 1 of 3 serotypes for which incidence has substantially increased (by 60%) in the past 10 years (2). Salmonella Infantis has been identified in a variety of foods, animals, and environmental settings.
Salmonellosis usually causes a self-limited gastroenteritis; however, current guidelines recommend that antimicrobial therapy be considered for groups of persons at increased risk for invasive infection. For those patients, treatment with ceftriaxone, ciprofloxacin, trimethoprim/sulfamethoxazole, or amoxicillin is recommended (3). Extended-spectrum β-lactamases (ESBLs) confer resistance to most third-generation cephalosporins and penicillins, including ampicillin.
Some Enterobacteriaceae produce CTX-M ESBLs, which are encoded by blaCTX-M genes that were discovered in 1989 (4). Since then, their prevalence has increased dramatically (5) and they have been isolated worldwide, primarily from Escherichia coli (6). Identification of ESBLs, including the CTX-M types, in Salmonella in the United States is relatively rare (7,8).
In the United States, the National Antimicrobial Resistance Monitoring System (NARMS) is a surveillance system that tracks changes in the antimicrobial susceptibility of certain enteric bacteria isolated from ill persons, retail meats, and food animals. In July 2015, the US Food and Drug Administration (FDA) notified the Centers for Disease Control and Prevention (CDC) of a CTX-M-65–producing Salmonella Infantis strain isolated from retail chicken meat in December 2014 (9). The isolate had a rare pulsed-field gel electrophoresis pattern, JFXX01.0787 (pattern 787). To clarify the sources and potential effects of this strain on human health, we analyzed data from several CDC surveillance systems to describe the prevalence, epidemiology, antimicrobial drug resistance, and molecular phylogenetics of Salmonella Infantis pattern 787 isolates from humans.
Methods
Background Rates
To determine the expected demographics, rates of hospitalization, and international travel among patients with Salmonella Infantis infections compared with patients with infections caused by other common nontyphoidal Salmonella serotypes, we analyzed data collected through the Foodborne Disease Active Surveillance Network (FoodNet; https://www.cdc.gov/foodnet/index.html) during 2012–2015. Begun in 1996, FoodNet has conducted active, population-based surveillance for culture-confirmed cases of infection caused by 9 pathogens transmitted commonly through food, including Salmonella. FoodNet is a collaboration of CDC, 10 state health departments, the US Department of Agriculture Food Safety and Inspection Service (USDA-FSIS), and the FDA. The FoodNet surveillance area includes 15% of the US population; these data are used to estimate the burden of US foodborne illnesses, hospitalizations, and deaths (1). We defined other common nontyphoidal Salmonella as the top 20 S. enterica serotypes (excluding Infantis) isolated from humans: Typhimurium, Enteritidis, Newport, Heidelberg, Javiana, Saintpaul, Montevideo, Agona, Oranienburg, Muenchen, Thompson, Hadar, Braenderup, Derby, I 4,[5],12:i:-, Paratyphi B var. L(+) tartrate+, Blockley, Anatum, Mississippi, and Panama. These 20 serotypes represented 69% of nontyphoiodal Salmonella isolates reported to FoodNet in 2015.
Case Finding
We looked for pattern 787 isolates reported to the National Molecular Subtyping Network for Foodborne Disease Surveillance (PulseNet; https://www.cdc.gov/pulsenet/index.html) through 2017. The PulseNet database contains pulsed-field gel electrophoresis patterns from state and local public health laboratories and food regulatory agencies. Only the first isolate from each patient was included in case counts.
CDC also requested patient data and clinical isolates from state and local public health departments for any case with pattern 787 reported through October 2015. Patient data included age, sex, date of symptom onset, hospitalization, and recent international travel (defined as travel outside of the United States in the 7 days before symptom onset). Isolate data included specimen collection date(s) and source.
Isolate Characterization
We used the NARMS standard broth microdilution protocol (Sensititer; Thermo Fisher Scientific, Oakwood Village, OH, USA) to determine the MICs for 14 antimicrobial agents: gentamicin, streptomycin, ampicillin, amoxicillin/clavulanic acid, ceftiofur, ceftriaxone, cefoxitin, azithromycin, sulfasoxazole, trimethoprim/sulfamethoxazole, chloramphenicol, ciprofloxacin, nalidixic acid, and tetracycline (10). These agents were categorized into 9 classes defined by the Clinical and Laboratory Standards Institute guidelines (https://clsi.org/); where available, CLSI interpretive criteria were used to define resistance. Transformation studies to confirm plasmid-associated genes were conducted by using plasmid purification and electroporation as previously described (11).
