Biofilm-residing bacteria embedded in an extracellular matrix are protected from diverse physicochemical insults. In addition to the general recalcitrance of biofilm bacteria, high bacterial loads in biofilm-associated infections significantly diminish the efficacy of antimicrobials due to a low per-cell antibiotic concentration.
KEYWORDS: Pseudomonas aeruginosa, biofilm regrowth, biofilm treatment, killing kinetics, treatment optimization
ABSTRACT
Biofilm-residing bacteria embedded in an extracellular matrix are protected from diverse physicochemical insults. In addition to the general recalcitrance of biofilm bacteria, high bacterial loads in biofilm-associated infections significantly diminish the efficacy of antimicrobials due to a low per-cell antibiotic concentration. Accordingly, present antimicrobial treatment protocols that have been established to serve the eradication of acute infections fail to clear biofilm-associated chronic infections. In the present study, we applied automated confocal microscopy on Pseudomonas aeruginosa to monitor dynamic killing of biofilm-grown bacteria by tobramycin and colistin in real time. We revealed that the time required for surviving bacteria to repopulate the biofilm could be taken as a measure for effectiveness of the antimicrobial treatment. It depends on the (i) nature and concentration of the antibiotic, (ii) duration of antibiotic treatment, (iii) application as monotherapy or combination therapy, and (iv) interval of drug administration. The vicious cycle of killing and repopulation of biofilm bacteria could also be broken in an in vivo model system by applying successive antibiotic dosages at intervals that do not allow full reconstitution of the biofilm communities. Treatment regimens that consider the important aspects of antimicrobial killing kinetics bear the potential to improve control of biofilm regrowth. This is an important and underestimated factor that is bound to ensure sustainable treatment success of chronic infections.
INTRODUCTION
Biofilm formation is widely found in natural environments and is considered an adaptation of microbes to hostile habitats. Protected in their own matrix, the bacteria efficiently withstand physical and chemical stressors of their natural surroundings. Due to the ongoing progress of medical sciences, more and more indwelling devices for medical treatment as well as foreign-body implants are used in humans. Biofilm-associated infections are a major complication associated with their use. Once a bacterial biofilm infection is established, it becomes very difficult, if not impossible, to eradicate it (1, 2). Biofilm infections are not necessarily associated with foreign bodies only. Chronic wound infections, chronic obstructive pulmonary diseases, endocarditis, and chronic infections of the middle ear and the sinuses, as well as infections of the lungs of cystic fibrosis (CF) patients, are all due to bacteria residing in biofilms (3). Such chronically infected patients may carry very high bacterial loads (up to 109 cells ml−1) associated with ongoing inflammation and changes in the structure and function of the affected organs (4–6). These infections therefore largely determine morbidity and mortality of the patients.
In this context, an exceedingly problematic Gram-negative pathogen is Pseudomonas aeruginosa. This opportunistic pathogen has emerged as one of the most important pathogens involved in acute nosocomial infections (7). It also plays a dominant role as the causative agent of chronic biofilm-associated infections, especially in the lungs of patients suffering from cystic fibrosis (1, 8–10).
A number of factors may explain biofilm-dependent bacterial survival despite intensified antibiotic treatment (11). These include (i) restricted penetration of antimicrobials into a biofilm (12–14), (ii) decreased growth rate of bacteria in biofilms (13, 15), (iii) biofilm-specific expression of possible resistance and stress response genes (16–24), (iv) metabolic heterogeneity (20, 25), and (v) the evolution of persister cells (26, 27). However, there also appears to be a functional relationship between the density of bacteria and the pharmacodynamics (PD) of antibiotics. A substantial inoculum effect can be observed on the efficacy of antibiotics under high-cell-density conditions (28). This density effect is primarily associated with a decrease in antibiotic concentration per cell and can partly be explained by the production of extracellular enzymes that degrade antimicrobials. Although it is debated whether the inoculum effect plays a role in vivo, standard protocols for antibiotic treatment fail to clear high-density-biofilm-associated infections.
One way to counteract this inoculum effect is to increase dosages of the antimicrobials as is done in inhalative antimicrobial therapy for the treatment of, e.g., CF lung infections. Colistin (CST) or tobramycin (TOB) aerosol inhalation is generally well tolerated and commonly applied, and these drugs are also well tolerated in combination (29). More recently, aztreonam and levofloxacin have become available as aerosols, and further antimicrobials may be characterized that could be developed into inhalative drugs (30, 31). Nevertheless, treatment schedules have largely been developed based on pharmacokinetics (PK) and pharmacodynamics of antibiotic activity against planktonic bacterial cells. Whether these schedules are equally applicable for the treatment of chronic biofilm-associated infections is currently unknown.
Initial results on pharmacodynamics within biofilms have been described previously (32, 33). In the present study, we applied automated confocal laser scanning microscopy (CLSM) to monitor the dynamics of various concentrations of two inhalative antibiotics, colistin and tobramycin, on the killing of P. aeruginosa biofilm cells in flow chambers in real time. We extended these studies to an in vivo biofilm system which we have recently established. It consists of a transplantable subcutaneous tumor which the bacteria efficiently target and colonize. Within 2 days, the bacteria produce mature biofilms, which protect them from antibiotics. Apparently, for the bacteria this niche is very similar to the niche they colonize in the lungs of humans with CF. We can conclude this from their gene expression profile, which is almost identical to the profile of bacteria isolated from CF lungs (34–36).
Our results suggest that treatment regimens should consider the important aspects of antimicrobial killing kinetics which exhibit substantial interantibiotic variation. Not only the choice of the antibiotic but also their specific combinations, concentrations, and especially the timing of application are of profound importance to enhance efficacy. Optimized treatment schemata, which are adapted to the conditions of high-cell density infections, promise to increase the efficiency of antibiotic treatment strategies that target chronic biofilm-associated infections.
RESULTS
Killing of biofilm-grown cells depends on the nature and concentration of the antibiotic.
