Treating high-density bacterial infections is a challenging clinical problem. We have a paucity of new agents that can address this problem.
KEYWORDS: Pseudomonas aeruginosa, fosfomycin, meropenem, pharmacodynamics
ABSTRACT
Treating high-density bacterial infections is a challenging clinical problem. We have a paucity of new agents that can address this problem. Pseudomonas aeruginosa is a particularly difficult pathogen to treat effectively because of the plethora of resistance mechanisms it carries. Fosfomycin is an agent discovered circa 40 years ago. Recently, it has been resurrected in the United States and studied for intravenous therapy. We hypothesized that, to maximize its utility, it would require combination chemotherapy when used in a clinical circumstance in high-bacterial-burden infections. We chose to examine the combination of meropenem plus fosfomycin. These agents were studied in the hollow-fiber infection model. We utilized a fully factorial study design, looking at 2 doses of meropenem alone (1 and 2 g 8-hourly) and two doses of fosfomycin alone (6 and 8 g 8-hourly), as well as all possible combinations plus a no-treatment control. We used a high-dimensional model of 5 inhomogeneous differential equations with 5 system outputs to analyze all data simultaneously. Combination therapy outperformed all monotherapy regimens, with all combinations driving >6 log10 CFU/ml of bacterial killing. Combination therapy was able to counterselect resistance emergence (meropenem mutants being killed by the combination, as well as fosfomycin mutants being killed by the combination) in all regimens studied. The analysis demonstrated that the combination was significantly synergistic for bacterial cell killing and resistance suppression. Meropenem plus fosfomycin is a promising combination for therapy of high-burden Pseudomonas aeruginosa infections and requires further study.
INTRODUCTION
Fosfomycin is the only epoxide antimicrobial. While it was discovered approximately 4 decades ago, it has only been available in the United States as an oral preparation approved for use in uncomplicated urinary tract infections. In many places in the world, it has had a long history of intravenous use.
Recently, Zavante Therapeutics (now acquired by Nabriva Therapeutics) began studying this agent as an intravenous product. Ultimately, it is important to understand the effect of fosfomycin against serious pathogens such as Pseudomonas aeruginosa. We had previously examined fosfomycin alone in the hollow-fiber infection model (HFIM) against Escherichia coli (1). Three isolates were evaluated, with MIC values of 1 mg/liter, 1 mg/liter, and 64 mg/liter. Exposures of 12 g/day through 36 g/day administered in a dose fractionation fashion were examined. All 12 g/day arms failed with resistance emergence. In some instances, larger doses suppressed resistance for the isolates with an MIC of 1 mg/liter. No activity was seen against the isolate with an MIC of 64 mg/liter. In addition, the combination of meropenem plus fosfomycin was significantly synergistic for the wild-type isolate and additive for the fosfomycin-resistant subpopulation.
Pseudomonas aeruginosa is a major hospital-acquired pathogen. We wished to evaluate fosfomycin for possible therapy of serious infections with this organism. We had previously found rapid emergence of resistance to fosfomycin as a single agent for P. aeruginosa PAO1 (2). We were able to identify the area under the concentration-time curve (AUC)/MIC ratio as the dynamically linked index for bacterial cell killing but also identified time > MIC (or, equivalently, minimum concentration of drug in serum [Cmin]/MIC ratio) as the dynamically linked index for resistance suppression.
In this evaluation, we wished to examine combination therapy with fosfomycin plus meropenem against P. aeruginosa PAO1 to see whether the prior synergistic effects against E. coli (1) could be demonstrated for Pseudomonas species. We employed a fully factorial design for this evaluation, with two doses of fosfomycin, 2 doses of meropenem, all possible combinations, and a no-treatment control, for 9 arms in total.
RESULTS
Organism MIC and mutational frequency to resistance.
We studied Pseudomonas aeruginosa PAO1. The broth microdilution fosfomycin MIC for this isolate was 64 mg/liter. The mutational frequency to resistance was 1/4.71 log10 CFU/ml (1/51,286 CFU/ml), with fosfomycin at a concentration of 3× baseline MIC incorporated into the selecting agar. For meropenem, the MIC was 0.5 mg/liter, and the mutational frequency to resistance was 1/6.32 log10 CFU/ml (1/292,050 CFU/ml).
Pseudomonas aeruginosa bacterial killing and resistance emergence for monotherapy.
