Like many taxa, the Zetaproteobacteria lack well-defined taxonomic divisions, making it difficult to compare them between studies. We designed ZetaHunter to reproducibly assign 16S rRNA gene sequences to previously described operational taxonomic units (OTUs) based on a curated database.
ABSTRACT
Like many taxa, the Zetaproteobacteria lack well-defined taxonomic divisions, making it difficult to compare them between studies. We designed ZetaHunter to reproducibly assign 16S rRNA gene sequences to previously described operational taxonomic units (OTUs) based on a curated database. While ZetaHunter can use any given database, we included a curated classification of publicly available Zetaproteobacteria.
ANNOUNCEMENT
Taxonomic groups with limited cultivated representatives are often lumped into a single taxonomic label encompassing multiple distinct ecological units. This makes it difficult to explicitly discuss the abundance, significance, and niche preference of these units between studies. One group with poor taxonomic resolution is the class Zetaproteobacteria, which is only accurately classified to this taxonomic level by standard small subunit rRNA (16S rRNA) gene classification tools. ZetaHunter allows for reproducible, higher-resolution comparisons across studies by using a curated data set with a defined taxonomy based on operational taxonomic units (OTUs). ZetaHunter comes with a curated database to identify the members of the Zetaproteobacteria, though it may be used with any curated 16S rRNA gene database. (This article was submitted to an online preprint archive [1].)
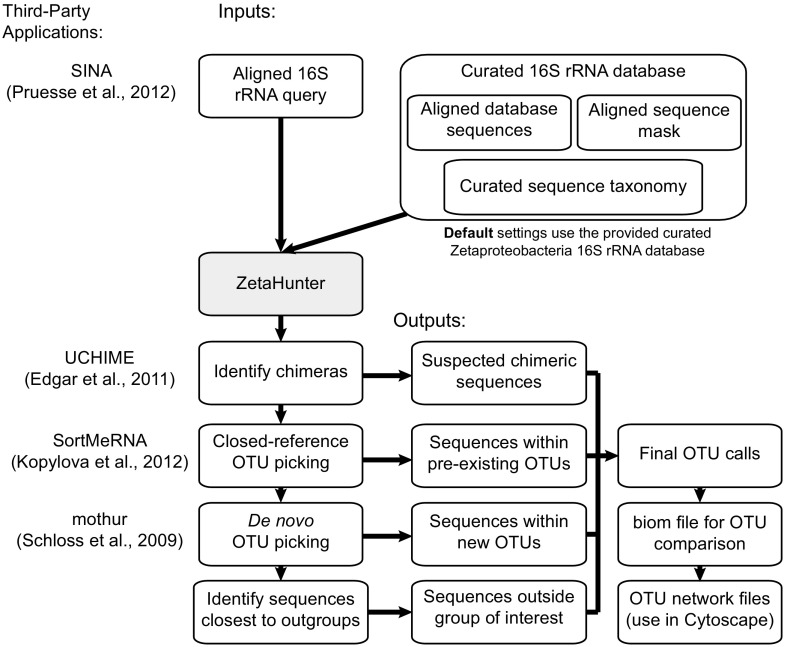
ZetaHunter is a command line program written in Ruby designed to assign user-supplied 16S rRNA gene sequences to OTUs defined by a reference sequence database. ZetaHunter can be used on Linux, Mac OSX, and Windows platforms through a Docker container or through installation from source (Linux and Mac OSX only). By default, ZetaHunter uses a curated database of Zetaproteobacteria 16S rRNA genes from ARB SILVA (release 128) (2) and Zetaproteobacteria genomes from the Joint Genome Institute (JGI) Integrated Microbial Genomes (IMG) database (3). Zetaproteobacteria OTU (ZOTU) definitions include those reported by McAllister et al. (4) at 97% identity, maintaining ZOTU number order from ZOTU1 to ZOTU28 for ease in comparisons across studies. Numbered ZOTUs from ZOTU29 upward were discovered after 2011.
