Methicillin-resistant Staphylococcus aureus (MRSA) infection has increased in recent years among cystic fibrosis (CF) patients. Linezolid (LZD) is one of the antistaphylococcal antibiotics widely used in this context.
KEYWORDS: 23S rRNA, MRSA, Staphylococcus aureus, cystic fibrosis, linezolid, oxazolidinones, ribosomal resistance, WGS
ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) infection has increased in recent years among cystic fibrosis (CF) patients. Linezolid (LZD) is one of the antistaphylococcal antibiotics widely used in this context. Although LZD resistance is rare, it has been described as often associated with long-term treatments. Thirteen MRSA strains isolated over 5 years from one CF patient were studied for LZD resistance emergence and subjected to whole-genome sequencing (WGS). Resistance emerged after three 15-day LZD therapeutic regimens over 4 months. It was associated with the mutation of G to T at position 2576 (G2576T) in all 5 rrl copies, along with a very high MIC (>256 mg/liter) and a strong increase in the generation time. Resistant strains isolated during the ensuing LZD therapeutic regimens and until 13 months after LZD stopped harbored only 3 or 4 mutated rrl copies, associated with lower MICs (8 to 32 mg/liter) and low to moderate generation time increases. Despite these differences, whole-genome sequencing allowed us to determine that all isolates, including the susceptible one isolated before LZD treatment, belonged to the same lineage. In conclusion, LZD resistance can emerge rapidly in CF patients and persist without linezolid selective pressure in colonizing MRSA strains belonging to the same lineage.
INTRODUCTION
With 70.6% of patients colonized/infected and a median age at the first infection of 3.6 years, Staphylococcus aureus is one of the major and earliest bacteria detected in infants and children with cystic fibrosis (CF) (1). Despite antibiotic intervention, S. aureus colonization or chronic infection persists in the lung for many years. Longitudinal studies have shown that in the majority of cases, patients are chronically colonized/infected with the same clone (2–4). With age, a decreasing incidence of S. aureus has been shown to coincide with an increased incidence of Pseudomonas aeruginosa colonization in CF patients (1). However, S. aureus is still present in 50% of patients with P. aeruginosa infection (5). CF lung colonization with methicillin-resistant S. aureus (MRSA) has increased in recent years, with MRSA being detected in 26% of the CF patients in the United States in 2015 (1). Several studies have reported that MRSA infection is associated with an increased rate of lung function decline and worse clinical outcomes (6, 7). The airways of CF patients provide a niche for bacteria in a hostile environment where the challenges include host immune response, antibiotics, interspecies competition, hypoxia, and starvation, which trigger various forms of S. aureus adaptations (8). These adaptations include the emergence of small-colony variants (SCVs), hypermutator phenotypes, multidrug resistance, and increased ability to form biofilm (4, 9, 10).
Linezolid (LZD) is widely used in CF patients for the treatment of MRSA infections (11). It is the first member of the oxazolidinone class of antibiotics, which has been available in France since 2002. LZD disrupts the beginning of the protein synthesis by binding to domain V of the 23S rRNA in the 50S subunit of the bacterial ribosome, specifically in the peptidyl transferase center (PTC) at site A (12). Clinical LZD-resistant S. aureus (LRSA) is still rarely isolated globally (<1%) (13). However, resistance in S. aureus emerged in CF patients several years ago, often associated with long-term treatments. The mutations implicated are mostly alterations in the 23S rRNA, especially the mutation G2576U (Escherichia coli numbering) (14). Alterations in ribosomal proteins L3 and L4, as well as the cfr gene, were also reported (15–20). Suboptimal dosing has been demonstrated to play an important role in resistance emergence due to the variation of the pharmacokinetic profile of LZD among children with CF, which requires dosing adjustment. LZD resistance has also emerged in patients without LZD exposure, which is probably due to transmission of LRSA between CF patients (20).
Our objective was to explore the dynamic of LZD resistance in MRSA isolates recovered over a long period of time from one CF patient and, in particular, to describe the isolates’ genetic evolution and relatedness.
RESULTS
LZD treatment, MRSA isolates, and emergence of LZD resistance.
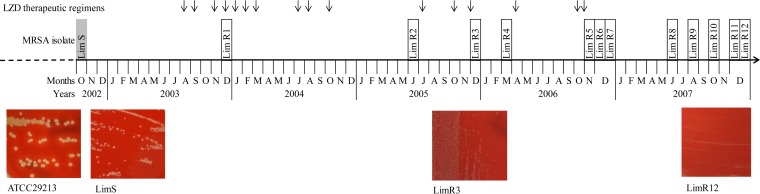
One CF patient was followed up at the CF unit of Limoges teaching hospital. Chronic MRSA colonization was diagnosed at 3 years of age. Thirteen frozen MRSA strains (LimS, which was LZD susceptible, and LimR1 to R12, which were LZD resistant) isolated from sputum samples over a 5-year period (October 2002 to December 2007) were included. The timeline of LZD treatments and the concomitant MRSA isolates are presented in Fig. 1. An initial therapeutic regimen of LZD (600 mg twice daily) was introduced in August 2003 over a 15-day period. After this one, successive therapeutic regimens of LZD (600 mg twice daily) were used alternately with intravenous glycopeptides until November 2006. Thirteen MRSA strains isolated either before (LimS), during (LimR1 to -R5), or after LZD treatment (LimR6 to -R12) were studied (Fig. 1). LZD resistance was detected in December 2003 after three 15-day LZD therapeutic regimens over 4 months (LimR1). Resistance was determined to be associated with the mutation of G to T at position 2576 (G2576T) in the 23S rRNA gene (rrl) by Sanger sequencing. All isolates harbored 5 rRNA operons (rrn), and the mutation was observed in 3 to 5 copies of rrl. Moreover, rplC and rplD genes were not mutated and the cfr gene was absent as determined by PCR amplification. LZD resistance levels were different during persistence, with MICs between 8 and >256 mg/liter. Those MIC variations were correlated with the number of mutated copies of the rrl gene (Table 1; Table S1 in the supplemental material). The LZD resistance was still unchanged 1 year after the last LZD therapeutic regimen, with MICs of 8 to 32 mg/liter, and was associated with mutations in 4 rrl copies.
