Six genome sequences of papillomavirus were determined from oral and skin swabs and fecal samples collected from wild rats. Three genomes were 7,722 bp, two genomes were 7,716 bp, and one was 7,730 bp, displaying typical papillomavirus genome organizations.
ABSTRACT
Six genome sequences of papillomavirus were determined from oral and skin swabs and fecal samples collected from wild rats. Three genomes were 7,722 bp, two genomes were 7,716 bp, and one was 7,730 bp, displaying typical papillomavirus genome organizations. Phylogenetic analysis revealed that these six genomes belonged to two different clusters.
ANNOUNCEMENT
Papillomaviruses (PVs) are known to infect a wide variety of mammals, birds, and reptiles (1) and are classified into genera based on the open reading frame (ORF) L1 (2). While the overwhelming majority of PVs only infect the epithelium (2, 3), the bovine papillomaviruses have the ability to infect both epithelial and mesenchymal cells and to infect multiple species (4). PVs are believed to have coevolved with their vertebrate host species, a hypothesis supported by the fact that PVs of closely related host species are generally closely related themselves (5, 6). Recently, more complete genomes of PVs were reported in nonhuman species (7–14).
Here, using viral metagenomics, we investigated the viral nucleic acid sequences from oral swabs, skin swabs, and feces. All samples were from 20 wild rats collected in Zhenjiang City, China, in 2016. Viral metagenomic analysis was performed as previously reported (15, 16). Briefly, 60 sample supernatants were pooled into 5-sample pools according to the 3 sample types and generated 12 pools where each type of sample included 4 pools. Total nucleic acid was then isolated using the QIAamp MinElute virus spin kit (Qiagen) according to manufacturer’s protocol. Twelve libraries were then constructed using the Nextera XT DNA sample preparation kit (Illumina) and sequenced using the MiSeq Illumina platform with 250-bp paired-end reads with dual barcoding for each pool. The total numbers of sequence reads generated for the 12 libraries were 3,204,222 (feces01), 2,009,444 (feces02), 2,330,836 (feces03), 1,674,810 (feces04), 203,916 (oral01), 212,296 (oral02), 125,674 (oral03), 39,314 (oral04), 292,992 (skin01), 87,252 (skin02), 81,526 (skin03), and 61,134 (skin04). An in-house analysis pipeline running on a 32-node Linux cluster was used to process the data. Reads were considered duplicates if bases 5 to 55 were identical and only one random copy of duplicates was kept. Clonal reads were removed, and low sequencing quality tails were trimmed using Phred quality score 10 as the threshold. Adaptors were trimmed using the default parameters of VecScreen, which is NCBI BLASTn with specialized parameters designed for adapter removal. The cleaned reads from each library were de novo assembled within each barcode using the Ensemble assembler (17). Contigs and singlets reads were then matched against a customized viral proteome database using BLASTx with an E value cutoff of <10−5.
We acquired six complete genomes by assembling the viral reads showing significant similarity to papillomavirus and conducting common PCR bridging the gaps between contigs using primers designed based on their closest relative genomes in GenBank, which are hereafter referred to as Ne-or02-zj, Or01-zj, Or02-zj, Sk01-zj, Sk02-zj, and Fe02-zj. Ne-or02-zj was found in the oral swab sample (containing 434 reads in library oral02) and shared 86.25% nucleotide sequence identity with a papillomavirus strain (GenBank accession no. KY370097) over the complete genome based on a BLASTn search, which was detected in Rattus norvegicus according to the sequence annotation in GenBank. Or01-zj (containing 25,285 reads in library oral01) and Or02-zj (containing 1,402 reads in library oral02) were detected in two different oral swab pools. Sk01-zj (containing 3,187 reads in library skin01) and Sk02-zj (containing 1,393 reads in library skin02) were found in two different skin swab sample pools. Fe02-zj (containing 81 reads in library feces02) was acquired from a fecal sample pool. A BLASTn search in GenBank based on the complete genome sequences of Or01-zj, Or02-zj, Fe02-zj, Sk01-zj, and Sk02-zj showed their best match was Rattus norvegicus papillomavirus 2 (GenBank accession no. HQ625441) (18). Five distinct ORFs on the same coding strand in the genomes of Or01-zj, Or02-zj, Fe02-zj, Sk01-zj, and Sk02-zj were identified using Geneious version 11.0, including the early genes E6, E1, and E2 and the late genes L2 and L1, while there were six ORFs in the genome of Ne-or02-zj which had an additional E7 gene before E6.
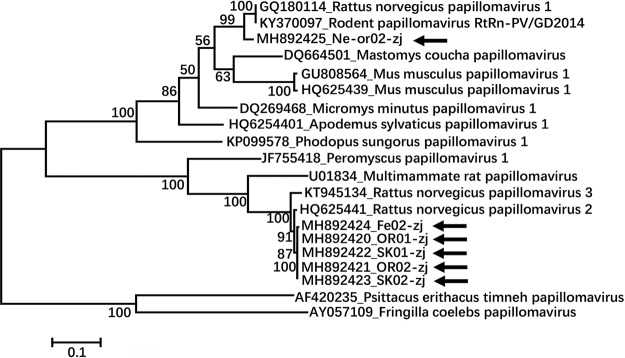
To determine the divergence in sequence between the PVs found here and other PVs, amino acid sequence alignment of the complete L1 protein was performed using Clustal W version 2.0 (19), and a phylogenetic tree was generated using the maximum-likelihood method in MEGA7 (20) with 1,000 bootstrap resamples of the alignment data sets. Phylogenetic analysis indicated that the six papillomavirus genomes fell into two different clusters, where Ne-or02-zj clustered with Rodent papillomavirus isolate RtRn-PV/GF2014 (GenBank accession no. KY370097), and the other five strains clustered with R. norvegicus papillomavirus 2 (GenBank accession no. HQ625441) (Fig. 1).