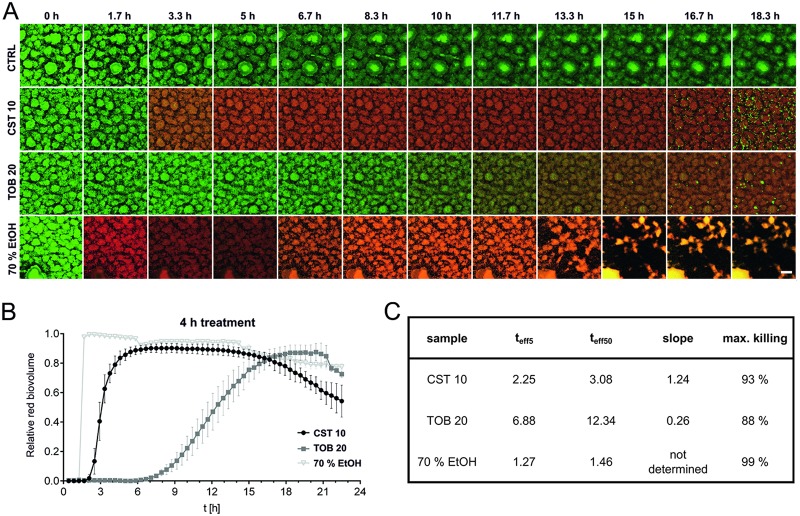
Besides genetic determinants of resistance, structured bacterial communities are known to exhibit tolerance against various antimicrobials. In this study, we evaluated the dynamics of antibiotic killing of biofilm-grown cells as well as the distribution of killing zones within the biofilm population over the course of treatment. By using established automated, time-lapse confocal laser scanning microscopy, we monitored the activity of two commonly used antimicrobials with different mode of actions on biofilm-grown green fluorescent protein (GFP)-tagged P. aeruginosa. We tested colistin, which disrupts the outer membrane, and tobramycin, a well-known translational inhibitor. The biofilms were allowed to establish in flow chambers under continuous-flow conditions, and the antibiotic activity was monitored by following uptake of propidium iodide (PI), which is not permeant to live cells and thus has proved to be a reliable indicator of cell death (37–40). For Fig. 1, established biofilms were subjected to antimicrobial treatment by adding tobramycin or colistin to the flow medium at concentrations corresponding to 10 to 20 times the MIC (TOB MIC, 1 µg ml−1; CST MIC, 0.5 µg ml−1). In addition, 70% ethanol was used as a killing control. After 4 h of treatment, the supply was changed to normal medium, which fully replaced the antibiotic-containing medium within approximately 120 min. Time-lapse microscopy clearly demonstrated that colistin (10 µg ml−1) kills the biofilm-grown cells almost immediately and almost as fast as ethanol (Fig. 1; see also Movie S1C and F in the supplemental material). Kinetics of cell death appears to reflect antibiotic penetration of the biofilm when treated with higher concentrations (10 µg ml−1 and 25 µg ml−1). Outer layers of bacteria in the biofilm are killed first, followed by a progression of killing as the antibiotic proceeds into deeper layers (Movie S1F and G). In contrast, killing of biofilm cells by high tobramycin concentrations (20 µg ml−1 and 50 µg ml−1) is significantly delayed (Fig. 1; see also Movie S1I and J). Furthermore, differences in the distribution of the killing zones can be observed. Tobramycin-mediated killing appears to occur most effectively in the periphery of the biofilms and to a much lesser extent in the deeper layers. A more detailed image analysis (Fig. 1C; see also Table S1) revealed that the overall levels of effectiveness of killing were comparable for the two antibiotics. Approximately 90% killing was achieved, determined as percent increase in propidium iodide-stained red biovolume.
FIG 1.
Killing dynamics of P. aeruginosa biofilm cells. Biofilms were cultivated for 48 h and treated with 10 µg ml−1 of colistin, 20 µg ml−1 of tobramycin, or 70% EtOH (killing control) for a period of 4 h. The first image series was acquired at the time of the flow start. The maximum antibiotic concentration was reached after approximately 90 min. Following a 4-h administration of the drug, the antibiotics were washed off the system within approximately 2 h. (A) Maximum-intensity projections of untreated control (CTRL, first row), colistin-treated (second row), tobramycin-treated (third row), and ethanol-treated (fourth row) biofilms at given intervals are shown. Green, GFP fluorescence; red, PI fluorescence. Scale bar: 50 µm. (B and C) Increase of the relative red biovolume (PI signal) over time (B) and relevant parameters describing the killing curve (C). teff5 and teff50, time points when 5% and 50%, respectively, of the biofilm cells are killed (slope of the sigmoidal curve and maximal killing rate). Data are means ± SDs for two positions in two independent experiments.
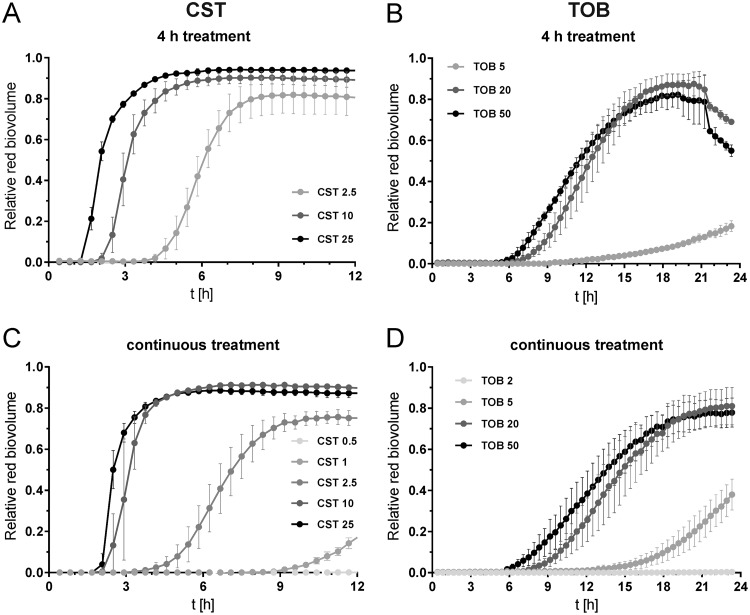
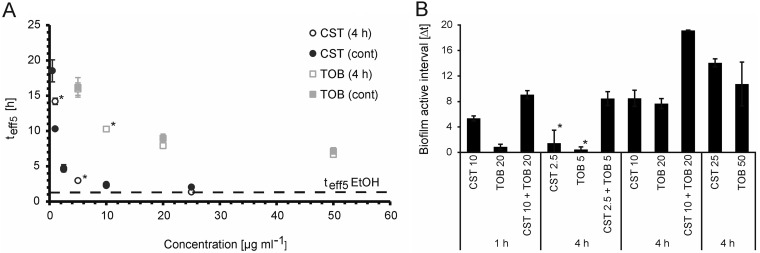
We also observed for both antibiotics that an increase in concentration led to an earlier onset of killing and higher maximum killing rates. In addition, maximum killing was achieved earlier (Fig. 2; see also Movie S1F versus Movie S1G and Movie S1I versus Movie S1J). This could be observed for both biofilms treated for 4 h (Fig. 2A and B; see also Movie S1F, G, I, and J) and biofilms treated continuously (Fig. 2C and D; see also Movie S1L, M, O, and P). When the antibiotic concentration versus time to effectively kill 5% of the biofilm population (teff5) is plotted for colistin and tobramycin, it can be seen that high colistin concentrations (25 µg ml−1) reached the teff5 after 2.3 h, which is close to the 1.3 h needed for 70% ethanol to reach maximum (Fig. 1C and Fig. 3A). In contrast, even high concentrations of tobramycin were not able to kill 5% of the biofilm bacterial population faster than 6 h (Fig. 2B and D and Fig. 3A). Low concentrations (approximately the MIC) were tested only in continuous treatment within the given intervals. The teff5 values were hardly reached by colistin (0.5 µg ml−1) or not reached at all by tobramycin (2 µg ml−1). Apparently, such concentrations are insufficient to kill biofilm bacteria efficiently. In conclusion, the lower the antibiotic concentration is, the longer it takes to kill 5% of the bacteria in the biofilm. This is true for both antibiotics and only when an antibiotic concentration above the MIC is applied.