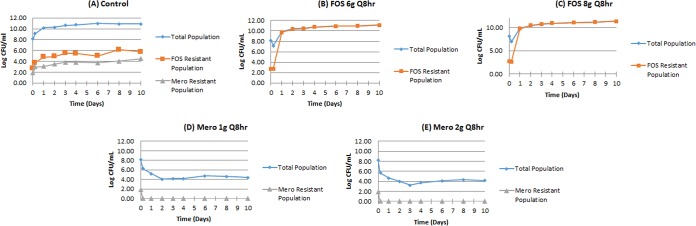
For fosfomycin, drug concentration-time profiles in the HFIM that approximated the kinetics of adult humans administered 6 g every 8 h and 8 g every 8 h were studied. For meropenem, concentration-time profiles for doses of 1 and 2 g every 8 h were examined. The initial bacterial inoculum was 8.18 log10 CFU/ml. There were 2.69 log10 CFU/ml of less-susceptible organisms in the population for fosfomycin at baseline and 1.86 log10 CFU/ml at initiation for meropenem. The presence of less-susceptible organisms at baseline is understandable when one compares the baseline bacterial burden to the mutational frequency to resistance. The regimen impact on the total population of P. aeruginosa and on the amplification of less susceptible, preexisting subpopulations is displayed in Fig. 1A to E.
FIG 1.
Monotherapy evaluations of fosfomycin and meropenem against P. aeruginosa PAO1 in the HFIM. Panels A to E.
Both fosfomycin monotherapy regimens rapidly selected for resistant isolates. In contrast, both of the meropenem doses rapidly suppressed resistance amplification, even though there were 72 CFU/ml of less-susceptible (MIC = 2 mg/liter versus 0.5 mg/liter for the wild-type isolate) organisms present at baseline. It is likely that this relatively small baseline change in MIC for the isolate less susceptible to meropenem was insufficient to allow amplification. For fosfomycin, the resistant isolates had an MIC for fosfomycin that increased from 64 mg/liter to 512 to 1,028 mg/liter. This change in susceptibility could not be counterselected by the exposures achieved at the doses studied. We had seen this in our previous experiment of fosfomycin monotherapy against this organism (2). Consequently, to evaluate the utility of fosfomycin for therapy of P. aeruginosa in the setting of a high-bacterial-burden infection, it was necessary to examine combination therapy and to model the results to determine whether the interaction between the agents was synergistic, additive, or antagonistic.
Pseudomonas aeruginosa bacterial killing and resistance emergence for combination therapy.
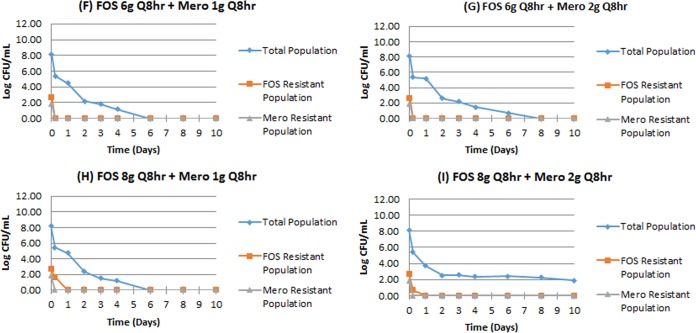
The effect of combination therapy is shown in Fig. 2F to I. In all instances, combination therapy suppressed all amplification of less-susceptible populations, even though these subpopulations existed at baseline for both meropenem and fosfomycin. All less-susceptible populations were reduced to unmeasurable by 24 h of therapy. Of interest, arms F to H reduced the total burden to unmeasurable by day 6 to 8. In arm I, the lowest counts were reached at day 10, at 1.88 log10 CFU/ml, representing a 6.3 log10 CFU/ml reduction from baseline. This arm had the largest meropenem and fosfomycin doses. The profile resembles that of having nonreplicative persister phenotype organisms present, but there are no direct data to address this.
FIG 2.
Combination therapy with fosfomycin and meropenem against P. aeruginosa PAO1. Panels F to I.
Mathematical analysis of drug interaction.
One of the reasons to examine this combination is to determine the drug-drug interaction for both bacterial cell killing and suppression of resistance emergence. We have previously described a system of inhomogeneous differential equations to look at these issues (1, 3, 4). The model was fitted to all system outputs from all arms simultaneously. The estimates of the mean and median parameter estimates, as well as the standard deviations, are displayed in Table 1. It is of note that all α values (drug-drug interaction terms) were strongly positive, indicating synergy.
TABLE 1.