FIG 1.
Timeline of MRSA strain isolation over the 5-year period, along with the dates of the 15-day periods of the 600-mg twice-daily (b.i.d.) LZD therapeutic regimen (black arrows). A total of 13 MRSA isolates were included in the study. The name of the MRSA isolate susceptible to LZD is highlighted in gray, and those resistant to LZD are framed in black. Images of isolates cultured on Colombia blood agar plates are presented for 3 isolates, with S. aureus reference strain ATCC 29213 shown for comparison.
TABLE 1.
Characterization of LZD resistance mechanisms and correlation with generation times
| Isolate | LZD MIC (mg/liter)a |
LZD resistance mechanismb |
Minimum generation time (mean ± SD) |
|||
|---|---|---|---|---|---|---|
| 23S rRNA gene (no. of mutated rrl alleles/total no. of rrl alleles) |
L3 protein (rplC gene) |
L4 protein (rplD gene) |
cfr gene | |||
| LimS | 0.75 | WT | WT | WT | — | 49.9 ± 0.7 |
| LimR1 | >256 | G2576T (5/5) | WT | WT | — | 260.7 ± 38.5c |
| LimR3 | 8 | G2576T (3/5) | WT | WT | — | 54.9 ± 1.9c |
| LimR6 | 8 | G2576T (4/5) | WT | WT | — | 158.0 ± 11.4 |
| LimR8 | 32 | G2576T (4/5) | WT | WT | — | 158.9 ± 16.4 |
| LimR9 | 24 | G2576T (4/5) | WT | V142A | — | 65.5 ± 4.2c |
| LimR12 | 16 | G2576T (4/5) | WT | WT | — | 67.8 ± 4.8c |
LZD resistance is defined by a MIC of >4 mg/liter by EUCAST and CLSI for S. aureus.
Determined with WGS data. WT, wild type; —, absent.
P ≤ 0.05, Student’s test in comparison with the results for the first isolate LimS.
Multilocus sequence type (MLST) and spa type analysis classified 10 isolates as sequence type 72 (ST72) and spa type t148 (Table S1). No classification could be obtained for the remaining 3 isolates (LimR6, -R8, and -R11) due to point mutations leading to changes in alleles for MLST or in repeats for spa type. Consequently, new ST and spa types were submitted in the corresponding databases (Table S1). Nevertheless, LimR6, which belongs to the new ST4898, also belongs to spa type t148, and conversely, LimR8 and -R11, with new spa types, belong to ST72. Accordingly, these data suggested that all isolates belong to the same lineage.
Whole-genome comparison.
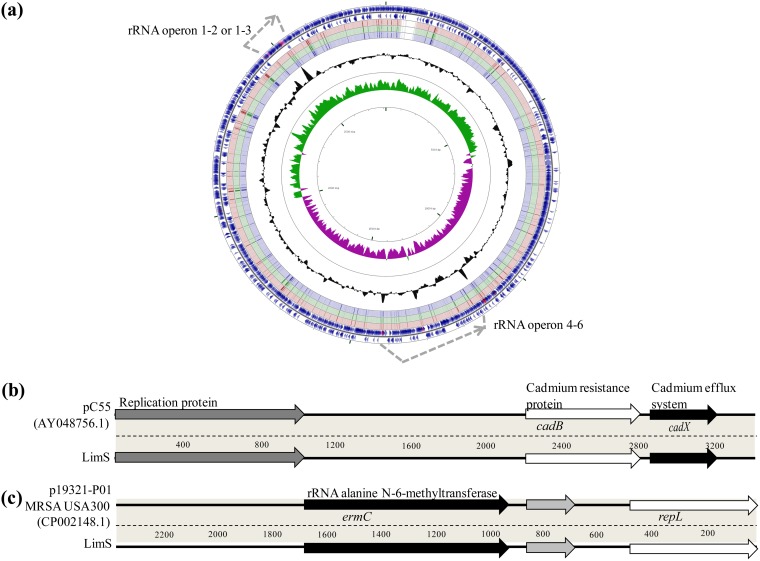
Among the 13 isolates, 7 (LimS, LimR1, LimR3, LimR6, LimR8, LimR9, and LimR12) were selected according to LZD resistance and isolation timeline for whole-genome sequencing (WGS). All had a genomic content compatible with S. aureus species with draft genomes size of 2.74 Mb and a mean GC content of 32.72%. Genomic content comparison of LimS, LimR1, and LimR12 with the FORC_012 MRSA strain (the closest ST72 strain in the database) is represented in Fig. 2a. The virulomes of all isolates were identical, similar to those of other ST72 strains (FORC_012 and 2148) with the absence of the Panton-Valentine leukocidin (PVL) and TSST-1-coding genes (Table S2). All Lim isolates contained a small plasmid of 3,332 bp, similar to pC55 (GenBank accession number AY048756.1), carrying 2 genes responsible for cadmium resistance (cadB and cadX, coding, respectively, a cadmium resistance protein and an efflux system) (Fig. 2b). Another plasmid (2,473 pb), identical to p19321-P01 (GenBank accession number CP002148.1) and coding for the ermC gene, was detected in all isolates except LimR6 (Fig. 2c).
FIG 2.
