The capsid of the hepatitis B virus is an attractive antiviral target for developing therapies against chronic hepatitis B infection. Currently available core protein allosteric modulators (CpAMs) mainly affect one of the two major types of protein-protein interactions involved in the process of capsid assembly, namely, the interaction between the core dimers.
KEYWORDS: 20-deoxyingenol, Arbidol, hepatitis B virus, cell model, compound screen, core protein, dimer, split luciferase
ABSTRACT
The capsid of the hepatitis B virus is an attractive antiviral target for developing therapies against chronic hepatitis B infection. Currently available core protein allosteric modulators (CpAMs) mainly affect one of the two major types of protein-protein interactions involved in the process of capsid assembly, namely, the interaction between the core dimers. Compounds targeting the interaction between two core monomers have not been rigorously screened due to the lack of screening models. We report here a cell-based assay in which the formation of core dimers is indicated by split luciferase complementation (SLC). Making use of this model, 2 compounds, Arbidol (umifenovir) and 20-deoxyingenol, were identified from a library containing 672 compounds as core dimerization regulators. Arbidol and 20-deoxyingenol inhibit the hepatitis B virus (HBV) DNA replication in vitro by decreasing and increasing the formation of core dimer and capsid, respectively. Our results provided a proof of concept for the cell model to be used to screen new agents targeting the step of core dimer and capsid formation.
INTRODUCTION
Hepatitis B infection represents a significant public health burden worldwide. An estimated 2 billion people have been infected, and more than 350 million are chronic carriers of the virus (1). Two arms of treatment, interferon alpha (IFN-ɑ) and nucleos(t)ide analogues (NUCs), have been approved for the therapy of HBV infection. IFN-ɑ has an immunomodulatory and a modest antiviral effect, and can cause significant side effects in over 30% of patients (2). NUCs directly and potently inhibit the reverse transcriptase activity of the HBV polymerase to suppress virus replication but do not eliminate the viral covalently closed circular DNA (cccDNA) from infected cells (3). The persistence of cccDNA causes virological relapse after discontinuation of NUC treatment. There is an unmet need for novel anti-HBV agents with new antiviral target(s), since the drugs currently available rarely cure chronic hepatitis B (3).
The nucleocapsid is an attractive antiviral target distinct from those of current drugs. Upon infection of the hepatocyte, the viral genomic relaxed circular DNA (rcDNA) is released into the nucleus, where the rcDNA is converted into cccDNA. Viral RNAs are transcribed using the cccDNA as the template. The pregenomic RNA (pgRNA) is translated to produce both the core protein and DNA polymerase. Core proteins interact with the DNA polymerase, which binds to the epsilon sequence within the 5′ portion of pgRNA, initiating nucleocapsid assembly. The encapsidated pgRNA is then reverse transcribed into viral DNA by the polymerase. The matured nucleocapsid can be enveloped and secreted out of the cell or enter into a recycling process to deliver its rcDNA back into the nucleus to amplify the cccDNA pool (4). Since the nucleocapsid hosts the HBV replication complex and exerts multifunction during the viral life cycle, agents affecting the formation of nucleocapsid should have the potential to suppress the viral replication.
The formation of the capsid involves two kinds of interactions, namely, the interaction between two core monomers and that between two core dimers (5). Dimers are the major intermediate during the formation of capsid in vitro. Recombinant Hepatitis B virus core protein (HBc) expressed from bacteria forms stable, noncovalent dimers in solution (6). The capsid assembly starts with a slow nucleation step from dimers (formation of trimers of dimers), followed by a rapid elongation phase (7–9). Mass spectrometry analysis has demonstrated the presence of a number of species of up to 12 dimers, including a dimer of dimers and trimer of dimers (10). A combination of mass spectrometry and cryo-electron microscopy (cryo-EM) has revealed kinetically trapped incomplete capsids, which may represent intermediates later in assembly (11).
Several lines of core protein allosteric modulators (CpAMs) have been demonstrated to possess an effect that deviates the process of HBV capsid formation. Heteroaryldihydropyrimidines (HAPs) were found to affect virus production in vitro and in vivo (12, 13). In vitro studies revealed that HAPs misdirected capsid assembly to form aberrant noncapsid polymers (14, 15). High-resolution crystal structure analysis demonstrated that a potent inhibitor from the HAP series, NVR-010–001-E, binds at the dimer-dimer interface of the core proteins, forms a new interaction surface promoting protein-protein interaction, induces protein assembly, and increases stability (16). The second class of CpAMs, phenylpropenamides (PPAs; AT130), were also shown to inhibit HBV replication in cell cultures (17, 18). AT130 was found to bind a hydrophobic pocket that also accommodates HAP compounds and to induce tertiary and quaternary structure changes in the capsid (19, 20). The third class of CpAMs includes the sulfanilamides and the sulfamoylbenzamides, which have been shown to disrupt normal capsid formation by decreasing at least the dimer-dimer interaction (21, 22). Together, these kinds of compounds are all believed to affect the dimer-dimer interaction.
In this study, we aimed to screen compounds that affect the core protein dimerization, in order to reduce the building blocks for capsid assembly. A technical obstacle for doing this is the lack of an assay that detects the formation of core dimers in a rapid and quantitative manner. HBV replication cell lines, such as HepG2.2.15 (23), HepAD38 (24), AML12HBV10 (25), and HepDES19 (26) are the commonly used models to screen compounds with inhibitory effects on HBV replication and have scored capsid inhibitors, but the assay readouts are normally for viral DNA and antigens. Zlotnick et al. have developed an in vitro screening model based on fluorescence quenching of dye-labeled core protein (27). This model helped identify small molecules that prevent or misdirect capsid formation; however, the molecular details of the capsid formation by bacterially expressed core proteins are different from what happen in cells (28). For example, at physiologically low HBc concentrations, the N-terminal domain is insufficient for capsid assembly, the C-terminal domain (CTD) is also required, and the phosphorylation and dephosphorylation of the CTD regulate capsid assembly (28). Thus, we set out to develop a cell-based assay for screening of compounds targeting core protein dimerization, which is quantified by the split luciferase complementation assay. Making use of this screening system, we identified two compounds that affect core dimerization and capsid formation from a library containing 672 agents, demonstrating the feasibility of the assay.
RESULTS
Rationale of assay design.
To establish a screening model for identifying compounds that target HBV core dimerization, we harnessed split luciferase complementation (SLC) technology, which has been used to indicate the interaction between two proteins (29, 30). In our case, a luciferase was split into two fragments and each was fused to the core protein (Fig. 1B). The split luciferase will be complemented when a heterodimer forms, and the recovered luciferase activity can then be detected conveniently. If a compound that affects the formation of core dimer is present, an alteration (a decrease or an increase) in the luciferase activity can be expected, and the extent of alteration may reflect the potency of an agent for interfering in the interaction between two core monomers.
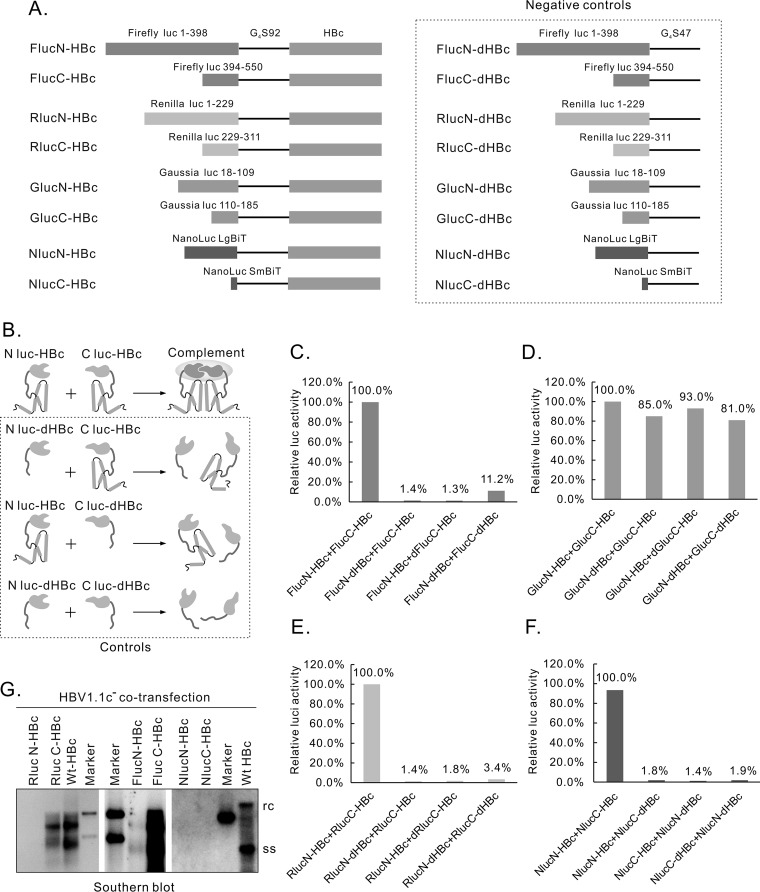
FIG 1.
