Carbapenem-resistant Enterobacteriaceae (CRE) represent a health threat, but effective control interventions remain unclear. Hospital wastewater sites are increasingly being highlighted as important potential reservoirs.
KEYWORDS: antimicrobial resistance, carbapenemase-producing Enterobacteriaceae, genome sequencing, infection control, molecular epidemiology
ABSTRACT
Carbapenem-resistant Enterobacteriaceae (CRE) represent a health threat, but effective control interventions remain unclear. Hospital wastewater sites are increasingly being highlighted as important potential reservoirs. We investigated a large Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli outbreak and wider CRE incidence trends in the Central Manchester University Hospital NHS Foundation Trust (CMFT) (United Kingdom) over 8 years, to determine the impact of infection prevention and control measures. Bacteriology and patient administration data (2009 to 2017) were linked, and a subset of CMFT or regional hospital KPC-producing E. coli isolates (n = 268) were sequenced. Control interventions followed international guidelines and included cohorting, rectal screening (n = 184,539 screens), environmental sampling, enhanced cleaning, and ward closure and plumbing replacement. Segmented regression of time trends for CRE detections was used to evaluate the impact of interventions on CRE incidence. Genomic analysis (n = 268 isolates) identified the spread of a KPC-producing E. coli outbreak clone (strain A, sequence type 216 [ST216]; n = 125) among patients and in the environment, particularly on 2 cardiac wards (wards 3 and 4), despite control measures. ST216 strain A had caused an antecedent outbreak and shared its KPC plasmids with other E. coli lineages and Enterobacteriaceae species. CRE acquisition incidence declined after closure of wards 3 and 4 and plumbing replacement, suggesting an environmental contribution. However, ward 3/ward 4 wastewater sites were rapidly recolonized with CRE and patient CRE acquisitions recurred, albeit at lower rates. Patient relocation and plumbing replacement were associated with control of a clonal KPC-producing E. coli outbreak; however, environmental contamination with CRE and patient CRE acquisitions recurred rapidly following this intervention. The large numbers of cases and the persistence of blaKPC in E. coli, including pathogenic lineages, are of concern.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) represent a global public health threat (1). Major carbapenemases include the metallo-β-lactamases, some oxacillinases, and the Klebsiella pneumoniae carbapenemase (KPC) (encoded by blaKPC), one of the most common carbapenemases globally (2). Transfer of carbapenemase genes on mobile genetic elements has resulted in rapid interspecies dissemination of carbapenem resistance (3, 4). Since few therapeutic options remain for CRE infections (5, 6), effective control is critical.
Escherichia coli is a major human pathogen, but it also a gastrointestinal commensal and can be transmitted between humans and the environment. Carbapenem resistance in E. coli, including that encoded by blaKPC, is increasing (7, 8) but is uncommon, and KPC-producing E. coli outbreaks have not been observed to date. The emergence and persistence of carbapenem resistance in E. coli in human and/or environmental reservoirs are of concern.
CRE detections in England have increased since 2008 (9) and are approximately 10 times the national average in Greater Manchester (10). Central Manchester University Hospital NHS Foundation Trust (CMFT) has experienced an ongoing, multispecies, blaKPC-associated CRE outbreak since 2009. Intensive infection prevention and control (IPC) measures, in line with national and international recommendations (11–13), have been implemented in response.
In 2015, a sudden increase in cases of fecal colonization with KPC-producing E. coli was detected in the Manchester Heart Centre (MHC) at the Manchester Royal Infirmary (part of CMFT). We retrospectively investigated the genomic epidemiology and evidence for nosocomial transmission of KPC-producing E. coli and KPC plasmids isolated from patients and the environment in this context, and we assessed the impact of guideline-compliant IPC bundles on CRE and KPC-producing E. coli incidence.
RESULTS
High prevalence of CRE colonization in the MHC.
Between 1 April 2014 and 30 December 2014, 23 new CRE-colonized individuals were detected in the MHC, including 2 with E. coli (Fig. 1A). A CRE outbreak was declared on 2 January 2015, when 6 new CRE-colonized individuals were identified (4 with blaKPC and 2 with blaNDM; no E. coli). Consequently, intensified IPC measures were implemented (Fig. 1B; also see Table S1 in the supplemental material), and wards 3 and 4 were closed (on 6 January 2015), terminally cleaned (with hypochlorite), and decontaminated (with hydrogen peroxide vapor). Ward 3 was reopened on 11 January 2015, and ward 4 was reopened on 23 January 2015. High-risk patients (with previously detected CRE or a history of hospitalization abroad or in a UK hospital with known CRE transmission in the past 12 months) were screened; CRE-positive patients were transferred to a cohort ward or, if they required cardiac monitoring, to side rooms.
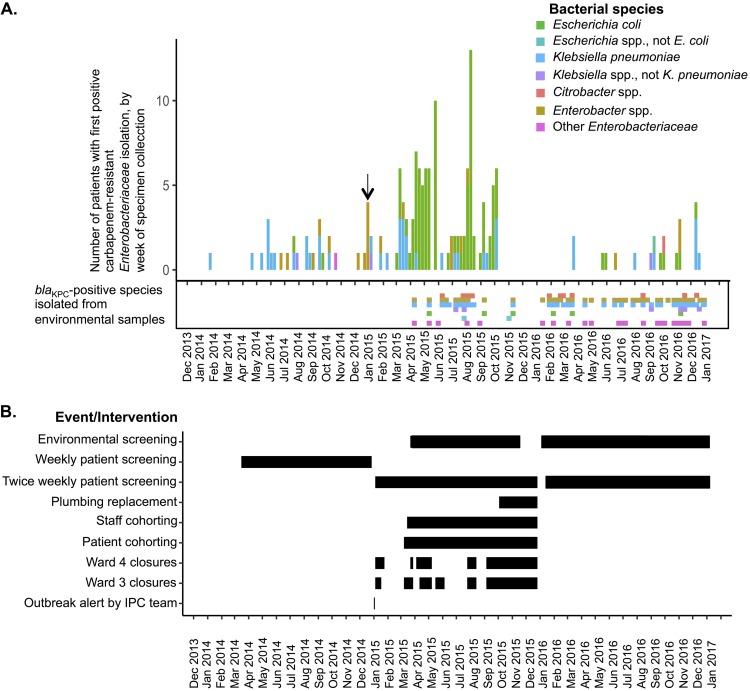
FIG 1.