Genome features of the chromosome (a) and the 2 plasmids (b and c) identified in Lim isolates. (a) Comparison by BLAST of the coding sequences (CDS) of the FORC_012 (blue arrows of the 2 outer rings) and LimS (red), LimR1 (green), and LimR12 (purple) genomes (corresponding to the 3rd, 4th, and 5th rings, respectively, from outside to inside) (CGview server [56]). GC content is represented on the plot inside the ring. The major difference between the FORC_012 genome and those of Lim isolates is the number of rRNA operons (gray dotted-line arrows), with 6 rrl genes for FORC_012 conversely to 5 for Lim isolates, corresponding to the loss of the 3rd operon. The other difference is the presence of a supplementary prophage in the FORC_012 genome. (b and c) Genes are labeled on the coding sequences for the 2 small cryptic plasmids of isolate LimS.
Antimicrobial resistance pattern and resistome.
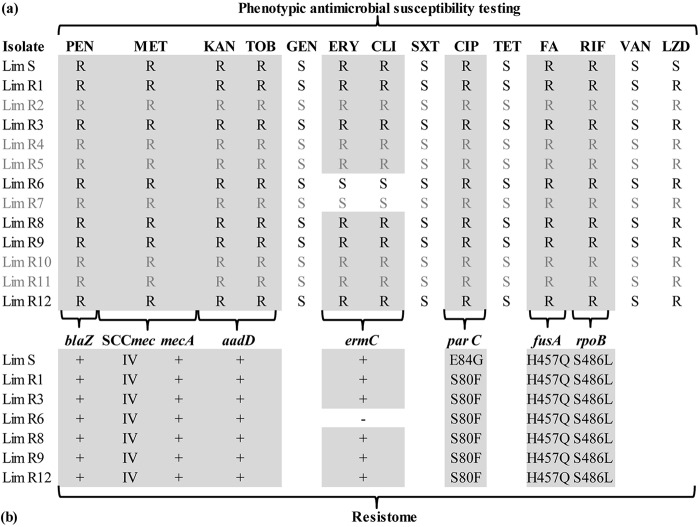
All Lim isolates shared the same antimicrobial resistance pattern and were resistant to methicillin, kanamycin, tobramycin, ciprofloxacin, rifampin, and fusidic acid. Almost all were also resistant to erythromycin and clindamycin (except LimR6 and LimR7) (Fig. 3a). Comparison of genomic resistance markers, obtained by WGS, correlated well with the antimicrobial susceptibility profiles and showed similar resistance genes or point mutations (Fig. 3b). Resistance to methicillin was conferred by a mecA gene in a staphylococcal cassette chromosome mec element (SCCmec) of type IV. The joining region J3 of the SCCmec element additionally encodes aminoglycosides (aaD gene) and bleomycin resistance on a pUB110 region. A constitutive macrolide-lincosamide-streptogramin B (MLSB) phenotype was present in all but 2 strains (LimR6 and -R7) due to the ermC gene on the p19321-P01-like plasmid. LimR6 susceptibility to erythromycin and clindamycin correlated with the loss of this plasmid. Resistance to fluoroquinolones, associated with a mean MIC of 2 mg/liter for ciprofloxacin, was linked to 2 different point mutations (E84G and S80F) previously reported in parC (21, 22). High-level resistance (MIC > 32mg/liter) was observed for rifampin associated with the point mutation S486L in rpoB (23). Finally, the H457Q mutation in fusA was responsible for low-level resistance to fusidic acid, as already described (24).
FIG 3.
Comparison of phenotypic antimicrobial susceptibility patterns and resistomes inferred from WGS data through the bioMérieux EpiSeq analysis pipeline. (a) Antimicrobial susceptibilities of the 13 isolates included in the study. Isolate names and susceptibility results (R, resistance; S, susceptibility) are displayed in black for sequenced isolates and gray for nonsequenced isolates. (b) Resistome associated with each antimicrobial drug shown in panel a (except for LZD, which is detailed in Table 1) for the 7 sequenced isolates. At the top are the antibiotic resistance genes. +, present; −, absent. The enzyme encoded by the aaD gene is ANT(4′)-Ia. Antibiotic resistance associated with single point mutations in the drug target is indicated by the observed amino acid change conferring resistance. PEN, penicillin; MET, methicillin; TOB, tobramycin; KAN, kanamycin; GEN, gentamicin; ERY, erythromycin; CLI, clindamycin; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; TET, tetracycline; FA, fusidic acid; RIF, rifampin; VAN, vancomycin; LZD, linezolid.
Phylogenetic relatedness and SNP analysis.