Evaluation of the 4 split luciferase pairs. (A) Diagram of the structure of different split-luc-HBcs and corresponding negative controls. Four pairs of split luciferases were fused at the N terminus of HBc, separated by a glycine-serine linker with a length of 92 amino acids (92G4S). The split site for each luciferase was indicated by amino acid numbers. Each negative control contains a G4S sequence with a length of 47 amino acids. (B) Working model of the complementation of split luciferase. The 2 split-luc-HBcs are expected to form dimers and reconstitute luciferase activity when cotransfected, while those negative controls with the interaction domains deleted will not complement each other. (C) Evaluation of split-Fluc-HBcs. Constructs expressing split-Fluc-HBcs or controls were cotransfected into HEK293 cells, as illustrated in panel B, and Fluc activities were detected. (D) Evaluation of split-Gluc-HBcs. (E) Evaluation of split-Rluc-HBcs. (F) Evaluation of split-Nluc-HBcs. (G) Evaluation of split-luc-HBcs for their ability to support HBV DNA replication. Different split-luc-HBcs constructs were cotransfected with plasmid HBV1.1c−, and core particle-associated HBV DNA was extracted and assayed by Southern blotting 6 days after transfection. Wt, wild type; WB, Western blot; EGFP, enhanced green fluorescent protein; anti-HA, anti-human influenza virus hemagglutinin; rc, relaxed circular DNA; ss, single-stranded DNA.
Evaluation of different split luciferase pairs.
We evaluated 4 pairs of split luciferases, including Firefly, Renilla, Gaussia, and NanoLuc luciferases, for their capability to reflect HBc dimer formation. Each split luciferase fragment was fused to the N terminus of the HBc through a glycine-serine linker (G4S) with a length of 92 amino acids (G4S92). This long linker was used because our previous results (31) showed that it helped maintain the functions of HBc fusion proteins. The split site of each luciferase, according to previous reports, is shown in Fig. 1A (32–35). Negative controls were constructed that express split luciferases only with the linker sequence G4S47 (Fig. 1A, right panel).
Cotransfection experiments were performed, with each pair of plasmids expressing different split luciferase HBcs (split-luc-HBcs) (Fig. 1B, upper panel) or controls (Fig. 1B, lower 3 panels). A desirable split-luc-HBc pair should present results consistent with the model shown in Fig. 1B, i.e., cotransfection with split-luc-HBcs produces significantly higher signals than those of the controls. Based on this criterion, the split Gaussia luciferase approach was terminated due to the signal strength being indistinguishable from that of the controls (Fig. 1D). Cotransfection of split Firefly (Fig. 1C), split Renilla (Fig. 1E), and split NanoLuc (Fig. 1F) luciferases fused with HBc produced signals significantly higher than those of the corresponding controls by about 10-fold, 20-fold, and 50-fold, respectively, suggesting that the complementation of the split-luc-HBcs did occur through the interaction between the 2 HBcs. As for the absolute value of the signal, that of the split-Nluc-HBcs was higher than that of the split-Rluc-HBcs by about 10-fold, while that of the split-Rluc-HBcs was higher than that of the split-Fluc-HBcs by about 100-fold (data not shown).
The formation of split-luc-HBc dimers indicated that the functions of the recombinant HBc were at least partially maintained. To further assess to what extent these split-luc-HBcs keep their functions, we tested these HBc fusion proteins for their capability to support HBV DNA replication. The 6 split-luc-HBcs were cotransfected with HBV1.1c−, a plasmid that expresses all the elements needed for HBV DNA replication except HBc (31). Southern blotting was used to detect the intracellular core particle-associated HBV DNA. As shown in Fig. 1G, RlucC-HBc, FlucC-HBc, and FlucN-HBc (very weakly) supported HBV DNA replication, while RlucN-HBc, NlucN-HBc, and NlucC-HBc did not in our experiments. These results suggested that the 2 split Nlucs and RlucN influenced the function of HBc more than did the RlucC and the 2 split Flucs.
Further analysis of the function of split Rluc-HBcs.
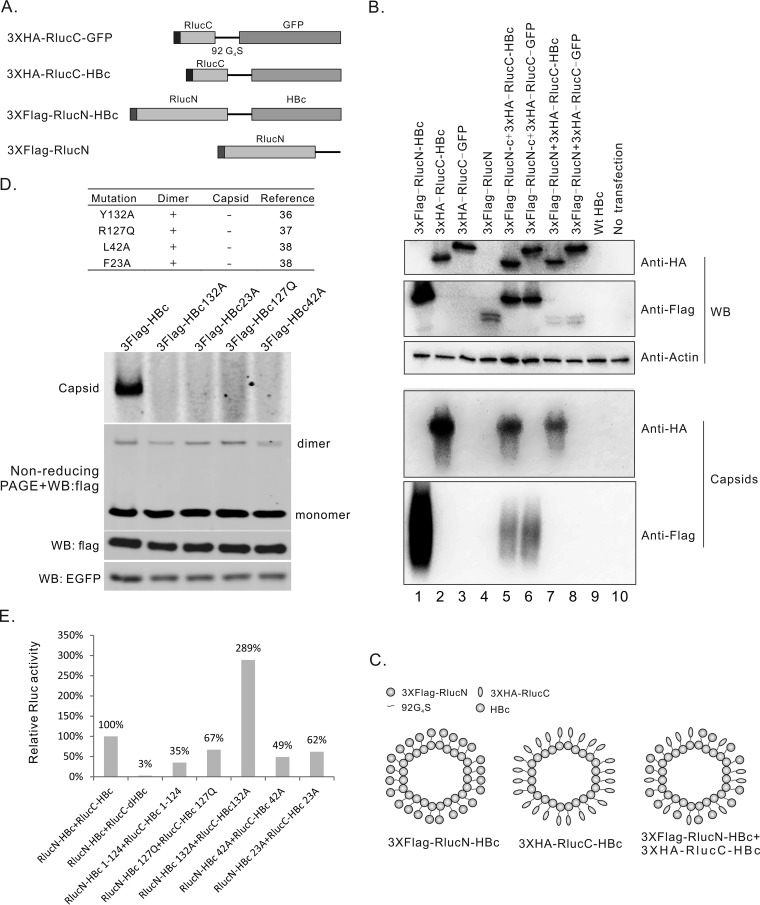
We chose split Rlucs for further tests, considering a balance between the efficiency of split luciferase complementation and the impact on the HBc function. The capability of split-Rluc-HBcs to form capsid-like particle was assayed. For the convenience of detection, we added a 3 × Flag tag to the N-terminus of RlucN-HBc, and a 3×HA tag to the N terminus of RlucC-HBc (Fig. 2A). As the control, the HBc portion was deleted from 3×Flag-RlucN-HBc or replaced with enhanced green fluorescent protein (EGFP) in 3×HA-RlucC-HBc (Fig. 2A). Proteins expressed from these plasmids were confirmed by Western blotting (Fig. 2B, upper panel). Capsid-like particles in the cell lysates were resolved in native agarose gels and detected with anti-Flag or anti-human influenza virus hemagglutinin (anti-HA) antibodies after blotting. As shown in Fig. 2B, 3×Flag-RlucN-HBc and 3×HA-RlucC-HBc formed capsid-like particles, while the 2 controls did not, either alone (lanes 3 and 4) or when cotransfected with 3×Flag-RlucN-HBc or 3×HA-RlucC-HBc (lanes 6 and 7). Together, we confirmed that both 3×Flag-RlucN-HBc and 3×HA-RlucC-HBc kept the ability to form capsids. This suggested that the RlucN fusion does not have a severe negative impact on capsid assembly, although RlucN-HBc does not support HBV DNA replication (Fig. 1G). Whether 3×Flag-RlucN-HBc and 3×HA-RlucC-HBc incorporated into one capsid, as in the model depicted in Fig. 2C, is not clear from our assay. We have attempted to perform a coimmunoprecipitation (coIP) experiment using anti-Flag and anti-HA antibodies but were not successful due to the cross-reaction between these two antibodies in our experiment (Fig. S1B).
FIG 2.
Further evaluation of split-Rluc-HBcs in supporting dimerization, capsid assembly, and luciferase reconstitution. (A) Diagram of the structures of the tagged split-Rluc-HBcs and controls. (B) capsid-like particle formed by split-Rluc-HBcs. Constructs indicated were transfected into HEK293 cells. Western blotting was used to confirm the expression of proteins, and capsid-like particles were detected by particle gel assay. (C) Modeling of capsid-like particles formed by tagged split-Rluc-HBcs. 3×Flag-RlucN-HBc and 3×HA-RlucC-HBc can form capsid-like particles; and cotransfection of these 2 constructs may generate mosaic capsid-like particles. (D) Specific mutations in HBc abolished the formation of the capsid. Recombinant HBc with mutations Y132A, R127Q, L42A, or F23A was reported to form dimers but not capsids in vitro (references for these mutants are listed). Constructs expressing 3×Flag-HBc containing single mutations Y132A, R127Q, L42A, or F23A, were transfected into HEK293 cells. 3×Flag-HBc dimers were detected by nonreducing PAGE and Western blotting, and capsids were detected by particle gel assay. (E) Split luciferase complementation assay of split-Rluc-HBc mutants. The indicated split-Rluc-HBc mutant pairs were transfected into HEK293 cells, and Rluc activity was tested 48 h after transfection.