(A) Numbers of individuals in MHC wards with first CRE-positive detection, by week, stratified by genus group and species of the organism isolated. The blaKPC-positive Enterobacteriaceae strains detected in environmental samples over the same time frame are also shown. The MHC outbreak was declared by the IPC team in the first week in 2015 (arrow). (B) Timeline of IPC measures instituted.
By January 2015, CMFT was operating a Trustwide CRE screening program (>110 screens/day) (Table S2). Between 1 September 2014 and 30 December 2014, screening transitioned from culture-based methods to PCR-based methods; during this period, 16,612 samples from 7,239 inpatients were screened using either culture (9,808 samples) or PCR and culture (6,804 samples), with an overall CRE prevalence of 3.8% (438 positive samples from 272 patients). Molecular mechanism data for 135/163 PCR-positive samples (83%) indicated that blaKPC accounted for most carbapenem resistance (97%).
KPC-producing E. coli outbreak despite IPC interventions.
Following the implementation of enhanced IPC activity, there was a further sharp increase in the number of CRE-colonized patients detected from 9 March 2015 (carbapenem-resistant [CR]E. coli and other species, mostly containing blaKPC and a few with blaNDM) (Fig. 1A). Ward 3 was again closed to admissions (from 11 March 2015 to 28 March 2015), and environmental decontamination was repeated; the following week, ward 4 was closed after detection of additional CRE-colonized patients (Fig. 1A and B). From 1 April 2015, KPC-producing E. coli predominated in the outbreak (Fig. 1A).
From April to September 2015, wards 3 and 4 were closed repeatedly, with 2 peaks in KPC-producing E. coli patient colonization (in April to May and in August) (Fig. 1B). Ward 3 capacity was reduced to 10 day-case beds (on 12 August 2015; day-case patients were not screened for CRE) and ward 4 capacity to 12 inpatient beds. Between 10 August 2015 and 28 September 2015, there were 27 new KPC-producing E. coli colonizations detected in the MHC (Fig. 1A) and 2 cases with other KPC-producing Enterobacteriaceae species. Of 88 KPC-producing E. coli cases between 24 February 2015 and 28 September 2015, 86 (98%) represented colonizations only; 1 individual additionally had a urinary tract infection and 1 a sternal wound infection (treated with gentamicin and ciprofloxacin, respectively, to which the isolates were susceptible).
CR E. coli cases in CMFT.
CR E. coli had been isolated in CMFT prior to the 2015 MHC outbreak, with 514 CRE. coli cases (considering first positive results by patient from clinical/screening isolates) in 2010 to 2016 (inclusive), including a separate outbreak on the gerontology wards (wards 45 and 46) in late 2012 (Fig. 2A and B). Of those, 434 cases were detected on day ≥2 of admission and 80 on day 0 or 1 of admission. Case peaks were not related to screening policy changes or rates (Fig. S6). CRE. coli strains were detected almost invariably from rectal screening samples (420/434 cases [97%]).
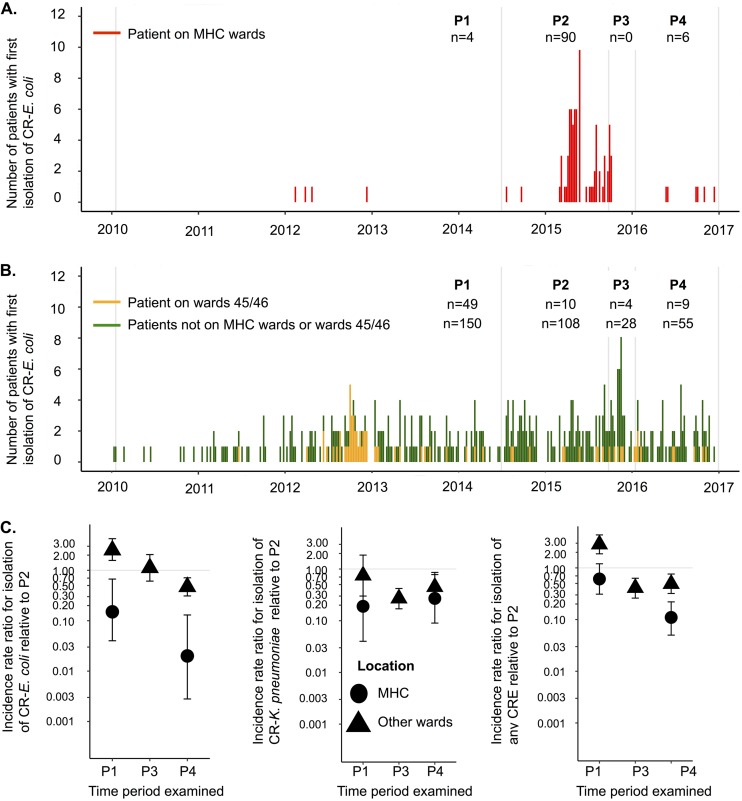
FIG 2.
(A and B) Counts of individuals with first CR E. coli detection by ward location. Detections on days 0 and 1 of admission are excluded. Faint vertical lines correspond to the boundaries of the 4 time periods, as follows: period 1 (P1), prior to implementation of a systematic CPE rectal screening policy; period 2 (P2), implementation of a CPE rectal screening policy consistent with national guidance; period 3 (P3), closure of wards 3 and 4 and replacement of plumbing infrastructure; period 4 (P4), reopening of wards 3 and 4 to patient admissions. (C) Incidence rate ratios for rates of first positive CR E. coli detection, carbapenem-resistant K. pneumoniae detection, and any CRE detection ≥2 days postadmission, relative to period 2 in the same location (MHC versus the rest of CMFT). An incidence rate ratio is not shown for period 3 in the MHC due to unit closure during this period, to facilitate plumbing replacement.
Environmental sampling yielding CRE from sinks and drains.