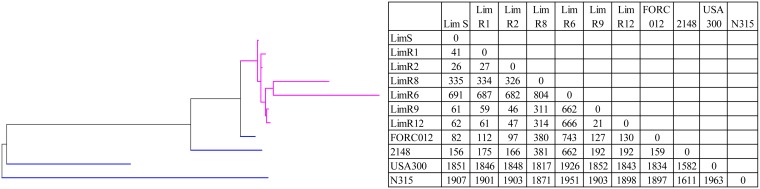
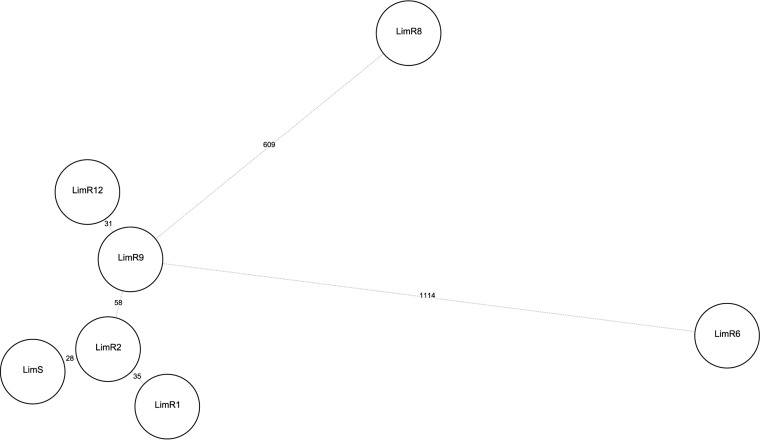
Phylogenetic relatedness was first evaluated with whole-genome MLST (wgMLST) analysis on 3,897 loci (core and accessory genes). It confirms a clonal relationship between the isolates (Fig. 4), with pairwise distances (MLST loci of wgMLST) of 21 to 46 different alleles between LimS, -R1, -R3, -R9, and -R12. ST72 isolates FORC_012 and 2148 were more distant, with 82 and 156 different loci, respectively, compared to those of LimS. On the other hand, LimR6 and -R8 were genetically divergent, with 662 and 311 different alleles, respectively, compared with LimR9 (the closest Lim isolate). To further assess phylogenetic links, whole-genome single-nucleotide polymorphism (wgSNP) analysis was performed and indicated that the isolates had a common ancestor. Comparison with FORC_012 indicated a total of 91 common strain-specific SNPs among the 7 isolates, indicating clonal diversification. Comparison with LimS (the first isolate) showed that pairwise distances varied between 28 and 58 SNPs over 5 years. Moreover, 21 and 36 SNPs were shared among the first 6 LZD-resistant isolates (LimR1, -R3, -R6, -R8, -R9, and -R12) and the last 4 isolates (LimR6, -R8, -R9, and -R12), respectively. Pairwise distances were not identical throughout the 5 years, and the mutation rate was more elevated between LimS and LimR1, as well as between LimR9 and -R12. LimR6 and -R8 showed more unique SNPs than the remaining 5 isolates (Fig. 5). A hypermutator phenotype was suspected. A premature termination of the mutL gene, part of the pathway for DNA proofreading mismatch repair, was observed in the last 4 isolates (LimR6, -R8, -R9, and -R12). This was linked to a 4-bp deletion which resulted in the frameshift Tyr501stop. Additionally, in isolates LimR1 and LimR8, a stop codon appeared in the recQ gene, involved in DNA replication and repair, following a 1-bp deletion.
FIG 4.
Results of phylogenetic analysis of the 7 MRSA isolates analyzed by wgMLST. A maximum parsimony tree (Bionumerics) with default parameters was used for the analysis of all loci (wgMLST and MLST alleles). The root position in the tree was assigned to the deepest branch measured by average branch length. S. aureus strains FORC_012 (ST72, t664), 2148 (ST72), N315 (ST5), and USA300-FPR3757 (ST8) were used for reference genomes (blue). Lim isolate branches are depicted in purple. Numbers in the body of the table represent the pairwise wgMLST allelic differences between isolates.
FIG 5.
Evolution of the pairwise distances (SNPs) in comparison with the LZD-susceptible Lim isolate, LimS, based on SNP analysis. A minimum spanning tree (BioNumerics version 7.6) with default parameters (priority rule1, maximum number of N locus variants [N = 1]; weight, 10,000) was used. Numbers on the tree branches indicate SNP distances between genomes (circles). Genome codes refer to Lim strain numbers, which are chronologically ordered as shown in Fig. 1.
Mutations that could be associated with LZD resistance, linked to ribosomal modification, were also sought in LimR isolates compared to LimS. No potentially compensatory mutation was observed in 23S rRNA genes. The mutation Lys716Glu in the C-terminal domain, for ribosomal fixation of the relA/spoT enzyme, could be a consequence of the ribosomal modification, as well as the mutation Gly57Glu observed in the SSU ribosomal protein S10p.
Phenotype.
As all isolates shared a common ancestor, the phenotypical consequences of LZD resistance acquisition were investigated. The first isolate, LimS, presented SCV features, with pinpoint-size colonies without beta hemolysis and absence of pigmentation observed after 24 h of growth on a blood agar plate at 37°C in ambient atmosphere (Fig. 1). However, no auxotrophy for thymidine, hemin, menadione, or CO2 dependency was observed and, concordantly, no previously described mutations in corresponding genes were present. LZD-resistant isolates presented even smaller colonies than LimS, with LimR6 and -R8 being the smallest even after 48 h of growth. However, they contained various SNPs that could be responsible for this phenotype and for the high generation time increase. The generation times of all other LZD-resistant isolates correlate well with the number of rrl copies with the G2576T mutation, as already described for the G2576T mutation in vitro (Table 1) (25).
