Currently available therapies for chronic hepatitis B virus (HBV) infection can efficiently reduce viremia but induce hepatitis B surface antigen (HBsAg) loss in very few patients; also, these therapies do not greatly affect the viral covalently closed circular DNA (cccDNA). To discover new agents with complementary anti-HBV effects, we performed a drug repurposing screen of 1,018 Food and Drug Administration (FDA)-approved compounds using HBV-infected primary human hepatocytes (PHH).
KEYWORDS: HBV, RAR agonist, tazarotene, antiviral agents, metabolic pathway
ABSTRACT
Currently available therapies for chronic hepatitis B virus (HBV) infection can efficiently reduce viremia but induce hepatitis B surface antigen (HBsAg) loss in very few patients; also, these therapies do not greatly affect the viral covalently closed circular DNA (cccDNA). To discover new agents with complementary anti-HBV effects, we performed a drug repurposing screen of 1,018 Food and Drug Administration (FDA)-approved compounds using HBV-infected primary human hepatocytes (PHH). Several compounds belonging to the family of retinoic acid receptor (RAR) agonists were identified that reduced HBsAg levels in a dose-dependent manner without significant cytotoxicity. Among them, tazarotene exhibited the most potent anti-HBV effect, with a half-maximal inhibitory concentration (IC50) for HBsAg of less than 30 nM in PHH. The inhibitory effect was also observed in HBV-infected differentiated HepaRG (dHepaRG) models, but not in HepG2.215 cells, and HBV genotypes A to D were similarly inhibited. Tazarotene was further demonstrated to repress HBV cccDNA transcription, as determined by the levels of HBV cccDNA and RNAs and the activation of HBV promoters. Moreover, RNA sequence analysis showed that tazarotene did not induce an interferon response but altered the expression of a number of genes associated with RAR and metabolic pathways. Inhibition of RARβ, but not RARα, by a specific antagonist significantly attenuated the anti-HBV activity of tazarotene, suggesting that tazarotene inhibits HBV in part through RARβ. Finally, a synergistic effect of tazarotene and entecavir on HBV DNA levels was observed. Therefore, RAR agonists as represented by tazarotene were identified as potential novel anti-HBV agents.
INTRODUCTION
Hepatitis B virus (HBV) is a small hepatotropic partially double-stranded DNA virus. Although there have been effective vaccines against it, chronic hepatitis B (CHB) virus infection, affecting approximately 250 million people worldwide, remains a serious health problem and is the main risk factor for the development of liver cirrhosis and hepatocellular carcinoma.
At present, two classes of antiviral therapies have been approved for the treatment of hepatitis B, alpha interferon (IFN-α) and nucleos(t)ide analogues. The antiviral efficacy of IFN-based therapy is about 20% to 40%, as determined by HBe antigen (HBeAg) loss. The nucleos(t)ide analogs, including lamivudine, adefovir, entecavir, tenofovir, and telbivudine, suppress HBV by inhibiting the viral reverse transcriptase. Although they are able to induce viral suppression in the majority of patients, sustained HBeAg and HBV surface antigen (HBsAg) loss, which is considered a "functional cure," is only achieved in very few patients (1, 2). There have been reports suggesting that HBsAg contributes to the impaired innate immune responses (3), as well as to the suppression of antiviral T-cell activation (4, 5). Furthermore, HBsAg was reported to play a role in the pathogenesis of viral hepatitis B (6, 7). Besides, since the viral covalently closed circular DNA (cccDNA) in the nucleus, serving as the template for the transcription of viral RNAs and the viral reservoir (8), is persistent for long periods of time and cannot be directly targeted, most patients require long-term and even lifelong therapies, and this may result in the selection of drug-resistant mutations and increase the cost and potential risk of side effects (1, 9). There is thus a need for new therapeutic approaches that can reduce HBsAg and target the viral cccDNA.
Here, using HBV-infected primary human hepatocytes (PHH), we performed a drug repurposing screen of 1,018 Food and Drug Administration (FDA)-approved compounds for which bioactivity and cytotoxicity information has been well characterized. Several compounds belonging to the family of retinoic acid receptor (RAR) agonists were identified to efficiently reduce HBsAg levels in a dose-dependent manner without significant cytotoxicity. Among them, tazarotene exhibited the most potent anti-HBV activity, actively repressing HBV cccDNA transcription and HBsAg production. We further analyzed the alterations in gene expression induced by tazarotene, and a synergistic antiviral effect was observed when tazarotene and entecavir were combined. These findings identified RAR agonists as a novel class of anti-HBV agents with complementary mechanisms of action.
RESULTS
Screening of an FDA-approved drug library identifies retinoic acid compounds as potent anti-HBV agents.
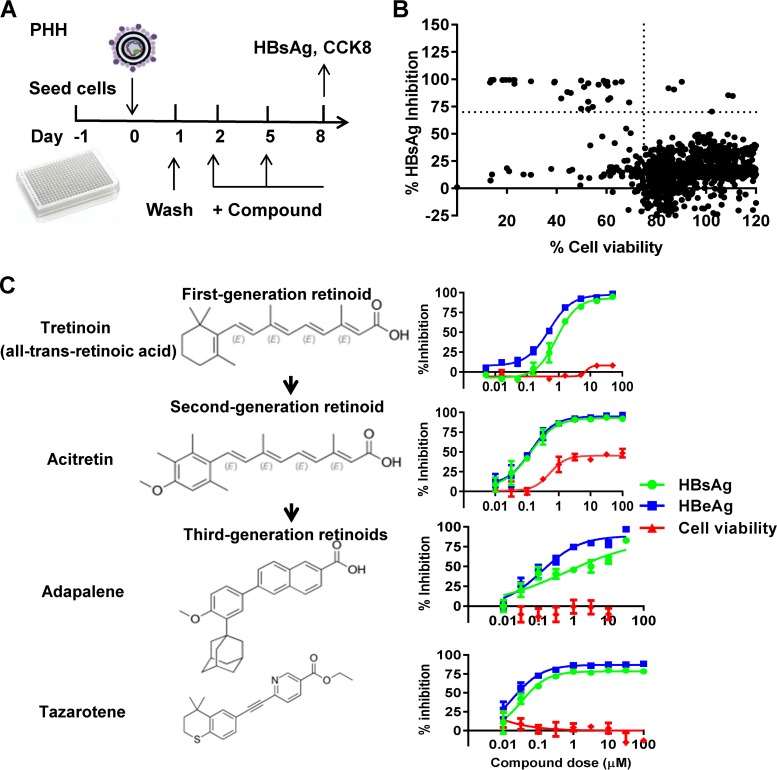
In order to discover new anti-HBV compounds in the context of HBV infection, we performed a drug repurposing screen of 1,018 Food and Drug Administration (FDA)-approved compounds in HBV-infected PHH cells (Fig. 1A). The high-throughput screening (HTS) assay conditions, including cell density and HBV inoculum, were preoptimized for a sufficient assay window. The Z-factor, which quantifies the assay quality, was calculated to characterize the assay performance for HTS. Here, the coefficient of variation (CV) value and the Z-factor were 14.1% and 0.57, respectively, showing that the assay was robust and ready for HTS. A total of 6 compounds that reduced HBsAg levels by more than 70% under the conditions of cell viability over 75% were identified through screening (Fig. 1B). Their inhibitory effects on HBsAg and HBeAg were further evaluated and confirmed in an independent experiment (see Table S1 in the supplemental material). Among the 6 compounds, 3 of them were approved for the treatment of cancer. Interestingly, the other 3 compounds, including adapalene, tretinoin, and tazarotene, belong to a family of RAR agonists. We further tested the inhibitory effects of retinoids against HBV. All the retinoids tested here showed a significant inhibitory effect on HBV antigen production that repressed HBsAg and HBeAg production in a dose-dependent manner without significant cytotoxicity (Fig. 1C). The half-maximal inhibitory concentrations (IC50s) for the tested retinoids and RAR antagonist are summarized in Table S2. Tazarotene, the third-generation retinoid which was approved for treatment of psoriasis, acne, and sun-damaged skin (photodamage), showed the most potent anti-HBV activity, with an IC50 of 30 nM to HBsAg. We also have evaluated the anti-HBV effect of RAR agonist acitretin, the retinoic acid, and the RXR agonist phytanic acid. Phytanic acid showed some anti-HBV activity but not as potent as that of the RAR agonists (Fig. S1).