Intermittent environmental sampling was undertaken to identify potential reservoirs. Overall, 927 samples from 833 sites were obtained between 9 April and 17 November 2015; 355 samples (38%) from 333 sites (40%) were from ward 3 or ward 4, and the remainder were from 11 other wards. A total of 850 samples were from sink/drain/shower/bath sites, 18 from toilets, hoppers, or sluices, and 33 from high-touch sites (including keyboards, door handles, and sponges; the labeling was unclear for 26 samples). Eighty-five samples (9%) and 72 sites (9%) were CRE positive, including 26/355 samples (7%) from 21/333 sites (6%) in wards 3 and 4. CRE-positive sites included shower drains (n = 19), sink taps (n = 7), sink drain tailpieces (n = 10), sink drain strainers (n = 8), sink trap water (n = 1), toilet bowls (n = 1), and other sites (n = 26). Common isolates cultured included Klebsiella spp. (n = 34), Enterobacter spp. (n = 25), and E. coli (n = 11) (Fig. 1A). All CRE-positive cultures were from wastewater/plumbing-associated sites; no other sites tested were CRE positive.
Of 10 sites yielding 11 KPC-producing E. coli isolates, 5 were in the ward 3/ward 4 kitchen (14 to 18 May 2015 [n = 4] and 10 September 2015 [n = 1]), 1 was a ward 4 staff sink (14 May 2015), and 4 were kitchen sinks or drains on wards 31 and 32 (sampling in response to a separate ward 31/32 outbreak, 12 to 17 November 2015). Ward 3/ward 4 sink-specific interventions included sink trap replacement for CRE-colonized sinks (16 April 2015, 31 July 2015, and 11 August 2015) and horizontal pipework cleaning with a brush to try to remove biofilms (11 August 2015).
Cardiac service relocation and decline in CRE colonization incidence.
Given the ongoing difficulty in preventing KPC-producing E. coli acquisitions and the isolation of KPC-producing E. coli from sink and drain sites, wards 3 and 4 were closed from 25 September 2015 and patients were relocated to another ward to allow replacement of the plumbing infrastructure back to the central drainage stacks. Replaceable sink plughole devices designed to prevent water aerosolization in the sink U-bend and to limit biofilm formation (HygieneSiphon; Aquafree) were installed.
Controlling for screening and compared to the period immediately before intervention (when screening policies were the same), the incidence of first detection of any CRE or E. coli strain decreased significantly following the plumbing intervention, both in the MHC and elsewhere in the hospital (Fig. 2C and Table 1). The decline in incidence was significantly greater in the MHC (heterogeneity P < 0.001), where incidence fell by 89% for any CRE strain and by 98% for CR E. coli. The incidence of CR K. pneumoniae also decreased significantly in both settings, but there was no evidence that the declines differed between the two settings (heterogeneity P = 0.31) (Table 1). However, when patients were transferred back to wards 3 and 4 (from 18 January 2016), CR E. coli continued to be detected in patients (6 first detections in 2016) (Fig. 2A). Patient colonization with other CRE strains was also observed, in numbers similar to those for 2014 (Fig. 1A); environmental contamination with CRE in sink and wastewater sites recurred rapidly (Fig. 1A), and 2 environmental sites (both ward utility room sink drains) were CRE positive even prior to patient readmissions to the ward, suggesting residual contamination after the plumbing replacement or reintroduction following the plumbing replacement but prior to patient readmissions.
TABLE 1.
Incidence rate ratios for detection from screening swabs ≥2 days after admission (a proxy marker of acquisition) in CMFT for all CRE cases, CR E. coli cases, and CR K. pneumoniae cases, modeling the impact of the ward 3 and ward 4 closures and plumbing replacement on acquisition
| Location and perioda | All CRE (3,086 cases) |
CR E. coli (502 cases) |
CR K. pneumoniae (1,134 cases) |
|||
|---|---|---|---|---|---|---|
| IRR (95% CI) | P | IRR (95% CI) | P | IRR (95% CI) | P | |
| MHC | ||||||
| Week 3, 2010, to week 26, 2014 (period 1) | 0.61 (0.31–1.20) | 0.15 | 0.15 (0.04–0.67) | 0.012 | 0.19 (0.04–0.82) | 0.026 |
| Week 27, 2014, to week 39, 2015 (period 2; reference period) | 1.00 | 1.00 | 1.00 | |||
| Week 40, 2015, to week 2, 2016 (period 3; wards 3 and 4 closed) | ||||||
| Week 3, 2016, to week 52, 2016 (period 4) | 0.11 (0.05–0.22) | <0.001 | 0.02 (0.00–0.14) | <0.001 | 0.27 (0.09–0.78) | 0.015 |
| Other hospital locations | ||||||
| Week 3, 2010, to week 26, 2014 (period 1) | 2.85 (1.87–4.34) | <0.001 | 2.51 (1.57–4.03) | <0.001 | 0.75 (0.30–1.86) | 0.53 |
| Week 27, 2014, to week 39, 2015 (period 2; reference period) | 1.00 | 1.00 | 1.00 | |||
| Week 40, 2015, to week 2, 2016 (period 3) | 0.41 (0.26–0.63) | <0.001 | 1.12 (0.61–2.05) | 0.71 | 0.27 (0.17–0.42) | <0.001 |
| Week 3, 2016, to week 52, 2016 (period 4) | 0.49 (0.32–0.76) | 0.002 | 0.47 (0.31–0.71) | <0.001 | 0.47 (0.28–0.77) | 0.003 |
| MHC vs other location in reference period (period 2) | 1.69 (0.81–3.50) | 0.16 | 9.05 (3.98–20.55) | <0.001 | 0.45 (0.24–0.86) | 0.015 |
| Heterogeneity for reduction in MHC vs other location | ||||||
| Week 3, 2010, to week 26, 2014 (period 1) | <0.001 | 0.001 | 0.098 | |||
| Week 40, 2015, to week 2, 2016 (period 3) | ||||||
| Week 3, 2016, to week 52, 2016 (period 4) | <0.001 | 0.003 | 0.31 | |||
Four time periods were evaluated, as follows: period 1, prior to implementation of a systematic CPE rectal screening policy; period 2, implementation of a CPE rectal screening policy consistent with national guidance; period 3, closure of wards 3 and 4 and replacement of plumbing infrastructure; period 4, reopening of wards 3 and 4 to patient admissions. Period 2 was chosen as the reference period because of the change in screening policies between period 1 and period 2 (see Table S2 and Fig. S6 in the supplemental material), meaning that a greater incidence would be expected in period 2, due to more patients being screened every week. IRR, incidence rate ratio; CI, confidence interval.