DISCUSSION
We studied the dynamics of LZD resistance among related MRSA isolates over a 5-year period in one CF patient who had been colonized for 15 years. LZD was used for the first time in 2003 when the patient was 18 to treat multidrug-resistant S. aureus infection. Unfortunately, LZD resistance emerged rapidly after three 15-day therapeutic regimens. LZD resistance emergence in CF patients has been mainly associated with more LZD therapeutic regimens, longer duration of treatment, and transmission of a resistant isolate from another patient (16, 18). In the study by Endimiani et al., over 10% of patients developed resistance after prolonged treatment (18). However, as in our patient, rapid acquisition of resistance has also been described. It has been suggested that reduced bioavailability of LZD in CF could be responsible (17, 20). However, in contrast with other reports, resistance emerged immediately and with a high level (MIC > 256 mg/liter) in our patient (17). Resistance was conferred by the G2576T mutation in domain V of the 23S rRNA gene, which is the most frequently described mutation in Staphylococcus clinical strains (26). The high resistance level was linked to the mutation of all rrl copies. All isolates were shown to have 5 copies of the ribosomal operon, which is more frequent in hospital strains, favoring easier antibiotic pressure adaptation, while community strains often have 6 copies (27). Thus, the presence of only 5 rrn copies could have facilitated the mutation of all copies. Concordantly, so far, no isolate with the G2576T mutation in 6 rrl copies has been described. Moreover, very few clinical S. aureus strains with mutation in all 5 copies have been mentioned (28). We looked for other events that could have facilitated the mutation of all rrn copies. The DNA helicase RecQ is involved in DNA replication and repair. It has been hypothesized that a mutation in this gene could contribute to rapid spread of domain V mutations after acquisition of the first mutation by increasing the frequency of short sequence recombination and, thus, potentially facilitating recombination among rrn loci (29). Isolate LimR1 has a truncated recQ gene, so we hypothesize that recQ deletion could have facilitated the mutation of all of the rrn copies. The G2576T mutation in 5 copies is associated with a high fitness cost in our isolate, as already described in vitro (25). This high increase in generation time could also explain the disappearance of this isolate over time. The ensuing isolates had 3 or 4 mutated copies, as generally described in clinical strains. Those isolates could have emerged from another subpopulation, as the presence of variants of a same S. aureus clone has been described in CF patients (30). They persisted without LZD treatment for 1 year, as was described for this mutation in vitro (17, 31, 32).
Further epidemiological comparison showed that all isolates belonged to ST72 and/or spa type t148. All strains carried a type IV SCCmec element commonly found in community-acquired MRSA (CA MRSA), which correlates with the ST72 lineage, also known as a CA MRSA (33). Contrary to the high rifampin resistance level observed in our isolates, the mutation S486L has been previously described as conferring low-level resistance and is not frequent in clinical strains because of its high fitness cost (23). Two different mutations in parC were observed, E84G for LimS (first strain) and S80F for all other isolates. The first has already been described but is infrequent, unlike the second (22). Finally the loss of the constitutive MLSB phenotype observed in 2 strains, along with the ermC gene, has already been described during acquisition of LZD resistance (34).
The duration and dynamics of S. aureus persistence in the airways of individual CF patients over extended periods have been conclusively attributed to one single clone in 63% to 80% of patients by different typing methods (pulsed-field gel electrophoresis [PFGE] and variable-number tandem repeat–multilocus variable-number tandem-repeat analysis [VNTR-MLVA]) (2, 3). However, variation in the same lineage has been observed over the years with the MLVA technique (2). WGS- and SNP-based methods surpass all previous methods used for typing in terms of discriminatory power and were used to precisely analyze our isolates’ relatedness (35). Moreover, wgMLST has recently been reported as the method of choice for surveillance of food-borne bacterial pathogens by PulseNet International (36). In our study, wgMLST analysis confirmed the clonality of all the isolates. Strain-specific SNPs indicated clonal diversification of our isolates compared with other ST72 strains. The pairwise distance between isolates varied over time between 28 and 58 SNPs (for LimS, -R1, -R3, -R9, and -R12). If changes were accumulating according to a molecular clock (i.e., at a constant mutation rate of 3.10−6 per site per year as described for S. aureus), we could expect about 8.4 mutations per year (37). However, the mutation rates varied over time between 17.5 and 93 SNPs/year, with phylogeny showing LimR2 to be closer to other isolates than to the first susceptible isolate (LimS). The explanation could be that at sampling times, there were multiple strains in the population, as already proposed (38). Moreover, the maximum SNP distance between 2 isolates of the same strain is still debated and will likely depend on the organism and even the strain under investigation, as well as on the setting. SNP analysis could be very useful for the follow-up of persistent infection in patients over several years and the evolution of strains under different pressures (39–41). However, the sampling of one S. aureus colony per sputum sample for SNP analysis may be considered a limitation of our study. Indeed during long-term colonization, different evolutions of a same isolate may be found (42). In a recent study with intra-CF patient SNP comparison, isolates with 74 SNPs or fewer were considered to be the same strain (38), which is higher than the 40 SNPs considered in some outbreak studies but could permit us to hypothesize that our isolates have the same ancestor (43). In CF patients, the ability of S. aureus colonizing strains to present a high mutation rate in response to environmental stress arises through mutations in mutator genes and could lead to high levels of SNP differences (44). Accordingly, mutations in recQ (LimR1 and LimR6) and mutL (LimR6, -R8, -R9, and -R12) could have increased the mutation rate and, thus, SNP differences. However, this cannot by itself explain the very high numbers of unique SNPs of LimR6 and LimR8 compared to the sequences of LimR9 and -R12, which harbored the same truncated mutL gene. Of note, this high mutation rate could also explain the unexpected appearance of point mutations in 2 MLST alleles of LimR6, leading to a new gmk allele and, consequently, to a new ST. The same applies to the new spa type for LimR8 and LimR11. However, this adaptation, with minor changes in spa type, has already been reported in CF patients (5, 45).
In conclusion, physicians should be aware that LZD resistance can emerge rapidly and at a high level in CF patients and persist for a long time without linezolid selective pressure in MRSA colonizing strains belonging to the same lineage. All in all, this also reflects the strong bacterial adaptability over years to minimize fitness cost and keep resistance at the required level.
MATERIALS AND METHODS
Bacterial growth conditions and antimicrobial susceptibility testing.
Bacterial colonies were grown on Columbia blood agar plates (bioMérieux, Marcy l’Étoile, France) for 24 to 48 h at 37°C in ambient atmosphere. Antimicrobial susceptibility testing was performed using the agar disk diffusion method, and MICs were determined by Etest (bioMérieux) on Mueller-Hinton medium (MH; Bio-Rad, Hercules, CA) with or without blood at 5% and NAD (MHF; Bio-Rad) (46).