Next, we asked whether the complementation of the split Rlucs came only from heteromonomers (a RlucN-HBc monomer and a RlucC-HBc monomer), or partly from two different kinds of homodimers (a RlucN-HBc dimer and a RlucC-HBc dimer) aligned adjacently on a capsid. To address this question, two plasmids were constructed, which expressed 3×Flag-G4S92-HBc and 3×HA-G4S92-HBc, respectively. The 3×Flag-G4S92-HBc plasmid maintains the major functions of wild-type HBc, supporting the formation of capsids and the replication of HBV DNA, while the 3×HA-G4S92-HBc plasmid exhibits deficiency in capsid assembly and HBV DNA replication (Fig. S1D).
Mutations Y132A, R127Q, L42A, and F23A were introduced into the HBc sequence of the 3×Flag-G4S92-HBc. All of these mutations have been reported to make the Escherichia coli-expressed Cp149 not form capsids but to support dimer formation in vitro (36–38). The characteristics of these mutants and references are summarized in the upper panel of Fig. 2D. These mutants provide us a chance to determine whether the complementation of the split Rlucs depends on the formation of capsids or not. To analyze the function of these mutants in cells, the plasmids were cotransfected with pEGFP-N1 (as the control) into HEK293 cells. Formation of capsids and dimers was determined by particle gel assay and nonreducing PAGE. As shown in Fig. 2D, all of these mutants were deficient in the formation of capsids but allowed core dimerization, which is consistent with previous reports (36–38). The same mutations were then introduced into RlucN-HBc and RlucC-HBc. Each pair of Rluc-HBc mutants, such as RlucN-HBc132A and RlucC-HBc132A, were cotransfected into HEK293 cells to determine the influence of these mutations on Rluc activity. As shown in Fig. 2E, all of these mutants retained about 50% or higher Rluc activity compared with that of the wild type. The mutants with 132A even showed a higher Rluc activity than that of the wild type. These data supported a notion that at least a considerable part of Rluc activity was from the complemented heterodimers.
Establishment of the stable cell line SRluc-HBc6.
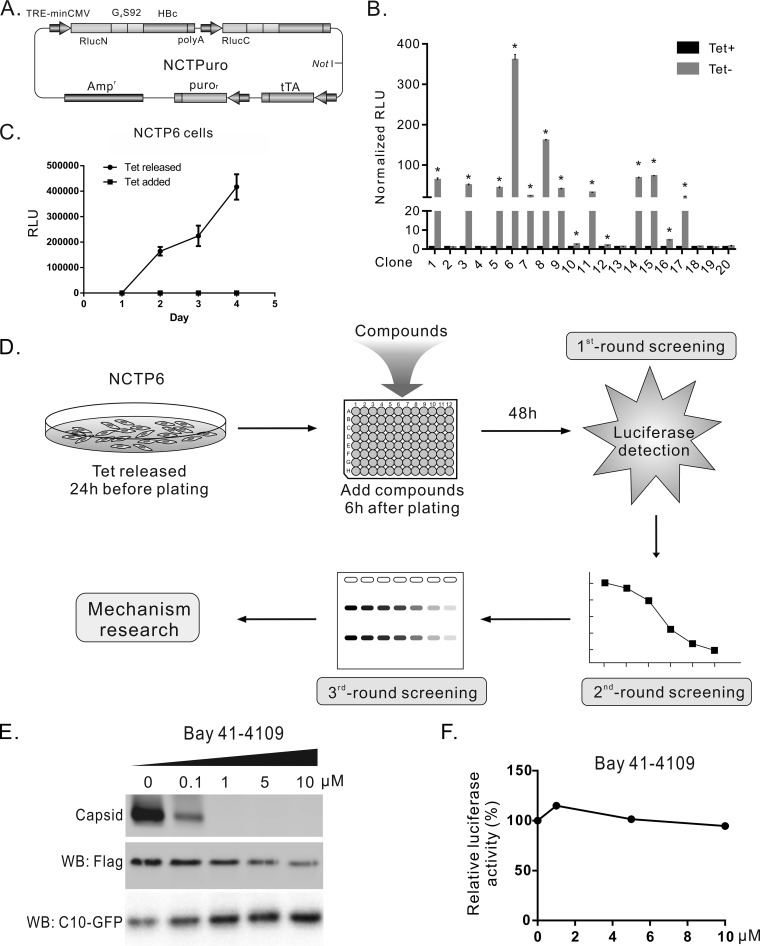
To establish the stable cell line that expresses the split-Rluc-HBcs, a tetracycline-inducible plasmid named NCTPuro was constructed, which contained a tandem RlucN-HBc and RlucC-HBc open reading frame driven by the tetracycline response element (TRE)-minimal cytomegalovirus (miniCMV) promoter, a tetracycline-controlled transactivator (tTA) gene, and a puromycin-resistant gene (Fig. 3A). NCTPuro was linearized with Not I and transfected into HEK293 cells. After puromycin selection, twenty clones were obtained and characterized by comparing the Rluc activity under tetracycline-containing or tetracycline-free conditions. Among them, clone 6 showed the strongest Rluc activity in a tet-off manner (Fig. 3B) and exhibited a gradual increase of Rluc activity in a time course study (Fig. 3C). This cell line was named SRluc-HBc6 and used thereafter to screen compounds.
FIG 3.
Establishment of stable cell line SRluc-HBc6 and compound screening procedure. (A) Schematic illustration of plasmid NCTPuro. The plasmid contains 4 open reading frames, which express an RlucN-HBc, an RlucC-HBc, a puromycin resistance gene, and a tetracycline-controlled transactivator (tTA). (B) Selection and characterization of stable cell lines. Twenty clones were obtained after puromycin selection. Rluc activities of these clones were detected in the presence and absence of tetracycline. (C) Time course measurement of Rluc activity in SRluc-HBc6 cells. (D) The diagram of the compound screening procedure. SRluc-HBc6 cells cultured in a 10-cm diameter dish were seeded into 96-well plates with the tetracycline-free medium. Six hours after seeding, compounds were added, and Rluc activity was assayed 48 h after treatment for the first-round screening. Compounds identified from the first-round screening were tested for dose-effect curves in SRluc-HBc6 cells as the second-round screening. For the third-round screening, compounds were tested in HepAD38 cells for their effects on HBV DNA replication by Southern blotting. (E) Bay 41-4109 inhibited capsid formation. HepG2 cells cotransfected with 3×Flag-G4S92-HBc and C10-GFP were treated with Bay 41-4109 for 6 days. Intracellular capsids were detected by particle gel assay. The expression of the recombinant core protein and C10-GFP (control) was detected by Western blotting. (F) Bay 41-4109 did not affect the Rluc activity of SRluc-HBc6 cells. SRluc-HBc6 cells were treated for 2 days with Bay 41-4109 of different concentrations, and the Rluc activity was assayed.
Compound screening.
The screening procedures are outlined in Fig. 3D. Briefly, (i) The tetracycline-free medium was used to culture SRluc-HBc6 cells the day before seeding. On the next day, SRluc-HBc6 cells were seeded into 96-well plates, and compounds were added to the medium 6 h later. Rluc activity in the cell lysate of each well was assayed after 48 h. (ii) Those compounds significantly affecting the Rluc activities were tested for their dose-dependent effects in the second-round screening. (iii) The confirmed compound hits were analyzed in HepAD38 cells for their biological effects on the replication of HBV DNA.
We first tested the reference CpAM compound Bay 41–4109 in SRluc-HBc6. Bay 41-4109 potently inhibited the formation of capsid in HepG2 cells transfected with 3×Flag-G4S92-HBc at 1 μM (Fig. 3E), but no inhibition of Rluc activity was observed in SRluc-HBc6 cells treated even with 10 μM Bay 41-4109 (Fig. 3F). The known mechanism of action (MoA) of heteroaryldihydropyrimidine (HAP) compounds, including Bay 41-4109 is misdirecting capsid assembly to form aberrant noncapsid polymers (14, 15, 39, 40). Thus, our assay would help to identify compounds with different mechanisms from those of the classic CpAMs.
