Ashy storm-petrels (order Procellariiformes) are seabirds that are found along the coast of California to Baja Mexico. A novel gyrovirus was identified from a cloacal swab of an ashy storm-petrel, which is the second gyrovirus to be identified in sea birds, the first being found in the related northern fulmar.
ABSTRACT
Ashy storm-petrels (order Procellariiformes) are seabirds that are found along the coast of California to Baja Mexico. A novel gyrovirus was identified from a cloacal swab of an ashy storm-petrel, which is the second gyrovirus to be identified in sea birds, the first being found in the related northern fulmar.
ANNOUNCEMENT
The ashy storm-petrel (Oceanodroma homochroa) is a seabird of conservation concern that is endemic to the California Current between western Baja California, Mexico, and northern California (1), with breeding populations concentrated at the South Farallon and Channel Islands (2, 3). The South Farallon Islands, 42 km west of San Francisco in the United States and part of the Farallon Islands National Wildlife Refuge, represent the largest colony, with ∼40% to 50% of the world population (3) and high visitation during the spring and summer (1, 2).
No viruses have been identified in ashy storm-petrels to date. As part of a viral discovery project, 40 individual cloacal swabs were collected in 2012 from adult ashy storm-petrels inhabiting the Farallon Islands and stored in RNAlater and guanidinium-isothiocyanate buffer. A 100-µl aliquot from each sample was used for viral DNA extraction as previously described (4, 5), and circular molecules were enriched by rolling-circle amplification using TempliPhi 100 amplification (GE Healthcare, USA). The resulting DNA was used to construct a 2 × 150-bp library using the Illumina TruSeq Nano DNA library prep kit and sequenced on an Illumina HiSeq 4000 platform at Macrogen, Inc. (South Korea). The raw reads (13,328,366 paired-end reads) were trimmed using Trimmomatic (6) and then de novo assembled using ABySS 2.0 (7). In the resulting 227,835 contigs (N50, 5,545 nucleotides [nt]), which were predominately Pseudomonas spp., an 818-nucleotide contig (with 15× coverage) was identified as having similarities to gyrovirus sequences using BLASTx (8). Gyroviruses (family Anelloviridae, genus Gyrovirus) are small, circular, negative-sense, single-stranded DNA viruses that have GC-rich noncoding regions, high sequence variability, and conserved genome organization (9). Relatively few gyroviruses have been identified, and little is known about their impacts on host organisms. Chicken anemia virus has been shown to cause immunosuppression, anemia, and hemorrhaging in young chickens (10). Using metagenomic approaches, various novel gyroviruses have been identified from chickens, human feces (in Chile, China, France, Hong Kong, South Africa, and Tunisia), a northern fulmar (in spleen and uropygial gland tissue [United States]), and ferret feces (in Hungary); however, no direct disease correlations have been demonstrated (11–16).
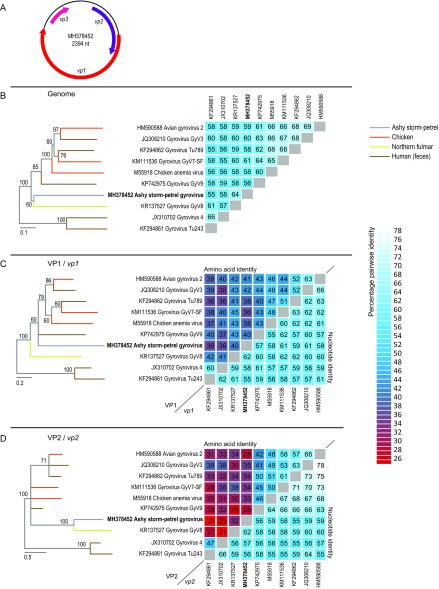
Based on the gyrovirus-like de novo-assembled contigs, a set of back-to-back (for recovery of circular genomes) primers (5′-GTTACTTTCCAAGGTATTATTCTCATCCCC-3′, 5′-TCCGAGTGAGTTGTATGGTTTGGTAAC-3′) was designed and used to amplify the full genome of the gyrovirus. The cloned and Sanger-sequenced genome of ashy storm-petrel-associated gyrovirus (ASPaGyV) is 2,365 nucleotides in length, containing three large open reading frames (ORFs; VP1, VP2, VP3) (Fig. 1A). Representative genome sequences of gyroviruses were analyzed with that of ASPaGyV. The genome of ASPaGyV1 is most closely related to the gyrovirus GyV8 (GenBank accession number KR137527) from a northern fulmar, sharing 64% genome-wide pairwise identity (Fig. 1B). The VP1 of ASPaGyV shares 36% to 42% amino acid and 57% to 61% nucleotide identities with the VP1s of other gyroviruses (Fig. 1C). The VP2 shares 20% to 38% amino acid and 56% to 59% nucleotide identities with the VP2s of other gyroviruses (Fig. 1D). No homologues for the VP3 of ASPaGyV were identified. The pathology in ashy storm-petrels associated with ASPaGyV is unknown, and further work is necessary to determine the incidence rate and diversity of these viruses in these wild birds.