Whole-genome sequencing was performed according to the PulseNet protocol for the Illumina MiSeq (Illumina, La Jolla, CA, USA) (12). Closed PacBio genomes were generated as part of a previous study and used as references where indicated (8). Genome assemblies for short-read data were generated de novo and analyzed for acquired antimicrobial drug–resistance determinants and plasmid replicons by using ResFinder and PlasmidFinder (13,14). To confirm the absence of certain genes, we performed read mapping in CLC Genomics Workbench version 8.5 (https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/). To identify mutational resistance, we extracted the gyrA and parC genes from genome assemblies by using a perl script (https://github.com/lskatz/lskScripts/blob/master/blastAndExtract.pl). To identify mutations in the quinolone resistance–determining regions of these genes, we aligned gene sequences in CLC Workbench. To assess isolate relatedness, we generated high-quality single-nucleotide polymorphism (hqSNP) phylogenies. In brief, isolates were aligned to the closed chromosomal sequence of 2014AM-3028 (omitting the plasmid contig) by using Lyve-SET-v1.1.4f (https://github.com/lskatz/lyve-SET/). Genome alignments were processed by using Gubbins (https://sanger-pathogens.github.io/gubbins/) to omit areas of recombination, uninformative sites were removed, and the resulting SNP alignment was used to calculate pairwise differences and generate hqSNP trees by using scripts bundled with Lyve-SET (15,16). We performed a phylogeographic analysis (a type of molecular clock analysis) by using Bayesian Evolutionary Analysis Sampling Trees (BEAST; https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003537).
To sample more diverse Salmonella Infantis isolates, we obtained sequence data on isolates from sources other than CDC NARMS assigned to the same National Center for Biotechnology Information (NCBI) SNP cluster PDS000003955.192 as our study isolates on the NCBI Pathogen Detection page (17). These additional genomes were from isolates recovered from hospitalized patients, meat, and environmental samples from Peru during 2010–2014 and collected by the Center for Food Safety and Applied Nutrition at FDA and the US Naval Medical Research Unit (18), and isolates from chicken samples obtained at slaughter by USDA-FSIS. These isolates were used to produce a time-measured maximum clade credibility tree to estimate the dates that study isolates in addition to all isolates (study isolates and isolates from Peru and cecal isolates from USDA-FSIS) shared most recent common ancestors (MRCAs), and the geographic location of these MRCAs (18). The date of the MRCA was estimated by reporting the height of the most ancestral node of the maximum clade credibility tree and the 95% highest posterior density (HPD) intervals for these estimates. More details on genetic analyses are available in the Technical Appendix.
Statistical Analyses
We used the χ2 test (or Fisher exact test when cell counts were ≤5) for statistical comparisons. All denominators exclude persons for whom data were missing. We considered p<0.05 to be significant. All p values were 2-tailed. We used SAS 9.3 or 9.4 (SAS Institute, Cary, NC, USA) to conduct our analyses.
Results
The first pattern 787 Salmonella Infantis isolate from a human in the United States was collected from a patient in June 2012. By the end of 2017, PulseNet contained 312 Salmonella Infantis pattern 787 isolates from persons living in 43 states; Washington, DC; and Puerto Rico. The number of cases detected each year increased from 5 in 2012 to 174 in 2017 and represented 8.4% of all Salmonella Infantis isolated that year (Figure 1).
Figure 1.
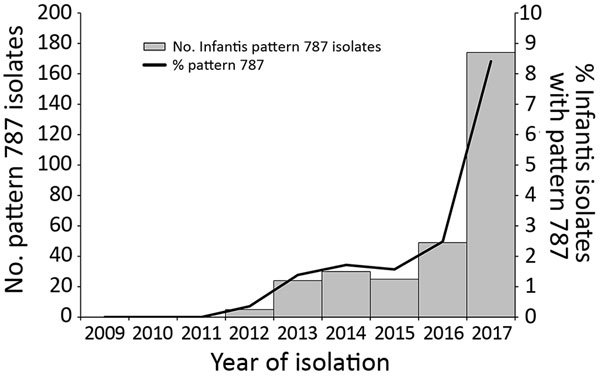
Total of 312 Salmonella enterica serotype Infantis isolates from humans with pattern JFXX01.0787 as a percentage of all Salmonella Infantis isolates by year, United States, 2012–2017. Source: PulseNet (https://www.cdc.gov/pulsenet/index.html).