FIG 1.
Screening for FDA-approved compounds with anti-HBV activity. (A) Schematic representation of the compound screening. PHH cells seeded in 384-well plates were infected with HBV and treated with compounds (10 μM) for 6 days. The levels of HBsAg in the supernatant were determined. (B) Scatter plot of the % HBsAg inhibition versus % cell viability for all tested compounds. Each dot represents the percent inhibition of compounds at 10 µM. Dots located in the upper-right box represent an inhibition of >70% with viability of >75%. The results were normalized to the DMSO control. (C) PHH cells infected with HBV were treated with the indicated retinoids with multiple doses 2 days postinfection. Cells were cultured with medium in the presence of compounds for 6 days before the levels of HBsAg and HBeAg and the cell viability were examined.
Tazarotene potently reduces the production of HBV antigens and viral DNA in HBV-infected cells.
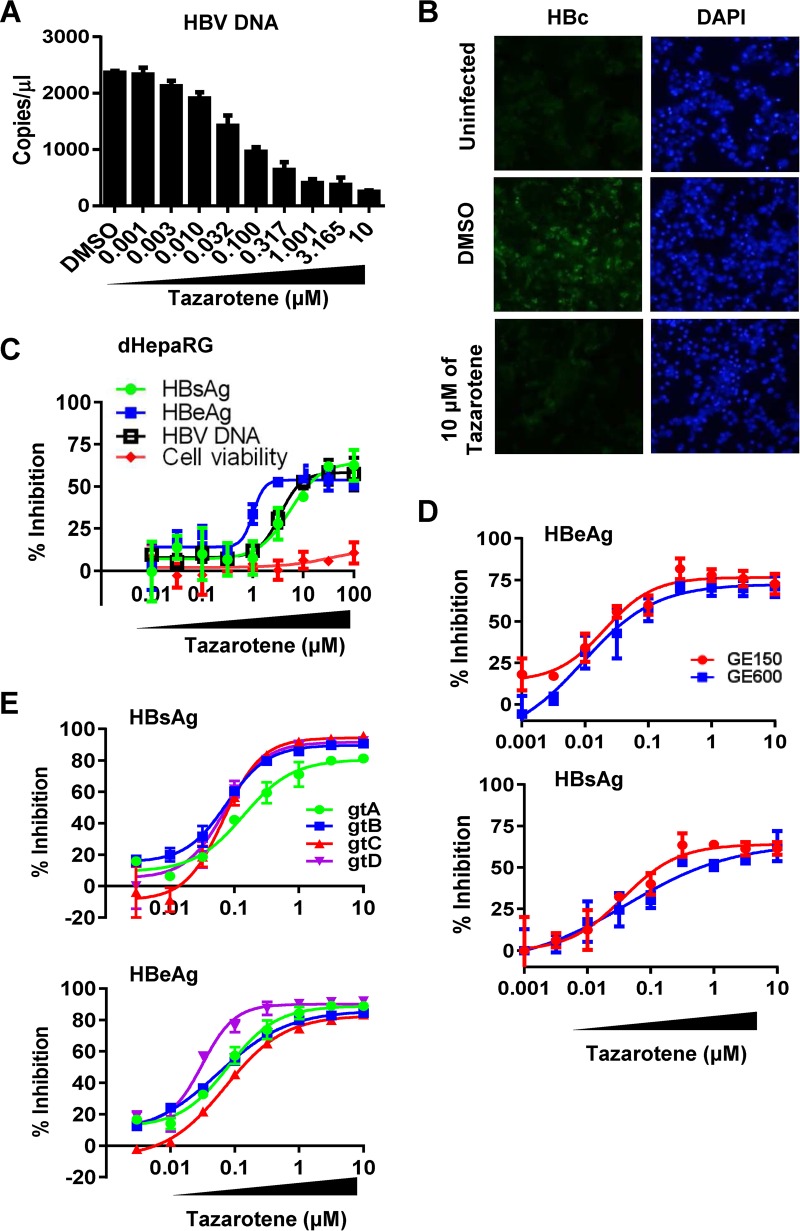
We further confirmed the inhibitory effect of tazarotene on HBV antigen expression and DNA production in cell culture-based HBV infection models. Considering the high donor-to-donor variability of PHH, we first repeated the experiment using PHH derived from other two donors and obtained similar results (Fig. S2). Moreover, we showed that tazarotene reduced the levels of HBV DNA in the cell culture supernatant in a dose-dependent manner (Fig. 2A) and strongly inhibited the intracellular HBc expression at a concentration of 10 µM (Fig. 2B). In dHepaRG cells treated with tazarotene, the supernatant HBsAg, HBeAg, and HBV DNA levels were also significantly inhibited (Fig. 2C). The inhibitory effect of tazarotene on HBeAg production was also observed in the HBV-infected HepG2-sodium taurocholate cotransporting polypeptide (HepG2-NTCP) systems (Fig. S3). Moreover, we found that the HBV replication was inhibited when the cells were pretreated with tazarotene one time before infection, indicating that tazarotene has a prophylactic effect on HBV infection (Fig. S4A). Three days of treatment showed an inhibitory effect similar to that of 6 days of tazarotene treatment, and the effect lasted for 6 days after the cessation of treatment in PHH cells (Fig. S4B). In addition, the inhibitory effect of tazarotene on HBV remained potent when the HBV inoculum was increased to 600 genome equivalents (GE) per cell instead of 150 GE (Fig. 2D). Since clinical observations have suggested that HBV genotype plays a role in therapy response to IFN-α (10), we also investigated the anti-HBV effect of tazarotene across different HBV genotypes. As determined by the levels of HBsAg and HBeAg in the culture supernatant of infected cells, HBV genotypes A to D were similarly inhibited by tazarotene (Fig. 2E and Table S3), suggesting that tazarotene has a pangenotypic anti-HBV effect.
FIG 2.
Effect of tazarotene on the production of HBV antigens and DNA in HBV-infected cells. (A) HBV-infected PHH cells were treated with tazarotene with multiple doses 2 days postinfection. Cells were cultured with medium in the presence of compounds for 6 days before the levels of supernatant HBV DNA were examined. (B) The intracellular HBcAg (HBc) expression (green) in PHH cells treated or not treated with tazarotene were examined. DAPI, 4′,6-diamidino-2-phenylindole. (C) dHepaRG cells infected with HBV were treated with tazarotene from day 3 to day 12 postinfection. The levels of HBsAg, HBeAg, and HBV DNA in the cell culture medium and the cell viability were then analyzed. (D and E) PHH cells infected with indicated viral inoculum dose (D) or with different genotypes of HBV (E) were treated with multiple doses of tazarotene for 6 days starting on day 2 postinfection. The levels of HBsAg and HBeAg in the cell culture medium were then analyzed.