Parameter estimates for all parameters for the combination of meropenem plus fosfomycin against Pseudomonas aeruginosa
| Parametera | Mean | Median | SD |
|---|---|---|---|
| VMero (liter) | 17.7 | 18.2 | 0.827 |
| VFos (liter) | 27.7 | 28.0 | 1.87 |
| CLMero (liter/h) | 11.5 | 11.8 | 0.736 |
| CLFos (liter/h) | 6.84 | 7.02 | 0.524 |
| Kg-s (h−1) | 0.229 | 0.134 | 0.221 |
| Kkill-s (h−1) | 8.44 | 9.92 | 2.25 |
| E50-Mero-s (mg/liter) | 359 | 258 | 336 |
| E50-Fos-s (mg/liter) | 5,085 | 2,585 | 5,775 |
| αs | 8.67 | 9.87 | 2.34 |
| Kg-Mero-r (h−1) | 0.174 | 0.0474 | 0.267 |
| Kkill-Mero-r (h−1) | 8.82 | 8.64 | 0.803 |
| E50-Mero-r (mg/liter) | 1,834 | 279 | 2,467 |
| αmero-r | 8.35 | 9.73 | 1.96 |
| Kg-Fos-r (h−1) | 0.168 | 0.0591 | 0.207 |
| Kkill-Fos-r (h−1) | 8.35 | 8.65 | 1.53 |
| E50-Fos-r (mg/liter) | 3,608 | 3,379 | 1,101 |
| αFos-r | 8.44 | 9.1 | 1.17 |
| HMero-s | 3.05 | 2.05 | 1.55 |
| HFos-s | 2.55 | 1.97 | 0.885 |
| HMero-r | 2.19 | 1.35 | 1.30 |
| HFos-r | 1.54 | 1.24 | 0.658 |
V, volume of the central compartment; CL, drug clearances; Kg, growth rate constant for organisms; Kkill, kill rate constants for a subpopulation; E50, drug concentration at which the kill rate is half maximal; α, drug interaction constant; H, Hill’s constant for a subpopulation; Fos, fosfomycin; Mero, meropenem; s, susceptible; r, resistant.
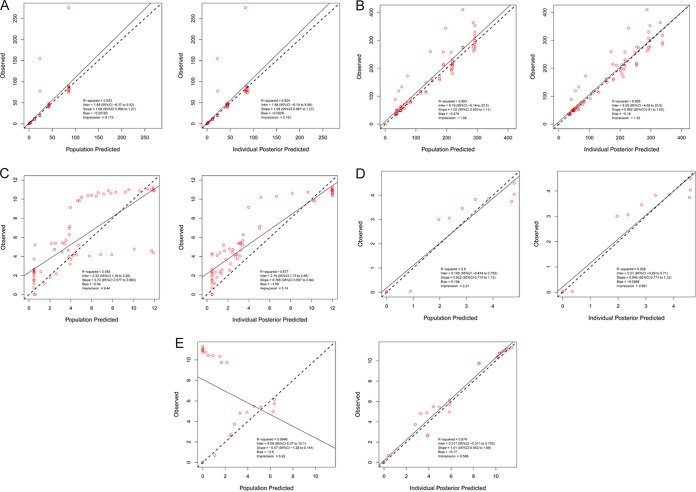
In Fig. 3, we show the predicted-observed regressions for the pre-Bayesian (population) estimates and the Bayesian (individual) estimates for all system outputs (meropenem concentrations, fosfomycin concentrations, total Pseudomonas population burden, population burden for meropenem-resistant isolates, and population burden for fosfomycin-resistant isolates). The fit of the models to the data were acceptable, even for the pre-Bayesian analyses. It should be noted that for the fosfomycin-resistant population, there were a subset of observations which were substantially underpredicted (Fig. 3E). These poorly predicted observations were from resistance emergence from the fosfomycin monotherapy arms. These exposures were predicted by the pre-Bayesian model not to have had substantial resistance amplification. In the case of the Bayesian analyses, the lowest r2 value (coefficient of determination, the amount of variance explained by the model system) was 0.877, or 87.7% of the variance. The poorly predicted values in the pre-Bayesian set were quite well predicted by the Bayesian posterior estimates (overall r2 = 0.974).
FIG 3.
Predicted-observed regressions for pre-Bayesian (population) and Bayesian (individual) analyses for (A) meropenem concentrations, (B) fosfomycin concentrations, (C) total bacterial burden, (D) meropenem-resistant bacterial burden, and (E) fosfomycin-resistant bacterial burden.