State health departments submitted 34 pattern 787 isolates from humans to CDC; 29 (85%) had resistance phenotypes consistent with ESBL-conferred resistance to ceftriaxone, ceftiofur, and ampicillin (Tables 1, 2). All 29 isolates with ESBL-resistant phenotypes had the blaCTX-M-65 gene. In addition to ESBL-conferred resistance, these isolates were also resistant to chloramphenicol, sulfisoxazole, tetracycline, nalidixic acid, and trimethoprim/sulfamethoxazole and intermediately susceptible to ciprofloxacin and gentamicin. Resistance to these drugs was plasmid mediated and transferable to E. coli, except for nalidixic acid and ciprofloxacin, for which resistance was caused by a chromosomal mutation in gyrA (D87Y). Transferable resistance resulted from a large IncFIB-like plasmid containing the resistance genes aph(3′)-Ic, aph (4)-Ia, aadA1, aac (3)-IVa, blaCTX-M-65, fosA, floR, sul1, tetA, and dfrA14. These observations are consistent with the organization of these genes on a single IncF1B-like plasmid closed by using long read sequencing, which was completed as part of a previous study by Tate et al., who recently published a further detailed description of this plasmid and a comparative genomics analysis of blaCTX-M-65–positive IncFIB-like plasmids from Salmonella Infantis in the United States (9).
Table 1. Characteristics of patients and 30 Salmonella enterica serotype Infantis pattern JFXX01.0787 isolates associated with CTX-M-65, United States*.
| Accession no.† | Year | Specimen source | Resistance profile | Plasmids | gyrA | parC | Hospitalized | Recent travel | Prolonged infection‡ |
|---|---|---|---|---|---|---|---|---|---|
| SRR2485281 |
2012 |
Urine |
ACSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
Yes |
Peru |
|
| SRR2485278 |
2012 |
Blood |
ACSuTCxTioFoxCip(I)NalCotGen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
Peru |
Yes |
| SRR4019593 |
2013 |
Feces |
ASuTCxTioCip(I)NalCotGen |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
|
| SRR4019592 |
2013 |
Feces |
ACSuTCxTioCip(I)NalCotGen |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
|
| SRR2485284 |
2013 |
Feces |
ACSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
Yes |
| SRR4025935 |
2013 |
Feces |
ACSuTCxTioCip(I)Nal |
IncFIB-like |
D87Y |
Wt |
No |
Peru |
|
| SRR4025936 |
2013 |
Feces |
ACSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No data |
Peru |
Yes |
| SRR3178069 |
2013 |
Feces |
ACSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
Peru |
Yes |
| SRR4019589 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot
Gen |
IncFIB-like |
D87Y |
Wt |
No |
Ecuador |
|
| SRR2407791 |
2014 |
Retail chicken meat |
ACTCxTioCip(I)NalGen(I) |
IncFIB-like |
D87Y |
Wt |
NA |
NA |
NA |
| SRR2353201 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
|
| SRR4025938 |
2014 |
Feces |
ACSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
|
| SRR2485287 |
2014 |
Feces |
ACSSuTAug(I)CxTioFox
Cip(I)NalCotGen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
Yes |
| SRR4019601 |
2014 |
Feces |
ASSuTCxTioCip(I)NalCotGen |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
Yes |
| SRR4019595 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
No travel |
|
| SRR2485288 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No data |
No travel |
|
| SRR2485282 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No |
Peru |
Yes |
| SRR2485286 |
2014 |
Feces |
ASSuTCxTioCip(I)NalCotGen |
IncFIB-like |
D87Y |
Wt |
Yes |
Peru |
|
| SRR3178071 |
2014 |
Feces |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
Yes |
Peru |
|
| SRR4019602 |
2015 |
Feces |
ACSSuTCxTioFox(I)Cip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
Ecuador |
|
| SRR4019594 |
2015 |
Feces |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No data |
No data |
|
| SRR3185043 |
2015 |
Urine |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
No data |
Yes |
| SRR4019591 |
2015 |
Feces; urine |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
Yes |
No data |
Yes |
| SRR4019588 |
2015 |
Feces |
ACSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like, colE |
D87Y |
Wt |
Yes |
No travel |
Yes |
| SRR4019587 |
2015 |
Feces |
ACSSuTCxTioCip(I)Nal
Gen(I) |
IncFIB-like |
D87Y |
Wt |
No |
No travel |
|
| SRR3184311 |
2015 |
Feces |
ACSSuTCxTioCip(I)NalCot
Gen(I) |
IncFIB-like |
D87Y |
Wt |
Yes |
No travel |
Yes |
| SRR4019590 |
2015 |
Feces |
ACSuTCxTioCip(I)NalCot |
pXuzhou21, IncFIB-like |
D87Y |
Wt |
Yes |
No travel |
|
| SRR4019603 |
2015 |
Urine |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
No travel |
|
| SRR3178070 |
2015 |
Urine |
ACSSuTCxTioCip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
Yes |
Peru |
|
| SRR4019600 | 2015 | Feces | ACSuTCxTioCip(I)NalCot | IncFIB-like | D87Y | Wt | No | Peru | Yes |
*A, ampicillin; Aug, amoxicillin-clavulanic acid; C, chloramphenicol; Cip, ciprofloxacin; Cot, trimethoprim/sulfamethoxazole; Cx, ceftriaxone; Cx, ceftriaxone; Fox, cefoxitin; Gen, gentamicin; (I), intermediate susceptibility; NA, not applicable; Nal, nalidixic acid; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Tio, ceftiofur. †Accession numbers are the sequence read archive run identification numbers from the National Center for Biotechnology Information. ‡Documented illness >2 wk after symptom onset. Blank cells indicate no evidence of prolonged infection (either no additional data were available or additional data indicated no clear evidence of prolonged infection).