FIG 2.
Antibiotic concentration-dependent increase of the red (dead) biovolume of biofilms. Biofilms were cultivated for 48 h and exposed to different concentrations of colistin (A and C) and tobramycin (B and D) for 4 h (A and B) and continuously (C and D). Data are means ± SDs of two positions in two independent experiments. Data for biofilms treated for 4 h with 10 µg ml−1 of colistin and 20 µg ml−1 of tobramycin are shown in Fig. 1.
FIG 3.
Concentration- and time-dependent time point of killing and regrowth. (A) Relationship between antibiotic concentration and the time to kill 5% of the biofilm population (teff5). The dashed line represents the teff5 of the killing control, ethanol. Asterisks indicate data of one single experiment (see Table S1). (B) Biofilm-active interval (interval [in h] between the time point when the antibiotic starts to kill and the time point when regrowth becomes visible) of 48-h-old PA14 biofilms treated for 1 h and 4 h with tobramycin and/or colistin at the indicated concentrations (in micrograms per milliliter). Asterisks indicate that for CST at 2.5 µg ml−1 and TOB at 5 µg ml−1 (4-h treatment), a BAI could not be determined for all experiments. Data are means ± SDs of two positions in two independent experiments.
Surviving bacteria repopulate biofilms very efficiently.
As soon as the administration of antibiotics is discontinued, surviving bacteria start to repopulate the biofilm structures in the flow chambers (Movie S2). For instance, in a biofilm that had been treated for 4 h with 10 µg ml−1 of colistin, repopulation had already begun after 18 h (Movie S1F). The repopulating clusters of biofilm bacteria grow very fast, reaching doubling times of less than 60 min (Table S1). Interestingly, this is as fast as the initial growth of biofilm microcolonies within the flow cells (55 to 60 min [data not shown]). Detailed information about image analysis, including start of killing and regrowth, is provided in Table S1 and Movie S2.
It has been reported that regrowth in colistin-treated biofilms was mainly found close to the substratum (39). In this study, we observed that regrowth after 1 h of colistin treatment (10 µg ml−1) appeared more or less randomly in the vertical axis of the biofilm (Fig. S1). However, regrowth after the combined treatment with colistin (10 µg ml−1) and tobramycin (20 µg ml−1) for 1 h was observed only close to the substratum and at later time points.
The interval between the time point when the antibiotic starts to kill and the time point when regrowth becomes visible was defined as the biofilm-active interval (BAI). As demonstrated in Fig. 3B, the BAI increases with increasing concentrations of either of the two antibiotics. This indicates that effective killing delays regrowth of the biofilm bacteria. When the two antibiotics colistin and tobramycin were combined to kill biofilm bacteria, regrowth was further delayed. Thus, antibiotic combinations showed a longer BAI than the sum of the single drugs (Fig. 3B). This indicates that the two drugs exhibit synergistic antibiofilm activities.
Of note, when biofilms were treated for 4 h with low antibiotic concentrations (CST, 2.5 µg ml−1, and TOB, 5 µg ml−1), killing was slower and less pronounced than with higher concentrations (Movie S1E and H). Moreover, in parallel to the inwards-proceeding killing, (re)growth of biofilm cells took place at the biofilm top layer (Movie S1E and H). This might be due to the washout of the antibiotics. Interestingly, colistin-treated bacterial cells also grew on the surface layer despite continuous treatment with 2.5 µg ml−1 (Movie S1K), which was not observed for continuous treatment with 5 µg ml−1 of tobramycin (Movie S1N). Apparently, the colistin-treated cells at the top layers face conditions which allow them to adapt to the activity of colistin. They grow despite antibiotic concentrations that exceed the MIC. The detailed killing and regrowth pattern can be seen in Fig. S2 and Movie S3. Importantly, the bacteria growing in the presence of the antibiotic did not exhibit mutations conferring resistance. When restreaked, bacteria showed the same susceptibility to colistin as control samples (data not shown).
Expanding the time of exposure to antibiotics delays regrowth.
We also exposed the P. aeruginosa biofilms continuously to the antimicrobial agents (Fig. 2C and D; see also Movie S1K to P). Killing dynamics of the biofilm cells under such conditions compared very well with the 4-h exposure to the same concentration of the antibiotic as long as the concentration was clearly higher than the MIC (>10×) (Fig. 2; see also Fig. S2 and Movie S1F and G versus Fig. S1L and M [CST] and Fig. S1I and J versus Fig. S1O and P [TOB]). On the other hand, treatment with an antibiotic concentration close to the MIC (1 µg ml−1 of colistin) killed biofilm bacteria only when given continuously. In that case, almost no killing was observed when the exposure time was reduced to 4 h (Table S1). In addition, we could show that when the time of exposure of the biofilm cells to the antibiotics was shortened from 4 h to 1 h, bacterial regrowth in biofilms also started earlier (Fig. S3A and B and Movie S1A and D). Detailed information about killing and regrowth parameters of all experiments is found in Table S1.
