Ceftobiprole is a fifth-generation cephalosporin with activity against methicillin-resistant Staphylococcus aureus (MRSA). One-year surveillance at the Regional Hospital of Ancona (Italy) disclosed a 12% ceftobiprole resistance rate (12/102 isolates; MIC, ≥4 mg/liter).
KEYWORDS: MRSA, cephalosporins, ceftobiprole, penicillin-binding protein, mecA gene
ABSTRACT
Ceftobiprole is a fifth-generation cephalosporin with activity against methicillin-resistant Staphylococcus aureus (MRSA). One-year surveillance at the Regional Hospital of Ancona (Italy) disclosed a 12% ceftobiprole resistance rate (12/102 isolates; MIC, ≥4 mg/liter). Epidemiological characterization demonstrated that the resistant isolates all belonged to different clones. Penicillin-binding protein (PBP) analysis showed substitutions in all PBPs and a novel insertion in PBP2a. The mecB and mecC genes were not detected. Ceftobiprole susceptibility screening is essential to avoid therapeutic failure and the spread of ceftobiprole-resistant strains.
INTRODUCTION
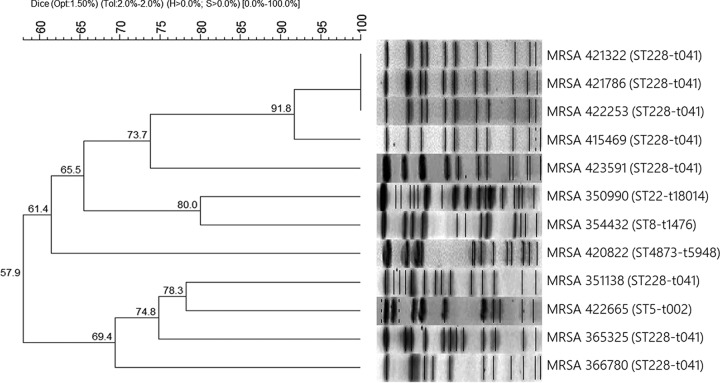
Methicillin-resistant Staphylococcus aureus (MRSA) is a pathogen with a wide diffusion in Europe (1), as well as in Italy, accounting for about 30% of all invasive S. aureus strains described in Italy to date (2). Resistance to β-lactamases is often due to the mecA gene, which encodes the low-affinity penicillin-binding protein 2a (PBP2a). Ceftobiprole, a fifth-generation broad-spectrum cephalosporin, shows activity against Gram-positive and Gram-negative bacteria and is also active against MRSA (3); in particular, it has demonstrated high affinity not only for the common PBPs but also for PBP2a (4). The antibiotic which has recently been approved has been shown to display relative stability against β-lactamases and a low propensity to develop resistance (3), as confirmed by the low rates of resistant staphylococcal isolates found in surveillance studies (5–8). Nonetheless, some papers have described ceftobiprole resistance among MRSA strains (5, 6, 9–11). This resistance is probably due to mutations in pbp genes, especially mecA or pbp4 (9–12). During a recent MRSA survey, we decided to test ceftobiprole against 102 strains isolated from February 2017 to February 2018 from a variety of specimens collected at Ospedali Riuniti in Ancona, Italy. MIC determination by broth microdilution (13) showed that 88% of MRSA strains (n = 90) were susceptible to ceftobiprole, with MICs ranging from 0.03 to 2 mg/liter. The resistance rate was 12% (12 isolates; MIC, ≥4 mg/liter), which is considerably higher than the rates (1.7 to 3.5%) detected in surveillance studies in Europe (5–6). Similar resistance rates (15%) have been found only in an African surveillance study (11). The 12 ceftobiprole-resistant MRSA strains were recovered from wounds (n = 4) and pulmonary secretions (n = 8) of patients admitted to different departments, except for 3 strains, which were collected from general medicine patients. The MIC50 and MIC90 of the 102 isolates were 1 mg/liter and 4 mg/liter, respectively. The resistant isolates were further characterized for pbp gene mutations and to determine the epidemiological relationships among them (Table 1). According to spa typing (14) and multilocus sequence typing (MLST) (15), 8 isolates shared the same spa type (t041) and MLST (sequence type 228 [ST228], clonal complex 5 [CC5]) and were classified as “South German” or “Italian” clones, i.e., nosocomial strains that are widespread in central and southern Europe (16). SmaI–pulsed-field gel electrophoresis (SmaI-PFGE) analysis (17) demonstrated that the 8 strains showed 4 different PFGE patterns (B, D, E, and F), with three identical strains (pattern F2) and two closely related isolates (patterns F1 and F3) belonging to pulsotype F (Fig. 1). These correlations suggested the possibility