Tazarotene significantly reduces the levels of HBV RNAs transcribed from cccDNA.
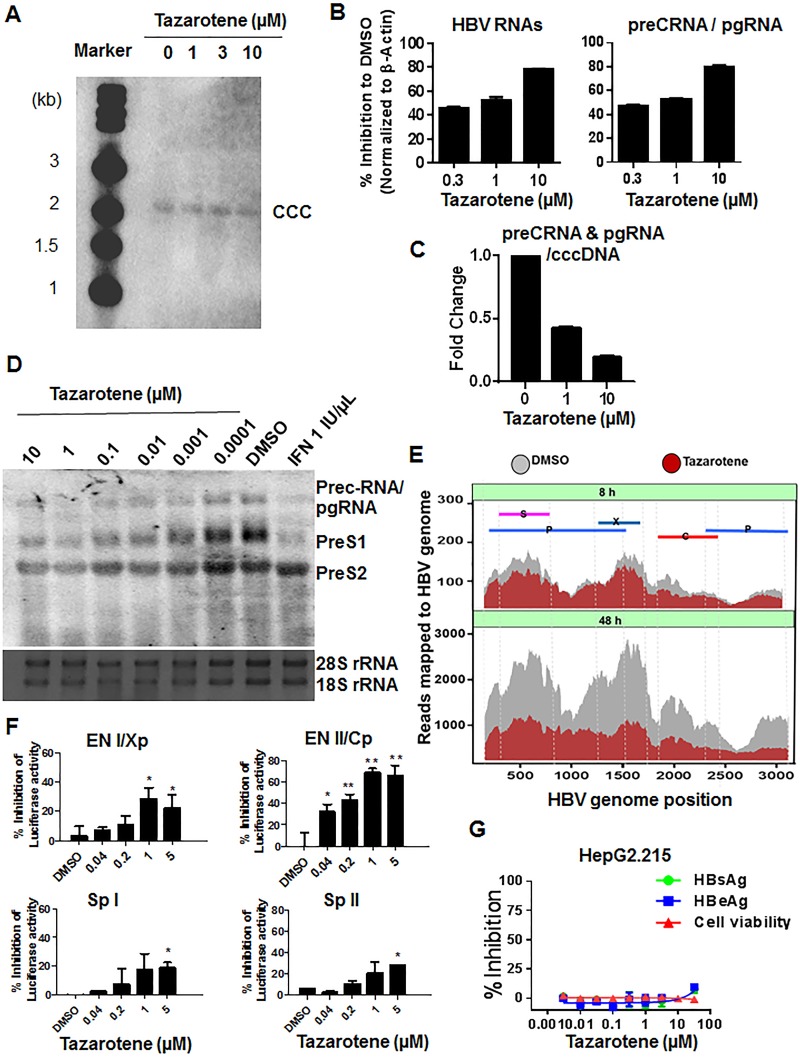
To determine at what level(s) tazarotene inhibits viral antigen expression, we next determined the effect of tazarotene on the cccDNA level, HBV RNA production, and HBV promoter activation. While the cccDNA level was not significantly affected by tazarotene treatment (Fig. 3A), the results of quantitative PCR (qPCR) with specific primers for HBV total RNAs or pregenomic RNA (pgRNA)/precore RNA showed that tazarotene treatment significantly reduced the intracellular HBV RNA levels (Fig. 3B), as well as the ratio of pgRNA/precore RNA to cccDNA (Fig. 3C). Also, the IC50s of tazarotene to RNAs were similar to those of the antigens, as shown in Fig. S5. We next confirmed that the HBV 3.5- and 2.4/2.1-kb mRNAs were inhibited by tazarotene using Northern blot (Fig. 3D). The RNA sequencing (RNA-seq) analysis further revealed that tazarotene induced a reduction in HBV RNA expression as early as 8 h posttreatment, and the reduction became more significant at 48 h posttreatment. By mapping the sequencing reads to the HBV genome, tazarotene was found to inhibit the expression of both pgRNA and subgenomic HBV RNAs (Fig. 3E). To further determine whether this inhibition occurred transcriptionally or posttranscriptionally, we employed reporter plasmids in which the luciferase reporter gene was under the control of HBV promoters/enhancers. The result showed that the activation of the four transcription elements within the HBV genome, including En I/Xp, En II/CP, Sp I, and Sp II, was inhibited in the presence of tazarotene, though the extent of inhibition varied (Fig. 3F). However, tazarotene had little effect on the cytomegalovirus (CMV) promoter-driven HBV RNA expression (data not shown). Furthermore, tazarotene did not greatly affect the expression levels of HBV RNAs and antigens in the Hep2.2.15 cell line (Fig. 3G), in which the HBV genome is integrated into host genome; thus, the majority of HBV RNAs in this cell model are not transcribed from the cccDNA. However, the suppression of HBsAg expression by tazarotene was observed in another HepG2-derived cell line that was recently established by us as a model for studying cccDNA (Fig. S6), in which HBV recombinant cccDNA serves as the template supporting pgRNA transcription and viral replication (11). These results suggested that the tazarotene-mediated reduction in HBV RNAs mainly occurred transcriptionally and that the inhibitory effect of tazarotene on HBV transcription is relatively HBV promoter and cccDNA specific.
FIG 3.
Effect of tazarotene on cccDNA levels, HBV RNA expression, and HBV promoter activation. (A) PHH cells infected with HBV were treated with tazarotene for 9 days. The Hirt DNA was extracted and analyzed by Southern blot. (B) The RNAs were extracted for determination of the levels of total HBV RNA and pregenomic/precore mRNA. (C) The ratio of precore RNA/pgRNA to cccDNA were calculated. (D) Northern blot analysis of 1 μg of RNA extracted from HBV-infected PHH cells treated with tazarotene for 6 days. (E) HBV transcript-level quantification by nucleotide mapping in the presence or absence of tazarotene after 8 h (top) and 48 h (bottom) of treatment was analyzed by RNA sequencing. Each of the HBV RNA transcripts localized to the HBV genome is shown. (F) Huh7 cells transfected with the indicated reporter plasmids were mock treated or treated with the indicated dose of tazarotene. The cells were extracted for luciferase analysis 48 h posttreatment. (G) HepG2.215 cells were treated with tazarotene for 5 days. The HBsAg and HBeAg in the culture supernatant were then measured. Data were analyzed using Student’s t test and are shown as the mean ± SD (n = 3). **, P < 0.01; *, P < 0.05.
Tazarotene does not induce an interferon response but alters the expression of a number of genes associated with RAR and metabolism pathways.