Colistin plays a dominant role in tobramycin-colistin combinations.
The effects of combinations of colistin and tobramycin given continuously at various concentrations are shown in Fig. S3C. Apparently, an increase in the concentration of colistin acting on the membrane directly has greater effects on the activity of the antibiotics than an increase of tobramycin concentrations. Killing was more advanced, i.e., faster with a steeper slope of the death curve when 10 µg ml−1 instead of 2.5 µg ml−1 of colistin was combined with 5 µg ml−1 of tobramycin. In contrast, no change in the killing curve was observed when the concentration of tobramycin was raised to 50 µg ml−1 and colistin was kept at 2.5 µg ml−1 or 10 µg ml−1 (Fig. S3C).
In addition, the killing activity of colistin was not influenced by pretreatment with tobramycin. When we exposed the biofilm bacteria which had been pretreated for 1 h with tobramycin (once, twice, or three times using a 12-h interval) to colistin, the fast and effective killing by colistin was found not to have been altered by the prior treatment. This highlights the strong potential for killing of biofilm-residing P. aeruginosa by colistin (Fig. S3D).
Shortening the interval between effective treatment doses delays biofilm regrowth.
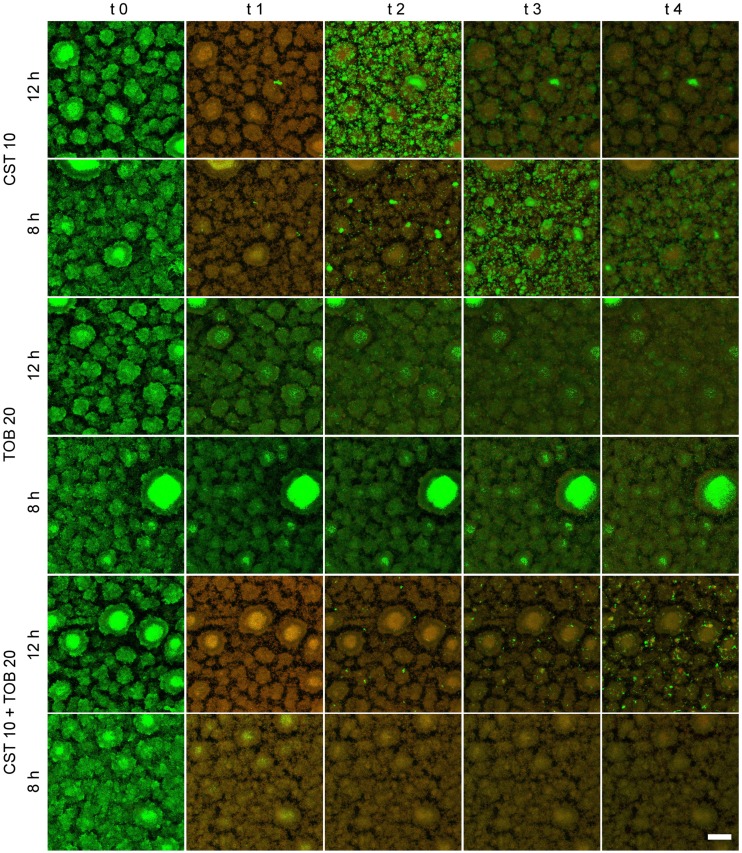
With the aim to mimic therapeutic treatment regimens used in the clinic, we tested the effects of repeated exposures to one or a combination of the two antibiotics. We exposed established biofilms for 1 h to antibiotics and repeated the exposure every 12 h over the course of 2 days (total of four applications). We had shown before that regrowth of biofilms that had been treated with a combination of colistin (10 µg ml−1) and tobramycin (20 µg ml−1) does not occur before 9 h (Fig. 3B). We therefore also tested an 8-h interval in order to evaluate whether avoidance of regrowth of the biofilm population between the treatments is critical for long-term success of antibiotic-mediated killing of biofilm bacteria. As observed before, the application of colistin killed the biofilm very efficiently and fast at both the 8-h and the 12-h intervals (Fig. 4; see also Movie S4B and E). However, the strong regrowth of surviving bacteria in the interval between the treatments also resulted in a very fast repopulation of the biofilm. Similarly, in tobramycin-treated biofilms, fast repopulation was observed in the 12-h regimen due to slower killing and incomplete penetration of the drug (Movie S4D), the latter also visible in the 8-h interval (Movie S4A).
FIG 4.
Killing dynamics in biofilms treated over a period of 48 h. The biofilms were exposed four times for 1 h to 10 µg ml−1 of colistin, 20 µg ml−1 of tobramycin, or a combination of the two. Two treatment intervals were tested: the first set was treated every 8 h and the second set every 12 h. Representative maximum-intensity projections of biofilms before treatment and 1 h after the time points of antibiotic addition are shown. Green, GFP fluorescence; red, PI fluorescence. Scale bar: 50 µm.
The combination of both antibiotics is clearly the most efficient strategy to kill biofilm bacteria, as seen in Fig. 4 and Movie S4C and F. Nevertheless, such treatment at a 12-h interval was not sufficient to abolish bacterial regrowth. After 24 h, clusters of bacteria start to repopulate the biofilm (Movie S4F).
This can, however, be prevented when the interval of antibiotic exposure is shortened to 8 h. We did not detect regrowth anymore (Movie S4C). Thus, shortening the intervals between two antibiotic administrations might allow complete clearance of the infection because regrowth of biofilm bacteria is suppressed.
Surviving bacteria repopulate biofilms very efficiently in an in vivo biofilm model.
The results of our in vitro studies revealed that the time surviving bacteria require to repopulate biofilms following antibiotic exposure is dependent on the overall concentration of the antibiotic, the duration of antibiotic treatment, monotreatment or combination treatment, and—if given as a bolus—the interval at which the drug is administered. The time of regrowth thus is a good measure for the effectiveness of antimicrobial therapy. Since modifications of treatment regimens could also have a major impact on antibiotic effectiveness in a clinical situation, we wanted to evaluate whether regrowth of surviving bacteria also occurs in vivo. We therefore used our previously established murine tumor model for biofilm infections. Mice bearing CT26 tumors colonized with biofilm-residing P. aeruginosa were treated intratumorally with both antibiotics. Mice were sacrificed 6 h and 24 h after antibiotic exposure, and CFU numbers in the tumors were determined by plating tumor homogenates. As demonstrated in Fig. S4, despite the fact that the bacteria grew within biofilm structures in the tumor tissue (34, 35), high concentrations of both drugs significantly reduced the bacterial load at the 6-h time point. However, when the mice were scarified 24 h after treatment, the CFU counts were comparable to those of the untreated controls. This clearly demonstrates that surviving bacteria are capable of efficiently repopulating the tumor tissue, similar to what had been observed in vitro.