of an intrahospital outbreak of a ceftobiprole-resistant clone. The remaining 4 isolates showed different PFGE pulsotypes (A, C, G, and H, respectively) and molecular types, as follows: MRSA 350990 was assigned to a novel spa type (t18014) and to ST22 (CC22), MRSA 354432 was assigned to spa type t1476 and ST8 (CC8), MRSA 420822 was assigned to spa type t5948 and the new ST4873 (CC59), and MRSA 422665 was assigned to spa type t002 and ST5 (CC5) (Table 1). CC22, CC8, and CC5 are associated with nosocomial infections (18–19), whereas CC59 is associated with community-acquired infections (20). PBP sequences of the resistant isolates were obtained (21–22) and compared with the susceptible strain S. aureus Mu50. The PBP mutations explained the epidemiological context in the following ways. First, spa type t041 and ST228 strains harbored the N146K mutation in PBP2a. This mutation, which has been reported in association with two other substitutions, as N146K-N204K-G246E (11), is found in the non-penicillin-binding domain (non-PBD), which mediates resistance through interactions with other proteins (23). Three of these 4 strains, MRSA 351138, 365325, and 366780, also bore the C197Y mutation in PBP2. Mutations in PBP1 (S194N) and PBP4 (N337D) were also detected in MRSA 365325. Second, MRSA 350990 (spa type t18014, ST22) exhibited mutations in all PBPs, with S225R in PBP2a; S629T in PBP1; C197Y, L256V, P285A, and T439V in PBP2; R504K and K584F in PBP3; and D98E, S189T, and E398A in PBP4. Despite the numerous mutations, the ceftobiprole MIC was only slightly above the breakpoint, suggesting that they do not all affect resistance. Third, MRSA 354432 (spa type t1476, ST8) lacked the mecA, mecB, and mecC genes and showed mutations in all the PBPs tested (D118N in PBP1, C197Y in PBP2, P233L and S438T in PBP3, and S189T in PBP4), which are probably responsible for cefoxitin resistance; in contrast, ceftobiprole resistance may be due to substitutions in PBP4, which seems to play a key role in ceftobiprole-resistant strains lacking PBP2a (24). Fourth, MRSA 420822 (spa type t5948, ST4873) harbored a wild-type mecA gene and carried several mutations in the other pbp genes: 3 mutations in PBP1 (A329V, E499D, and G515S), 4 mutations in PBP2 (C197Y, P285A, Q358H, and T439V), a mutation in PBP3 (A330S), and 2 mutations in PBP4 (S189T and V210I). Notably, this community-associated strain had an elevated MIC (32 mg/liter), which to the best of our knowledge is the highest ceftobiprole MIC reported so far in clinical strains, despite its wild-type PBP2a. Moreover, it showed the highest number of PBP mutations, which may be responsible for the high MIC, even though the involvement of other resistance mechanisms cannot be excluded (15). Fifth, MRSA 422665 (spa type t002, ST5) showed a 5-amino-acid insertion (VQHED) in the non-PBD at 259 to 260 in PBP2a, as well as a mutation in PBP2 (V607M). The insertion has the potential to affect interactions with other proteins (23), inducing ceftobiprole resistance, but it does not affect β-lactamase resistance. This is the first report of an amino acid insertion in PBP2a in a ceftobiprole-resistant strain. None of the strains carried the mecB or mecC gene. The present surveillance study, although limited to isolates recovered from a single center, showed a high ceftobiprole resistance rate and PBP mutations that were not confined to amino acid substitutions. Notably, since ceftobiprole became available at Ospedali Riuniti only in early 2017, selective pressure can be excluded. This suggests that mutations conferring ceftobiprole resistance can be induced not only by selective pressure but also arise independently. The present findings highlight the need to perform ceftobiprole screening before treatment to avoid therapeutic failure and the spread of resistant strains.
TABLE 1.