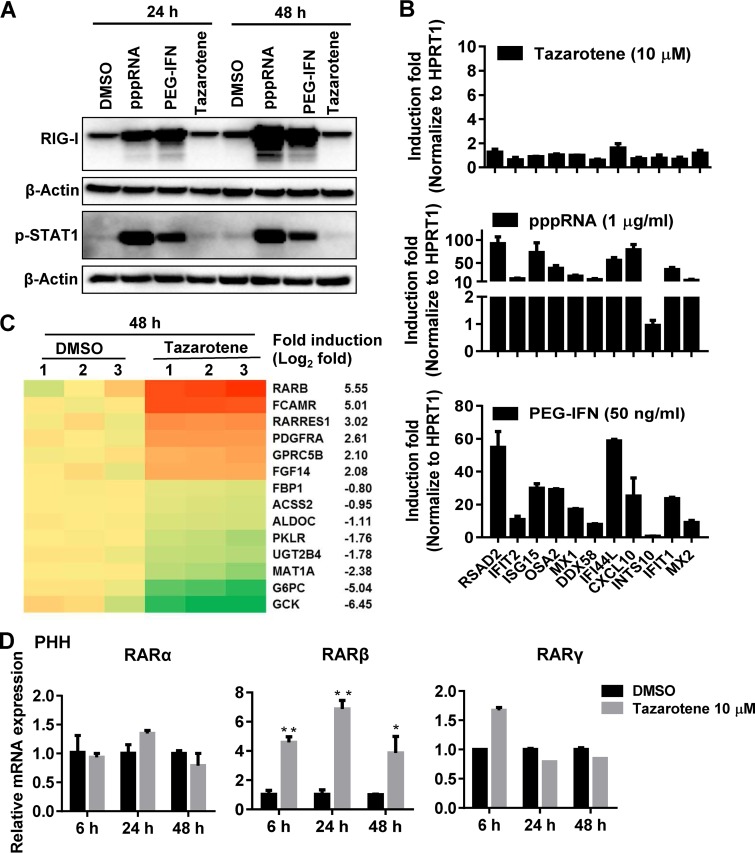
It has been known that the retinoic acid-inducible gene I (RIG-I), a pattern recognition receptor sensor for sensing virus (12, 13), plays an important role in inducing type I interferons and in the activation of intracellular innate antiviral immune responses. We thus further tested whether tazarotene, a retinoic acid, could induce RIG-I expression and result in the phosphorylation of signal transducer and activator of transcription 1 (STAT1) in PHH, which is a key step for IFN-mediated induction of IFN-stimulated genes (ISGs) and antiviral responses. The Western blot analysis showed that tazarotene hardly induced RIG-I expression and STAT1 phosphorylation compared to the cells treated with 5′-triphosphate double-stranded RNA (pppRNA; a potent RIG-I agonist) or pegylated IFN-α, which were used as positive controls (Fig. 4A). We also measured a number of ISGs using the QuantiGene Plex (QGP) method. None of the tested ISGs were significantly induced by tazarotene (Fig. 4B), suggesting that the anti-HBV effect of tazarotene observed in the current system did not rely on the activation of IFN responses.
FIG 4.
Effect of tazarotene treatment on cellular gene expression. (A) PHH cells infected with HBV were transfected with 0.5 µg/ml pppRNA or treated with 100 ng/ml pegylated IFN (PEG-IFN) or 10 μM tazarotene for 24 h and 48 h. The cells were then extracted for Western blot analysis. (B) ISGs were measured by QGP after 24 h stimulation by tazarotene, pppRNA, or PEG-IFN. (C) Heatmap analysis of cellular gene expression regulated by tazarotene treatment (48 h) compared to the mock-treated cells. The samples were triplicated (labeled 1, 2, and 3). (D) The PHH cells treated with tazarotene for the indicated times were extracted for analyzing the expression of RARα, RARβ, and RARγ at the mRNA level. Data were analyzed using Student’s t test and are shown as the mean ± SD (n = 3). **, P < 0.01; *, P < 0.05.
To determine the mechanism by which tazarotene inhibits HBV, we further characterized the tazarotene-mediated changes of cellular gene expression on a transcriptome-wide scale by the high-throughput RNA-seq technique. A series of genes and cellular pathways were found to be altered upon tazarotene treatment (Fig. 4C and S7). Among them, the genes related to RAR, such as RARβ, and genes associated with cellular metabolic pathway, such as G6PC, were the most significant differentially expressed genes. We further confirmed the effect of tazarotene on the expression of RAR-related genes in PHH and HepG2 cells. The RARβ gene, but not the RARα and RARγ genes, was shown to be significantly upregulated by tazarotene both in PHH cells (Fig. 4D) and HepG2 cells (Fig. S8A) at the mRNA level. Moreover, significant induction of intracellular RARβ at the protein level was observed after 72 h of tazarotene treatment (Fig. S8B).
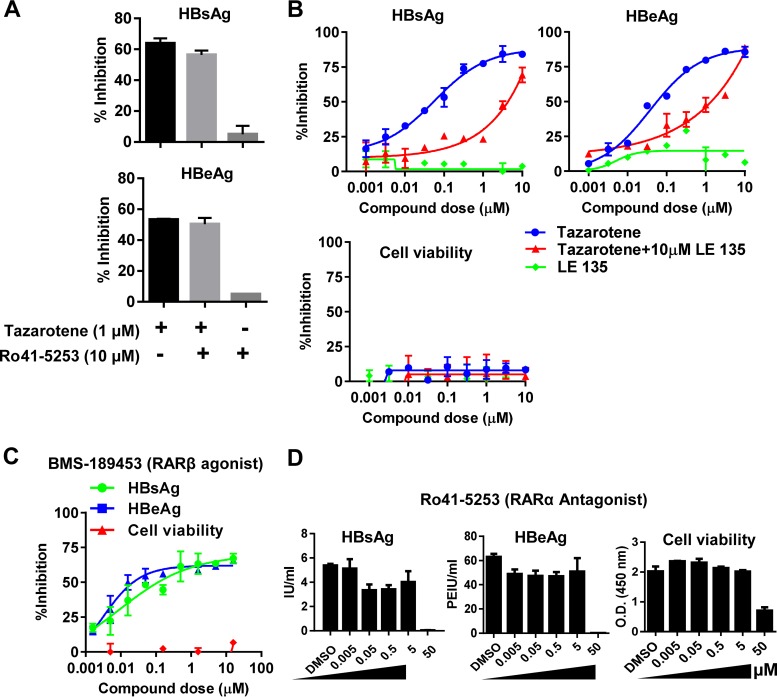
The anti-HBV action of tazarotene is partially through RARβ.
Since the biological activity of the retinoid acids is reported to be exhibited through different RAR receptors, we next examined whether the inhibitory effect of tazarotene was through the RAR receptors. We cotreated PHH cells with tazarotene and the RARα-specific antagonist Ro41-5253 or RARβ-specific antagonist LE135. The results showed that the RARα antagonist did not affect the tazarotene-mediated reduction of HBsAg and HBeAg levels (Fig. 5A), while the RARβ-specific antagonist LE135 partially abrogated the anti-HBV activity of tazarotene (Fig. 5B). Moreover, as expected, a potent RARβ agonist, which also acts as an antagonist against RARα and RARγ, BMS-189453 (Fig. 5C), but not Ro41-5253 (Fig. 5D), showed potent anti-HBV effect in PHH. Taken together, these results suggested that the anti-HBV action of tazarotene occurs partially via RARβ.
FIG 5.
The anti-HBV action of tazarotene is partially through RARβ. (A) PHH cells infected with HBV were treated with tazarotene in the presence or absence of the RARα antagonist Ro41-5253. The supernatant was collected for HBsAg and HBeAg analysis after 6 days of treatment. (B) PHH cells infected with HBV were treated with multiple doses of tazarotene in the presence or absence of the RARβ antagonist LE135. The supernatant was collected for HBsAg and HBeAg analysis after 6 days of treatment, and cell viability was determined by CCK-8. (C and D) PHH cells infected with HBV were treated with multiple doses of BMS-189453 (C) or Ro41-5253 (D), as indicated. The cell culture medium was collected for HBsAg and HBeAg analysis after 6 days of treatment.