Two libraries containing 304 natural products and 368 synthesized compounds were tested in the first-round screening. We arbitrarily chose for the second-round screening those agents that inhibited RLuc activity by more than 3-fold or increased it by more than 2-fold compared to that of the controls. A total of 36 compounds were identified according to this criterion. In the second-round screening, 12 out of the 36 compounds induced a dose-dependent response of Rluc activity in SRluc-HBc6 cells, 9 of which decreased Rluc activities, and 3 of which increased them (Fig. S2). These 12 compounds were tested in HepAD38 cells for their ability to inhibit the replication of HBV DNA. Four compounds, HBV002 (sinapine), HBV005 (20-deoxyingenol), HBV009 (Arbidol [umifenovir]), and HBV011 (sulfanilamide), showed a dose-dependent inhibition of HBV DNA replication in HepAD38 cells (Fig. S3). Further characterization indicated that sinapine was a false positive that suppressed the transcription from the TRE-miniCMV promoter (data not shown). Sulfanilamide, which has been reported to inhibit capsid formation of Cp149 in vitro (22), failed to exhibit significant antiviral activity in subsequent assays (data not shown). Thus, we prioritized Arbidol and 20-deoxyingenol for further studies.
Characterization of biological effects of the compound candidate Arbidol.
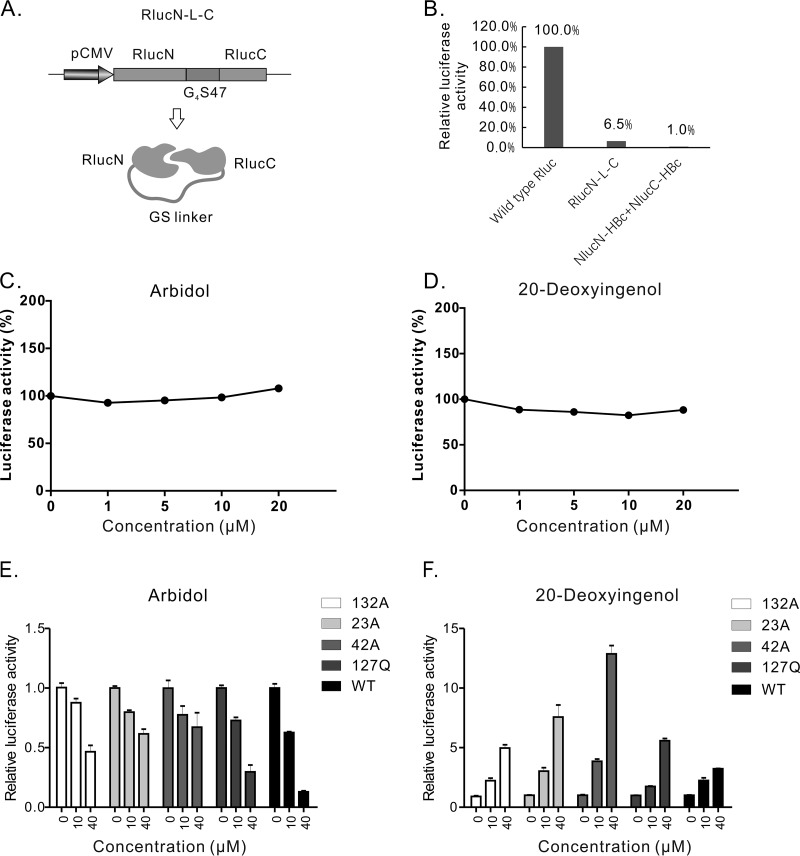
Arbidol showed a 50% effective concentration (EC50) of 2.3 ± 1.2 μM and a 50% cytotoxic concentration (CC50) of 58.4 ± 4.2 μM in SRluc-HBc6 cells (Table 1). To test whether Arbidol influences the complementation of the split Rluc, a plasmid was constructed that expresses the RlucN and RlucC linked by a G4S47 linker, driven by the cytomegalovirus-immediate early (CMV-IE) promoter (Fig. 4A). This construct (named pRlucN-L-C) produced 6.5% of the Rluc activity of the wild-type Rluc (Fig. 4B). HepG2 cells transfected with pRlucN-L-C were treated with Arbidol, and no significant change in the Rluc activity was observed under 20 μM Arbidol (Fig. 4C). All of this indicated that Arbidol did not affect the transcription activity of the CMV-IE promoter or the interaction between RlucN and RlucC.
TABLE 1.
Activity of Arbidol and 20-deoxyingenol in different cell models
| Compound | SRluc-HBc6 |
HepAD38 |
HepG2.2.15 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| EC50a (µM) | CC50b (µM) | SIc | EC50 (µM) | CC50 (µM) | SI | EC50 (µM) | CC50 (µM) | SI | |
| Arbidol | 2.3 ± 1.2 | 58.4 ± 4.2 | 25.9 | 5.3 ± 0.6 | 64.6 ± 3.0 | 12.1 | 5.6 ± 0.8 | 59.9 ± 4.1 | 10.7 |
| 20-Deoxyingenol | 8.9 ± 1.56 | >100 | >11.3 | 31.2 ± 3.1 | >100 | >3.2 | 14.5 ± 1.6 | >100 | >6.9 |
EC50, fifty percent effective concentration.
CC50, fifty percent cytotoxicity concentration.
SI, selectivity index calculated by the ratio of CC50 and EC50.
FIG 4.
Arbidol and 20-deoxyingenol did not affect the activity of RlucN-L-C but did affect dimerization of split-Rluc-HBc mutants. (A) Diagram of plasmid pRlucN-L-C and RlucN-L-C protein. (B) Activity test of RlucN-L-C. The indicated constructs were transfected into HEK293 cells. Rluc activities were tested 48 h posttransfection. (C) Arbidol did not significantly affect the Rluc activity produced by pRlucN-L-C transfection. (D) 20-Deoxyingenol did not significantly influence the Rluc activity produced by pRlucN-L-C transfection. (E) Arbidol decreased dimerization of split-Rluc-HBc mutants. split-Rluc-HBc pairs with mutations Y132A, R127Q, L42A, or F23A were transfected into HEK293 cells, and Arbidol of indicated concentrations were used to treat the cells. Rluc activity was assayed after 2 days of treatment. (F) 20-Deoxyingenol increased dimerization of split-Rluc-HBc mutants.
To test the influence of Arbidol on the dimerization of split-Rluc-HBc mutants, four pairs of mutants, such as RlucN-HBc132A and RlucC-HBc132A, were transfected into HEK293 cells. Arbidol with different concentrations were used to treat these cells, and Rluc activity was assayed 48 h posttransfection. As shown in Fig. 4E, Arbidol inhibited the Rluc activity of all the mutant pairs in a dose-dependent manner, indicating that the dimerization of these mutants was also suppressed by Arbidol.
Next, we tested the influence of Arbidol on the capsid formation. The construct 3×Flag-G4S92-HBc was transfected into HepG2 cells, and the cells were treated with Arbidol with different concentrations for 6 days. Intracellular capsid particles were assayed by native agarose gel electrophoresis and immunoblotting. As shown in Fig. 5A, Arbidol inhibited the capsid formation in a dose-dependent manner, with an EC50 of 25 ± 3.6 μM. For dimerization assay, the cells were lysed with NP-40 lysis buffer and loaded directly onto SDS-PAGE gels in the absence of SDS and β-mercaptoethanol. Bands with molecular weights equal to those of dimers or different polymers can be detected from 24 h to 96 h after transfection (Fig. S5). We chose 48 h of treatment posttransfection as the endpoint of the assay. As shown in Fig. 5A, Arbidol dose-dependently reduced the level of core dimers, but the ratios between dimer and monomer were not significantly changed (data not shown). In the meantime, the amount of 3×Flag-G4S92-HBc protein also showed a slight decrease. This decrease cannot be explained by a decrease in protein expression level, since Arbidol did not affect the level of C10-green fluorescent protein (GFP) expressed from a cotransfected plasmid with a similar construction (Fig. S4B). A plausible explanation is that HBc deficient in the formation of dimer or capsid is unstable in cells and prone to degradation.
FIG 5.