Characteristics of the ceftobiprole-resistant MRSA strains analyzed in this study
| MRSA strain |
Isolation date |
Departmenta | Infection typeb |
CFB MIC (mg/liter)c |
Mutation(s) by PBPd |
Molecular typinge |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP2a | PBP1 | PBP2 | PBP3 | PBP4 | Pulsotype | spa type | MLST (CC) | |||||
| 350990 | Feb 2017 | Spinal surgery | HAI | 4 | S225R | S629T | C197Y, L256V, P285A, T439V |
R504K, K584F |
D98E, S189T, E398A |
A | t18014 | ST22 (CC22) |
| 351138 | Feb 2017 | Dermatology | HAI | 4 | N146K | WT | C197Y | WT | WT | B | t041 | ST228 (CC5) |
| 354432 | Feb 2017 | Infectious diseases | HAI | 4 | ND | D118N | C197Y | P233L, S438T |
S189T | C | t1476 | ST8 (CC8) |
| 365325 | Apr 2017 | General medicine | HAI | 8 | N146K | S194N | C197Y | WT | N337D | D | t041 | ST228 (CC5) |
| 366780 | Apr 2017 | Neurosurgery | HAI | 4 | N146K | WT | C197Y | WT | WT | E | t041 | ST228 (CC5) |
| 415469 | Dec 2017 | Subintensive medicine | HAI | 4 | N146K | WT | WT | WT | WT | F1 | t041 | ST228 (CC5) |
| 420822 | Jan 2018 | Rehabilitation | CAI | 32 | WT | A329V, E499D, G515S |
C197Y, P285A, Q358H, T439V |
A330S | S189T, V210I | G | t5948 | ST4873 (CC59) |
| 421322 | Jan 2018 | ICU | HAI | 4 | N146K | WT | WT | WT | WT | F2 | t041 | ST228 (CC5) |
| 421786 | Jan 2018 | Otolaryngology | HAI | 4 | N146K | WT | WT | WT | WT | F2 | t041 | ST228 (CC5) |
| 422253 | Jan 2018 | Subintensive medicine | HAI | 4 | N146K | WT | WT | WT | WT | F2 | t041 | ST228 (CC5) |
| 422665 | Jan 2018 | LTC | HAI | 4 | INS 259 VQHED 260 | WT | V607M | WT | WT | H | t002 | ST5 (CC5) |
| 423591 | Jan 2018 | Subintensive medicine | HAI | 8 | N146K | WT | WT | WT | WT | F3 | t041 | ST228 (CC5) |
ICU, intensive care unit; LTC, long-term care.
HAI, hospital-associated infection; CAI, community-associated infection.
CFB, ceftobiprole.
WT, wild type; ND, not detected; INS, insertion.
MLST, multilocus sequence type; CC, clonal complex.
FIG 1.
SmaI-PFGE pattern and dendrogram of the ceftobiprole-resistant MRSA strains. Profiles were analyzed with BioNumerics software version 7.0 (Applied Maths Scientific Software Development, Sint-Martens-Latem, Belgium). The dendrogram was built by applying the Dice similarity coefficient, with 1.5% optimization and 2.0% tolerance. Clustering was obtained using the unweighted pair group method with arithmetic mean. Opt, optimization; Tol, tolerance; H, minimum height; S, minimum surface.
Data availability.
The nucleotide sequences of mutated PBPs were deposited in GenBank under accession numbers MH798847 to MH798870.
ACKNOWLEDGMENTS
We are grateful to Daniela Bencardino and Elisa Ponzio for their technical assistance.
This work was financially supported by Progetto Strategico di Ateneo 2016–Università Politecnica delle Marche, “Study of new compounds and innovative strategies to control complicated bacterial skin infections.”