Table 2. Characteristics of patients and 5 Salmonella enterica serotype Infantis pattern JFXX01.0787 isolates without CTX-M-65, United States*.
| Accession no.† | Year | Specimen source | Resistance profile | Plasmids | gyrA | parC | Hospitalized | Recent travel |
|---|---|---|---|---|---|---|---|---|
| SRR2485280 |
2012 |
Feces |
No resistance detected |
No replicons detected |
None |
Wt |
No |
No travel |
| SRR4025941 |
2012 |
Feces |
CSuTCip(I)NalCotGen(I) |
IncFIB-like |
D87Y |
Wt |
No |
Peru, Bolivia, Ecuador, Chile |
| SRR4019596 |
2014 |
Urine |
CSSuTFox(I)Cip(I)NalCot |
IncFIB-like |
D87Y |
Wt |
No |
No data |
| SRR3185042 |
2014 |
Feces |
No resistance detected |
No replicons detected |
None |
Wt |
Yes |
No travel |
| SRR2485289 | 2015 | Feces | No resistance detected | No replicons detected | None | Wt | No | No data |
*A, ampicillin; Aug, amoxicillin-clavulanic acid; C, chloramphenicol; Cip, ciprofloxacin; Cot, trimethoprim/sulfamethoxazole; Cx, ceftriaxone; Fox, cefoxitin; Gen, gentamicin; (I), intermediate susceptibility; Nal, nalidixic acid; S, streptomycin; Su, sulfisoxazole; T, tetracycline; Tio, ceftiofur. †Accession numbers are the sequence read archive run identification numbers from the National Center for Biotechnology Information.
We focused our analysis on the 29 patients with Salmonella Infantis isolates containing the blaCTX-M-65 gene (hereafter called CTX-M-65 Infantis). We compared patients infected with CTX-M-65 Infantis with patients infected with all strains of Salmonella Infantis and with patients infected with common Salmonella serotypes other than Infantis. The median age of patients with CTX-M-65 Infantis was 25 years (interquartile range [IQR] 15–50 years), and 69% were female (Table 3). The median age of patients with any strain of Salmonella Infantis was 37 years (IQR 12–58 years), and 56% were female. The median age of patients infected with other common Salmonella serotypes was 29 years (IQR 6–54 years), and 52% were female.
Table 3. Comparison of demographic and clinical characteristics of patients with CTX-M-65–positive isolates with patients infected with all Salmonella enterica serotype Infantis strains or other common Salmonella serotypes, FoodNet, United States, 2012–2015* .
| Characteristic |
CTX-M-65-positive Salmonella Infantis, pattern JFXX01.0787, n = 29* |
|
All Salmonella Infantis, n = 723† |
|
Other Salmonella, n = 21,285‡ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proportion§ |
% |
p value |
Proportion§ |
% |
p value |
Proportion§ |
% |
p value |
|||
| Female sex | 20/29 | 69 | Referent | 403/722 | 56 | 0.16 | 11,067/21,258 | 52 | 0.07 | ||
| Urine isolation | 5/29 | 17 | Referent | 66/723 | 9 | 0.14 | 1,076/21,214 | 5 | <0.01 | ||
| Hospitalization | 8/18 | 44 | Referent | 199/695 | 29 | 0.14 | 5,951/20,619 | 29 | 0.14 | ||
| International travel | 12/19 | 63 | Referent | 51/611 | 8 | <0.01 | 1,431/16,496 | 9 | <0.01 | ||
*Patient age range 0–82 y; median age (interquartile range) 25 (15–50) y. †Patient age range 0–95 y; median age (interquartile range) 37 (12–58) y. ‡Patient age range 0–101 y; median age (interquartile range) 29 (6–54) y. Other Salmonella defined as the top 20 Salmonella enterica serotypes (excluding Infantis): Typhimurium, Enteritidis, Newport, Heidelberg, Javiana, Saintpaul, Montevideo, Agona, Oranienburg, Muenchen, Thompson, Hadar, Braenderup, Derby, I 4,[5],12:i:-, Paratyphi B var. L(+) tartrate+, Blockley, Anatum, Mississippi, Panama. §Unknowns were excluded from the denominator.