Shortening the interval between effective treatment doses reduces bacterial burden in the in vivo biofilm model.
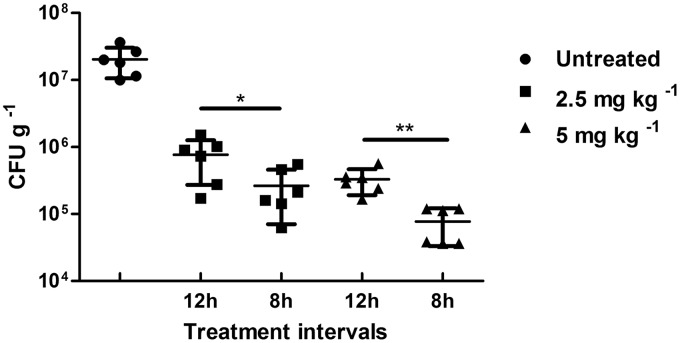
A fast repopulation of biofilm by the bacteria was observed in the murine tumors after the termination of antibiotic treatment (Fig. S4). Therefore, we wondered whether the data obtained in vitro could be confirmed in toto in our in vivo model. Tumor-bearing mice were infected with P. aeruginosa. This leads to stable biofilm formation within 2 days postinfection. These mice were then treated with a combination of colistin and tobramycin. We had demonstrated before that monotherapies with such antibiotics independent of the application interval led to inferior antimicrobial activity compared to that of combination therapies (34). When the animals bearing tumors colonized by biofilm-producing bacteria received the mixture of antibiotics in four doses at a 12-h interval, a significant reduction of bacterial load was observed at 2.5 mg kg−1 of body weight, which was even more pronounced at 5 mg kg−1 (Fig. 5). However, when the application regimen was altered to four applications with an 8-h interval, the antibiotic effect was strongly increased at both concentrations. At 5 mg kg−1, CFU counts were reduced almost to the detection limit. Thus, as predicted from the in vitro data when antibiotics were repeatedly administered at intervals that interfere with regrowth of biofilm bacteria, efficacy of combination therapy can be strongly enhanced. Possibly complete clearance may be achieved with this treatment regimen.
FIG 5.
In vivo killing is dependent on the treatment interval. Mice were treated with a combination of 2.5 mg kg−1 of colistin and 2.5 mg kg−1 of tobramycin (squares) or 5 mg kg−1 of colistin and 5 mg kg−1 of tobramycin (triangles). In total, four doses were injected intravenously either every 8 h or every 12 h. CFU counts per gram of homogenized tumor tissue are shown. Single antibiotics were less effective than combinatorial treatment (34). Each group contained 5 mice.
DISCUSSION
Despite intensified antimicrobial treatment, eradication of chronic persistent infections is almost impossible nowadays (1, 2). The urgent medical need for new therapy options against biofilm-associated infections spurred global research to uncover the mechanisms of biofilm formation and antimicrobial tolerance. However, we are only at the very beginning of understanding biofilm recalcitrance. In addition, although initial results using small molecules with antibiofilm activity have been published (31, 41–44), currently no antibiofilm compound is in clinical use.
This emphasizes the need to optimize current antimicrobial therapies for the treatment of chronic biofilm-associated infections. However, most established antimicrobial treatment protocols only consider the collective contribution of pharmacokinetics and pharmacodynamics (PK/PD) of antibiotics for the treatment of planktonic bacteria, responsible for acute infections. Whether such treatment schemes are equally well applicable to bacteria residing within biofilms at very high cell densities is under debate (6, 45). It appears that PK/PD values for the treatment of biofilm infections differ (46, 47) and thus that concentrations, combinations, and dosing intervals have to be optimized for the treatment of biofilm infections.
In this study, we took advantage of recent technological advancements in time-lapse imaging. Very precise automatic x, y, and z focusing has become possible, and this enables comparison of several samples in a single experiment over extended intervals. The spatial organization of complex biofilm systems as well as the spatiotemporal relationships of the bacterial cells that constitute the biofilm can now be analyzed in great detail and real time. This is of particular interest when it comes to monitoring the dynamic effect of antibiotic killing of biofilm bacteria. Dynamic monitoring of the death and regrowth kinetics of biofilm cells can lead to more precise conclusions. For example, it has been repeatedly described that colistin treatment results in effective killing of cells in the inner parts of the biofilm, while the surrounding cells at the biofilm surface layer are less affected (20, 25, 39, 48). Now, by dynamic monitoring of the death and regrowth kinetics, we are able to extend this interpretation. The profile observed is not due to preferential killing of metabolic inactive bacteria in the inner parts of the biofilm since high concentrations of colistin efficiently kill bacteria in the outer layers of the biofilm as well. However, bacteria at the surface layers apparently face conditions which allow them to tolerate colistin even when present at concentrations above the MIC. These bacteria actively repopulate the surface of the biofilm and thus quickly build a new surface population. The gradual increase of antibiotic concentration within the flow cell until the final peak concentration is reached (90 min) might facilitate the adaptation process. Interestingly, Chua and colleagues demonstrated that colistin-tolerant subpopulations migrate from lower parts of the biofilm to the upper parts via type IV pilus-driven motility (39) and thus contribute to the repopulation at the surface.
Bacterial adaptation to increased colistin concentrations has been described before. It appears to involve increased expression of various two-component systems, including PhoP-PhoQ, PmrA-PmrB, and ParR-ParSas, as well as the MexAB-oprM efflux pump (25, 39, 49–52). All of them have an impact on lipopolysaccharide (LPS) modifications.