DISCUSSION
Pseudomonas aeruginosa remains a therapeutic challenge for clinicians. When there is an infection with a large bacterial burden, such as that seen with ventilator-associated bacterial pneumonia (VABP), rates of emergence of resistance during therapy have been seen in the realms of 40 to 60% when carbapenems were employed as monotherapy (3–5) and approximately 33% when fluoroquinolones were used as monotherapy (4). Consequently, when considering the options for chemotherapy, it is critical to have both substantial bacterial cell killing and to select a combination that has a high likelihood of suppressing resistance.
We have used the approach here in several previous investigations (1, 6, 7). In two of these analyses, the combination produced good bacterial killing and suppressed amplification of less-susceptible subpopulations (1, 7). However, the use of two drugs in combination does not guarantee that there will be suppression of resistance emergence. In one study (6), the combination of rifampin plus linezolid was employed against Mycobacterium tuberculosis. Of all (n = 9) different drug doses in combination studied, only two were able to counterselect resistance amplification, and these were at the two highest exposures to the two agents. This may be because both rifampin and linezolid inhibit organism growth within the same pathway. Nonetheless, it is critical to choose combinations and then study them under conditions somewhat permissive for resistance emergence. The easiest path forward is to start with a total bacterial burden substantially larger than the inverse of the mutational frequency to resistance. This will allow less-susceptible populations to be present at the beginning of therapy and will test whether the combination will suppress resistance amplification.
In this evaluation, we started with 8.18 log10 CFU/ml. This allowed both meropenem and fosfomycin less-susceptible isolates to be present at therapy initiation.
The primary goal of anti-infective chemotherapy is to kill the pathogen to a degree that will allow the patient to survive the infection. Here, the pharmacodynamics of meropenem plus fosfomycin show that this combination would likely achieve this end. For the four combination arms evaluated, three of the four drove the colony counts below 1.0 log10 CFU/ml between days 6 and 8 of therapy. This was a higher degree of organism killing than that of either monotherapy arm. In the fourth arm, the bacterial burden declined below 2.0 log10 CFU/ml. Interestingly, this occurred in the arm with the largest simulated doses of both drugs. The reason for this is unclear, but we suggest that these organisms may have entered into a nonreplicative persister (NRP) phenotype state. It will be important to explore this, and we intend to study a Pseudomonas isolate that expresses an unstable green fluorescent phenotype reporter. In this way, we can directly confirm or deny the presence of NRP-phase organisms as explaining the loss of cell killing in this circumstance.
We were also able to demonstrate that the combination of meropenem plus fosfomycin was able to counterselect amplification of less-susceptible subpopulations (8, 9). This was seen in all four combination regimens studied. Consequently, this combination produced both rapid and extensive bacterial kill and complete suppression of resistance for this isolate of Pseudomonas aeruginosa.
The analysis we performed provides insight into why these salutary outcomes both occurred. Our system of differential equations allows us to estimate the way in which the two drugs interact, shown in the value of the interaction parameter α. In Table 2, we demonstrate how these two agents interact for isolates susceptible to both drugs and for isolates resistant to one agent but susceptible to the other. In all instances, the values of α were quite positive, and this represents at least a tendency toward synergy. To ascertain the statistical significance of this, we performed a bootstrapping exercise to generate a 95% credible interval around the point estimate of the α values. As seen in Table 2, the lower bound of the credible interval is still substantially positive, indicating that the α terms were all significantly synergistic. Given the interaction, we have insight into the good outcomes for bacterial killing and resistance suppression.
TABLE 2.
α-Interaction parameters (medians) and 95% credible intervals to determine type of drug-drug interaction
| α-Interaction parameter | α Estimate | 95% credible interval |
|---|---|---|
| α-s | 9.9 | 4.3–10.0 |
| α-Mero-r | 9.7 | 5.9–10.0 |
| α-Fos-r | 9.0 | 6.8–9.6 |
The mechanism of this synergistic interaction is currently unknown. We speculate that the mechanism of synergy is that the two agents act on different points in the pathway for cell wall synthesis. Fosfomycin inhibits the first committed enzymatic step in peptidoglycan synthesis. This is accomplished by binding to the enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and inhibiting the formation of N-acetylmuramic acid by N-acetylglucosamine and phosphoenolpyruvate (10). β-Lactams mainly act on a late stage in cell wall synthesis by preventing the transpeptidation of peptidoglycan. It is important to recognize that ampC overexpression results in a change in susceptibility to fosfomycin (11). Hamou-Segarra demonstrated this clearly (11), but it should be recognized this demonstration was with an isolate with inactivation of the three ampD amidases. Meropenem and other carbapenems are strong inducers of ampC production, and this may be another mechanism by which there is meropenem/fosfomycin synergy.