Whole-genome sequencing.
DNA libraries were prepared using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA) and Nextera XT index kit (Illumina). WGS was performed with a MiSeq (Illumina) instrument to generate 200-bp paired-end reads. An average sequencing depth of 204.0 (106.2 to 258.8) was achieved. De novo assembly was performed with a SPAdes de novo assembler on the BioNumerics (version 7.6) (Applied Maths, Sint-Martens-Latem, Belgium) cloud-based calculation engine (47). Gene prediction and annotation of the contigs were done using BioNumerics. Molecular resistance and virulence determinants were obtained from WGS data through the bioMérieux EpiSeq knowledge base and with VirulenceFinder (https://cge.cbs.dtu.dk/services/VirulenceFinder/) for reference strains (48).
Phylogenetic relatedness.
MLST and spa typing were inferred from WGS through the bioMérieux EpiSeq data analysis workflow for sequenced isolates. For other isolates and unknown ST or spa type from WGS data, Sanger sequencing was performed for determination or verification, respectively. New ST assignment was obtained by submission to the MLST online database (https://pubmlst.org/saureus/), and for the new spa type, submission to the RIDOM web server (http://spaserver.ridom.de/) was performed (49, 50).
To assess phylogenetic relatedness, whole-genome MLST (wgMLST) was performed on 3,897 loci (core and accessory genes) with the BioNumerics version 7.6 plugin (47). Two independent algorithms were used, an assembly-free k-mer-based method (k = 35) and a second, BLAST-based allele detection algorithm on de novo assemblies. Only the second algorithm was applied to reference strains. The closest ST72 strains, chosen among publicly available genomes by alignment of the largest contigs using BLAST, were FORC_012 and 2148 (GenBank accession no. CP010998.1 and CP016856.1, respectively). Strains with more distant genomes, MRSA N315 (ST5) and USA300-FPR3757 (ST8) (GenBank accession no. NC_002745.2 and CP000255.1, respectively), were also used. To further determine variation among isolates, whole-genome SNP analysis was performed. Two references were used for mapping with a Bowtie2 algorithm (BioNumerics). The de novo assembled and annotated genome of the first isolate (LimS) was used to assess distances between isolates, and the FORC_012 strain was used to assess clonal diversification. Strict SNP filtering was applied with the following conditions: extraction at positions shared by all strains, at least one variant within the strain set detected, minimum total coverage of 5 reads, at least 1 supporting read in each direction, and minimum distance between retained SNP positions of 12 bp. Clustering of the wgMLST and wgSNP results was performed with a maximum parsimony tree and a minimum spanning tree, respectively, based on pairwise wgMLST allelic differences between isolates using BioNumerics version 7.6.
Linezolid resistance determinants.
Specific LZD resistance determinants (23S rRNA genes rrl, rplC, rplD, and cfr) were searched by PCR and Sanger sequencing as previously described, confirmed from WGS data, and compared with reference S. aureus strain N315 using BLAST (51–53). For mutations in 23S rRNA genes, the mutated copy number was estimated by the coverage, with mutated reads divided by the total coverage (alternate and reference allele) according to the rRNA gene copy number determined. The latter was determined with a specific PCR as previously described (27).
Auxotrophy complementary testing and growth studies.
Auxotrophy was assessed by disc diffusion on MH agar using hemin disks (Oxoid, Waltham, MA, USA), 1.5 μg thymidine (Sigma-Aldrich, St. Louis, MO), or 1.5 μg menadione (Sigma-Aldrich), as already described (54). Growth studies were performed in triplicate by inoculating brain heart infusion (BHI) broth with an overnight BHI culture on a microplate. The optical density (OD) at 620 nm was measured every hour for 14 h. The generation times were calculated from the growth rates in the exponential growth phase as previously described (55).
Accession number(s).
The sequencing raw data for each isolate were submitted to GenBank with BioProject record number PRJNA434495.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by bioMérieux S.A. (Marcy l’Etoile, France).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Remy Bonnin, Katleen Vranckx, Gaël Kaneko, and Bruno H. Muller for WGS data analysis assistance. We are also grateful to François Vandenesh for fruitful discussion about genomic data and to Peggy Haneman-Castex for English editing.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00720-18.