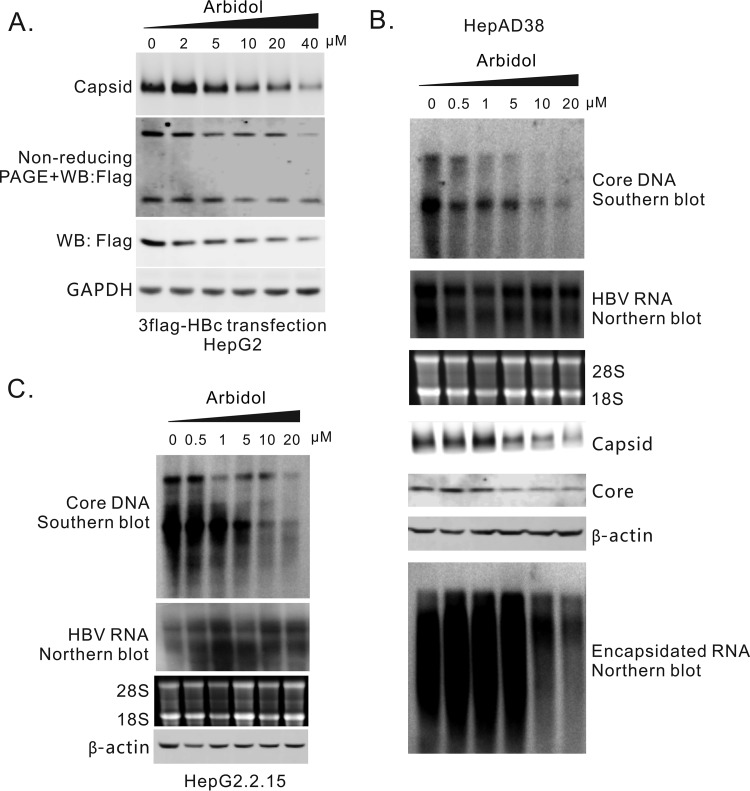
Antiviral effects of Arbidol on HBV replication in cell cultures. (A) Arbidol reduced capsid formation in HepG2 cells transiently transfected with 3×Flag-G4S92-HBc. HepG2 cells transfected with 3×Flag-G4S92-HBc were treated with Arbidol at different concentrations. Intracellular capsids were detected by particle gel assay. Core protein expression was detected by Western blotting, and the core dimer was assayed by nonreducing PAGE. (B) Arbidol suppressed HBV DNA replication in HepAD38 cells by inhibiting capsid formation. Different concentrations of Arbidol were used to treat HepAD38 cells for 6 days. Intracellular core DNA was then extracted and assayed by Southern blotting. Intracellular total RNA and encapsidated RNA were extracted, and HBV RNA was assayed by Northern blotting. Capsid was analyzed by particle gel assay. Core protein levels were detected by Western blotting. (C) Arbidol inhibited HBV DNA replication in HepG2.2.15 cells. Different concentrations of Arbidol were used to treat HepG2.2.15 cells for 6 days. Intracellular core DNA was extracted and detected by Southern blotting. The levels of β-actin in the lysates (the first step of core DNA extraction) were used as controls for cell numbers. Total RNA was extracted, and HBV RNA was assayed by Northern blot.
To test the antiviral activity of Arbidol against HBV replication, intracellular core DNA was assayed in HepAD38 and HepG2.2.15 cells treated with Arbidol. As shown in Fig. 5B and C and Table 1, HBV DNA replication levels were significantly inhibited in both HepAD38 (EC50, 5.3 ± 0.6μM) and HepG2.2.15 cells (EC50, 5.6 ± 0.8μM). The CC50 values were 64.6 ± 3.0 μM and 59.9 ± 4.1 μM, respectively (Table 1). The HBV RNA levels in both cell models were unchanged, indicating that Arbidol did not influence the transcription activity of the TRE-miniCMV promoter or HBV core promoter. Capsid, core protein, and encapsidated pgRNA in HepAD38 cells were suppressed in a dose-dependent manner (Fig. 5B). Together, these results suggested that Arbidol inhibits HBV DNA replication by blocking the formation of capsids.
Characterization of biological effects of the compound candidate 20-deoxyingenol.
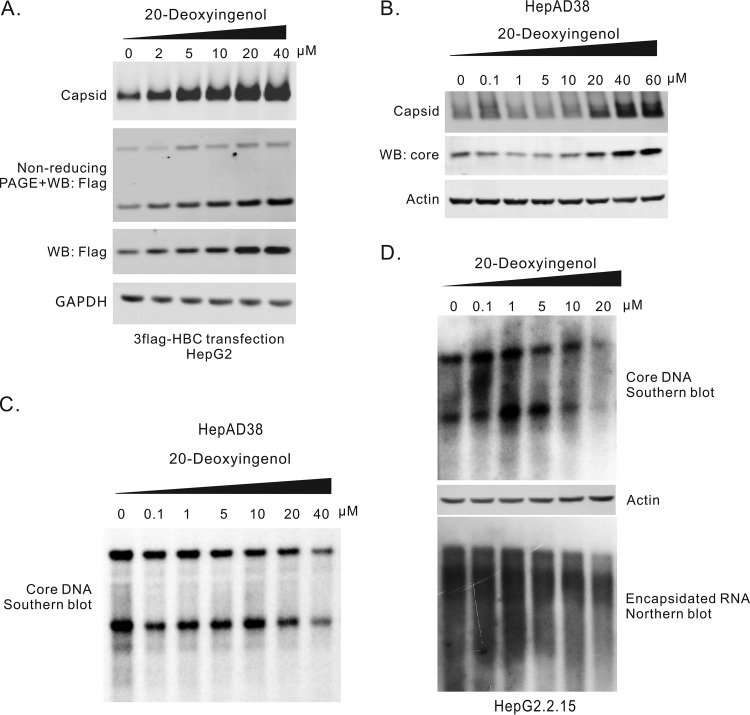
20-Deoxyingenol has an EC50 of 8.9 ± 1.56 μM, and no significant cytotoxicity was observed at 100 μM on SRluc-HBc6 cells (Table 1). It did not alter the Rluc activity in HepG2 cells transfected with RlucN-L-C (Fig. 4D), indicating that 20-deoxyingenol did not affect the transcription activity of the CMV-IE promoter or the interaction between RlucN and RlucC. In the HepG2 cells transfected with 3×Flag-G4S92-HBc, 20-deoxyingenol increased the formation of dimers and capsids in a dose-dependent manner. The amount of 3×Flag-G4S92-HBc protein also increased, likely due to stability enhancement in dimer and/or capsid formation (Fig. 6A), since the expression of the cotransfected C10-GFP did not change (Fig. S4C). In HepAD38 cells, treatment of 20-deoxyingenol also resulted in an increase of both capsid and core protein levels (Fig. 6B), consistent with the results from the transfection system with 3×Flag-G4S92-HBc (Fig. 6A). Interestingly, 20-deoxyingenol slightly suppressed HBV DNA replication in both 20-deoxyingenol HepAD38 and HepG2.2.15 cells, with EC50 values of 31.2 ± 3.1μM (CC50 > 100 μM) and 14.5 ± 1.6 μM (CC50 > 100 μM), respectively (Table 1), suggesting that 20-deoxyingenol-induced enhancement of capsid formation possesses a negative impact on pgRNA encapsidation and DNA replication (Fig. 6D).
FIG 6.
Antiviral effects of 20-deoxyingenol on HBV replication in cell cultures. (A) 20-Deoxyingenol increased capsid formation in HepG2 cells transfected with 3×Flag-G4S92-HBc. HepG2 cells transfected with 3×Flag-G4S92-HBc were treated with 20-deoxyingenol for 6 days. Intracellular capsids were detected by particle gel assay. Core protein expression was detected by Western blotting, and the core dimer was assayed by nonreducing PAGE. (B) 20-Deoxyingenol promoted capsid formation in HepAD38 cells. Different concentrations of 20-deoxyingenol were used to treat HepAD38 cells for 6 days. Capsids and core proteins were detected by particle gel assay and Western blotting, respectively. (C) 20-Deoxyingenol inhibited HBV DNA replication in HepAD38 cells. Intracellular core DNA was extracted from HepAD38 cells treated with 20-deoxyingenol for 6 days and assayed by Southern blotting. (D) 20-Deoxyingenol inhibited HBV DNA replication in HepG2.2.15 cells. Different concentrations of 20-deoxyingenol were used to treat HepG2.2.15 cells for 6 days. Intracellular core DNA was extracted and detected by Southern blotting. The levels of β-actin in the lysates (the first step of core DNA extraction) were used as controls for cell numbers.
DISCUSSION
In this study, we developed a cell-based assay for screening compounds against the HBV capsid formation step. Specifically, this model can, at least partly, predict the interaction between two HBc monomers by utilizing SLC technology. Since the first publication of the principle of SLC by the group of Umezawa et al. in 2001 (41), this technology has been widely used to study protein-protein interactions (for reviews, see references 29 and 30). However, this powerful technology has not been used in HBV research. To our best knowledge, this is the first successful attempt to establish an in-cell model using SLC technology for identifying compounds against the dimerization of HBV core protein during capsid assembly.
We have evaluated 4 luciferase species for their ability to indicate the interaction between the split-luc-HBcs, with two essential criteria, as follows: (i) HBc should not significantly disturb the structure, and in turn the function, of a fused split-luc, and (ii) a split-luc fusion should minimally affect the structure, and in turn the function, of HBc. An ideal split-luc-HBcs system would completely meet these 2 criteria. The function of a split-luc can be indicated by luciferase activity when complemented with the other piece of the split-luc. The function of a split-luc-HBc can be assessed on three levels for convenience of detection. Level 1 is maintaining the interaction between 2 HBc monomers (dimer formation). Level 2 is forming capsid-like particles, and level 3 is supporting the synthesis of HBV DNA in the capsid. As summarized in Table 2, our results showed that the split-Fluc-HBcs maintained both the functions of split Flucs and HBc the best (Fig. 1G), while presenting low signal intensity when complemented. split-Gluc did not work for our purpose. split-Nluc-HBcs showed the best signal intensity when complemented, but neither of the 2 fusion proteins supported HBV DNA replication (Fig. 1G). split-Rluc-HBcs had a relatively balanced performance between the signal intensity and the maintenance of HBc function.