Of 18 patients infected with CTX-M-65 Infantis for whom outcome data were available, 8 (44%) were hospitalized, compared with a hospitalization rate of 29% among patients in both comparison groups (those with Infantis and those with other Salmonella serotypes; Table 3). Patients with CTX-M-65 Infantis infection were more likely (17%) to have the organism isolated from urine compared with all patients with Salmonella Infantis (9%, p = 0.14) and patients with other common serotypes (5%, p<0.01).
For 20 CTX-M-65 Infantis–infected patients with information about symptom onset date and isolation date(s), Salmonella Infantis was recovered from 12 who provided fecal samples >2 weeks after their reported date of symptom onset. A total of 8 patients were still reporting symptoms and had positive cultures >30 days after symptom onset, including 3 patients who sought care and had multiple isolates recovered from 50 days to 8 months after symptom onset.
Twelve (63%) patients with CTX-M-65 Infantis infections reported international travel in the 7 days before symptom onset. All reported travel to South America, 10 to Peru, and 2 to Ecuador (Table 3). Travel was less common among all Salmonella Infantis–infected patients (7%, p<0.01) and those infected with other common serotypes (8%, p<0.01; Table 3).
A maximum-likelihood hqSNP phylogeny of the 34 isolates from humans and that from retail chicken meat revealed that isolates from patients with travel-associated infections formed a well-supported clade (clade A) with isolates from patients with infections not associated with travel (Figure 2). Isolates in clade A differed by only 2–47 pairwise hqSNPs, suggesting that these isolates recently evolved from a common ancestor. Included in clade A was the isolate from retail chicken meat collected in 2014.
Figure 2.
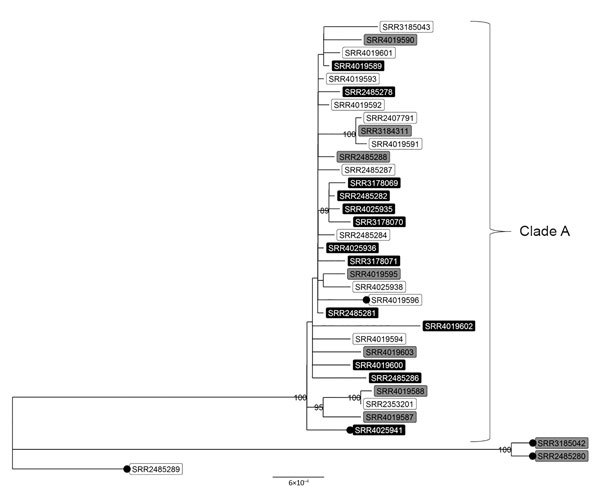
High-quality single-nucleotide polymorphism–based phylogenetic tree of clinical and retail meat isolates of Salmonella enterica serotype Infantis with pulsed-field gel electrophoresis pattern JFXX01.0787 collected in the United States and submitted to the National Antimicrobial Resistance Monitoring System for whole-genome sequencing. Tree tips are labeled with National Center for Biotechnology Information accession numbers (sequence read archive run identification numbers); shading indicates patients’ international travel history for clinical isolates (black, recent international travel; gray, no recent international travel; white, travel data missing). Black circles indicate isolates that are missing the blaCTX-M-65 gene. Isolates in top clade differed by 2–47 high-quality single-nucleotide polymorphisms. Numbers displayed on nodes are bootstrap support values, an indication of the reliability of the tree. Only bootstrap values >80 are displayed. More information on patient and isolate characteristics are provided in Tables 1 and 2. Scale bar indicates nucleotide substitutions per site.
Of the 5 CTX-M-65-negative isolates, 2 grouped within clade A and contained a multidrug-resistance plasmid that was lacking the blaCTX-M-65 gene. An additional 3 CTX-M-65–negative isolates lacked the IncFIB-like plasmid replicon, lacked resistance determinants, and differed by 96–273 pairwise hqSNPs from isolates in clade A. Absence of the blaCTX-M-65 gene in these 5 isolates was confirmed by read-mapping procedures and supported by the observed phenotypic susceptibility to β-lactam antimicrobial drugs.