In this experiment, we demonstrated synergy at both the level of cell killing and resistance suppression. The PAO1 isolate examined is wild type (WT) for all major Pseudomonas resistance mechanism (no efflux upregulation, oprD+, ampC WT). To be optimally useful, these agents would ideally remain synergistic for Pseudomonas isolates expressing these resistance mechanisms. As a next step, we intend to look at the combination of meropenem plus fosfomycin against three isogenic less meropenem-susceptible isolates (oprD−, MexAB overexpression, stably derepressed ampC). We have previously published experiments with carbapenems for all these isolates (12, 13). If the hypothesis regarding highly stably derepressed ampC production driving fosfomycin hypersusceptibility (11) is correct, the utility of the combination should be maintained. Furthermore, it is important to examine other mechanisms of meropenem resistance to identify the utility of this combination against non-WT Pseudomonas aeruginosa isolates.
The major limitations of this study are two. First, more isolates need to be studied, both in isogenic mutant with defined resistance mechanisms and also in other clinically derived wild-type isolates. Second, these findings need to be validated in animal systems, both with and without granulocytes. In this way, a database can be built that will allow a judgement to be made regarding testing this regimen in patients.
MATERIALS AND METHODS
Microorganisms.
Pseudomonas aeruginosa PAO1, a well-studied organism in our laboratory, was the isolate examined in these studies.
Drugs.
Fosfomycin disodium salt (fosfomycin for injection, Zolyd; Zavante Therapeutics, recently acquired by Nabriva Therapeutics) was used for the susceptibility testing, preparation of resistance-selecting plates, bioanalytical methods (liquid chromatography/tandem mass spectrometry [LC-MS/MS]) and as medium for the hollow-fiber infection model studies. Fosfomycin was supplied by Zavante Therapeutics. Meropenem was purchased from CuraScript (Orlando, FL).
In vitro susceptibility testing.
The in vitro susceptibility to meropenem and fosfomycin was measured using both broth dilution and agar dilution according to CLSI methodology (14). Cation-adjusted Mueller-Hinton agar (Mueller-Hinton II [MH II]) plates (Becton, Dickinson, Sparks, MD) containing meropenem or fosfomycin (3× baseline MIC) were prepared. Plates were incubated for 18 to 24 h in ambient air at 35°C.
Mutation frequency.
An overnight incubation in Mueller-Hinton II (MH II) broth of P. aeruginosa PAO1 was subsequently serially diluted and plated on drug-free Ca-Mueller Hinton agar (MHA) plates to estimate the total bacterial burden and also on drug-containing (3× baseline MIC) Ca-MHA plates to estimate the less-susceptible subpopulation burden. To investigate whether the mutants that grew on drug-containing plates had elevated meropenem or fosfomycin MICs, 3 colonies were selected, and the meropenem or fosfomycin MICs were reestimated using agar dilution and the broth microdilution method, as previously described.
Hollow-fiber infection model.
An HFIM was used to investigate the pharmacodynamics of meropenem and fosfomycin alone and in combination against P. aeruginosa PAO1. Mueller-Hinton II broth was pumped from a central compartment through a hollow-fiber cartridge (FiberCell Systems, Frederick, MD) before being returned to the central compartment. A peristaltic pump was employed. Meropenem and/or fosfomycin was administered into the central compartment by using a programmable syringe pump. Fresh Mueller-Hinton II broth was pumped from a reservoir into the central compartment, and the same volume of drug-containing medium was removed as waste. The rate was controlled to simulate pharmacokinetic profiles for meropenem (15) and/or fosfomycin (modern data for the fosfomycin pharmacokinetic parameter values in volunteers were kindly provided by E. J. Ellis-Grosse of Zavante Therapeutics). The extracapillary space of each HFIM was inoculated with 12 ml of bacterial suspension. The desired inoculum was confirmed with quantitative cultures. The HFIM was incubated at 37°C in ambient air. Bacterial densities were determined by removing 0.4 ml from the extracapillary space via a sampling port. Serial dilutions in 0.1 ml volumes were then plated on both drug-free and drug-containing Ca-MHA plates to enumerate total and resistant subpopulations, respectively.
Study design.
There were nine treatment arms examined, as follows: a no-treatment control; monotherapy arms of 1 and 2 g 8-hourly for meropenem and 6 and 8 g 8-hourly for fosfomycin; all possible combinations of meropenem plus fosfomycin were studied. We sampled the system at baseline, 0.21, 1, 2, 3, 4, 6, 8, and 10 days after therapy initiation for microbiological endpoints (total counts, meropenem-resistant counts, and fosfomycin-resistant counts). For drug concentration determinations, we sampled at baseline and 1, 3, 6, 8, 9, 24, 25, 27, 30, 32, 33, 48, and 49 h postinitiation of a 1 h infusion.