FIG 1.
(A) Organization of the genome of ashy storm-petrel-associated gyrovirus VP1 (putative capsid protein; 1,368 nucleotides), VP2 (unknown function; 699 nucleotides), and VP3 (unknown function; 273 nucleotides). (B) Neighbor-joining phylogenetic tree of representative sequences (NCBI RefSeq) of gyroviruses with 1,000 bootstrap replicate branch support and pairwise identity matrix. (C) Maximum likelihood phylogenetic tree of the VP1 amino acid sequences and the pairwise identities of the VP1 protein and VP1 nucleotide sequences of representative gyroviruses. (D) Maximum likelihood phylogenetic tree of the VP2 amino acid sequences and the pairwise identities of the VP2 protein and VP2 nucleotide sequences of representative gyroviruses. The maximum likelihood phylogenetic trees were inferred using PHYML (17) with the WAG+G substitution model, determined as the optimal model using ProtTest (18), and the pairwise identities were inferred using SDT v1.2 (19).
Data availability.
The complete genome sequence of the ashy storm-petrel-associated gyrovirus isolate was deposited at GenBank under the accession number MH378452.
ACKNOWLEDGMENTS
This study was supported by seed funding from the Center of Fundamental and Applied Microbiomics (Arizona State University, USA) awarded to A.V.
Sampling on the Farallon Islands National Wildlife Refuge was conducted under cooperative agreement with the U.S. Fish and Wildlife Service (F14AC00237) and Bird Banding Laboratory permit number 9316.
REFERENCES
- 1.Spear LB, Ainley DG. 2007. Storm-petrels of the Eastern Pacific Ocean: species assembly and diversity along marine habitat gradients. Ornithol Monog 1–77. doi: 10.2307/40166847. [DOI] [Google Scholar]
- 2.Carter H, McIver W, McChesney G. 2008. Ashy storm-petrel (Oceanodroma homochroa), p 117–124. In California bird species of special concern: a ranked assessment of species, subspecies, and distinct populations of birds of immediate conservation concern in California. Studies of western birds 1. Western Field Ornithologists, Camarillo, CA, and California Department of Fish and Game, Sacramento, CA. [Google Scholar]
- 3.Carter HR, Ainley GG, Wolf SG, Weinstein AM. 2016. Range-wide conservation and science of the ashy storm-petrel Oceanodroma homochroa. Mar Ornithol 44:53–62. [Google Scholar]
- 4.Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, Eilers AA, Shero MR, Beltran R, Kirkham A, McCorkell R, Berngartt RK, Male MF, Ballard G, Ainley DG, Breitbart M, Varsani A. 2017. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol 3:vex017. doi: 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smeele ZE, Burns JM, Van Doorsaler K, Fontenele RS, Waits K, Stainton D, Shero MR, Beltran RS, Kirkham AL, Berngartt R, Kraberger S, Varsani A. 2018. Diverse papillomaviruses identified in Weddell seals. J Gen Virol 99:549–557. doi: 10.1099/jgv.0.001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27:768–777. doi: 10.1101/gr.214346.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, Varsani A. 2017. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 10.Schat KA. 2009. Chicken anemia virus. Curr Top Microbiol Immunol 331:151–183. [DOI] [PubMed] [Google Scholar]
- 11.Chu DKW, Poon LLM, Chiu SSS, Chan KH, Ng EM, Bauer I, Cheung TK, Ng IHY, Guan Y, Wang D, Peiris JSM. 2012. Characterization of a novel gyrovirus in human stool and chicken meat. J Clin Virol 55:209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehér E, Pazár P, Kovács E, Farkas SL, Lengyel G, Jakab F, Martella V, Bányai K. 2014. Molecular detection and characterization of human gyroviruses identified in the ferret fecal virome. Arch Virol 159:3401–3406. doi: 10.1007/s00705-014-2203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Pesavento PA, Gaynor AM, Duerr RS, Phan TG, Zhang W, Deng X, Delwart E. 2015. A gyrovirus infecting a sea bird. Arch Virol 160:2105–2109. doi: 10.1007/s00705-015-2468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rijsewijk FA, Dos Santos HF, Teixeira TF, Cibulski SP, Varela AP, Dezen D, Franco AC, Roehe PM. 2011. Discovery of a genome of a distant relative of chicken anemia virus reveals a new member of the genus Gyrovirus. Arch Virol 156:1097–1100. doi: 10.1007/s00705-011-0971-6. [DOI] [PubMed] [Google Scholar]
- 15.Sauvage V, Cheval J, Foulongne V, Gouilh MA, Pariente K, Manuguerra JC, Richardson J, Dereure O, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol 85:7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao S, Gao X, Tuo T, Han C, Gao Y, Qi X, Zhang Y, Liu C, Gao H, Wang Y, Wang X. 2017. Novel characteristics of the avian gyrovirus 2 genome. Sci Rep 7:41068. doi: 10.1038/srep41068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 18.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of the ashy storm-petrel-associated gyrovirus isolate was deposited at GenBank under the accession number MH378452.