REFERENCES
- 1.Köck R, Becker K, Cookson B, Van Gemert-Pijnen JE, Harbarth S, Kluytmans J, Mielke M, Peters G, Skov RL, Struelens MJ, Tacconelli E, Navarro Torné A, Witte W, Friedrich AW. 2010. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15:pii=19688. [DOI] [PubMed] [Google Scholar]
- 2.European Centre for Disease Prevention and Control. 2017. Surveillance of antimicrobial resistance in Europe. 2016. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control, Solna, Sweden: https://ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net. [Google Scholar]
- 3.Zhanel GG, Lam A, Schweizer F, Thomson K, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Noreddin AM, Karlowsky JA. 2008. Ceftobiprole: a review of a broad-spectrum and anti-MRSA cephalosporin. Am J Clin Dermatol 9:245–254. doi: 10.2165/00128071-200809040-00004. [DOI] [PubMed] [Google Scholar]
- 4.Davies TA, Page MGP, Shang W, Andrew T, Kania M, Bush K. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob Agents Chemother 51:2621–2624. doi: 10.1128/AAC.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrell DJ, Flamm RK, Sader HS, Jones RN. 2014. Ceftobiprole activity against over 60,000 clinical bacterial pathogens isolated in Europe, Turkey, and Israel from 2005 to 2010. Antimicrob Agents Chemother 58:3882–3888. doi: 10.1128/AAC.02465-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Flamm RK, Duncan LR, Streit JM, Castanheira M, Sader HS. 2018. Antimicrobial activity of ceftobiprole and comparator agents when tested against contemporary Gram-positive and -negative organisms collected from Europe (2015). Diagn Microbiol Infect Dis 91:77–84. doi: 10.1016/j.diagmicrobio.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 7.Hischebeth GTR, Gravius S, Molitor E, Kohlhof H, Hoerauf A, Hilgers C, Randau TM. 2018. Activity of ceftobiprole against Staphylococcus spec. isolates derived from foreign body associated infections. Diagn Microbiol Infect Dis 91:175–178. doi: 10.1016/j.diagmicrobio.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Henriksen AS, Smart J, Hamed K. 2018. Comparative activity of ceftobiprole against coagulase-negative staphylococci from the BSAC Bacteraemia Surveillance Programme, 2013–2015. Eur J Clin Microbiol Infect Dis 37:1653–1659. doi: 10.1007/s10096-018-3295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan LC, Basuino L, Diep B, Hamilton S, Chatterjee SS, Chambers HF. 2015. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 59:2960–2963. doi: 10.1128/AAC.05004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greninger AL, Chatterjee SS, Chan LC, Hamilton SM, Chambers HF, Chiu CY. 2016. Whole-genome sequencing of methicillin-resistant Staphylococcus aureus resistant to fifth-generation cephalosporins reveals potential non-mecA mechanisms of resistance. PLoS One 11:e0149541. doi: 10.1371/journal.pone.0149541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaumburg F, Peters G, Alabi A, Becker K, Idelevich EA. 2016. Missense mutations of PBP2a are associated with reduced susceptibility to ceftaroline and ceftobiprole in African MRSA. J Antimicrob Chemother 71:41–44. doi: 10.1093/jac/dkv325. [DOI] [PubMed] [Google Scholar]
- 12.Fishowitz J, Hermoso JA, Chang M, Mobashery S. 2014. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66:572–577. doi: 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Committee on Antimicrobial Susceptibility Testing. 2018. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0. European Committee on Antimicrobial Susceptibility Testing, Vaxjo, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf. [Google Scholar]
- 14.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol 37:3556–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mato R, Campanile F, Stefani S, Crisostomo MI, Santagati M, Sanches SI, de LH. 2004. Clonal types and multidrug resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) recovered in Italy during the 1990s. Microb Drug Resist 10:106–113. doi: 10.1089/1076629041310109. [DOI] [PubMed] [Google Scholar]
- 17.Brenciani A, Morroni G, Pollini S, Tiberi E, Mingoia M, Varaldo PE, Rossolini GM, Giovanetti E. 2016. Characterization of novel conjugative multiresistance plasmids carrying cfr from linezolid-resistant Staphylococcus epidermidis clinical isolates from Italy. J Antimicrob Chemother 71:307–313. doi: 10.1093/jac/dkv341. [DOI] [PubMed] [Google Scholar]
- 18.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tristan A, Ferry T, Durand G, Dauwalder O, Bes M, Lina G, Vandenesch F, Etienne J. 2007. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect 65:105–109. doi: 10.1016/S0195-6701(07)60025-5. [DOI] [PubMed] [Google Scholar]
- 20.Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. 2005. Successful multiresistance community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type V or SCCmec type IV. J Clin Microbiol 43:4719–4730. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argudín AM, Dodémont M, Taguemount M, Roisin S, de Mendonça R, Deplano A, Nonhoff C, Denis O. 2017. In vitro activity of ceftaroline against clinical Staphylococcus aureus isolates collected during a national survey conducted in Belgian hospitals. J Antimicrob Chemother 72:56–59. doi: 10.1093/jac/dkw380. [DOI] [PubMed] [Google Scholar]
- 22.Becker K, van Alen S, Idelevich EA, Schleimer N, Seggewiss J, Mellman A, Kaspar U, Peters G. 2018. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242–248. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 52:2089–2096. doi: 10.1128/AAC.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequences of mutated PBPs were deposited in GenBank under accession numbers MH798847 to MH798870.