Combination therapy was accomplished according to the methods described by Blaser (16).
Mathematical modeling.
The system of differential equations employed for modeling the interaction of antimicrobial agents has been described previously (6).
The model was implemented in the program PMetrics (17). We simultaneously modeled 5 system outputs for the analysis of the data, using the same model as previously published for tuberculosis (6). These were (i) concentration of meropenem, (ii) concentration of fosfomycin, (iii) total Pseudomonas colony counts, (iv) colony counts resistant to meropenem, and (v) colony counts resistant to fosfomycin. No isolates in any experiment were found to be resistant to both drugs.
We used the nonparametric adaptive grid (NPAG) program within the Pmetrics package for R (17). Our modeling approach allowed us to apply classical properties of drug interaction for effect (synergy, additivity, and antagonism) to multiple populations simultaneously. The enabling equations of our Drusano-Greco model for the interaction are published in our previous report (6).
Pmetrics partitions model error into assay (fixed) and residual (random). The assay error is calculated as an output-dependent standard deviation, SD = A0 + A1 × [C] + A2 × [C]2 + A3 × [C]3, where [C] is the measured output of drug concentration or the log10-transformed colony count. There is one set of coefficients, A, for each of the five output equations.
Additionally, we used a fitted multiplicative term, γ, such that each observation in the fitting process was weighted by the Fisher information, i.e., 1/(γ × SD2). Values of γ from 1 to 3 indicate reasonable model and data appropriateness, and the γ for the final model was 1.73.
To summarize population parameter values, we used a bootstrapping procedure to calculate median values and 95% credibility intervals. Briefly, using the 6 support points, which each contain a vector of values for every parameter in the model and an associated probability of that parameter set, we generated 1,000 sets of 6 random weighted samples (with replacement) for any parameter, e.g., αr1 for the interaction term of the meropenem-resistant population. From these 1,000 sets, we calculated the median, 2.5th percentile, and 97.5th percentile.
We selected the final model based on fit of the observed versus predicted data using the median Bayesian posterior parameter values to generate predictions.
Pre-Bayesian (population) regressions are generated employing the population median parameter vector for generating the predicted output values. Bayesian (individual) regressions are generated using the Bayesian posterior parameter values for each of the HFIM experiment tubes.
The α values (see Results) are indicative of whether the drugs interact synergistically, additively or antagonistically. Whether statistically significant synergy is present is determined by the credible interval around the point estimate of α. If α is positive and the lower bound of the credible interval does not cross zero, then the interaction is statistically significant synergy. If the α is negative and the upper bound of the credible interval does not cross zero, then the interaction is statistically significant antagonism. Any α where the credible interval crosses zero is accorded to be an additive interaction.
Meropenem LC-MS/MS assay for Mueller-Hinton II broth.
Mueller-Hinton II (MH II) broth samples were stored at −80°C until analysis. After thawing at room temperature, 0.020 ml of each sample and 20 µl of internal standard (ceftazidime) was diluted using 1 ml of water. Resulting sample was transferred to a liquid chromatography/mass spectrometry (LC/MS) vial, and 2 µl was used as injection volume for analysis.
Determination of meropenem was performed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) consisting of a Prominence high-performance liquid chromatography (HPLC) system (Shimadzu) and an API5000 triple quadrupole mass spectrometer (AB Sciex). Separation was achieved using a Gemini NX C18 HPLC column (150 × 4.6 mm, 5 µm; Phenomenex) at 40°C with a run time of 5 min. Mobile phases consisted of 0.1% fluorescent antibody (FA) in water (A) and acetonitrile (B) at a flow rate of 0.750 ml/min in isocratic mode using 30% B.
The mass spectrometer was operated in positive ion mode using the turbo ion spray (TIS) probe interface. Multiple reaction monitoring (MRM) at m/z 384.3/141 (quantifier) and m/z 384.257/68 (qualifier) was used for meropenem, as well as at m/z 547.1/468.1 for internal standard ceftazidime. API5000 parameters (arbitrary units unless specified) were as follows: collision cell gas setting (CAD), 6; curtain plate glass setting (CUR), 30; nebulizer gas (gas 1) setting (GS1), 60; auxiliary gas (gas 2) setting (GS2), 60; ion spray voltage (IS), 5,500; temperature of heater gas (TEM), 650°C; MRM, m/z 384.3/141: declustering potential (DP), 141; collision cell energy (CE), 23; collision cell exit potential (CXP), 20; dwell, 200 ms; MRM m/z 384.257/68: DP, 141; CE, 77; CXP, 16; dwell time, 200 ms; MRM m/z 547.1/468.1: DP, 111; CE, 19; CXP, 18; dwell, 200 ms. Concentration calculations were performed using Analyst software v 1.6.2 (AB Sciex).