A phylogeographic analysis of the 32 isolates in the main clade and additional genomes from NCBI generated a time-measured phylogenetic tree of isolates from the United States and Peru (Technical Appendix Figure 1). These isolates last shared a common ancestor around 2006 (95% HPD interval 2003–2008). This analysis also suggests that the MRCA of these isolates existed in Peru with a probability of 98.7%. The probability of the tree being rooted in the United States was ≈1.3%. Clinical isolates from humans in the United States sequenced as part of our study last shared a common ancestor sometime around 2009 (95% HPD interval 2008–2009), before the first isolation of pattern 787 in the United States in 2012.
Discussion
A new strain of Salmonella Infantis, which has pattern 787 and frequently carries a multidrug-resistant plasmid with a CTX-M-65 ESBL, has emerged in the United States. This strain possesses clinically important resistance associated with higher hospitalization rates. Using epidemiologic and phylogenetic methods, we demonstrated that the earliest cases of CTX-M-65 Infantis infection were among travelers returned from South America, whereas subsequent infections were acquired domestically.
The demographic and clinical characteristics among patients with CTX-M-65 Infantis infections differed from those of patients infected with all strains of Salmonella Infantis or other common Salmonella serotypes. CTX-M-65 Infantis–infected patients were younger and more frequently female, and rates of hospitalization were 50% higher for these patients than for those in the other 2 groups. Among patients with CTX-M-65 Infantis infections, 63% reported recent travel to South America, predominantly to Peru, compared with <10% in the comparison groups reporting travel. This finding is consistent with those of other studies that found foreign travel to be a risk factor for CTX-M–type ESBLs (19–21). Studies have shown that CTX-M-65 has recently emerged in commensal E. coli in Bolivia (22) and in Salmonella Infantis in Ecuador (23), 2 countries to which patients in our study also traveled.
Our phylogeographic analysis, which included many isolates from Peru, suggested that isolates from patients in the United States last shared a common ancestor that existed in Peru in 2009. This finding makes sense, given the high proportion of patients in our study who reported travel to Peru and the known circulation of CTX-M-65–positive Enterobacteriaceae in South America. In our study, patients with CTX-M-65 Infantis infection and a history of travel were reported every year from 2012 through 2015; patients with no history of travel were first reported in 2014.
One caveat is that genetic information for CTX-M Infantis from countries in South America other than Peru was not available; therefore, we could not distinguish the role that other countries in South America may have played in the spread of this Salmonella Infantis strain to the United States. We cannot definitively determine how patients with this strain of Salmonella Infantis who did not report travel to South America became infected; however, our analysis does show a close genetic relationship between clinical isolates from these patients and isolates collected by USDA-FSIS from chickens in the United States.
Poultry consumption may be a source of CTX-M-65 Infantis infection in the United States and abroad. In our study, more than one third of patients with CTX-M-65 Infantis infections did not report recent international travel and thus were exposed via a domestic source. CTX-M genes have been linked to poultry, particularly broiler chickens (24–26). A study of 14 chicken farms in Henan Province, China, conducted during 2007–2008, was the first to describe detection of CTX-M-65–producing E. coli in chickens (27). Recently, CTX-M-65 Infantis was found to be transmitted from broiler chickens and chicken meat to humans in Italy (28).
The results of our initial hqSNP analysis demonstrated that the original isolate collected from retail chicken in 2014 was genetically related to isolates from humans. In addition, our data show a clear partition between the dates of specimen collection from the first travel-associated infections detected in the United States (2012) and the dates of the first domestically acquired infections detected (2014) (Technical Appendix Figure 2). Sampling in poultry production plants and sequence data from USDA-FSIS demonstrate that this strain was present in domestic food processing plants during the latter part of the study period (2014–2015) (29). We cannot, however, determine precisely when or how this strain was introduced into poultry stock in the United States because enhanced poultry plant sampling was not conducted before 2014. International distribution of infected breeder stocks, chicken feed, or feed additives contaminated with CTX-M-65 Infantis could help explain how these broiler-associated infections have spread globally during the same period.
CTX-M-65, in comparison with more well-characterized CTX-M enzymes, differs by only 2 substitutions (A77V, S272R) from CTX-M-14, one of the more commonly detected CTX-M variants worldwide (30,31). The blaCTX-M-65 gene was transmitted on a large IncFIB-like plasmid containing multiple resistance genes. The presence of multidrug resistance in CTX-M-65 Infantis isolates in our study suggests that a variety of antimicrobial drugs could provide positive selection pressure and thus promote persistence of this strain. These characteristics suggest that the potential for spread of the gene is high. Research conducted in Bolivia has shown that even in the absence of selective pressure from antimicrobial drug use, plasmid transfer of CTX-M-65 from E. coli to other pathogens was achieved at high frequency and shown to be stable (22).