TABLE 2.
Function test of split-luc-HBcs
| Fusion protein | Luciferase function |
Signal intensity |
HBc function |
||
|---|---|---|---|---|---|
| Monomer interaction |
Capsid formation |
DNA replication |
|||
| FlucN-HBc | + | + | + | ND | + (weak) |
| FlucC-HBc | + | + | + | ND | + |
| RlucN-HBc | + | ++ | + | + | − |
| RlucC-HBc | + | ++ | + | + | + |
| GlucN-HBc | − | − | NDa | ND | ND |
| GlucC-HBc | − | − | ND | ND | ND |
| NlucN-HBc | + | +++ | + | ND | − |
| NlucC-HBc | + | +++ | + | ND | − |
ND, not determined.
Theoretically, cotransfection of the 2 split-Rluc-HBcs could generate 3 kinds of dimers, 2 homodimers and 1 heterogeneous dimer. These 2 homodimers produced apparently only ignorable luciferase signals (Fig. 1E). Therefore, the signal detected mainly reflects the formation of heterogeneous dimers, at least for those mutants with the deficiency in capsid formation (Fig. 2E). It is possible that part of the signal might be from the complementation of 2 homodimers aligned adjacently on the same capsid as some of the mutations abolishing capsid formation reduced the signal intensity (Fig. 2E). What is more important is that a considerable part (over 50%) of the signal is produced from the complemented heterogeneous dimers, as estimated from the results of mutants. That means the compound screening would not miss those effects on the complementation of split Rlucs by interfering with the interaction between the 2 heterogeneous monomers.
Making use of the established cell model (SRluc-HBc6), we have identified 2 compounds with anti-HBV activity from 2 libraries. One of the 2 compounds, Arbidol, has been reported to inhibit a broad spectrum of pathogenic viruses, including respiratory viruses, nonenveloped Reoviridae and hepatitis viruses, etc. (for a review, see reference 42). The reported antiviral mechanisms of Arbidol include inhibiting viral entry by binding to both lipid and protein residues and by inhibiting viral replication, assembly, and budding (42). Although Arbidol has been reported to inhibit HBV DNA replication in HepG2.2.15 cells (43, 44), its mechanism of this action has not been elucidated. Our results confirmed that Arbidol inhibited HBV DNA replication in both HepAD38 and HepG2.2.15 cells (Fig. 5). Arbidol reduced the complemented Rluc activities of those mutants deficient in capsid formation (Fig. 4E), suggesting that Arbidol targeted the dimerization step. The ratio of dimers and monomers seemed not to be significantly altered by Arbidol treatment (Fig. 5A and data not shown), suggesting that Arbidol may not prevent the formation of core dimers but may induce an allosteric effect on core proteins. How Arbidol imposes this influence on core dimers warrants further study.
Transfection experiments with 3×Flag-G4S92-HBc showed that Arbidol suppressed formation of the capsid and reduced the protein levels of 3×Flag-G4S92-HBc (Fig. 5A). A similar pattern has been seen in HepAD38 cells treated with Arbidol (Fig. 5B). This raised the question of whether the suppression of capsid formation is a result of the decrease of the 3×Flag-G4S92-HBc levels or vice versa. The evidence arguing for the latter possibility includes the following: (1) Arbidol did not influence the activity of RlucN-L-C, the expression of which was driven by a CMV-IE promoter (Fig. 4C), suggesting that the expression of RlucN-L-C was not affected; (2) Arbidol decreased the levels of 3×Flag-G4S92-HBc, wheras it did not affect the levels of C10-GFP expressed from a similar construct in the cotransfection experiments (Fig. S4B); and (3) the HBV RNA levels in the HepAD38 cells did not decrease upon Arbidol treatment (Fig. 5B), suggesting that HBV RNA transcription was not the compound target. In line with this notion, it has been reported that heteroarylpyrimidines (HAP), a capsid protein allosteric modulator (CpAM), blocks capsid formation and reduces core protein levels, likely due to a reduction of the core protein half-life after blocking capsid assembly (13).
The other identified compound with anti-HBV activity is 20-deoxyingenol, a natural product compound. Interestingly, 20-deoxyingenol significantly increased Rluc activity in SRluc-HBc6 cells (Fig. S2) but did not affect Rluc activity in the HepG2 cells transfected with RlucN-L-C (Fig. 4D). Consistently, the capsid formation in HepG2 cells transfected with 3×Flag-G4S92-HBc and that in HepAD38 cells were promoted by 20-deoxyingenol, together with an increased level of core protein (Fig. 6A and B). As mentioned above, the observed increase of HBc protein levels is supposed to be a consequence of enhanced capsid formation. On the other hand, 20-deoxyingenol treatment marginally decreased the replication of HBV DNA in both HepAD38 and HepG2.2.15 cells (Fig. 6C and D). The differential effect of 20-deoxyingenol on HBV capsid formation and DNA replication indicates that this compound influences the function of capsid through promotion of the capsid formation process. As a mechanism of action proposed by Zlotnick et al., CpAMs enhance but misdirect the kinetics of capsid assembly, preventing the packaging of viral pregenomic RNA and polymerase complex into the capsid (5). However, none of the reported CpAMs has shown enhanced capsid formation in particle gel assays (13, 18, 21, 45–51), and thus 20-deoxyingenol represents a novel reference compound to demonstrate that a CpAM could promote capsid formation but inhibit HBV DNA replication.
In conclusion, we have established a new in-cell model for screening compounds that target HBV core protein dimerization and capsid formation. Two compounds, Arbidol and 20-deoxyingenol, have been identified from the pilot screen and functionally validated, thus providing a proof of principle for the usage of this assay to screen capsid targeting candidates. These two compounds are also worthy of further investigations to dissect in detail their mechanisms of action and to develop efficacious agents helping to achieve a cure for HBV infection.
MATERIALS AND METHODS
Plasmids.
Plasmid pCH-9/3091, which expresses an HBV 1.1× genome governed by CMV-IE promoter (52), was kindly provided by Lin Nan (Southwest Hospital, Chongqing, China). pHBV1.1C− was constructed previously (31) in which the 40th amino acid of the core protein was mutated to a stop codon (GAA to TAA). wtHBc was a plasmid expressing wild-type HBc of genotype D from PCH9/3091. Plasmids FlucN-HBc, FlucC-HBc, RlucN-HBc, RlucC-HBc, GlucN-HBc, GlucC-HBc, NlucN-HBc, and NlucC-HBc were constructed by cloning the 4 pairs of luciferases to the N termini of HBcs, separated by the G4S92 linker. The split sites of different luciferases are shown in Fig. 1A. FlucN-dHBc, FlucC-dHBc, RlucN-dHBc, RlucC-dHBc, GlucN-dHBc, GlucC-dHBc, NlucN-dHBc, and NlucC-dHBc were plasmids obtained by deleting the respective HBc sequences on the above plasmids. 3×Flag-RlucN-HBc and 3×HA-RlucC-HBc were generated by adding a 3×Flag and a 3×HA tag sequence, respectively, to the N termini of RlucN-HBc and RlucC-HBc. 3×Flag-RlucN was constructed by deleting the HBc sequence from 3×Flag-RlucN-HBc. 3×HA-RlucC-GFP was generated by replacing the HBc sequence on 3×HA-RlucC-HBc with the GFP sequence. A plasmid expressing C10-GFP was constructed by fusing the HiBit tag (Promega) at the N terminus of GFP with the G4S47 linker (Fig. S4A). Plasmid NCTPuro was constructed with the following steps. First, the TRE-miniCMV fragment was amplified from plasmid pTRE-HBV1.1 and used to generate plasmids pTRE-RlucN-HBc and pTRE-RlucN-HBc. Second, the 2 fragments, TRE-RlucN-HBc and TRE-RlucN-HBc, were placed onto one plasmid, leading to the plasmid pTRE-RN+RC. Third, the puromycin resistance gene was amplified from pXPR and ligated with the backbone of PCH9/3091, leading to PCH-Puro. Fourth, the tTA gene was amplified from the genomic DNA of HepAD38 cells and ligated with the backbone of PCH9/3091, leading to PCH-tTA. Fifth, the puromycin resistance gene and tTA gene were placed on one plasmid (PCH-tTA-puro). Finally, the tTA-puro fragment was transferred onto pTRE-RN+RC, producing the plasmid NCTPuro (Fig. 3A).
Cell culture and transfection.