Genomic epidemiology of KPC-producing E. coli.
A total of 268 clinical and environmental CR E. coli isolates were sequenced, including 82 isolates from the MHC (2015 to 2016 [16 environmental isolates]), 36 from wards 45 and 46 (2010 to 2016), 109 from other CMFT wards or units, and 41 from other regional hospitals (Table S3). Nine isolates were blaKPC negative on sequencing; 5 of those isolates contained blaOXA-48, 1 blaOXA-181, and 1 blaNDM-5, with no known carbapenem resistance mechanisms identified for the remaining 2. The 259 KPC-producing E. coli isolates included all 16 environmental CR E. coli isolates, 158 isolates that were the first CR E. coli isolates cultured from patients, and 38 sequentially cultured CR E. coli isolates from patients (longitudinal cultures from 12 patients); sequencing and patient epidemiological identifiers could not be linked for 47/259 isolates.
Forty sequence types (STs), including known pathogenic lineages (e.g., ST131), occurred among the KPC-producing E. coli isolates (Fig. 3; also see Table S3), highlighting regional KPC-producing E. coli diversity. In contrast, 67/80 MHC isolates (84%) were ST216, compared with 59/179 (33%) elsewhere. ST216 has rarely been reported in other settings.
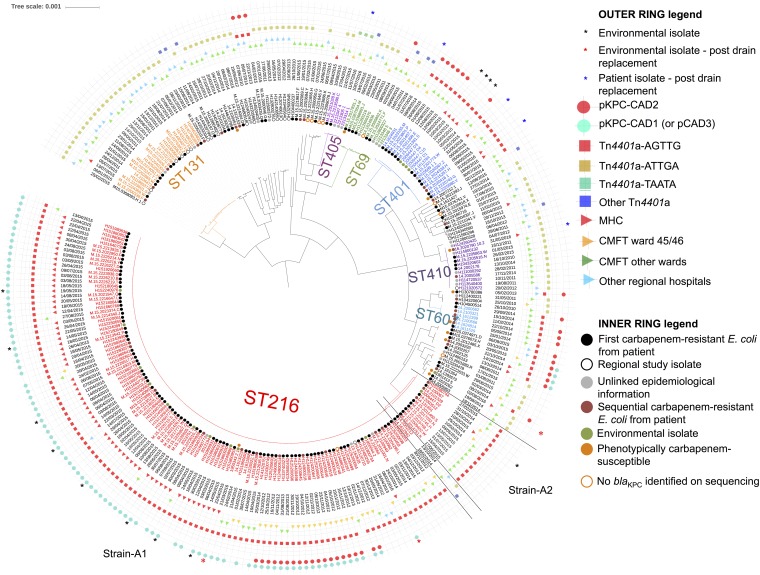
FIG 3.
Recombination-corrected phylogeny of 259 sequenced KPC-producing E. coli isolates (and 9 E. coli isolates that were blaKPC negative on sequencing) from CMFT and other regional hospitals in northwest England, annotated with collection date, ward/center location, Tn4401 type, and outbreak plasmid types. The earliest available sequences per patient are denoted “first carbapenem-resistant E. coli from patient” if the stored isolate collection date was ≤7 days from the first isolation date in the TRACE database or “sequential carbapenem-resistant E. coli from patient” if the stored isolate date was after that. KPC-producing E. coli isolates from a Public Health England (PHE) project that sequenced the first 10 KPC-producing Enterobacteriaceae strains from hospitals in northwest England (2009 to 2014) are denoted “regional study isolate.” “Environmental isolate” denotes KPC-producing E. coli strains cultured during an initial environmental prevalence survey on wards 3 and 4 (10 March 2015), any KPC-producing E. coli strain isolated as part of subsequent, intermittent, IPC-associated environmental sampling (9 April 2015 to 17 November 2015), and isolates available at the time of analysis from environmental and patient samples from a separate ongoing study (commenced January 2016).
ST216 KPC-producing E. coli.
The ST216 KPC-producing E. coli group (n = 126, including 1 blaKPC-negative isolate [H134880341]; 9,118 variable sites) was represented by 2 main genetic subgroups, consisting of 112 isolates (the main outbreak strain [strain A1 in Fig. 3]; ≤65 single-nucleotide variations [SNVs] among isolates in this cluster [2012 to 2016]) and 12 isolates (the secondary outbreak strain [strain A2 in Fig. 3]; ≤25 SNVs among isolates in this cluster and >7,800 SNVs divergent from strain A1 isolates [2012 to 2015]). Although the SNV-based distances between strains A1 and A2 were large, review of the ClonalFrameML output suggested that these differences represented a single “mega-recombination event” affecting ∼1 Mb of the genome (Fig. S7).
All except 3 ST216 isolates carried blaKPC-2 in a Tn4401a transposon (14), which is typically associated with high-level blaKPC expression (15), flanked by a 5-bp target site duplication, AGTTG, which was previously observed only with the Tn4401b isoform in an isolate from Colombia (Fig. 3; also see Table S3). This relatively unique transposon-flanking sequence unit was also observed in other lineages within CMFT (e.g., ST401) (Fig. 3). However, plasmid and resistance gene profiles varied considerably, even to some extent within the ST216 KPC-producing E. coli outbreak strains (Fig. 3; also see Fig. S8). Overall, these results demonstrated clonal expansion of specific KPC-producing E. coli strains, with significant accessory genome mobility. Most notable were the emergence and persistence of ST216 KPC-producing E. coli strain A1, which was isolated from patients and the environment over 4 years and caused outbreaks in wards 45 and 46 (2012) and the MHC (2015).