Previous studies had demonstrated synergy of the two antibiotics colistin and tobramycin on P. aeruginosa biofilm-residing cells (53, 54). In addition, Pamp and colleagues combined colistin with tetracycline and ciprofloxacin (25). They showed that tetracycline and ciprofloxacin, which target the metabolically active bacteria, and colistin, which targets less active biofilm cells, complemented each other in terms of spatial killing profiles. Indeed, monitoring the dynamic killing patterns in the present work confirmed that the activity of colistin efficiently killed metabolically inactive cells in the center of the biofilms. In addition, tobramycin prevented the adaptive resistance of the metabolically more active cells on the top layers of the biofilm toward colistin. Thus, it inhibited repopulation of the biofilm top layer, which most likely explains the synergistic effect against P. aeruginosa biofilms.
Studying the dynamics of spatiotemporal killing patterns of different antimicrobials provides important information which may help to design new synergistic drug combinations. It is essential to determine (i) which antibiotics, (ii) which concentrations, and (iii) which combinations have to be employed to obtain the best effect on the reduction of the biofilm populations. However, as has to be concluded from our study now, it is equally important to prevent regrowth of the biofilm bacteria. Indeed, the most effective antibiotic therapy would be worthless when surviving bacteria fully reconstitute the biofilm population during the treatment intervals.
It appears that for a successful reduction of the bacterial load in chronically infected CF patients, the timing of the antibiotic therapy as well as the time point of termination will be very critical. Otherwise, due to the incomplete reduction of the biofilm biomass, surviving bacteria will find optimal conditions and very rapidly reconstitute the population until a balanced biomass is reached again.
The more efficient an antimicrobial therapy is, the more space it leaves for surviving bacteria to repopulate the empty niche. A similar fast repopulation of the biofilms might be the reason why in the clinic colistin treatment, despite being more effective in killing of biofilm cells (demonstrated here and before [37]), is not superior to an inhalative therapy with tobramycin (55–57). One might even argue that repeated killing and fast regrowth of the bacteria leave bacterial debris that may trigger a strong inflammatory response contributing to tissue destruction. Indeed, it was previously shown that the supernatant of colistin-treated biofilms is more cytotoxic to macrophages compared to the supernatant of nontreated biofilms or colistin solved in medium (39).
Importantly, not only was fast regrowth of bacterial biofilm populations observed after effective killing, but also we demonstrated that shortening the treatment intervals between two antibiotic doses reduced the overall bacterial burden in both our in vitro and in vivo model systems. Thus, if treatment intervals are short enough to interfere with full reconstitution of the biofilm population, the overall burden can be reduced in an additive or even synergistic manner. In the present study, the reduction from a 12-h to an 8-h treatment interval was sufficient to reduce the overall bacterial load in the murine tumors and might even lead to a complete clearance of the biofilm infection. However, reduction of the overall bacterial burden was dependent not only on the application interval. In addition, an increase of the overall dose of the combination of the two antibiotics improved the efficacy.
One could argue that under the condition of shortened application intervals, antibiotics simply accumulate and the increased efficacy is due to a raised concentration. However, we find this highly unlikely. It is well established that the antibiotics are rapidly cleared from circulation and tissues (46, 47, 58). Thus, timing of the individual doses appears to be the most critical factor and seems to depend on the effectiveness of the individual doses, as they determine when the biofilm population starts to regrow. Under these circumstances, it might be even conceivable that intervals between individual treatment doses are handled dynamically and might be extended with every cycle of antibiotic administration. Due to the gradual reduction of the bacterial load, the per-cell antibiotic concentration will increase with every cycle of antibiotic administration. Thus, regrowth should be progressively delayed.
New strategies to break the vicious cycle of killing and regrowth of biofilm-associated bacteria are essential. In the future, fine-tuned time-lapse microscopy combined with in silico model building (59) might prove to become a very valuable tool that serves to optimize antibiotic therapy of chronic biofilm-associated infections. The in silico approach will allow the prediction of various combinations and applications of different antibiotics applied at different intervals to maximize efficacy. Such in silico and subsequently in vitro-optimized protocols should lead to favorable outcomes in the in vivo model (35). This is prone to pave the way for successful antibiotic therapies, which would involve repeated antibiotic administration and clearance of the bacterial population in a stepwise manner.
MATERIALS AND METHODS
Biofilm cultivation and treatment.
Biofilms of a chromosomally GFP-tagged PA14 strain were cultivated in FAB medium with citrate (0.3 mM) at 30°C (inoculum, 5 × 107 cells ml−1) as previously described using modified 6-channel flow cells (µ-Slide VI0.4; ibidi, Germany) (60). After a 30-min settling period, flow was started at a constant rate (∼3.3 ml h−1 per channel) using a peristaltic pump (205S; Watson Marlow, United Kingdom). After 48 h of incubation, biofilms were treated for 1 h, 4 h, or continuously with medium containing tobramycin (sulfate salt) (TOB; Sigma-Aldrich, Germany) and colistin (sulfate) (CST; Carl Roth, Germany) at various concentrations and combinations as specified above. To add the antibiotics, the pump was paused, reflow was stopped with clamps, and the tubes and bubble traps upstream of the clamps were emptied and refilled with FAB minimal medium containing antibiotics. Clamps were removed and the flow was restarted. Due to the distance between bubble trap and flow cell (tube length, approximately 175 cm), antibiotic-containing FAB medium reached the flow cell about 15 to 20 min after restart. Pumping dyes (crystal violet and methylene blue) through the system, we measured the optical density (OD) of the flowthrough and could detect that both dyes reached their maximum concentration (=OD of stock concentration) in the flow cell approximately 90 min after flow restart. For the dilution of dyes after switching back to FAB-only medium (washing off period), more time was needed (approximately 120 min) due to the bubble trap volume (data not shown). Accordingly, both drugs could be detected in the flowthrough at the same time (Fig. S5). However, while tobramycin concentrations were found to be unchanged from the input, the concentration of colistin decreased to approximately one-third in the flowthrough. This indicates that colistin was most likely absorbed by the bacterial material. As a killing control, 70% ethanol (EtOH) was used. Propidium iodide (PI) was constantly supplied in the medium at a final concentration of 6.7 µM to monitor bacterial killing. All experiments were performed at least in duplicates. Images were taken at two positions within the flow cell.
Image acquisition via automated microscopy.
