We report the isolation and genomic characterization of a Sapelovirus A strain, or porcine sapelovirus (PSV), from a diarrheic Corsican piglet in France. It shares 87% nucleotide identity with a 2014 German isolate.
ABSTRACT
We report the isolation and genomic characterization of a Sapelovirus A strain, or porcine sapelovirus (PSV), from a diarrheic Corsican piglet in France. It shares 87% nucleotide identity with a 2014 German isolate.
ANNOUNCEMENT
Porcine sapelovirus (PSV; family Picornaviridae, genus Sapelovirus) is a 7.5- to 8.3-kb single-stranded, positive-sense polyadenylated and nonenveloped RNA virus. The genus is closely related to the genus Enterovirus and includes three species, Avian sapelovirus, Sapelovirus A (porcine sapelovirus), and Sapelovirus B (simian sapelovirus). It consists of a single serotype. PSV is transmitted by the fecal-oral route, and infection can be asymptomatic or associated with diarrhea, respiratory distress, encephalitis, skin lesions, and reproductive disorders (1–3). The virus has been detected worldwide in Asia (China [2], India [1], and South Korea [4, 5]), the Americas (United States [6, 7] and Brazil [8]), and Europe (Germany, United Kingdom [9], and Spain [10]).
During a campaign dedicated to identifying pigs infected by the hepatitis E virus (HEV), stool samples with positive reverse transcription-quantitative PCR (qRT-PCR) detection of the HEV genome were inoculated onto cell cultures (PLC/PRF/5 cells). Unexpectedly, a gross cytopathic effect (CPE) was observed for one specimen. It was sampled in 2017 from a 3- to 4-month-old female Nustrale (Corsican breed) piglet in a farm located in the northeast of Corsica, France. While CPE was maintained after passaging the culture supernatant, HEV was not detected, suggesting isolation of a different virus. Viral RNA was extracted from the cell culture supernatant at passage 1, using the EZ1 device and EZ1 virus minikit 2.0 (Qiagen), following the manufacturer’s instructions. Reverse transcription, nonspecific amplification, and library building were performed as previously described (11), followed by sequencing using Ion S5 technology (Thermo Fisher).
Data were treated as previously reported (12). In brief, the 95,906 reads obtained were cleaned, using CLC Genomics Workbench software v11.0.1, according to quality score (limit, 0.01) and length (reads shorter than 100 bp were removed). De novo assembly (requirements included the following: map reads back to contigs, mismatch cost, 2 occurrences; insertion cost, 3 occurrences; deletion cost, 3 occurrences; length fraction, 50%; similarity fraction, 80%; and minimum contig length, 350 bp) was realized, and a 7,532-nucleotide (nt)-long PSV consensus sequence, including the complete open reading frame (ORF) (7,014 nt, determined from the CLC Genomics Workbench v11.0.1 software pipeline: start codon ATG, and standard genetic code), was identified from 49,761 reads. Partial 5′ and 3′ noncoding region sequences were obtained (423/480 and 95/106 nt long, respectively, with reference to previously published PSV sequences (GenBank accession no. KJ821019, KX574284, LC326555, JX286666, and KF539414). According to current knowledge (4, 6, 13), the ORF encodes four structural proteins (VP4, VP3, VP2, and VP1) and seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D). The strain was assigned the name PSV OPY-1-Corsica-2017 and made available in the European Virus Archive collection (and under GenBank accession no. MH513612).
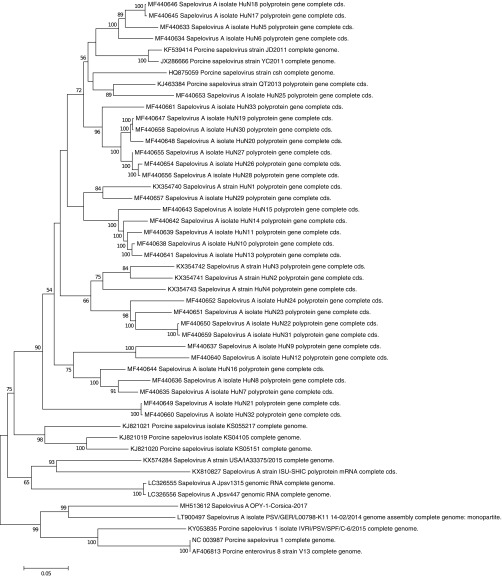
A maximum likelihood phylogenetic reconstruction (GTR+G+I model chosen from the best model option on MEGA6 software, determined from the data set using the MEGA6 program [14]) was obtained using full ORF sequences and indicates that PSV OPY-1-Corsica-2017 is close to a 2014 German isolate (GenBank accession no. LT900497), with ca. 87% nt (6,561/7,527) and 96% amino acid (aa) (2,238/2,337) identities (Fig. 1). Phylogenetically, PSV OPY-1-Corsica-2017 clusters with Indian (KY053835) and German (LT900497, NC_003987, and AF406813) PSV isolates and is clearly separated from other strains isolated from China and the United States, among others.
FIG 1.
Phylogenetic tree reconstructed from PSV ORF nucleotide sequences using the maximum likelihood algorithm, 500 resampling bootstraps (GTR+G+I model, determined from the data set), and the MEGA 6.06 software.