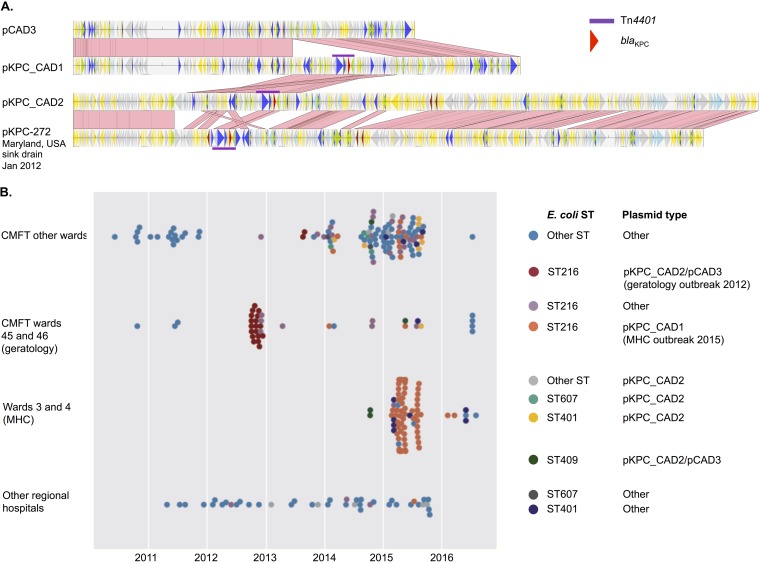
Long-read sequencing demonstrated that the ST216 KPC-producing E. coli strain A1 isolate H124200646 (ward 46 [2012]) contained 2 plasmids, pKPC-CAD2 (307 kb; IncHI2/HI2A, with blaKPC present) and pCAD3 (152 kb; IncFIB/FII, with blaKPC absent); 83% of pKPC-CAD2 was highly similar (99% sequence identity) to pKPC-272 (282 kb, Enterobacter cloacae; GenBank accession no. CP008825.1), which was identified in a sink drain in the National Institutes of Health Clinical Centre in 2012 (16). The other long-read sequence, H151860951 (ward 4 [April 2015]), which was also an ST216 KPC-producing E. coli strain A1 isolate, contained a blaKPC plasmid, pKPC-CAD1 (200 kb; IncFIB/FII), which had 99% sequence identity to pCAD3 over 76% of its length, together with a 48-kb contiguous region including blaKPC that was 99% identical to part of pKPC-CAD2 (Fig. 4A). These results suggest the evolution of a blaKPC plasmid similar to pKPC-272 in CMFT within ST216 KPC-producing E. coli strain A from 2012 to 2015, including recombination between pKPC-CAD2 and pCAD3, giving rise to pKPC-CAD1. Although plasmid typing based on mapping of short-read data to plasmid references should be interpreted cautiously, sequence comparisons with the outbreak plasmids pKPC-CAD1 and pKPC-CAD2 were consistent with the emergence of pKPC-CAD1 and its domination within ST216 KPC-producing E. coli strain A after 2014, as well as exchange of pKPC-CAD1, pKPC-CAD2, and pCAD3 with other E. coli STs (Fig. 3 and 4B).
FIG 4.
(A) Alignments of the 2012 MHC outbreak KPC plasmid pKPC-CAD2 (wards 45 and 46; Tn4401a plus blaKPC) and the 2015 MHC KPC plasmid pKPC-CAD1 (Tn4401a plus blaKPC), highlighting the recombination of the Tn4401a- and blaKPC-harboring 48-kb segment from pKPC-CAD2 with pCAD3 to generate pKPC-CAD1. Regions of sequence homology are represented by pink links drawn between alignments. pKPC-272 (GenBank accession no. CP008825.1), a plasmid identified in an isolate from a sink drain at the National Institutes of Health Clinical Centre in Maryland in 2012, demonstrates significant sequence homology with pKPC-CAD2. (B) Incidence plot of different E. coli STs and likely MHC-related KPC plasmid types across hospital locations.
Environmental CRE isolates.
Thirty environmental CRE isolates from wards 3 and 4 were sequenced, 27 of which were isolated prior to the plumbing replacement and 16 of which were CR E. coli, as described above (13 isolated prior to the plumbing replacement). Eleven of the 16 E. coli isolates were ST216 KPC-producing E. coli strains (10 strain A1 and 1 strain A2), isolated on 8 separate days (in March, May, and September 2015 and February 2016), consistent with transmission between patients and the environment (Fig. 3) and persistence or reintroduction following the plumbing replacement. The other 14 isolates represented diverse KPC-producing CRE species, including K. pneumoniae (n = 7), Citrobacter freundii (n = 4), Klebsiella oxytoca (n = 1), Enterobacter cloacae (n = 1), and Kluyvera intermedia (n = 1). The KPC plasmids in these KPC-producing CRE isolates likely included the outbreak plasmids pKPC-CAD1 and pKPC-CAD2, pKpQIL, and other plasmids, consistent with the interspecies transfer of a diverse set of blaKPC plasmids.
DISCUSSION
Our detailed analyses of the largest institutional KPC-producing E. coli outbreak described to date demonstrate a complex genetic and epidemiological picture, including the emergence of ST216 KPC-producing E. coli strain A1 as a significant clone in CMFT, causing the major 2015 MHC outbreak, an antecedent outbreak in 2012, and sporadic cases and small clusters in other wards and regional health care settings. Plasmid-associated dissemination of blaKPC to other E. coli lineages, including recognized high-risk clones such as ST131, was evident and the problem was substantial, with 514 confirmed patient acquisitions of CR E. coli over a 6-year period.
Environmental sampling on wards 3 and 4 confirmed that sinks and drains were colonized by multiple CRE strains, including the ST216 KPC-producing E. coli strains A1 and A2 and other CRE strains containing the outbreak KPC plasmids (pKPC-CAD1 and pKPC-CAD2), potentially representing a persistent reservoir between patient-associated outbreaks and plausibly explaining why this large outbreak was refractory to standard IPC bundles. Supporting this, the incidence of new CR E. coli detections declined substantially after ward plumbing replacement and temporary relocation of patients (Fig. 1A and 2A and C), consistent with a major contribution from the ward environment. After wards 3 and 4 reopened, however, the environment was rapidly recontaminated, including with ST216 KPC-producing E. coli strain A1, and CRE strains were again detected in patients, suggesting that this type of intervention has limited durability. National and international guidelines on CRE management recommend rectal screening, strict contact precautions, isolation/cohorting of cases, and antimicrobial stewardship to limit transmission (12, 13, 17), all measures already being implemented in CMFT. Current guidelines do not address the control of large persistent outbreaks or provide advice on the sampling and management of environmental reservoirs, and there is limited evidence in support of any given measure (18). It is unclear why a particular strain of KPC-producing E. coli predominated in the outbreak described, as opposed to other CRE strains found contemporaneously in the environment; differences in the gastrointestinal colonization ability of species or an unidentified point source are potential hypotheses.