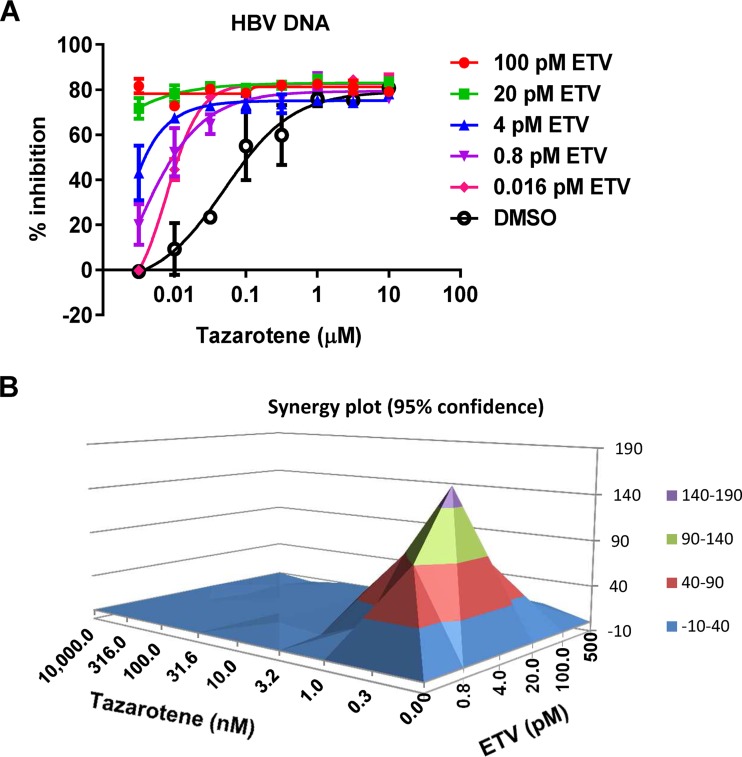
Combination of tazarotene and entecavir exhibits a synergistic effect.
We next studied whether the combination of tazarotene and the clinically used nucleoside analogue entecavir increases the anti-HBV efficacy. As shown in Fig. 6A, the dose-dependent inhibition of tazarotene to HBV DNA was dramatically shifted with multiple doses of entecavir (ETV). The MacSynergy II program calculated the theoretical additive interactions of the drugs based on the Bliss independence mathematical definition of expected effects for drug-drug interactions. The combination of tazarotene and ETV demonstrated significant synergy in inhibiting HBV DNA production, as assessed by the MacSynergy method (197 µM2% in MacSynergy in 95% confidence intervals) (Fig. 6B).
FIG 6.
Effect of combining tazarotene and ETV on HBV DNA production in PHH. PHH cells infected with HBV were treated by tazarotene or ETV alone or both of them. The supernatant was extracted after 9 days of incubation for HBV DNA-level quantitation. (A) The dose-response curve of tazarotene with multiple doses of ETV. (B) The raw data from the combination assays were imported into the MacSynergy II software program, and the compound interactions were calculated at the 95% confidence interval.
DISCUSSION
In this study, by screening an FDA-approved drug library in the PHH-based HBV infection model, we identified several compounds belonging to the family of RAR agonists as a class of potent anti-HBV agents. Among them, tazarotene, as the third-generation retinoid, exhibited the most potent anti-HBV effect that can strongly reduce the levels of HBsAg and HBeAg in a pangenotypic anti-HBV manner. Tazarotene was shown to potently reduce the HBV RNA levels transcribed from cccDNA. Since the current therapies for chronic HBV infection do not efficiently reduce the HBsAg levels and cannot directly target the viral transcription template cccDNA, our findings have provided new insights into the development of novel therapeutic approaches and strategies for chronic HBV infection.
The RAR is a type of nuclear receptor which is activated by retinoic acid and can heterodimerize with RXR to regulate the expression of specific genes, thereby controlling the development, homeostasis, and metabolism of the cells (14). Retinoid acids have been implicated in clinical applications, both as a potential antitumor agent and for the treatment of skin diseases (15). In addition, there have been a number of studies suggest that the retinoic acid or its analogues exhibit antiviral activities against a variety of pathogens, including human immunodeficiency virus type 1 (HIV-1) (16, 17), measles virus (18), human herpesvirus 8 (HHV-8) (19), hepatitis C virus (HCV) (20–22), and adenovirus (23). Particularly, acitretin has been used to treat patients with HIV who have psoriasis (17, 24), and the drug is well tolerated. Here, screening of an FDA-approved drug identified three compounds, adapalene, tretinoin, and tazarotene, which all belong to the RAR agonists, as potent HBV inhibitors. In addition, although we did not see a significant reduction in viral antigens when the RARα antagonist Ro41-5253 was added to the cells post-HBV infection, a previous study showed that pretreatment of the cells with Ro41-5253 could decrease host susceptibility to HBV infection through reducing the expression of HBV entry receptor NTCP (25). A very recently published paper using the HBV-replicating hepatoma cell line HepG2.215 indicated that peretinoin, an acyclic retinoid, could inhibit HBV replication (26). Moreover, our previous study suggested that all-trans retinoic acid (ATRA) is able to restore the HBV core-specific responses of T cells by targeting myeloid-derived suppressor cells in ex vivo peripheral blood mononuclear cells (PBMCs) from patients and in mouse models (5). Together, these results suggest that the regulation of RAR activity has a great impact on HBV transcription and replication and may also have some immunomodulatory effect.
Among the RAR agonists tested in this study, tazarotene exhibited the most potent anti-HBV effect. We showed that tazarotene treatment significantly reduced the HBsAg levels and suppressed the activation of HBV promoters and enhancers. Moreover, by mapping the RNA sequencing reads to the HBV genome, tazarotene was shown to reduce the expression levels of both viral pgRNA and subgenomic RNAs. Notably, considering that the retinoic acid response element has been reported to locate within the HBV genome (27–29), the activation of RAR by retinoic acid may theoretically activate rather than inhibit HBV transcription. This was actually supported by an early report using a transient expression system, which showed that the level of HBsAg in Hep3B/C16 cells increased after 24 h of retinoic acid treatment; however, HBsAg production was severely suppressed after long-term (120-h) retinoic acid treatment (30). Here, a single base mutation (G to A) was introduced into the retinoic acid responsive element (RARE) site, as described previously (31), and we found that tazarotene could still effectively inhibit the mutated HBV En II/Cp (Fig. S9). Considering that the RARE site located in the HBV genome has been suggested to be mainly activated by PPAR/RXR, but not RAR (27, 32, 33), we speculated that tazarotene does not inhibit HBV transcription by directly acting on an HBV promoter through RAR. Considering that retinoic acid regulates a variety of cellular gene expression levels and biological functions, it is possible that in addition to the inhibition of HBV transcription, tazarotene affects other steps within viral life cycles, such as viral replication, encapsidation, and secretion, which may also contribute to the suppression of viral antigen expression by tazarotene.
As indicated previously, retinoic acids have been reported to be able to induce the pattern recognition sensor RIG-I expression and activate a type I interferon-mediated innate immune response. Several reports have explained the antiviral effect of retinoic acid against HIV and HCV from the perspective of innate immune activation (17, 21, 34). Here, our results showed that compared to the cells treated with the positive controls, including pppRNA and pegylated IFN-α, tazarotene did not significantly induce genes of IFN responses, such as RIG-I, MX2, and IFIT1, suggesting that tazarotene inhibits HBV in our system in an IFN-independent manner. Through the detection of RAR expression and use of the relatively specific antagonists to RARα or RARβ and the agonist of RARβ, we identified that the anti-HBV action of tazarotene is, to some extent, selectively through RARβ. Considering that RARβ is closely linked to antiproliferation of the cells, it will be interesting to further investigate whether an RARβ agonist could also reduce the risk of development of chronic HBV infection-associated hepatocellular carcinoma (HCC). Moreover, HBV-encoded X protein (HBx) was reported to be able to overcome the growth-inhibitory potential of retinoic acid by downregulating RARβ2 (35), which might reflect an important role of retinoic acid and RARβ in the control of HBV persistence and pathogenesis in hepatocytes.