HEK293 and HepAD38 cells were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% (vol/vol) fetal calf serum. SRluc-HBc6 cells were maintained in the same way as HEK293 cells, with the addition of 2 μg/ml puromycin or 5 μg/ml tetracycline. For transfection, the cells were seeded into 12-well or 96-well plates and allowed to adhere overnight. On the following day, when the cells were about 80% confluent, the culture medium was replaced with fresh medium, and 1 μg (for 12-well plates) or 0.2 μg (for 96-well plates) of plasmid was transfected into the cells in each well using X-tremeGENE HP DNA transfection reagent (Roche Diagnostics, Mannheim, Germany), according to the instructions provided by the supplier.
Stable cell line establishment.
HEK293 cells were seeded in a 6-cm-diameter dish and grown for 18 h to achieve 80% confluence. Four micrograms of NCTPuro linearized by Not I (Promega, Madison, USA) digestion were transfected into the cells with 12 μl X-tremeGENE HP DNA transfection reagent. The cells were subcultured 48 h posttransfection, and medium containing 2 μg/ml of puromycin was used to screen resistant cells from the next day on. Two to three weeks afterwards, a portion of the surviving cells was seeded in 96-well plates by limiting dilution. The resulting cell clones were expanded and characterized by testing Rluc activity after tetracycline release.
Luciferase activity assay.
Firefly, Renilla, Gaussia, and NanoLuc luciferase activity assays were performed by using commercially available kits, including the Luciferase assay system (Promega, Madison, USA), Renilla luciferase assay system (Promega, Madison, USA), BioLux Gaussia luciferase assay kit (NEB, USA), and Nano-Glo luciferase assay system (Promega, Madison, USA), according to the manufacturers’ instructions. A GloMax-Multi Jr detection system (Promega, Madison, USA) or a Synergy H1 microplate reader (BioTek, USA) was used to detect bioluminescence.
HBV DNA and RNA assay.
(i) Intracellular HBV core DNA extraction. For extraction of intracellular viral core DNA, cells from one well of a 12-well plate were lysed for 10 min at room temperature with 200 μl of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.2% NP-40. Cell debris and nuclei were removed by centrifugation, and the supernatant was mixed with 10 μl of 100 mM CaCl2 and 2 μl micrococcal nuclease (NEB, USA) to digest the contaminating plasmid. After 1 h at 37°C, the reaction was terminated by adding 4 μl of 0.5 M EDTA. Ten microliters of 10% SDS and 10 μl of 10 mg/ml pronase (Roche Diagnostics, Germany) were added to digest (1 h at 37°C) core particles. The digestion mixture was extracted twice with phenol. The DNA was precipitated with ethanol and dissolved in Tris-EDTA (TE buffer) (10 mM Tris–HCl [pH 8.0], 1 mM EDTA).
(ii) Southern blot analysis. For Southern blot analysis, DNA samples were resolved in 1% agarose gels and transferred to positively charged nylon membranes (Roche Diagnostics, Germany). Hybridization and detection were performed with a DIG DNA labeling and detection kit (Roche Diagnostics, Germany), according to the manufacturer’s instructions.
(iii) RNA assay. Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. To extract encapsidated RNA, cells from one well of a 12-well plate were lysed with 250 μl of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1% NP-40. Cell debris and nuclei were removed by centrifugation, and the supernatant was mixed with 10 μl of 100 mM CaCl2 and 2 μl micrococcal nuclease (NEB, USA) to digest free nucleic acids for 30 min. The reaction was terminated by adding 4 μl of 0.5 M EGTA. TRIzol LS (750 μl; Life Technologies) was used to lyse the mixture for 5 min. Chloroform (200 μl) was added and vigorously mixed for 15 s. Then, after centrifugation at 13,000 × g for 15 min, the supernatant was precipitated with 500 μl isopropyl alcohol. The pellet was washed with 75% ethanol and precipitated again. The final pellet was dissolved in nuclease-free water.
For Northern blotting, 5 μg of total RNA of each sample was separated on a 1.2% agarose gel containing 2% formaldehyde and then transferred to a positively charged nylon membrane. HBV RNA was detected by a digoxigenin-labeled RNA probe prepared with a DIG Northern starter kit (Roche Diagnostics, Germany), according to the manufacturer’s instructions.
Particle gel assay.
Capsids and associated viral DNA were analyzed by a native agarose gel electrophoresis-based assay (28), with modification. Briefly, transfected cells cultured in 12-well plates were lysed by the addition of 200 μl of buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.2% NP-40 per well. Cell debris were removed by centrifugation at 5,000 × g for 10 min. Ten microliters of the clarified cell lysates were fractionated by electrophoresis through nondenaturing 1% agarose gels and transferred to a nitrocellulose filter by blotting with TNE buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, and 1 mM EDTA). Capsid particles were detected by probing the membrane with an antibody against GFP, red fluorescent protein (RFP), the Flag tag or the HA tag. Bound antibody was revealed by IRDye-conjugated secondary antibodies (Licor Biosciences, USA) and visualized by an Odyssey CLx system (Licor Biosciences, USA).
To detect capsid-associated HBV DNA, the membranes were treated for 5 min with a denaturing solution containing 0.5 M NaOH and 1.5 M NaCl, followed by neutralization for 5 min with a buffer containing 1 M Tris-HCl and 1.5 M NaCl. The viral DNA was then detected by hybridization.
Nonreducing PAGE and Western blotting.
Transfected or untransfected HEK293 cells were lysed with radioimmunoprecipitation assay (RIPA) buffer. Lysates were clarified by centrifugation, and the supernatants were mixed with Laemmli buffer without both SDS and β-mercaptoethanol. Samples were subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, followed by blotting onto polyvinylidene difluoride (PVDF) filters. Filters were blocked with phosphate-buffered saline with Tween 20 (PBST)-milk and incubated with an antibody. Filters were incubated with IRDye-conjugated secondary antibodies. Blots were revealed by an Odyssey CLx system. For C10-GFP detection, samples transferred onto membranes were directly detected by the Nano-Glo HiBiT blotting system (Promega) according to the manufacturer’s instructions and visualized by a Fusion FX5 system (Vilber Lourmat).
Compound screening.
SRluc-HBc6 cells were seeded into 96-well plates with 200 μl medium in each well in the absence of tetracycline. Six hours after seeding, when the cells reached a confluence of about 90%, 2 μl of 2 mM each compound dissolved in dimethyl sulfoxide (DMSO) was added into each well, and 2 μl of 20% DMSO (final concentration, 0.2% in medium) was used as the control. Each compound treatment was repeated 3 times on 3 different plates. Bay 41-4109 was from Medchem Express. Other compounds, including 304 natural products and the other 368 anti-infectious compounds, were from commercially available libraries (Medchem Express or Selleck, USA). Forty-eight h later, cells were lysed, and the lysates were assayed by using the Renilla luciferase assay system (Promega, Madison, USA) and a Synergy H1 microplate reader (BioTek, USA).
For dose-response effect assay, SRluc-HBc6 cells were seeded into 96-well plates as described above. Cells were treated with each compound with concentrations of 0.2, 0.5, 1, 2, 5, 10, and 20 μM, respectively, and 0.2% DMSO (final concentration) was used as a control. Renilla luciferase activity of each well was assayed as mentioned above.
To test compounds for their capability to inhibit HBV DNA replication, HepAD38 cells were seeded into 12-well plates with medium absent of tetracycline. The next day, compounds were added to the wells to reach final concentrations of 0, 0.1, 1, 5, 10, and 20 μM. The medium was replaced with fresh compounds 3 days later. On the 6th day of treatment, cells were lysed, and intracellular HBV DNA was extracted and assayed by Southern blotting.
Calculation of EC50 and CC50.
Core DNA samples, extracted from HepAD38 or HepG2.2.15 cells treated with different concentrations of agents in 3 independent experiments, were detected by Southern blotting. The volume of Core DNA of each sample was quantified using imageJ software (https://imagej.nih.gov/ij/). The volumes were standardized relative to that of the untreated controls. These relative values were used to generate a best-fit sigmoid dose-inhibition curve using Prism 6.0 software (GraphPad, San Diego, CA), with the drug concentration as the independent variable and the relative HBV DNA amount as the dependent variable. The EC50s were calculated from the concentration (inhibitor) versus inhibition curves. Cytotoxicity of the compounds was determined by using Cell counting kit-8 according to the procedure provided by the manufacturer (MedChemExpress). The cytotoxicity of a compound was expressed by CC50, calculated from concentration versus viability curves.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants 81871635, 81671997, 81471946, and 81661148057 from the National Natural Science Foundation of China, the Major National S&T program grants 2017ZX10202203 and 2017ZX10302201 from Science & Technology Commission of China, grant cstc2015jcyjA10023 from Chongqing Science & Technology Commission, grant CXTDX201601015 from the Program for Innovation Team of Higher Education in Chongqing, grant CSTCCXLJRC201719 from the Leading Talent Program of CQ CSTC, and NIH grants R01AI094474 and R01AI123271.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01302-18.