Linearity for meropenem in Mueller-Hinton II (MH II) broth, with a range of 0.0625 to 100 µg/ml, was demonstrated for each calibration curve over 2 separate runs with a correlation coefficient (R) of ≥0.9998 and linear regression (R2) of ≥0.9996. Within-run as well as between-run accuracies for each calibration curve were within ±9% of the nominal concentrations and <4% for the respective coefficients of variation of the mean values. Calibration curve precision within-run ranged from 0.7% to 4.7% and between-run 1.6% to 4.4%. The quality control (QC) sample within-run as well as between-run accuracies were within ±6% of the nominal concentrations and <5.4% for the respective coefficients of variation of the mean values. QC precision within-run ranged from 2.4% to 3.7% and between-run from 3.1% to 5.1%.
Fosfomycin LC-MS/MS assay for Mueller-Hinton II broth.
Mueller-Hinton II broth samples were stored at −80°C until analysis. After thawing at room temperature, 0.010 ml of each sample and 10 µl of internal standard (ethylphosphonic acid) was diluted using 1 ml of water. The resulting sample was transferred to an LC/MS vial, and 2 µl was used as injection volume for analysis. Determination of fosfomycin was performed using liquid chromatography-tandem mass spectrometry (LC-MS/MS), consisting of a Prominence HPLC system (Shimadzu) and an API5000 triple quadrupole mass spectrometer (AB Sciex). Separation was achieved using a Synergi Polar-RP HPLC column (150 × 4.6 mm, 4 µm; Phenomenex) at 40°C with a run time of 6 min. Mobile phases consisted of 10 mM ammonium acetate (pH 5) (A) and acetonitrile (B) at a flow rate of 0.750 ml/min in gradient mode.
The mass spectrometer was operated in negative ion mode using the turbo ion spray (TIS) probe interface. Multiple reaction monitoring (MRM) at m/z 137/63 (quantifier) and m/z 137/80 (qualifier) was used for fosfomycin, as well as at m/z 109/79 for the internal standard, ethylphosphonic acid. API5000 parameters (arbitrary units unless specified) were as follows: CAD, 6; CUR, 30; GS1, 60; GS2, 60; IS, −4500; TEM, 650°C; MRM m/z 137/63: DP, −15; CE −22; CXP, −11, dwell, 200 ms; MRM m/z 137/80: DP, −15, CE, −20; CXP, −9; dwell, 200 ms; MRM m/z 109/79: DP, −70; CE, −70, CXP, −5; dwell, 200 ms. Concentration calculations were performed using Analyst software v 1.6.2 (AB Sciex).
Linearity for fosfomycin in Mueller-Hinton II broth, with a range of 7.8125 to 500 mg/liter, was demonstrated for each calibration curve over 2 separate runs with a correlation coefficient (R) of ≥0.9990 and linear regression (R2) of ≥0.9979. Within-run as well as between-run accuracies for each calibration curve were within ±10% of the nominal concentrations and within <4.6% for the respective coefficients of variation of the mean values. Calibration curve precision within-run ranged from 0.1% to 7.8% and between-run from 1% to 6.8%. The QC sample within-run as well as between-run accuracies were within ±7% of the nominal concentrations and within <1.6% for the respective coefficients of variation of the mean values. QC within-run precision ranged from 2.2% to 5.1%, and between-run precision ranged from 3.5% to 4.3%.
ACKNOWLEDGMENT
These studies were supported by grant R01 AI121430 from NIAID.