The response to this outbreak caused major disruption to the hospital and regional cardiac services. Given that almost all cases represented colonizations and not infections, the risks of associated delays in cardiac interventions were debated, although the impacts were not formally quantified. The estimated cost to CMFT of CRE in the first 8 months of 2015 was £5.2 million (19), and the MHC outbreak contributed significantly to this, with approximately £240,000 being spent on the ward 3/ward 4 plumbing replacement.
The study has several limitations, including its observational nature, with only 1 year of follow-up monitoring after the ward 3/ward 4 plumbing replacement. Limited environmental sampling might have meant that the extent of contamination and the diversity of CRE in environmental niches were underestimated. Environmental sampling was restricted to wards in which CRE outbreaks had been detected, and it focused predominantly on sink/drain sites (because initial sampling suggested that those sites were most heavily contaminated); however, component parts of each sink drainage system were not sampled consistently due to resource issues, and the relative prevalence of CRE isolation from any given site type needs to be interpreted with caution. We sequenced only single isolates cultured from individuals at any given time point, due to resource limitations, and therefore might have underestimated the CRE strain diversity within patients. Other non-E. coli Enterobacteriaceae strains were not comprehensively sequenced, possibly underestimating dissemination of pKPC-CAD1 and pKPC-CAD2; however, even our limited sequencing of CRE strains from the environment in 2015 identified those plasmids (and other KPC plasmids) in multiple species. Although genetic overlap between environmental and patient isolates was consistent with transmission between these compartments (Fig. 3), the numbers were too small to infer directionality. Of the predominant KPC plasmid types present within the ST216 KPC-producing E. coli strain A1 outbreak clone, one (pKPC_CAD2) was transferred to multiple E. coli STs (Fig. 3 and 4B), and another (pKPC_CAD1) might have contributed to the clone’s success from 2014 (Fig. 4B), although the genetic and biological mechanisms underpinning this have not been explored.
Our experience highlights the limited evidence for managing large CRE outbreaks, including environmental sampling protocols and interventions, despite numerous centers reporting similar experiences with wastewater sites acting as CRE reservoirs (18, 20–23). Widespread colonization with KPC-producing E. coli is a concern, as E. coli is a common gastrointestinal colonizer and cause of infection, and any stable association between blaKPC and E. coli, particularly in pathogenic lineages such as ST131 (Fig. 3), represents a significant clinical and transmission threat. Although our analyses focused on CRE, similar wider environmental contamination and dissemination of carbapenem-susceptible Enterobacteriaceae seem plausible. A more robust evidence base delineating transmission networks (including initial contamination of sink sites), drivers, and effective control measures (including differential impacts of decontamination methods on particular species and strains) is needed to minimize the financial, clinical, and social impacts of CRE outbreaks.
MATERIALS AND METHODS
Setting.
CMFT is one of the largest hospital trusts in northwest England. The MHC manages >10,000 patients/year and in 2015 included two 28-bed inpatient wards (wards 3 and 4), an acute facility (ward 35), an intensive care unit, and a cardiac catheter laboratory. Ward 3 and ward 4 both included 3 bays and 4 single-patient side rooms, with a shared kitchen (see Fig. S1A and B in the supplemental material).
IPC measures.
CRE screening and IPC measures, based on UK guidelines (11), were implemented Trust-wide from mid-2014. Enhanced measures were introduced in April 2015 in response to the MHC KPC-producing E. coli outbreak (Table S1). In addition, wards 3 and 4 (where most KPC-producing E. coli cases were observed) were closed to replace the plumbing infrastructure back to the drainage stacks (Fig. S2) from September 2015. Staff screening was not undertaken, consistent with national guidelines (11).
Patient CRE screening.
Rectal swabs were screened for CRE using selective chromogenic agar, i.e., ChromID CARBA (bioMérieux) (published sensitivity, 89 to 100%; specificity, 95% [24–26]) to August 2014 and the Cepheid Xpert Carba-R assay (published sensitivity, 97 to 100%; specificity, 99% [27, 28]) from August 2014, along with an in-house multiplex PCR (blaKPC, blaNDM, and blaOXA-48) from November 2014. The Cepheid assay was used for specimens from patients with admissions to the Trust in the past 12 months, those admitted from overseas, or those due to be transferred to a district general hospital (to facilitate transfer planning). All other samples were tested using the multiplex PCR. Species identification of isolates was performed using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker).
Epidemiological analyses.
CMFT electronic bacteriology records were linked, based on NHS numbers, to patient administration data (1 January 2010 to 1 January 2017) and anonymized, and the first CRE-positive test result per patient (rectal screening or clinical specimen) was considered in the evaluation of CRE incidence trends. Trends and the impact of IPC interventions were analyzed retrospectively.
Because CMFT CRE screening rates changed over time in response to national guidance and local IPC interventions and a key aim was to evaluate specifically the impact of ward closure and a radical plumbing intervention in the MHC on CRE acquisition rates, we considered CRE detection rates in 4 periods delineated by 3 time points, namely, the implementation of national carbapenemase-producing Enterobacteriaceae (CPE) IPC policy in mid-2014 (which substantially increased the number of screens performed), the beginning of the MHC-specific intervention (patient relocation and plumbing infrastructure replacement in wards 3 and 4), and the end of the MHC intervention.
First CRE-positive screens were used as a pragmatic proxy for CRE acquisition (i.e., a “case”), given that 89% of patients with first CRE-positive results in the MHC had a negative rectal screen within the preceding 14 days (79% within 7 days) (Fig. S3 to S5). Information on specific carbapenemase mechanisms was not consistently available for all isolates, hampering our ability to perform these analyses specifically by carbapenemase gene family (Table S2).