Established biofilms were monitored by confocal laser scanning microscopy (CLSM) using an inverted Leica SP8 system (Leica Microsystems, Germany). Settings for fluorescence signal detection were as follows: GFP, excitation at 488 nm (multiargon laser) and emission at between 500 and 550 nm; PI, excitation at 561 nm and emission at between 650 and 720 nm. Scanning of each individual position was repeated every 25 min. The first stack/image series was acquired at the time of the flow start (between 0 to 25 min, dependent on chamber position). Z-stacks were acquired for a total height of 57 µm with a step size of 3 µm by the use of a 40×/numerical aperture (NA) 1.1 water immersion objective. Two positions per lane (technical replicate) were imaged at a distance of 1 mm within the first third of the lane.
Image analysis.
Acquired image stacks were processed with the software Developer XD (Definiens) and Imaris (version 7.6; Bitplane). A customized solution in the Developer software similar to the known biofilm quantification tool PHLIP (61) was used to determine specific parameters for the stained biofilm describing the structure and the fluorescence ratios of both dyes as well as maximum intensity projections. In detail, automated image thresholding with fixed settings was applied for each stack after empty image removal to determine biophysical parameters, as well as ratios of red, green, and overlapping events within the Developer software. The increase in red biovolume proportion over time was used as a readout for antibiotic killing (values of >90% did correlate with nearly full killing). As shown before, the increase in red biovolume correlates with the amount of dead cells (determined via CFU counts [38]). Curve sketching was divided into two main sections to separately analyze the killing (initial increase of red biovolume proportion determined with the Definiens software) and the regrowth interval (increase of strong GFP signals due to bacterial growth determined via calculated isosurfaces of regrowth clusters within in the biofilm population with the Imaris software). A correction factor to compensate signal bleaching could not be used since the declines in fluorescence intensities were different for the two fluorophores (due to fixed thresholds), not constant over time, and sample dependent (changes in intensity during killing and regrowth were different for each sample and dependent on the antibiotic/antibiotic concentration used). As result, a constant intensity reduction led to an overall drop in the red biovolume proportion (see, for example, Fig. 1, isopropanol control).
Imaris was used to visualize three-dimensional (3D) reconstruction of the biofilms and to prepare video/time series sequences. Furthermore, Imaris was used to calculate the increase of isosurface volumes over time to indirectly determine the doubling time of regrowth clusters and microcolonies as well as to determine the center of homogeneous mass in z-direction. Excel and GraphPad Prism were used to perform nonlinear regression of the sigmoid killing curves. Video processing was achieved with the software VSDC Free Video Editor (Flash-Integro LLC).
In vivo tumor mouse model.
Experiments were performed as described previously (34). In brief, 8-week-old female BALB/c mice (Janvier, Germany) were injected intradermally with 5 × 105 CT26 (colon carcinoma) or F1A11 (fibrosarcoma) murine tumor cells. Mice bearing tumors with a diameter of 150 to 200 mm3 were infected intravenously (i.v.) with 5 × 106 P. aeruginosa PA14 cells. After 48 h, bacterial biofilms are established within tumors (35, 36). Thus, mice were treated either i.v. or locally (intratumoral application) with the desired antibiotic concentrations at specific intervals. At the end of treatment, the mice were euthanized by CO2 asphyxiation and sacrificed to isolate the tumors. These tumors were homogenized in 0.1% (vol/vol) Triton X-100–phosphate-buffered saline (PBS) using gentle MACS M-tubes and a dissociator from Miltenyi Biotec. The samples were serially diluted and plated on Luria-Bertani (LB) agar plates containing ampicillin (0.1 mg ml−1).
Ethics statement.
All experiments were performed in compliance with the German animal welfare act (TierSchG BGBl. I S 1105; 25.05.1998) and were approved by the Lower Saxony Committee on the Ethics of Animal Experiments as well as the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety under permit number 33.9-42502-04-12/0713).
Supplementary Material
ACKNOWLEDGMENTS
We thank Volkhard Kaever (Research Core Unit Metabolomics, Hannover Medical School) for helpful discussion on the optimization of antibiotic concentration measurements.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01635-18.
REFERENCES
- 1.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O, Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Doring G, Flume P, Heijerman H, Elborn JS, Consensus Study G. 2012. Treatment of lung infection in patients with cystic fibrosis: current and future strategies. J Cyst Fibros 11:461–479. doi: 10.1016/j.jcf.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Lebeaux D, Ghigo J-M, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohman DE, Chakrabarty AM. 1982. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun 37:662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tümmler B, Bosshammer J, Breitenstein S, Brockhausen I, Gudowius P, Herrmann C, Herrmann S, Heuer T, Kubesch P, Mekus F, Römling U, Schmidt KD, Spangenberg C, Walter S. 1997. Infections with Pseudomonas aeruginosa in patients with cystic fibrosis. Behring Inst Mitt 98:249–255. [PubMed] [Google Scholar]
- 6.Høiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, Hall-Stoodley L, Holá V, Imbert C, Kirketerp-Møller K, Lebeaux D, Oliver A, Ullmann AJ, Williams C. 2015. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21:S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 8.Tümmler B, Kiewitz C. 1999. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol Med Today 5:351–358. doi: 10.1016/S1357-4310(99)01506-3. [DOI] [PubMed] [Google Scholar]
- 9.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratjen F, Döring G. 2003. Cystic fibrosis. Lancet 361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3:10.1128/microbiolspec.MB-0010-2014. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suci PA, Mittelman MW, Yu FP, Geesey GG. 1994. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 38:2125–2133. doi: 10.1128/AAC.38.9.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol 15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, Iglewski BH, Storey DG. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, ’O’Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 18.Gillis RJ, White KG, Choi K-H, Wagner VE, Schweizer HP, Iglewski BH. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 49:3858–3867. doi: 10.1128/AAC.49.9.3858-3867.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Mah TF. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang W-C, Pamp SJ, Nilsson M, Givskov M, Tolker-Nielsen T. 2012. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol Med Microbiol 65:245–256. doi: 10.1111/j.1574-695X.2012.00929.x. [DOI] [PubMed] [Google Scholar]