It should be noted that hepatocytes, as the host cells of HBV, are major sites for metabolism. The native role of the most identified HBV-bound transcriptional factors is the control and coordination of hepatic metabolism (36). Furthermore, recent reports indicated that HBV actively interferes with hepatic metabolic pathways (37, 38). Consequently, HBV gene expression is closely linked to hepatic metabolic processes, such as glucose and fat production and utilization, as well as bile acid production and secretion. Interestingly, we showed that tazarotene can reduce a series of genes related to metabolism pathways, such as the genes related to the glycolysis pathway. Considering that HBV was reported to be able to reprogram the glucose metabolism in hepatocytes, which may facilitate its persistence and be of importance in the development of HBV-associated HCC (39), the pattern of gene expression changes induced by tazarotene could, to some extent, reverse the HBV-mediated changes to normal hepatocyte gene expression. In spite of this, the understanding of the complex networks related to the regulation of HBV cccDNA transcription and HBV-induced metabolic changes in hepatocytes is still very limited; thus, it requires further investigation to comprehensively clarify the effect of RAR agonists on HBV expression, as well as the effect on host gene expression in different cell and animal models.
In summary, the results from this study suggest that the RAR agonists, as represented by tazarotene, have potent anti-HBV effect that especially affect HBsAg production and cccDNA transcription. Since it has been recognized that a combination of therapies targeting different steps of viral life cycles and persistence will likely be needed to achieve a functional hepatitis B virus cure (1, 40), our findings might be a valuable complement to the existing antiviral strategies.
MATERIALS AND METHODS
Reagents.
An FDA-approved drug library (1,018 compounds, 10 mM solution in dimethyl sulfoxide [DMSO], catalog no. L1300) was purchased from Selleckchem. All-trans retinoic acid (ATRA), tazarotene, adapalene, and tretinoin were obtained from Sigma, and Ro41-5253 was obtained from Abcam. HBsAg and HBeAg chemiluminescence immunoassay (CLIA) kits were obtained from Autobio. Anti-HBV core antigen antibody was purchased from Dako. Phospho-STAT1 rabbit monoclonal antibody (MAb) and RIG-I rabbit MAb were obtained from CST. Maxima H Minus first-strand cDNA synthesis kit was purchased from Thermo Fisher.
Cell culture and transfection.
Proliferating HepaRG cells were purchased from Biopredic International (Rennes, France) and were amplified and differentiated according to the manufacturer’s protocol. HepG2-NTCP (kindly provided by Yi Ni and Stephan Urban) and primary human hepatocytes (PHH; purchased from Bioreclamation IVT) were cultured as described previously (41). The human hepatoma-derived cell line HepG2 (purchased from ATCC) was cultured in Dulbecco’s modified Eagle medium (DMEM)/F12 supplemented with 10% fetal bovine serum (Invitrogen), 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C under humidified air that contained 5% CO2. The transfection of the luciferase reporter constructs of HBV promoters/enhancers (En II/Cp, Sp1, Sp2, and En I/Xp) were described previously (42).
HBV infection.
HBV was prepared from the HepAD38 cells (43), and the infection was performed with 150 GE per cell in the presence of 4% (vol/vol) polyethylene glycol 8000 (PEG 8000; Sigma-Aldrich) at 37°C for 24 h, as we described previously (41). The cells were washed and maintained in medium with a medium change every 2 to 3 days. We also used concentrated (100-fold) medium of HepG2 cells transfected with an expression plasmid for either HBV genotype A, B, C, or D and infected into PHH cells at 1,000 GE per cell in the presence of 4% (vol/vol) PEG 8000 at 37°C for 24 h, as described previously (41).
HBs, HBe antigen quantification, and cell viability.
The levels of HBsAg and HBeAg from cell culture supernatant were measured by HBsAg and HBeAg CLIA kits (Autobio), according to the manufacturer’s manual. Cell viability was determined using the Cell Counting kit-8 (CCK-8) from Dojindo, which is a sensitive colorimetric method for the determination of cell viability in cell proliferation and cytotoxicity assays. Dojindo’s highly water-soluble tetrazolium salt, WST-8, is reduced by dehydrogenase activities in cells to give a yellow formazan dye, which is soluble in the tissue culture medium. The amount of the formazan dye, generated by the activities of dehydrogenases in cells, is directly proportional to the number of living cells. The absorbance at 450 nm was measured using a microplate reader after 2 h of incubation of the cells with 10% (vol/vol) CCK-8 solution.
Quantification of viral DNA and RNA and cellular RNA.
HBV DNA from culture medium was extracted using the MagNA Pure 96 system (Roche) and then quantified by real-time PCR using the LightCycler 480 master kit (Roche). The forward primer 5′-GCTGGATGTGTCTGCGGC-3′ (positions 372 to 389), reverse primer 5′-GAGGACAAACGGGCAACATAC-3′ (positions 459 to 479), and probe 5′-CATCCTGCTGCTATGCCTCATCTTCTTG-BHQ-2-3′ (BHQ, black hole quencher) (positions 409 to 436) were used. The cycling conditions were 95°C for 10 min and then 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s for a total of 40 cycles. Total cellular HBV RNA was extracted with the TRIzol reagent (Invitrogen). Isolated RNA was reverse transcribed using the cDNA synthesis kit (Thermo Fisher) with random primers. The precore RNA and pgRNA were quantified with specific primers 5′-GTCTGCGCACCAGCACCA-3′ (positions 1797 to 1814) and 5′-GTCCATGCCCCAAAGCCACC-3′ (positions 1887 to 1906) and probe 5′-FAM-TCTTGTTCATGTCCTACTGTTCAAGCCTCCAA-BHQ1-3′ (FAM, 6-carboxyfluorescein) (positions 1844 to 1875). To detect HBV total RNA, primers 5′-TGTGCCTTCTCATCTGCCG-3′ (positions 1554 to 1572) and 5′-ATTCTCAGACCGTAGCACAC-3′ (positions 1650 to 1674) and probe 5′-FAM-CGTGTGCACTTCGCTTCACCTCTGC-BHQ1-3′ (positions 1579 to 1603) were used. The induction levels of RARα, RARβ, and RARγ were quantitative by real-time PCR using commercial primers and probes from Thermo Fisher.
Hirt DNA extraction and Southern blot detection of HBV cccDNA.
HBV cccDNA was extracted from HBV-infected cells by a modified Hirt method as previously described, with minor modification (11). The Hirt DNA was analyzed by Southern blot, as described previously (41); briefly, the samples were loaded on a 1.2% agarose gel in 1× Tris-acetate-EDTA (TAE) buffer for electrophoresis and transferred onto a Hybond-N + membrane (GE Healthcare). The membrane was probed with digoxigenin (DIG) labeled with HBV probe. After incubation of the membrane with an alkaline phosphatase-conjugated anti-DIG antibody, hybridization signals were quantified using a standard chemiluminescence reaction.
Northern blot analysis of HBV RNA.