REFERENCES
- 1.Cystic Fibrosis Foundation. 2016. Cystic Fibrosis Foundation patient registry, 2015 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 2.Vu-Thien H, Hormigos K, Corbineau G, Fauroux B, Corvol H, Moissenet D, Vergnaud G, Pourcel C. 2010. Longitudinal survey of Staphylococcus aureus in cystic fibrosis patients using a multiple locus variable number of tandem repeats analysis method. BMC Microbiol 10:24. doi: 10.1186/1471-2180-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahl BC, Duebbers A, Lubritz G, Haeberle J, Koch HG, Ritzerfeld B, Reilly M, Harms E, Proctor RA, Herrmann M, Peters G. 2003. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J Clin Microbiol 41:4424–4427. doi: 10.1128/JCM.41.9.4424-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone A, Saiman L. 2007. Update on the epidemiology and management of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, in patients with cystic fibrosis. Curr Opin Pulm Med 13:515–521. doi: 10.1097/MCP.0b013e3282efbbac. [DOI] [PubMed] [Google Scholar]
- 5.Hirschhausen N, Block D, Bianconi I, Bragonzi A, Birtel J, Lee JC, Dübbers A, Küster P, Kahl J, Peters G, Kahl BC. 2013. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int J Med Microbiol 303:685–692. doi: 10.1016/j.ijmm.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Dasenbrook EC, Merlo CA, Diener-West M, Lechtzin N, Boyle MP. 2008. Persistent methicillin-resistant Staphylococcus aureus and rate of FEV1 decline in cystic fibrosis. Am J Respir Crit Care Med 178:814–821. doi: 10.1164/rccm.200802-327OC. [DOI] [PubMed] [Google Scholar]
- 7.Dasenbrook EC, Checkley W, Merlo CA, Konstan MW, Lechtzin N, Boyle MP. 2010. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA 303:2386–2392. doi: 10.1001/jama.2010.791. [DOI] [PubMed] [Google Scholar]
- 8.Kahl BC, Becker K, Löffler B. 2016. Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin Microbiol Rev 29:401–427. doi: 10.1128/CMR.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasenbrook EC. 2011. Update on methicillin-resistant Staphylococcus aureus in cystic fibrosis. Curr Opin Pulm Med 17:437–441. doi: 10.1097/MCP.0b013e32834b95ed. [DOI] [PubMed] [Google Scholar]
- 10.Prunier A-L, Malbruny B, Laurans M, Brouard J, Duhamel J-F, Leclercq R. 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J Infect Dis 187:1709–1716. doi: 10.1086/374937. [DOI] [PubMed] [Google Scholar]
- 11.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, Ratjen FA. 2014. Antibiotic management of lung infections in cystic fibrosis. I. The microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Ann Am Thorac Soc 11:1120–1129. doi: 10.1513/AnnalsATS.201402-050AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulle S, Saini JS, Homeyer N, Gohlke H. 2015. Complex long-distance effects of mutations that confer linezolid resistance in the large ribosomal subunit. Nucleic Acids Res 43:7731–7743. doi: 10.1093/nar/gkv729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flamm RK, Mendes RE, Hogan PA, Streit JM, Ross JE, Jones RN. 2016. Linezolid surveillance results for the United States (LEADER Surveillance Program 2014). Antimicrob Agents Chemother 60:2273–2280. doi: 10.1128/AAC.02803-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, Venkataraman L, Moellering RC, Ferraro MJ. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 15.Gales A, Sader H, Andrade S, Lutz L, Machado A, Barth A. 2006. Emergence of linezolid-resistant Staphylococcus aureus during treatment of pulmonary infection in a patient with cystic fibrosis. Int J Antimicrob Agents 27:300–302. doi: 10.1016/j.ijantimicag.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Hill RLR, Kearns AM, Nash J, North SE, Pike R, Newson T, Woodford N, Calver R, Livermore DM. 2010. Linezolid-resistant ST36 methicillin-resistant Staphylococcus aureus associated with prolonged linezolid treatment in two paediatric cystic fibrosis patients. J Antimicrob Chemother 65:442–445. doi: 10.1093/jac/dkp494. [DOI] [PubMed] [Google Scholar]
- 17.Tazi A, Chapron J, Touak G, Longo M, Hubert D, Collobert G, Dusser D, Poyart C, Morand PC. 2013. Rapid emergence of resistance to linezolid and mutator phenotypes in Staphylococcus aureus isolates from an adult cystic fibrosis patient. Antimicrob Agents Chemother 57:5186–5188. doi: 10.1128/AAC.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endimiani A, Blackford M, Dasenbrook EC, Reed MD, Bajaksouszian S, Hujer AM, Rudin SD, Hujer KM, Perreten V, Rice LB, Jacobs MR, Konstan MW, Bonomo RA. 2011. Emergence of linezolid-resistant Staphylococcus aureus after Prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob Agents Chemother 55:1684–1692. doi: 10.1128/AAC.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke JB, Zuill DE, Scharn CR, Deane J, Sahm DF, Denys GA, Goering RV, Shaw KJ. 2014. Linezolid-resistant Staphylococcus aureus strain 1128105, the first known clinical isolate possessing the cfr multidrug resistance gene. Antimicrob Agents Chemother 58:6592–6598. doi: 10.1128/AAC.03493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D, Stach LM, Newland JG. 2015. Linezolid-resistant Staphylococcus aureus in children with cystic fibrosis. J Pediatr Infect Dis Soc 4:e163–e165. doi: 10.1093/jpids/piv071. [DOI] [PubMed] [Google Scholar]
- 21.de Matos PDM, de Oliveira TLR, Cavalcante FS, Ferreira DC, Iorio NLP, Pereira EM, Chamon RC, Dos Santos KRN. 2016. Molecular markers of antimicrobial resistance in methicillin-resistant Staphylococcus aureus SCCmec IV presenting different genetic backgrounds. Microb Drug Resist 22:700–706. doi: 10.1089/mdr.2015.0255. [DOI] [PubMed] [Google Scholar]