FIG 1.
Phylogenetic analysis based on amino acid sequence alignment of the complete L1 protein. The sequences in the phylogenetic tree included the 6 papillomaviruses identified in this study, 12 papillomaviruses with complete genomes from rodents available in GenBank, and 2 complete papillomavirus genomes from birds as an outgroup. The GenBank number of the referenced genomes are provided. The six papillomavirus strains identified in this study are marked by arrows.
Data availability.
The raw sequence reads were deposited in the Sequence Read Archive under accession no. SRX4791096 to SRX4791099, SRX4791100, SRX4791137, SRX4791138, SRX4791154, SRX4791673, SRX4791804, SRX4791859, and SRX4791861. The six genomic sequences were deposited in GenBank under accession no. MH892420 to MH892425.
ACKNOWLEDGMENTS
Y.L., F.M., Y.H., M.Z., and H.X. performed the experiments. Y.L. and W.Z. analyzed the sequence data and wrote the manuscript. All the authors read and approved the final manuscript.
This work was partly supported by Key Research and Development Plan of Zhenjiang no. SH2018032 and Jiangsu Maternal and Child Health Care Research Project no. F201717.
REFERENCES
- 1.Van Doorslaer K. 2013. Evolution of the Papillomaviridae. Virology 445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Bernard H-U, Burk RD, Chen Z, van Doorslaer K, Zur Hausen H, de Villiers E-M. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joh J, Jenson AB, King W, Proctor M, Ingle A, Sundberg JP, Ghim S. 2011. Genomic analysis of the first laboratory-mouse papillomavirus. J Gen Virol 92:692–698. doi: 10.1099/vir.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 4.Munday JS. 2014. Bovine and human papillomaviruses: a comparative review. Vet Pathol 51:1063–1075. doi: 10.1177/0300985814537837. [DOI] [PubMed] [Google Scholar]
- 5.Van Ranst M, Fuse A, Fiten P, Beuken E, Pfister H, Burk RD, Opdenakker G. 1992. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology 190:587–596. doi: 10.1016/0042-6822(92)90896-W. [DOI] [PubMed] [Google Scholar]
- 6.Tachezy R, Duson G, Rector A, Jenson AB, Sundberg JP, Van Ranst M. 2002. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology 301:313–321. doi: 10.1006/viro.2002.1566. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues TCS, Subramaniam K, Cortés-Hinojosa G, Wellehan JFX, Ng TFF, Delwart E, McCulloch SD, Goldstein JD, Schaefer AM, Fair PA, Reif JS, Bossart GD, Waltzek TB. 2018. Complete genome sequencing of a novel type of Omikronpapillomavirus 1 in Indian River Lagoon bottlenose dolphins (Tursiops truncatus). Genome Announc 6:e00240-18. doi: 10.1128/genomeA.00240-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Wood CE, Abee CR, Burk RD. 2018. Complete genome sequences of three novel Saimiri sciureus papillomavirus types isolated from the cervicovaginal region of squirrel monkeys. Genome Announc 6:e01400-17. doi: 10.1128/genomeA.01400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Wong MCS, Wong PY, Ho WCS, Liang M, Yeung ACM, Chan PKS. 2017. Complete genome sequence of a novel human papillomavirus isolated from oral rinse. Genome Announc 5:e01281-17. doi: 10.1128/genomeA.01281-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regnard GL, Matiso A, Mounir L, Embarki T, Hitzeroth II, Rybicki EP. 2017. Complete genome sequence of Bos taurus papillomavirus type 1, isolated in Morocco. Genome Announc 5:e00979-17. doi: 10.1128/genomeA.00979-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tisza MJ, Yuan H, Schlegel R, Buck CB. 2016. Genomic sequence of canine papillomavirus 19. Genome Announc 4:e01380-16. doi: 10.1128/genomeA.01380-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regnard GL, Baloyi NM, Bracher LR, Hitzeroth II, Rybicki EP. 2016. Complete genome sequences of two isolates of Canis familiaris oral papillomavirus from South Africa. Genome Announc 4:e01006-16. doi: 10.1128/genomeA.01006-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munday JS, Thomson N, Dunowska M, Knight CG, Laurie RE, Hills S. 2015. Genomic characterisation of the feline sarcoid-associated papillomavirus and proposed classification as Bos taurus papillomavirus type 14. Vet Microbiol 177:289–295. doi: 10.1016/j.vetmic.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Stevens H, Rector A, Bertelsen MF, Leifsson PS, Van Ranst M. 2008. Novel papillomavirus isolated from the oral mucosa of a polar bear does not cluster with other papillomaviruses of carnivores. Vet Microbiol 129:108–116. doi: 10.1016/j.vetmic.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, Minor PD, Delwart E. 2015. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods 213:139–146. doi: 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Li L, Deng X, Blümel J, Nübling CM, Hunfeld A, Baylis SA, Delwart E. 2016. Viral nucleic acids in human plasma pools. Transfusion 56:2248–2255. doi: 10.1111/trf.13692. [DOI] [PubMed] [Google Scholar]
- 17.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. 2015. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res 43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz E, Gottschling M, Ulrich RG, Richter D, Stockfleth E, Nindl I. 2012. Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS One 7:e47164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence reads were deposited in the Sequence Read Archive under accession no. SRX4791096 to SRX4791099, SRX4791100, SRX4791137, SRX4791138, SRX4791154, SRX4791673, SRX4791804, SRX4791859, and SRX4791861. The six genomic sequences were deposited in GenBank under accession no. MH892420 to MH892425.