Total RNA was extracted from PHH using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions, and RNA was detected by Northern blot according to the method described previously (11).
Immunofluorescence analysis.
Virus-infected cells were fixed with 4% paraformaldehyde for 20 min, washed three times with phosphate-buffered saline (PBS), and permeabilized with PBS containing 0.5% Triton X-100 for 30 min. After incubation for 1 h with 10% goat serum for blockade of nonspecific binding, immunofluorescence analysis was conducted by using an anti-HBc antibody (Dako) at a dilution of 1:1,000. The bound antibodies were visualized by incubation with secondary antibodies (Alexa Fluor 488 goat anti-rabbit IgG). Images were acquired using a Zeiss confocal microscope.
Western blot.
Cells were lysed in a radioimmunoprecipitation assay (RIPA) lysis buffer with protease inhibitor cocktail (Roche), and the proteins were separated in a SDS-PAGE gel and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). The membranes were incubated overnight at 4°C with primary antibodies and β-actin. The bound antibodies were detected using chemiluminescence reagents (Thermo Fisher). β-Actin was used as a loading control.
RNA-seq by Illumina HiSeq and data analysis.
The RNA-seq analyses were performed by Genewiz (Suzhou, China). Briefly, the total RNA of each sample was extracted using the TRIzol reagent (Invitrogen). RNA quality was monitored using a RNA 6000 Nano chip on a Bioanalyzer 2100 (Agilent Technologies). Next-generation sequencing library preparations were constructed according to the manufacturer’s protocol (NEBNext Ultra RNA library prep kit for Illumina). Library preparations were performed using strand-specific TruSeq RNA-seq library prep (Illumina), according to the manufacturer's instructions. Then, libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument, according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was carried out using a 2 × 150-bp paired-end (PE) configuration; image analysis and base calling were conducted using the HiSeq control software (HCS) + OLB + GAPipeline-1.6 (Illumina) on a HiSeq instrument. In order to remove technical sequences, including adapters, PCR primers, or fragments thereof, and those with a quality of bases lower than 20, pass filter data in fastq format were processed by Trimmomatic (version 0.30) to be high-quality clean data. First, reference genome sequences and gene model annotation files of relative species were downloaded from genome websites, such as UCSC, NCBI, and ENSEMBL. Second, Hisat2 (version 2.0.1) was used to index the reference genome sequence. Finally, clean data were aligned to the reference genome using the software Hisat2 (version 2.0.1). Functional annotation was performed using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
QuantiGene Plex assay.
To verify the relative expression of the IFN-inducible genes (ISGs) in PHH, the QuantiGene Plex assay (Affymetrix) was performed on a customized panel of ISGs according to the manufacturer’s protocol. The fluorescence intensity in the hybridized complex was measured using a Milliplex analyzer (Millipore).
Data analysis and statistics.
The dose-response curve was plotted by nonlinear regression model, and the half-maximal inhibitory concentration (IC50) and the 50% cytotoxic concentration (CC50) were generated using the GraphPad Prism 5.0 software. The error bars in the figures represent the mean ± the standard deviation (SD). Statistical analysis was performed using Student’s t test. A P value of <0.05 was considered statistically significant.
Compound-interaction analysis using the MacSynergy II software program.
The calculated theoretical additive interactions were determined from the dose-response curves of the individual drugs. The calculated additive surface, which represents the predicted additive interaction, was then subtracted from the observed surface to reveal regions of statistically significant greater-than-expected (synergy) or less-than-expected (antagonism) interactions. If the interactions were additive, the resulting surface appeared as a horizontal plane at 0% above the calculated additive surface in the resulting difference plots. Peaks above this plane in the difference plots were indicative of synergy, while depressions below the horizontal plane indicated antagonism.
Data availability.
All the sequence data reported in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO series accession number GSE114003.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sheng Zhu from the Roche Innovation Center Shanghai for assistance in RNA-seq data analysis.
This is a project based on using the material and technical means from Roche and is supported by the grants from the National Natural Science Foundation of China (grants 81772189 and 91542207), the National Science and Technology Major Project of China (grants 2018ZX10301208 and 2017ZX10202202), and the innovation program of the Shanghai Municipal Education Commission (grant 201701070007E00057).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00465-18.
REFERENCES
- 1.Lok AS, Zoulim F, Dusheiko G, Ghany MG. 2017. Hepatitis B cure: from discovery to regulatory approval. Hepatology 66:1296–1313. doi: 10.1002/hep.29323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trepo C, Chan HL, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 3.Revill P, Yuan Z. 2013. New insights into how HBV manipulates the innate immune response to establish acute and persistent infection. Antivir Ther 18:1–15. doi: 10.3851/IMP2542. [DOI] [PubMed] [Google Scholar]
- 4.Zhu D, Liu L, Yang D, Fu S, Bian Y, Sun Z, He J, Su L, Zhang L, Peng H, Fu YX. 2016. Clearing persistent extracellular antigen of hepatitis B virus: an immunomodulatory strategy to reverse tolerance for an effective therapeutic vaccination. J Immunol 196:3079–3087. doi: 10.4049/jimmunol.1502061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Z, Li J, Yu X, Zhang D, Ren G, Shi B, Wang C, Kosinska AD, Wang S, Zhou X, Kozlowski M, Hu Y, Yuan Z. 2015. Polarization of monocytic myeloid-derived suppressor cells by hepatitis B surface antigen is mediated via ERK/IL-6/STAT3 signaling feedback and restrains the activation of T cells in chronic hepatitis B virus infection. J Immunol 195:4873–4883. doi: 10.4049/jimmunol.1501362. [DOI] [PubMed] [Google Scholar]
- 6.Kondo Y, Ninomiya M, Kakazu E, Kimura O, Shimosegawa T. 2013. Hepatitis B surface antigen could contribute to the immunopathogenesis of hepatitis B virus infection. ISRN Gastroenterol 2013:935295. doi: 10.1155/2013/935295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian X, Zhao C, Zhu H, She W, Zhang J, Liu J, Li L, Zheng S, Wen YM, Xie Y. 2010. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J Virol 84:3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JT, Guo H. 2015. Metabolism and function of hepatitis B virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Testoni B, Levrero M, Zoulim F. 2017. Challenges to a cure for HBV infection. Semin Liver Dis 37:231–242. doi: 10.1055/s-0037-1606212. [DOI] [PubMed] [Google Scholar]
- 10.Perrillo R. 2009. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49:S103–S111. doi: 10.1002/hep.22956. [DOI] [PubMed] [Google Scholar]
- 11.Wu M, Li J, Yue L, Bai L, Li Y, Chen J, Zhang X, Yuan Z. 2018. Establishment of Cre-mediated HBV recombinant cccDNA (rcccDNA) cell line for cccDNA biology and antiviral screening assays. Antiviral Res 152:45–52. doi: 10.1016/j.antiviral.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, Watanabe T, Iijima S, Sakurai Y, Watashi K, Tsutsumi S, Sato Y, Akita H, Wakita T, Rice CM, Harashima H, Kohara M, Tanaka Y, Takaoka A. 2015. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42:123–132. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Yi Z, Chen J, Kozlowski M, Yuan Z. 2015. Innate detection of hepatitis B and C virus and viral inhibition of the response. Cell Microbiol 17:1295–1303. doi: 10.1111/cmi.12489. [DOI] [PubMed] [Google Scholar]