In conclusion, we identified and characterized for the first time a porcine sapelovirus in France, on the Mediterranean island of Corsica. Importantly, the PSV-infected piglet from which PSV OPY-1-Corsica-2017 was isolated was born and farmed in Corsica. This is strongly suggestive of local transmission of the endemic virus.
Data availability.
The genome sequence reported here has been deposited in the GenBank database under the accession no. MH513612.
ACKNOWLEDGMENTS
We are grateful to the farmers who provided the samples used in the current study.
This study was supported by the Corsica territorial collectivity and by the European Virus Archive goes Global (EVAg) project (European Union–Horizon 2020 program) under grant agreement no. 653316.
REFERENCES
- 1.Ray PK, Desingu PA, Kumari S, John JK, Sethi M, Sharma GK, Pattnaik B, Singh RK, Saikumar G. 2018. Porcine sapelovirus among diarrhoeic piglets in India. Transbound Emerg Dis 65:261–263. doi: 10.1111/tbed.12628. [DOI] [PubMed] [Google Scholar]
- 2.Lan D, Ji W, Yang S, Cui L, Yang Z, Yuan C, Hua X. 2011. Isolation and characterization of the first Chinese porcine sapelovirus strain. Arch Virol 156:1567. doi: 10.1007/s00705-011-1035-7. [DOI] [PubMed] [Google Scholar]
- 3.Kim D-S, Son K-Y, Koo K-M, Kim J-Y, Alfajaro MM, Park J-G, Hosmillo M, Soliman M, Baek Y-B, Cho E-H, Lee J-H, Kang M-I, Goodfellow I, Cho K-O. 2016. Porcine sapelovirus uses α2,3-linked sialic acid on GD1a ganglioside as a receptor. J Virol 90:4067–4077. doi: 10.1128/JVI.02449-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bak G-Y, Kang M-I, Son K-Y, Park J-G, Kim D-S, Seo J-Y, Kim J-Y, Alfajaro MM, Soliman M, Baek Y-B, Cho E-H, Kwon J, Choi J-S, Park S-I, Cho K-O. 2017. Occurrence and molecular characterization of Sapelovirus A in diarrhea and non-diarrhea feces of different age group pigs in one Korean pig farm. J Vet Med Sci 78:1911–1914. doi: 10.1292/jvms.16-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D-S, Kang M-I, Son K-Y, Bak G-Y, Park J-G, Hosmillo M, Seo J-Y, Kim J-Y, Alfajaro MM, Soliman M, Baek Y-B, Cho E-H, Lee J-H, Kwon J, Choi J-S, Goodfellow I, Cho K-O. 2016. Pathogenesis of Korean Sapelovirus A in piglets and chicks. J Gen Virol 97:2566–2574. doi: 10.1099/jgv.0.000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arruda PHE, Arruda BL, Schwartz KJ, Vannucci F, Resende T, Rovira A, Sundberg P, Nietfeld J, Hause BM. 2017. Detection of a novel sapelovirus in central nervous tissue of pigs with polioencephalomyelitis in the USA. Transbound Emerg Dis 64:311–315. doi: 10.1111/tbed.12621. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Zheng Y, Guo B, Zhang J, Yoon K-J, Harmon KM, Main RG, Li G. 2016. Complete genome sequence of porcine sapelovirus strain USA/IA33375/2015 identified in the United States. Genome Announc 4:e01055-16. doi: 10.1128/genomeA.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donin DG, de Arruda Leme R, Alfieri AF, Alberton GC, Alfieri AA. 2014. First report of Porcine teschovirus (PTV), Porcine sapelovirus (PSV) and Enterovirus G (EV-G) in pig herds of Brazil. Trop Anim Health Prod 46:523–528. doi: 10.1007/s11250-013-0523-z. [DOI] [PubMed] [Google Scholar]
- 9.Schock A, Gurrala R, Fuller H, Foyle L, Dauber M, Martelli F, Scholes S, Roberts L, Steinbach F, Dastjerdi A. 2014. Investigation into an outbreak of encephalomyelitis caused by a neuroinvasive porcine sapelovirus in the United Kingdom. Vet Microbiol 172:381–389. doi: 10.1016/j.vetmic.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Buitrago D, Cano-Gómez C, Agüero M, Fernandez-Pacheco P, Gómez-Tejedor C, Jiménez-Clavero MÁ. 2010. A survey of porcine picornaviruses and adenoviruses in fecal samples in Spain. J Vet Diagn Invest 22:763–766. doi: 10.1177/104063871002200519. [DOI] [PubMed] [Google Scholar]
- 11.Stang A, Korn K, Wildner O, Überla K. 2005. Characterization of virus isolates by particle-associated nucleic acid PCR. J Clin Microbiol 43:716–720. doi: 10.1128/JCM.43.2.716-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piorkowski G, Jacquot F, Quérat G, Carbonnelle C, Pannetier D, Mentré F, Raoul H, de Lamballerie X. 2017. Implementation of a non-human primate model of Ebola disease: infection of Mauritian cynomolgus macaques and analysis of virus populations. Antiviral Res 140:95–105. doi: 10.1016/j.antiviral.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Krumbholz A, Dauber M, Henke A, Birch-Hirschfeld E, Knowles NJ, Stelzner A, Zell R. 2002. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J Virol 76:5813–5821. doi: 10.1128/JVI.76.11.5813-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence reported here has been deposited in the GenBank database under the accession no. MH513612.