REFERENCES
- 1.Docobo-Pérez F, Drusano GL, Johnson A, Goodwin J, Whalley S, Ramos-Martín V, Ballestero-Tellez M, Rodriguez-Martinez JM, Conejo MC, van Guilder M, Rodríguez-Baño J, Pascual A, Hope WW. 2015. Pharmacodynamics of fosfomycin: insights into clinical use for antimicrobial resistance. Antimicrob Agents Chemother 59:5602–5610. doi: 10.1128/AAC.00752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louie A, Maynard M, Duncanson B, Nole J, Vicchiarelli M, Drusano GL. 2018. Determination of the dynamically-linked indices of fosfomycin for Pseudomonas aeruginosa in the hollow fiber infection model (HFIM). Antimicrob Agents Chemother :pii: AAC.02627–p17. doi: 10.1128/AAC.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calandra G, Ricci F, Wang C, Brown K. 1986. Cross resistance and imipenem. Lancet Aug 9 2:340–341. [DOI] [PubMed] [Google Scholar]
- 4.Fink MP, Snydman DR, Niederman MS, Leeper KV Jr, Johnson RH, Heard SO, Wunderink RG, Caldwell JW, Schentag JJ, Siami GA. 1994. Treatment of severe pneumonia in hospitalized patients: results of a multicenter, randomized, double-blind trial comparing intravenous ciprofloxacin with imipenem-cilastatin. The Severe Pneumonia Study Group. Antimicrob Agents Chemother 38:547–557. doi: 10.1128/AAC.38.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luyt CE, Aubry A, Lu Q, Micaelo M, Brechot N, Brossier F, Brisson H, Rouby JJ, Trouillet JL, Combes A, Jarlier V, Chastre J. 2014. Imipenem, meropenem, or doripenem to treat patients with Pseudomonas aeruginosa ventilator-associated pneumonia. Antimicrob Agents Chemother 58:1372–1380. doi: 10.1128/AAC.02109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drusano GL, Neely M, Van Guilder M, Schumitzky A, Brown D, Fikes S, Peloquin C, Louie A. 2014. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS One 9:e101311. doi: 10.1371/journal.pone.0101311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louie A, Liu W, VanGuilder M, Neely MN, Schumitzky A, Jelliffe R, Fikes S, Kurhanewicz S, Robbins N, Brown D, Baluya D, Drusano GL. 2015. Meropenem plus levofloxacin is synergistic for the therapy of Pseudomonas aeruginosa as tested in a murine model of pneumonia. J Infect Dis 211:1326–1333. doi: 10.1093/infdis/jiu603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drusano GL, Louie A, MacGowan A, Hope W. 2015. Suppression of emergence of resistance in pathogenic bacteria: keeping our powder dry—part 1. Antimicrob Agents Chemother 60:1183–1193. doi: 10.1128/AAC.02177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drusano GL, Hope W, MacGowan A, Louie A. 2015. Suppression of emergence of resistance in pathogenic bacteria: keeping our powder dry—part 2. Antimicrob Agents Chemother 60:1194–1201. doi: 10.1128/AAC.02231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. 1996. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure 4:1465–1474. doi: 10.1016/S0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 11.Hamou-Segarra M, Zamorano L, Vadlamani G, Chu M, Sanchez-Diener I, Juan C, Blazquez J, Hattie M, Stubbs KA, Mark BL, Oliver A. 2016. Synergistic activity of fosfomycin, β-lactams and peptidoglycan recycling inhibition against Pseudomonas aeruginosa. J Antimicrob Chemother 72:448–454. doi: 10.1093/jac/dkw456. [DOI] [PubMed] [Google Scholar]
- 12.Louie A, Bied A, Fregeau C, Van Scoy B, Brown D, Liu W, Bush K, Queenan AM, Morrow B, Khashab M, Kahn JB, Nicholson S, Kulawy R, Drusano GL. 2010. Impact of different carbapenems and regimens of administration on resistance emergence for three isogenic Pseudomonas aeruginosa strains with differing mechanisms of resistance. Antimicrob Agents Chemother 54:2638–2645. doi: 10.1128/AAC.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drusano GL, Liu W, Fregeau C, Kulawy R, Louie A. 2009. Differing effect of combination chemotherapy with meropenem and tobramycin on cell kill and suppression of resistance on wild-type Pseudomonas aeruginosa PAO1 and its isogenic MexAB efflux pump over-expressed mutant. Antimicrob Agents Chemother 53:2266–2273. doi: 10.1128/AAC.01680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed CLSI document M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Lodise TP, Sörgel F, Mason B, Melnick D, Kinzig M, Drusano GL. 2011. Penetration of meropenem into epithelial lining fluid in intubated patients with nosocomial pneumonia. Antimicrob Agents Chemother 55:1606–1610. doi: 10.1128/AAC.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaser J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 15:125–130. doi: 10.1093/jac/15.suppl_A.125. [DOI] [PubMed] [Google Scholar]
- 17.Neely MN, van Guilder MG, Yamada WM, Schumitzky A, Jelliffe RW. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]