Additional studies on antimicrobial drug use and management practices in food animals may help us understand which factors contribute most to the emergence, persistence, and spread of resistance genes such as blaCTX-M-65. New efforts to perform whole-genome sequencing on all Salmonella isolates at public health laboratories nationwide will help determine whether plasmid-mediated blaCTX-M-65 has spread to other Salmonella serotypes. One limitation of our study was that we were unable to obtain epidemiologic data for all 312 cases. The data available from FoodNet enabled us to compare variables such as patient demographics, travel, and hospitalizations; however, we did not have a control population for evaluating other variables of interest to determine potential domestic sources of transmission.
The spread of CTX-M-65 is concerning because the presence of ESBLs eliminates 2 recommended treatment options, ceftriaxone and ampicillin, for the management of salmonellosis. Given the multidrug-resistant profile of CTX-M-65 Infantis, potential for plasmid-mediated transmission, increased hospitalization rate, and evidence of this strain in domestic poultry, action is needed to prevent widespread dissemination in the United States. Enhanced surveillance and additional studies in humans and food animals may help pinpoint the sources of infection for implementation of prevention and control measures. Meanwhile, travelers and healthcare providers should be aware of the risks and implications of infection with this strain, including the potential for antimicrobial treatment failure.
Supplementary methods, tip-dated maximum clade credibility tree of 160 closely related Salmonella serotype Infantis isolates, and dates of specimen collection for isolates in clade A.
Acknowledgments
We thank Heather Carleton and Lee Katz for helpful discussion and guidance on some analyses, Beth Tolar for fulfilling repeated data requests, the USDA-FSIS Eastern Laboratory Microbiology Characterization Branch for their work on adding whole-genome sequencing data to NCBI, Lauren Ahart for manuscript preparation, and the FoodNet sites and public health departments in the following states for their collaboration on this investigation: Arizona, California, Colorado, Florida, Illinois, Kentucky, Louisiana, Massachusetts, Michigan, New York, Oregon, Pennsylvania, Texas, Utah, Virginia, and Wisconsin.
Biography
Dr. Brown is team lead for AR Capacities and Special Studies in the Division of Healthcare Quality Promotion, National Center for Emerging and Zoonotic Infectious Diseases, CDC. Her research interests include emerging mechanisms of antimicrobial drug resistance in healthcare-associated infections.
Footnotes
Suggested citation for this article: Brown AC, Chen JC, Francois Watkins LK, Campbell D, Folster JP, Tate T, et al. CTX-M-65 extended-spectrum β-lactamase–producing Salmonella enterica serotype Infantis, United States. Emerg Infect Dis. 2018 Dec [date cited]. https://doi.org/10.3201/eid2412.180500
Preliminary results from this analysis were presented at the International Salmonella and Salmonellosis Symposium, June 6–8, 2016, St. Malo, France, and at the International Association for Food Protection Annual Meeting, July 9–12, 2017, Tampa, Florida, USA.