We tested the hypothesis that CRE acquisitions (reflected by first CRE-positive screens) changed in the MHC more than in other hospital wards following the ward 3/ward 4 closure and plumbing intervention, using negative binomial regression models for the weekly counts of first (per person) CRE detection ≥2 days postadmission (i.e. cases), using weekly numbers of persons screened ≥2 days postadmission as an offset (i.e., adjusting for screening rates), and counting each patient as screened as long as they had ≥1 screen per week. Models were fitted (R v3.4.1) for CRE, CR E. coli, and CR K. pneumoniae. We included period and ward location (MHC versus other wards) as independent variables, with interaction terms for period and location (see the supplemental material for details).
Environmental sampling and sample processing.
In 2015, environmental samples were taken from ward sites using charcoal swabs and were cultured on ChromID CARBA for 18 h at 37°C. After January 2016, ∼20 ml of wastewater was aspirated from sink P-traps, shower drains, or toilets. Aspirates were centrifuged at 4,000 rpm for 10 min, 15 ml of supernatant was discarded, and the pellet was resuspended in the remaining 5 ml. One milliliter of sample was then incubated aerobically overnight at ∼37°C in 5 ml of trypticase soy broth with an ertapenem disc; the multiplex PCR (as above) was performed on broths to identify blaKPC-positive samples for subsequent culture on ChromID CARBA. Environmental sampling prior to January 2016 was not systematic; after January 2016, 75 wastewater sites on wards 3 and 4 were sampled fortnightly on rotation (one half of the sites 1 week and the other half the next); these sites included toilets, sink basins, and sink drains.
Genome sequencing and sequence data analysis.
To provide genetic context for the outbreak, we sequenced retrievable, archived, KPC-producing E. coli patient and environmental isolates from CMFT and patient isolates collected for regional public health surveillance (see the supplementary methods and Table S3 in the supplemental material). We also sequenced a small subset of non-E. coli environmental CRE isolates that had been stored (n = 14) ad hoc as part of outbreak sampling prior to the plumbing replacement.
For Illumina sequencing (HiSeq 2500; 150-bp PE reads), DNA was extracted using the QuickGene system (Fujifilm, Japan), with an additional mechanical lysis step following chemical lysis (FastPrep; MP Biomedicals, USA). Two outbreak isolates (H124200646 and H151860951) were selected for long-read sequencing based on Illumina data. For long-read sequencing (with a PacBio [n = 1] or MinION [n = 1] system), DNA was extracted using the Qiagen genomic tip 100/G kit (Qiagen, Netherlands) (see the supplementary methods).
In silico species identification was performed using Kraken (29). Illumina reads were then mapped to species-specific references (E. coli CFT073 [GenBank accession no. AE014075.1] and the ST216 reference H151860951), and base-calling was performed as described previously (30). De novo assembly was performed using SPAdes v3.6 (31), and resistance gene, blaKPC plasmid, and Tn4401 typing was performed using BLASTn and mapping-based approaches (see the supplementary methods and Table S3).
Two-dimensional reads were extracted from MinION sequence data using poretools (32); hybridSPAdes (31) and Canu (33) were used to generate de novo hybrid assemblies from MinION and Illumina data (see the supplementary methods in the supplemental material). PacBio sequence data were de novo assembled using HGAP3 (34). E. coli phylogenies were reconstructed using IQ-Tree (35) and ClonalFrameML (36) and were visualized in iTOL (37) (see the supplementary methods).
Ethics approval.
Because the investigations formed part of a Trust board-approved outbreak response, ethics approval was not required under NHS governance arrangements (see the supplementary methods).
Accession number(s).
Sequencing data are available under NCBI BioProject PRJNA379782.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for and acknowledge the contributions of the clinical and support staff working in the MHC, CMFT; the microbiology laboratory staff and infection control teams at CMFT; the staff of the Manchester Medical Microbiology Partnership; and the research laboratory (in particular, Ali Vaughan) and informatics and project management teams working as part of the Modernising Medical Microbiology consortium (Oxford). We thank Jeff Scott, Ashley Sharp, and Theresa Shryane (Public Health England [PHE] North West) and Suzan Trienekens (Field Epidemiology Service, PHE) for data collection and Karen Mathieson (CMFT), Jane Turton, and Claire Perry (PHE) for outbreak investigation and support. We thank the Health Protection Research Unit Steering Group for review of the draft manuscript.
The Transmission of Carbapenemase-producing Enterobacteriaceae (TRACE) study investigators are listed alphabetically, with those also included as named individuals in the author list in parentheses: (Zoie Aiken), (Oluwafemi Akinremi), (Julie Cawthorne), (Paul Cleary), (Derrick W. Crook), (Valerie Decraene), (Andrew Dodgson), Michel Doumith, Matthew Ellington, David W. Eyre, (Ryan George), (Malcolm Guiver), Robert Hill, Katie Hopkins, Rachel Jones, (Cheryl Lenney), (Amy J. Mathers), (Ashley McEwan), Ginny Moore, Sarah Neilson, Tim E. A. Peto, (Hang T. T. Phan), Mark Regan, (Anna C. Seale), (Nicole Stoesser), Jay Turner-Gardner, (Vicky Watts), Jimmy Walker, (A. Sarah Walker), (David H. Wyllie), (William Welfare), and (Neil Woodford).
This work was supported by the National Institute for Health Research, Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance, Oxford University, in partnership with PHE (grant HPRU-2012-10041). The report presents independent research funded by the National Institute for Health Research. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, the Department of Health, or PHE. N.S. is funded by a PHE-University of Oxford Clinical Lectureship. Contemporaneous outbreak investigation by CMFT and PHE was undertaken as part of routine activity.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01689-18.