Input FASTA sequences for use in ZetaHunter must be aligned first using SINA (either online or standalone) (5). The default pipeline of ZetaHunter (Fig. 1) takes these SINA-aligned 16S rRNA gene sequences and processes them as follows: (i) input sequences are masked to the 1,282 bp used by McAllister et al. (4); (ii) sequences are checked for chimeras using the mothur UCHIME algorithm (6, 7); (iii) SortMeRNA is used to cluster new sequences with the reference database, assigning a ZOTU based on genetic distance (closed reference binning) (8); (iv) the remaining sequences are clustered into novel OTUs (NewZetaOtus) with mothur (de novo binning, average neighbor, numbered by abundance) (6); and (v) summary files (including final ZOTU calls and closest database hits), a biological observation matrix (biom) table (9) showing counts for each ZOTU by sample, and OTU network files are exported for use. The ZetaHunter database includes 21 proteobacterial outgroup sequences, allowing ZetaHunter to flag sequences potentially outside the Zetaproteobacteria. Additionally, sequences that are short, chimeric, or singletons/doubletons or contain ambiguous bases are also flagged. This pipeline is an implementation of open-reference OTU picking similar to the one found in QIIME (10).
FIG 1.
Flow chart showing the ZetaHunter pipeline. Third-party tools used in the pipeline are indicated to the left.
To use ZetaHunter for non-Zetaproteobacteria, users need (i) a database of SINA-aligned 16S rRNA gene sequences, (ii) a sequence mask with asterisks at each informative alignment column to be used for taxonomic assignment, and (iii) a tab-delimited file assigning each sequence to a particular taxonomic group at the similarity threshold desired by the user.
Data availability.
ZetaHunter is available for download at (https://github.com/mooreryan/ZetaHunter) (11), where it will be supported and maintained for at least the next 10 years. GitHub documentation includes installation instructions, a description of all dependencies, details of the ZetaHunter curated database, descriptions of output files, and examples of classifications of Zetaproteobacteria and non-Zetaproteobacteria. Detailed example data sets are included in the ZetaHunter_examples GitHub repository (https://github.com/mooreryan/ZetaHunter_examples).
ACKNOWLEDGMENTS
We thank Dave Emerson for providing 16S rRNA sequences for the ZetaHunter database from novel Zetaproteobacteria isolates. We also thank K. E. Wommack and S. Polson for their guidance and support in implementing ZetaHunter. D. Emerson, B. Chiu, and A. Harrison are thanked for their comments on early versions of the manuscript. We also thank K. Hager, H. Fullerton, J. Scott, and J. Vander Roost for testing and troubleshooting ZetaHunter using multiple platforms and data sets. We also acknowledge the ARB/SILVA group for their work with stable 16S rRNA alignments using the SINA tool; this tool is key to ZetaHunter’s function.
S.M.M. and R.M.M. designed the ZetaHunter program. S.M.M. developed the Zetaproteobacteria database. R.M.M. wrote the ZetaHunter program. S.M.M., R.M.M., and C.S.C. wrote and edited the manuscript.
This work was funded by National Science Foundation (NSF) grant number OCE-1155290 (to C.S.C.) and Agriculture and Food Research Initiative grant number 2012-68003-30155 from the USDA National Institute of Food and Agriculture. Computational infrastructure support by the University of Delaware Center for Bioinformatics and Computational Biology Core Facility was made possible through funding from Delaware INBRE (grant number NIH P20 GM103446) and the Delaware Biotechnology Institute.
We declare no conflicts of interest.
REFERENCES
- 1.McAllister SM, Moore RM, Chan CS. 2018. ZetaHunter: a reproducible taxonomic classification tool for tracking the ecology of the zetaproteobacteria and other poorly-resolved taxa. bioRxiv. doi: 10.1101/359620. [DOI] [PMC free article] [PubMed]
- 2.Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W. 2017. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 3.Chen I-MA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, Varghese N, Hadjithomas M, Tennessen K, Nielsen T, Ivanova NN, Kyrpides NC. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McAllister SM, Davis RE, McBeth JM, Tebo BM, Emerson D, Moyer CL. 2011. Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol 77:5445–5457. doi: 10.1128/AEM.00533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 9.McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, Knight R, Caporaso JG. 2012. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience 1:7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG, Knight R. 2013. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAllister SM, Moore RM, Chan CS. 2018. ZetaHunter v.1.0.0. https://github.com/mooreryan/ZetaHunter. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ZetaHunter is available for download at (https://github.com/mooreryan/ZetaHunter) (11), where it will be supported and maintained for at least the next 10 years. GitHub documentation includes installation instructions, a description of all dependencies, details of the ZetaHunter curated database, descriptions of output files, and examples of classifications of Zetaproteobacteria and non-Zetaproteobacteria. Detailed example data sets are included in the ZetaHunter_examples GitHub repository (https://github.com/mooreryan/ZetaHunter_examples).