References
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Jervis R, et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2017. MMWR Morb Mortal Wkly Rep. 2018;66:397–403. 10.15585/mmwr.mm6615a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, et al. 2017 Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65:1963–73. 10.1093/cid/cix959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Casellas JM, Goldberg M, Holley M, Jungwirth R, Mangold P, et al. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–63. 10.1007/BF01704610 [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol. 2012;12:883–93. 10.1016/j.meegid.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 6.Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–74. 10.1093/jac/dkl483 [DOI] [PubMed] [Google Scholar]
- 7.Sjölund-Karlsson M, Howie R, Krueger A, Rickert R, Pecic G, Lupoli K, et al. CTX-M-producing non-Typhi Salmonella spp. isolated from humans, United States. Emerg Infect Dis. 2011;17:97–9. 10.3201/eid1701.100511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjölund-Karlsson M, Howie RL, Blickenstaff K, Boerlin P, Ball T, Chalmers G, et al. Occurrence of β-lactamase genes among non-Typhi Salmonella enterica isolated from humans, food animals, and retail meats in the United States and Canada. Microb Drug Resist. 2013;19:191–7. 10.1089/mdr.2012.0178 [DOI] [PubMed] [Google Scholar]
- 9.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, et al. Comparative analysis of extended-spectrum-beta-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother. 2017;61:e00488–17. 10.1128/AAC.00488-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for enteric bacteria (NARMS): human isolates surveillance report for 2015 (final report). Atlanta: The Centers; 2016. p. 22–5. [Google Scholar]
- 11.Folster JP, Pecic G, McCullough A, Rickert R, Whichard JM. Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007. Foodborne Pathog Dis. 2011;8:1289–94. 10.1089/fpd.2011.0944 [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. Laboratory standard operating procedure for PulseNet Nextera XT Library Prep and Run Setup for the Illlumina MiSeq. Code PNL32. 2015. [cited 2016 Feb 1]. https://www.cdc.gov/pulsenet/pdf/pnl32-miseq-nextera-xt.pdf
- 13.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, et al. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio. 2013;4:e00398–13. 10.1128/mBio.00398-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. 10.1093/nar/gku1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.US National Library of Medicine. Pathogen detection [cited 2018 Jan 23]. https://www.ncbi.nlm.nih.gov/pathogens/
- 18.Gopinath G, Chase H, Gangiredla J, Patel I, Addy N, Beaubrun JJG, et al. Comparative genomics of blaCTX-M-65-resistant clinical strains of Salmonella enterica serovar Infantis from Peru and resistant strains from chicken, cattle and humans. Poster presented at: International Association for Food Protection annual meeting; 2017. Jul 9–12; Tampa, FL, USA. [Google Scholar]
- 19.Tängdén T, Cars O, Melhus A, Löwdin E. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum β-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother. 2010;54:3564–8. 10.1128/AAC.00220-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins KL, Batchelor MJ, Liebana E, Deheer-Graham AP, Threlfall EJ. Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int J Antimicrob Agents. 2006;28:180–92. 10.1016/j.ijantimicag.2006.03.027 [DOI] [PubMed] [Google Scholar]
- 21.Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MC, van Genderen PJ, Goorhuis A, et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis. 2016. [DOI] [PubMed] [Google Scholar]
- 22.Riccobono E, Di Pilato V, Di Maggio T, Revollo C, Bartoloni A, Pallecchi L, et al. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum β-lactamase in the Bolivian Chaco region. Antimicrob Agents Chemother. 2015;59:5340–7. 10.1128/AAC.00589-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cartelle Gestal M, Zurita J, Paz Y Mino A, Ortega-Paredes D, Alcocer I, Alcocer I. Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz J Infect Dis. 2016;20:406–7. 10.1016/j.bjid.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maciuca IE, Williams NJ, Tuchilus C, Dorneanu O, Guguianu E, Carp-Carare C, et al. High prevalence of Escherichia coli-producing CTX-M-15 extended-spectrum β-lactamases in poultry and human clinical isolates in Romania. Microb Drug Resist. 2015;21:651–62. 10.1089/mdr.2014.0248 [DOI] [PubMed] [Google Scholar]
- 25.Silva KC, Fontes LC, Moreno AM, Astolfi-Ferreira CS, Ferreira AJ, Lincopan N. Emergence of extended-spectrum-β-lactamase CTX-M-2-producing Salmonella enterica serovars Schwarzengrund and Agona in poultry farms. Antimicrob Agents Chemother. 2013;57:3458–9. 10.1128/AAC.05992-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weill FX, Lailler R, Praud K, Kérouanton A, Fabre L, Brisabois A, et al. Emergence of extended-spectrum-beta-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J Clin Microbiol. 2004;42:5767–73. 10.1128/JCM.42.12.5767-5773.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan L, Liu JH, Hu GZ, Pan YS, Liu ZM, Mo J, et al. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolates from chickens in Henan Province, China. J Med Microbiol. 2009;58:1449–53. 10.1099/jmm.0.012229-0 [DOI] [PubMed] [Google Scholar]
- 28.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One. 2015;10:e0144802. 10.1371/journal.pone.0144802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Food and Drug Administration. NARMS Now [cited 2018 Jan 19]. https://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm416741.htm
- 30.Yin J, Cheng J, Sun Z, Ye Y, Gao YF, Li JB, et al. Characterization of two plasmid-encoded cefotaximases found in clinical Escherichia coli isolates: CTX-M-65 and a novel enzyme, CTX-M-87. J Med Microbiol. 2009;58:811–5. 10.1099/jmm.0.006007-0 [DOI] [PubMed] [Google Scholar]
- 31.Bush K. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci. 2013;1277:84–90. 10.1111/nyas.12023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, tip-dated maximum clade credibility tree of 160 closely related Salmonella serotype Infantis isolates, and dates of specimen collection for isolates in clade A.