Contributor Information
the TRACE Investigators’ Group:
Zoie Aiken, Oluwafemi Akinremi, Julie Cawthorne, Paul Cleary, Derrick W. Crook, Valerie Decraene, Andrew Dodgson, Michel Doumith, Matthew Ellington, David W. Eyre, Ryan George, Malcolm Guiver, Robert Hill, Katie Hopkins, Rachel Jones, Cheryl Lenney, Amy J. Mathers, Ashley McEwan, Ginny Moore, Sarah Neilson, Tim E. A. Peto, Hang T. T. Phan, Mark Regan, Anna C. Seale, Nicole Stoesser, Jay Turner-Gardner, Vicky Watts, Jimmy Walker, A. Sarah Walker, David H. Wyllie, William Welfare, and Neil Woodford
REFERENCES
- 1.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215(Suppl 1):S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Centre for Disease Prevention and Control. 2013. ECDC technical report: carbapenemase-producing bacteria in Europe: interim results from the European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) project 2013. European Centre for Disease Prevention and Control, Solna, Sweden. [Google Scholar]
- 6.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 7.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. doi: 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasevic AT, Canton R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL. 2016. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 9.Public Health England. 2014. Carbapenemase-producing Enterobacteriaceae: laboratory confirmed cases, 2003 to 2013. www.gov.uk/government/publications/carbapenemase-producing-enterobacteriaceae-laboratory-confirmed-cases/carbapenemase-producing-enterobacteriaceae-laboratory-confirmed-cases-2003-to-2013. Accessed 9 February 2016.
- 10.Donker T, Henderson KL, Hopkins KL, Dodgson AR, Thomas S, Crook DW, Peto TEA, Johnson AP, Woodford N, Walker AS, Robotham JV. 2017. The relative importance of large problems far away versus small problems closer to home: insights into limiting the spread of antimicrobial resistance in England. BMC Med 15:86. doi: 10.1186/s12916-017-0844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Public Health England. 2014. Carbapenemase-producing Enterobacteriaceae: early detection, management and control toolkit for acute trusts. Public Health England, London, United Kingdom: www.gov.uk/government/publications/carbapenemase-producing-enterobacteriaceae-early-detection-management-and-control-toolkit-for-acute-trusts. [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2015. Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE): November 2015 update. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 13.European Centre for Disease Prevention and Control. 2016. Rapid risk assessment: carbapenem-resistant Enterobacteriaceae. European Centre for Disease Prevention and Control, Stockholm, Sweden: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/carbapenem-resistant-enterobacteriaceae-risk-assessment-april-2016.pdf. [Google Scholar]
- 14.Cuzon G, Naas T, Nordmann P. 2011. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother 55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheruvanky A, Stoesser N, Sheppard AE, Crook DW, Hoffman PS, Weddle E, Carroll J, Sifri CD, Chai W, Barry K, Ramakrishnan G, Mathers AJ. 2017. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 61:e00025-17. doi: 10.1128/AAC.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Public Health England. 2013. Acute trust toolkit for the early detection, management and control of carbapenemase-producing Enterobacteriaceae. Public Health England, London, UK: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/329227/Acute_trust_toolkit_for_the_early_detection.pdf. [Google Scholar]
- 18.Kizny Gordon AE, Mathers AJ, Cheong EY, Gottlieb T, Kotay S, Walker AS, Peto TEA, Crook DW, Stoesser N. 2017. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections: a systematic review of the literature. Clin Infect Dis 64:1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 19.Central Manchester University Hospital NHS Foundation Trust. 2015. Financial performance for 2015/16. Central Manchester University Hospital NHS Foundation Trust, Manchester, UK. [Google Scholar]
- 20.Carling PC. 2018. Wastewater drains: epidemiology and interventions in 23 carbapenem-resistant organism outbreaks. Infect Control Hosp Epidemiol 39:972–979. doi: 10.1017/ice.2018.138. [DOI] [PubMed] [Google Scholar]
- 21.Kotsanas D, Wijesooriya WR, Korman TM, Gillespie EE, Wright L, Snook K, Williams N, Bell JM, Li HY, Stuart RL. 2013. “Down the drain”: carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med J Aust 198:267–269. doi: 10.5694/mja12.11757. [DOI] [PubMed] [Google Scholar]
- 22.Leitner E, Zarfel G, Luxner J, Herzog K, Pekard-Amenitsch S, Hoenigl M, Valentin T, Feierl G, Grisold AJ, Hogenauer C, Sill H, Krause R, Zollner-Schwetz I. 2015. Contaminated handwashing sinks as the source of a clonal outbreak of KPC-2-producing Klebsiella oxytoca on a hematology ward. Antimicrob Agents Chemother 59:714–716. doi: 10.1128/AAC.04306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergara-López S, Domínguez MC, Conejo MC, Pascual Á, Rodríguez-Baño J. 2013. Wastewater drainage system as an occult reservoir in a protracted clonal outbreak due to metallo-β-lactamase-producing Klebsiella oxytoca. Clin Microbiol Infect 19:E490–E498. doi: 10.1111/1469-0691.12288. [DOI] [PubMed] [Google Scholar]
- 24.Papadimitriou-Olivgeris M, Bartzavali C, Christofidou M, Bereksi N, Hey J, Zambardi G, Spiliopoulou I. 2014. Performance of chromIDR CARBA medium for carbapenemases-producing Enterobacteriaceae detection during rectal screening. Eur J Clin Microbiol Infect Dis 33:35–40. doi: 10.1007/s10096-013-1925-6. [DOI] [PubMed] [Google Scholar]
- 25.Simner PJ, Gilmour MW, DeGagne P, Nichol K, Karlowsky JA. 2015. Evaluation of five chromogenic agar media and the Rosco Rapid Carb screen kit for detection and confirmation of carbapenemase production in Gram-negative bacilli. J Clin Microbiol 53:105–112. doi: 10.1128/JCM.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simner PJ, Martin I, Opene B, Tamma PD, Carroll KC, Milstone AM. 2016. Evaluation of multiple methods for detection of gastrointestinal colonization of carbapenem-resistant organisms from rectal swabs. J Clin Microbiol 54:1664–1667. doi: 10.1128/JCM.00548-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, Hindler J, Veltman J, Wang H, Cantón R. 2016. Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol 54:1814–1819. doi: 10.1128/JCM.00341-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyos-Mallecot Y, Ouzani S, Dortet L, Fortineau N, Naas T. 2017. Performance of the Xpert® Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int J Antimicrob Agents 49:774–777. doi: 10.1016/j.ijantimicag.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loman NJ, Quinlan AR. 2014. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 34.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.