- 14.Aranda A, Pascual A. 2001. Nuclear hormone receptors and gene expression. Physiol Rev 81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 15.Theodosiou M, Laudet V, Schubert M. 2010. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci 67:1423–1445. doi: 10.1007/s00018-010-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciaszek JW, Coniglio SJ, Talmage DA, Viglianti GA. 1998. Retinoid-induced repression of human immunodeficiency virus type 1 core promoter activity inhibits virus replication. J Virol 72:5862–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P, Kaiser P, Lampiris HW, Kim P, Yukl SA, Havlir DV, Greene WC, Wong JK. 2016. Stimulating the RIG-I pathway to kill cells in the latent HIV reservoir following viral reactivation. Nat Med 22:807–811. doi: 10.1038/nm.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trottier C, Chabot S, Mann KK, Colombo M, Chatterjee A, Miller WH Jr, Ward BJ. 2008. Retinoids inhibit measles virus in vitro via nuclear retinoid receptor signaling pathways. Antiviral Res 80:45–53. doi: 10.1016/j.antiviral.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Caselli E, Galvan M, Santoni F, Alvarez S, de Lera AR, Ivanova D, Gronemeyer H, Caruso A, Guidoboni M, Cassai E, Dolcetti R, Di Luca D. 2008. Retinoic acid analogues inhibit human herpesvirus 8 replication. Antivir Ther 13:199–209. [PubMed] [Google Scholar]
- 20.Bocher WO, Wallasch C, Hohler T, Galle PR. 2008. All-trans retinoic acid for treatment of chronic hepatitis C. Liver Int 28:347–354. doi: 10.1111/j.1478-3231.2007.01666.x. [DOI] [PubMed] [Google Scholar]
- 21.Hamamoto S, Fukuda R, Ishimura N, Rumi MA, Kazumori H, Uchida Y, Kadowaki Y, Ishihara S, Kinoshita Y. 2003. 9-cis retinoic acid enhances the antiviral effect of interferon on hepatitis C virus replication through increased expression of type I interferon receptor. J Lab Clin Med 141:58–66. doi: 10.1067/mlc.2003.8. [DOI] [PubMed] [Google Scholar]
- 22.Shimakami T, Honda M, Shirasaki T, Takabatake R, Liu F, Murai K, Shiomoto T, Funaki M, Yamane D, Murakami S, Lemon SM, Kaneko S. 2014. The acyclic retinoid peretinoin inhibits hepatitis C virus replication and infectious virus release in vitro. Sci Rep 4:4688. doi: 10.1038/srep04688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Zhang Q, Zhou Z, Liu M, Chen Y, Li J, Xu L, Guo J, Li Q, Yang J, Wang S. 2018. Retinoic acid receptor beta, a potential therapeutic target in the inhibition of adenovirus replication. Antiviral Res 152:84–93. doi: 10.1016/j.antiviral.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Buccheri L, Katchen BR, Karter AJ, Cohen SR. 1997. Acitretin therapy is effective for psoriasis associated with human immunodeficiency virus infection. Arch Dermatol 133:711–715. doi: 10.1001/archderm.1997.03890420043005. [DOI] [PubMed] [Google Scholar]
- 25.Tsukuda S, Watashi K, Iwamoto M, Suzuki R, Aizaki H, Okada M, Sugiyama M, Kojima S, Tanaka Y, Mizokami M, Li J, Tong S, Wakita T. 2015. Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis B virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression. J Biol Chem 290:5673–5684. doi: 10.1074/jbc.M114.602540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai K, Shirasaki T, Honda M, Shimizu R, Shimakami T, Nakasho S, Shirasaki N, Okada H, Sakai Y, Yamashita T, Kaneko S. 2018. Peretinoin, an acyclic retinoid, inhibits hepatitis B virus replication by suppressing sphingosine metabolic pathway in vitro. IJMS 19:108. doi: 10.3390/ijms19020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quasdorff M, Protzer U. 2010. Control of hepatitis B virus at the level of transcription. J Viral Hepat 17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. [DOI] [PubMed] [Google Scholar]
- 28.Shi YX, Huang CJ, Yang ZG. 2016. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J Gastroenterol 22:8161–8167. doi: 10.3748/wjg.v22.i36.8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Mertz JE. 2001. Critical roles of nuclear receptor response elements in replication of hepatitis B virus. J Virol 75:11354–11364. doi: 10.1128/JVI.75.23.11354-11364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu SL, Lin YF, Chou CK. 1993. Retinoic acid biphasically regulates the gene expression of hepatitis B virus surface antigen in human hepatoma Hep3B cells. J Biol Chem 268:23093–23097. [PubMed] [Google Scholar]
- 31.Huan B, Siddiqui A. 1992. Retinoid X receptor RXR alpha binds to and trans-activates the hepatitis B virus enhancer. Proc Natl Acad Sci U S A 89:9059–9063. doi: 10.1073/pnas.89.19.9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raney AK, Johnson JL, Palmer CN, McLachlan A. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol 71:1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huan B, Kosovsky MJ, Siddiqui A. 1995. Retinoid X receptor alpha transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J Virol 69:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho NE, Bang BR, Gurung P, Li M, Clemens DL, Underhill TM, James LP, Chase JR, Saito T. 2016. Retinoid regulation of antiviral innate immunity in hepatocytes. Hepatology 63:1783–1795. doi: 10.1002/hep.28380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung JK, Park SH, Jang KL. 2010. Hepatitis B virus X protein overcomes the growth-inhibitory potential of retinoic acid by downregulating retinoic acid receptor-beta2 expression via DNA methylation. J Gen Virol 91:493–500. doi: 10.1099/vir.0.015149-0. [DOI] [PubMed] [Google Scholar]
- 36.Bar-Yishay I, Shaul Y, Shlomai A. 2011. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liver Int 31:282–290. doi: 10.1111/j.1478-3231.2010.02423.x. [DOI] [PubMed] [Google Scholar]
- 37.Lamontagne J, Mell JC, Bouchard MJ. 2016. Transcriptome-wide analysis of hepatitis B virus-mediated changes to normal hepatocyte gene expression. PLoS Pathog 12:e1005438. doi: 10.1371/journal.ppat.1005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oehler N, Volz T, Bhadra OD, Kah J, Allweiss L, Giersch K, Bierwolf J, Riecken K, Pollok JM, Lohse AW, Fehse B, Petersen J, Urban S, Lutgehetmann M, Heeren J, Dandri M. 2014. Binding of hepatitis B virus to its cellular receptor alters the expression profile of genes of bile acid metabolism. Hepatology 60:1483–1493. doi: 10.1002/hep.27159. [DOI] [PubMed] [Google Scholar]
- 39.Liu B, Fang M, He Z, Cui D, Jia S, Lin X, Xu X, Zhou T, Liu W. 2015. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis 6:e1980. doi: 10.1038/cddis.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Testoni B, Durantel D, Zoulim F. 2017. Novel targets for hepatitis B virus therapy. Liver Int 37:33–39. doi: 10.1111/liv.13307. [DOI] [PubMed] [Google Scholar]
- 41.Shen F, Li Y, Wang Y, Sozzi V, Revill PA, Liu J, Gao L, Yang G, Lu M, Sutter K, Dittmer U, Chen J, Yuan Z. 2018. Hepatitis B virus sensitivity to interferon-alpha in hepatocytes is more associated with cellular interferon response than with viral genotype. Hepatology 67:1237–1252. doi: 10.1002/hep.29609. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Lin S, Chen Q, Peng L, Zhai J, Liu Y, Yuan Z. 2010. Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs. J Virol 84:6387–6399. doi: 10.1128/JVI.00236-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequence data reported in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO series accession number GSE114003.