- 21.Stewart PS, Franklin MJ, Williamson KS, Folsom JP, Boegli L, James GA. 2015. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 59:3838–3847. doi: 10.1128/AAC.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilton M, Charron-Mazenod L, Moore R, Lewenza S. 2016. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:544–553. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 26.Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 27.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 28.Udekwu KI, Parrish N, Ankomah P, Baquero F, Levin BR. 2009. Functional relationship between bacterial cell density and the efficacy of antibiotics. J Antimicrob Chemother 63:745–757. doi: 10.1093/jac/dkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann G, Yang L, Wu H, Song Z, Wang H, Hoiby N, Ulrich M, Molin S, Riethmuller J, Doring G. 2010. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis 202:1585–1592. doi: 10.1086/656788. [DOI] [PubMed] [Google Scholar]
- 30.Falagas ME, Trigkidis KK, Vardakas KZ. 2015. Inhaled antibiotics beyond aminoglycosides, polymyxins and aztreonam: a systematic review. Int J Antimicrob Agents 45:221–233. doi: 10.1016/j.ijantimicag.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Waters V, Smyth A. 2015. Cystic fibrosis microbiology: advances in antimicrobial therapy. J Cyst Fibros 14:551–560. doi: 10.1016/j.jcf.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Haagensen JA, Verotta D, Huang L, Spormann A, Yang K. 2015. New in vitro model to study the effect of human simulated antibiotic concentrations on bacterial biofilms. Antimicrob Agents Chemother 59:4074–4081. doi: 10.1128/AAC.05037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haagensen J, Verotta D, Huang L, Engel J, Spormann AM, Yang K. 2017. Spatiotemporal pharmacodynamics of meropenem- and tobramycin-treated Pseudomonas aeruginosa biofilms. J Antimicrob Chemother 72:3357–3365. doi: 10.1093/jac/dkx288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawar V, Komor U, Kasnitz N, Bielecki P, Pils MC, Gocht B, Moter A, Rohde M, Weiss S, Haussler S. 2015. In vivo efficacy of antimicrobials against biofilm-producing Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:4974–4981. doi: 10.1128/AAC.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawar V, Crull K, Komor U, Kasnitz N, Frahm M, Kocijancic D, Westphal K, Leschner S, Wolf K, Loessner H, Rohde M, Haussler S, Weiss S. 2014. Murine solid tumours as a novel model to study bacterial biofilm formation in vivo. J Intern Med 276:130–139. doi: 10.1111/joim.12258. [DOI] [PubMed] [Google Scholar]
- 36.Crull K, Rohde M, Westphal K, Loessner H, Wolf K, Felipe-López A, Hensel M, Weiss S. 2011. Biofilm formation by Salmonella enterica serovar typhimurium colonizing solid tumours. Cell Microbiol 13:1223–1233. doi: 10.1111/j.1462-5822.2011.01612.x. [DOI] [PubMed] [Google Scholar]
- 37.Müsken M, Klimmek K, Sauer-Heilborn A, Donnert M, Sedlacek L, Suerbaum S, Häussler S. 2017. Towards individualized diagnostics of biofilm-associated infections: a case study. NPJ Biofilms Microbiomes 3:22. doi: 10.1038/s41522-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müsken M, Di Fiore S, Römling U, Häussler S. 2010. A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc 5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 39.Chua SL, Yam JKH, Hao P, Adav SS, Salido MM, Liu Y, Givskov M, Sze SK, Tolker-Nielsen T, Yang L. 2016. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat Commun 7:10750. doi: 10.1038/ncomms10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benoit MR, Conant CG, Ionescu-Zanetti C, Schwartz M, Matin A. 2010. New device for high-throughput viability screening of flow biofilms. Appl Environ Microbiol 76:4136–4142. doi: 10.1128/AEM.03065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storz MP, Maurer CK, Zimmer C, Wagner N, Brengel C, De Jong JC, Lucas S, Müsken M, Häussler S, Steinbach A, Hartmann RW. 2012. Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J Am Chem Soc 134:16143–16146. doi: 10.1021/ja3072397. [DOI] [PubMed] [Google Scholar]
- 43.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. 2013. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A 110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reffuveille F, de la Fuente-Núñez C, Mansour S, Hancock REW. 2014. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother 58:5363–5371. doi: 10.1128/AAC.03163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. 2003. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest 123:1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 46.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. 2011. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 55:4469–4474. doi: 10.1128/AAC.00126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. 2012. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob Agents Chemother 56:2683–2690. doi: 10.1128/AAC.06486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pamp SJ, Sternberg C, Tolker-Nielsen T. 2009. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry 75A:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 49.Fernández L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock REW. 2012. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob Agents Chemother 56:6212–6222. doi: 10.1128/AAC.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the ‘last hope’ antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McPhee JB, Lewenza S, Hancock REW. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 52.Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol 34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 53.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, Elkins M, Thompson B, MacLeod C, Aaron SD, Harbour C. 2005. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol 43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PØ, Nielsen AK, Parsek M, Wozniak D, Molin S, Tolker-Nielsen T, Høiby N, Givskov M, Bjarnsholt T. 2011. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One 6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuster A, Haliburn C, Döring G, Goldman MH. 2013. Safety, efficacy and convenience of colistimethate sodium dry powder for inhalation (Colobreathe DPI) in patients with cystic fibrosis: a randomised study. Thorax 68:344–350. doi: 10.1136/thoraxjnl-2012-202059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tappenden P, Harnan S, Uttley L, Mildred M, Carroll C, Cantrell A. 2013. Colistimethate sodium powder and tobramycin powder for inhalation for the treatment of chronic Pseudomonas aeruginosa lung infection in cystic fibrosis: systematic review and economic model. Health Technol Assess 17(56):1–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uttley L, Harnan S, Cantrell A, Taylor C, Walshaw M, Brownlee K, Tappenden P. 2013. Systematic review of the dry powder inhalers colistimethate sodium and tobramycin in cystic fibrosis. Eur Respir Rev 22:476–486. doi: 10.1183/09059180.00001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dudhani RV, Turnidge JD, Coulthard K, Milne RW, Rayner CR, Li J, Nation RL. 2010. Elucidation of the pharmacokinetic/pharmacodynamic determinant of colistin activity against Pseudomonas aeruginosa in murine thigh and lung infection models. Antimicrob Agents Chemother 54:1117–1124. doi: 10.1128/AAC.01114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghanbari A, Dehghany J, Schwebs T, Müsken M, Häussler S, Meyer-Hermann M. 2016. Inoculation density and nutrient level determine the formation of mushroom-shaped structures in Pseudomonas aeruginosa biofilms. Sci Rep 6:32097. doi: 10.1038/srep32097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 61.Mueller LN, de Brouwer JFC, Almeida JS, Stal LJ, Xavier JB. 2006. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol 6:1. doi: 10.1186/1472-6785-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.