REFERENCES
- 1.Trepo C, Chan HL, Lok A. 2014. Hepatitis B virus infection. Lancet 384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Perrillo R. 2009. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49:S103–S111. doi: 10.1002/hep.22956. [DOI] [PubMed] [Google Scholar]
- 3.Zoulim F, Lebosse F, Levrero M. 2016. Current treatments for chronic hepatitis B virus infections. Curr Opin Virol 18:109–116. doi: 10.1016/j.coviro.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Hong X, Kim ES, Guo H. 2017. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: implications for epigenetic therapy against chronic hepatitis B. Hepatology 66:2066–2077. doi: 10.1002/hep.29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. 2015. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res 121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingfield PT, Stahl SJ, Williams RW, Steven AC. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 7.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. 1999. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry 38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 8.Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2010. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katen S, Zlotnick A. 2009. The thermodynamics of virus capsid assembly. Methods Enzymol 455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJ. 2011. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat Chem 3:126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 11.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JC, Zlotnick A, Jarrold MF. 2014. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc 136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, Haebich D, Ruebsamen-Waigmann H, Wohlfeil S. 2002. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res 54:69–78. doi: 10.1016/S0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 13.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 14.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. 2005. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A 102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stray SJ, Zlotnick A. 2006. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit 19:542–548. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 16.Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, Espiritu C, Baydo R, Atkins K, Abendroth J, Liao G, Efimov A, Hartman G, Flores OA. 2015. High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci U S A 112:15196–15201. doi: 10.1073/pnas.1513803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaney WEt, Edwards R, Colledge D, Shaw T, Furman P, Painter G, Locarnini S. 2002. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrob Agents Chemother 46:3057–3060. doi: 10.1128/AAC.46.9.3057-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King RW, Ladner SK, Miller TJ, Zaifert K, Perni RB, Conway SC, Otto MJ. 1998. Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (−)beta-L-2',3'-dideoxy-3'-thiacytidine. Antimicrob Agents Chemother 42:3179–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katen SP, Tan Z, Chirapu SR, Finn MG, Zlotnick A. 2013. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure 21:1406–1416. doi: 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourne CR, Finn MG, Zlotnick A. 2006. Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol 80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho MH, Song JS, Kim HJ, Park SG, Jung G. 2013. Structure-based design and biochemical evaluation of sulfanilamide derivatives as hepatitis B virus capsid assembly inhibitors. J Enzyme Inhib Med Chem 28:916–925. doi: 10.3109/14756366.2012.694879. [DOI] [PubMed] [Google Scholar]
- 23.Sells MA, Chen ML, Acs G. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Guo H, Pan XB, Mao R, Yu W, Xu X, Wei L, Chang J, Block TM, Guo JT. 2010. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J Virol 84:9332–9340. doi: 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 81:12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stray SJ, Johnson JM, Kopek BG, Zlotnick A. 2006. An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nat Biotechnol 24:358–362. doi: 10.1038/nbt1187. [DOI] [PubMed] [Google Scholar]
- 28.Ludgate L, Liu K, Luckenbaugh L, Streck N, Eng S, Voitenleitner C, Delaney WEt, Hu J. 2016. Cell-free hepatitis B virus capsid assembly dependent on the core protein C-terminal domain and regulated by phosphorylation. J Virol 90:5830–5844. doi: 10.1128/JVI.00394-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato N, Jones J. 2010. The split luciferase complementation assay. Methods Mol Biol 655:359–376. doi: 10.1007/978-1-60761-765-5_24. [DOI] [PubMed] [Google Scholar]
- 30.Azad T, Tashakor A, Hosseinkhani S. 2014. Split-luciferase complementary assay: applications, recent developments, and future perspectives. Anal Bioanal Chem 406:5541–5560. doi: 10.1007/s00216-014-7980-8. [DOI] [PubMed] [Google Scholar]
- 31.Chen JY, Gan CY, Cai XF, Zhang WL, Long QX, Wei XF, Hu Y, Tang N, Chen J, Guo H, Huang AL, Hu JL. 2018. Fluorescent protein tagged hepatitis B virus capsid protein with long glycine-serine linker that supports nucleocapsid formation. J Virol Methods 255:52–59. doi: 10.1016/j.jviromet.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Remy I, Michnick SW. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 33.Paulmurugan R, Gambhir SS. 2007. Combinatorial library screening for developing an improved split-Firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem 79:2346–2353. doi: 10.1021/ac062053q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulmurugan R, Gambhir SS. 2003. Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal Chem 75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, Wood KV. 2016. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol 11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 36.Bourne CR, Katen SP, Fulz MR, Packianathan C, Zlotnick A. 2009. A mutant hepatitis B virus core protein mimics inhibitors of icosahedral capsid self-assembly. Biochemistry 48:1736–1742. doi: 10.1021/bi801814y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konig S, Beterams G, Nassal M. 1998. Mapping of homologous interaction sites in the hepatitis B virus core protein. J Virol 72:4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander CG, Jurgens MC, Shepherd DA, Freund SM, Ashcroft AE, Ferguson N. 2013. Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc Natl Acad Sci U S A 110:E2782–E2791. doi: 10.1073/pnas.1308846110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Hu T, Zhou X, Wildum S, Garcia-Alcalde F, Xu Z, Wu D, Mao Y, Tian X, Zhou Y, Shen F, Zhang Z, Tang G, Najera I, Yang G, Shen HC, Young JA, Qin N. 2017. Heteroaryldihydropyrimidine (HAP) and sulfamoylbenzamide (SBA) inhibit hepatitis B virus replication by different molecular mechanisms. Sci Rep 7:42374. doi: 10.1038/srep42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hacker HJ, Deres K, Mildenberger M, Schroder CH. 2003. Antivirals interacting with hepatitis B virus core protein and core mutations may misdirect capsid assembly in a similar fashion. Biochem Pharmacol 66:2273–2279. doi: 10.1016/j.bcp.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. 2001. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem 73:2516–2521. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- 42.Blaising J, Polyak SJ, Pecheur EI. 2014. Arbidol as a broad-spectrum antiviral: an update. Antiviral Res 107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai H, Zhao Y, Zhao C, Gong P. 2006. Synthesis and in vitro anti-hepatitis B virus activities of some ethyl 6-bromo-5-hydroxy-1H-indole-3-carboxylates. Bioorg Med Chem 14:911–917. doi: 10.1016/j.bmc.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 44.Pecheur EI, Borisevich V, Halfmann P, Morrey JD, Smee DF, Prichard M, Mire CE, Kawaoka Y, Geisbert TW, Polyak SJ. 2016. The synthetic antiviral drug Arbidol inhibits globally prevalent pathogenic viruses. J Virol 90:3086–3092. doi: 10.1128/JVI.02077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mani N, Cole AG, Phelps JR, Ardzinski A, Cobarrubias KD, Cuconati A, Dorsey BD, Evangelista E, Fan K, Guo F, Guo H, Guo JT, Harasym TO, Kadhim S, Kultgen SG, Lee ACH, Li AHL, Long Q, Majeski SA, Mao R, McClintock KD, Reid SP, Rijnbrand R, Snead NM, Micolochick Steuer HM, Stever K, Tang S, Wang X, Zhao Q, Sofia MJ. 2018. Preclinical profile of AB-423, an inhibitor of hepatitis B virus pregenomic RNA encapsidation. Antimicrob Agents Chemother 62:e00082-18. doi: 10.1128/AAC.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S, Zhao Q, Zhang P, Kulp J, Hu L, Hwang N, Zhang J, Block TM, Xu X, Du Y, Chang J, Guo JT. 2017. Discovery and mechanistic study of benzamide derivatives that modulate hepatitis B virus capsid assembly. J Virol 91:e00519-17. doi: 10.1128/JVI.00519-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 2017. Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother 61:e00560-17. doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam AM, Ren S, Espiritu C, Kelly M, Lau V, Zheng L, Hartman GD, Flores OA, Klumpp K. 2017. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother 61:e00680-17. doi: 10.1128/AAC.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai D, Wang X, Yan R, Mao R, Liu Y, Ji C, Cuconati A, Guo H. 2016. Establishment of an inducible HBV stable cell line that expresses cccDNA-dependent epitope-tagged HBeAg for screening of cccDNA modulators. Antiviral Res 132:26–37. doi: 10.1016/j.antiviral.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res 76:168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Lu D, Liu F, Xing W, Tong X, Wang L, Wang Y, Zeng L, Feng C, Yang L, Zuo J, Hu Y. 2017. Optimization and synthesis of pyridazinone derivatives as novel inhibitors of hepatitis B virus by inducing genome-free capsid formation. ACS Infect Dis 3:199–205. doi: 10.1021/acsinfecdis.6b00159. [DOI] [PubMed] [Google Scholar]
- 52.Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66:4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.