- 22.Kwak YG, Truong-Bolduc QC, Bin Kim H, Song K-H, Kim ES, Hooper DC. 2013. Association of norB overexpression and fluoroquinolone resistance in clinical isolates of Staphylococcus aureus from Korea. J Antimicrob Chemother 68:2766–2772. doi: 10.1093/jac/dkt286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Shan W, Ma X, Chang W, Zhou X, Lu H, Dai Y. 2012. Molecular characterization of rifampicin-resistant Staphylococcus aureus isolates in a Chinese teaching hospital from Anhui, China. BMC Microbiol 12:240. doi: 10.1186/1471-2180-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H-J, Hung W-C, Tseng S-P, Tsai J-C, Hsueh P-R, Teng L-J. 2010. Fusidic acid resistance determinants in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother 54:4985–4991. doi: 10.1128/AAC.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. 2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross resistances. Antimicrob Agents Chemother 52:1570–1572. doi: 10.1128/AAC.01098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendes RE, Deshpande LM, Jones RN. 2014. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat 17:1–12. doi: 10.1016/j.drup.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Fluit AC, Jansen M, Bosch T, Jansen WTM, Schouls L, Jonker MJ, Boel CHE. 2016. rRNA operon copy number can explain the distinct epidemiology of hospital-associated methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 60:7313–7320. doi: 10.1128/AAC.01613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pillai SK, Sakoulas G, Wennersten C, Eliopoulos GM, Moellering RC, Ferraro MJ, Gold HS. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J Infect Dis 186:1603–1607. doi: 10.1086/345368. [DOI] [PubMed] [Google Scholar]
- 29.Iguchi S, Mizutani T, Hiramatsu K, Kikuchi K. 2016. Rapid acquisition of linezolid resistance in methicillin-resistant Staphylococcus aureus: role of hypermutation and homologous recombination. PLoS One 11:e0155512. doi: 10.1371/journal.pone.0155512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran Losada P, Chouvarine P, Dorda M, Hedtfeld S, Mielke S, Schulz A, Wiehlmann L, Tümmler B. 2016. The cystic fibrosis lower airways microbial metagenome. ERJ Open Res 2:00096-2015. doi: 10.1183/23120541.00096-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meka VG, Pillai SK, Sakoulas G, Wennersten C, Venkataraman L, DeGirolami PC, Eliopoulos GM, Moellering RC, Gold HS. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J Infect Dis 190:311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- 32.Tsakris A, Pillai SK, Gold HS, Thauvin-Eliopoulos C, Venkataraman L, Wennersten C, Moellering RC, Eliopoulos GM. 2007. Persistence of rRNA operon mutated copies and rapid re-emergence of linezolid resistance in Staphylococcus aureus. J Antimicrob Chemother 60:649–651. doi: 10.1093/jac/dkm246. [DOI] [PubMed] [Google Scholar]
- 33.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 34.Rouard C, Aslangul E, Rivière A, Deback C, Butel M-J, Doucet-Populaire F, Bourgeois-Nicolaos N. 2017. Mutation in the L3 ribosomal protein could be associated with risk of selection of high-level linezolid-resistant Staphylococcus epidermidis strains. Microb Drug Resist 23:462–467. doi: 10.1089/mdr.2016.0137. [DOI] [PubMed] [Google Scholar]
- 35.Tagini F, Greub G. 2017. Bacterial genome sequencing in clinical microbiology: a pathogen-oriented review. Eur J Clin Microbiol Infect Dis 36:2007–2020. doi: 10.1007/s10096-017-3024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadon C, Walle IV, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, Gilpin B, Smith AM, Kam KM, Perez E, Trees E, Kubota K, Takkinen J, Nielsen EM, Carleton H, Panel F-NE. 2017. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill 22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ankrum A, Hall BG. 2017. Population dynamics of Staphylococcus aureus in cystic fibrosis patients to determine transmission events by use of whole-genome sequencing. J Clin Microbiol 55:2143–2152. doi: 10.1128/JCM.00164-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannavino CR, Mendes RE, Sader HS, Farrell DJ, Critchley IA, Biek D, Le J, Skochko SM, Jones RN, Bradley JS. 2016. Evolution of ceftaroline-resistant MRSA in a child with cystic fibrosis following repeated antibiotic exposure. Pediatr Infect Dis J 35:813–815. doi: 10.1097/INF.0000000000001171. [DOI] [PubMed] [Google Scholar]
- 40.van Mansfeld R, de Been M, Paganelli F, Yang L, Bonten M, Willems R. 2016. Within-host evolution of the Dutch high-prevalent Pseudomonas aeruginosa clone ST406 during chronic colonization of a patient with cystic fibrosis. PLoS One 11:e0158106. doi: 10.1371/journal.pone.0158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viberg LT, Sarovich DS, Kidd TJ, Geake JB, Bell SC, Currie BJ, Price EP. 2017. Within-host evolution of Burkholderia pseudomallei during chronic infection of seven Australasian cystic fibrosis patients. mBio 8:e00356-17. doi: 10.1128/mBio.00356-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. 2016. Within-host evolution of bacterial pathogens. Nat Rev Microbiol 14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long SW, Beres SB, Olsen RJ, Musser JM. 2014. Absence of patient-to-patient intrahospital transmission of Staphylococcus aureus as determined by whole-genome sequencing. mBio 5:e01692-14. doi: 10.1128/mBio.01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver A, Mena A. 2010. Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clin Microbiol Infect 16:798–808. doi: 10.1111/j.1469-0691.2010.03250.x. [DOI] [PubMed] [Google Scholar]
- 45.López-Collazo E, Jurado T, de Dios Caballero J, Pérez-Vázquez M, Vindel A, Hernández-Jiménez E, Tamames J, Cubillos-Zapata C, Manrique M, Tobes R, Máiz L, Cantón R, Baquero F, del Campo R. 2015. In vivo attenuation and genetic evolution of a ST247-SCCmecI MRSA clone after 13 years of pathogenic bronchopulmonary colonization in a patient with cystic fibrosis: implications of the innate immune response. Mucosal Immunol 8:362–371. doi: 10.1038/mi.2014.73. [DOI] [PubMed] [Google Scholar]
- 46.The European Committee on Antimicrobial Susceptibility. 2018. Breakpoint tables for interpretation of MICs and zone diameters, version 8.0, 2018.
- 47.Earls MR, Kinnevey PM, Brennan GI, Lazaris A, Skally M, O’Connell B, Humphreys H, Shore AC, Coleman DC. 2017. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing: implications for screening. PLoS One 12:e0175542. doi: 10.1371/journal.pone.0175542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourgeois‐Nicolaos N, Massias L, Couson B, Butel M, Andremont A, Doucet‐Populaire F. 2007. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J Infect Dis 195:1480–1488. doi: 10.1086/513876. [DOI] [PubMed] [Google Scholar]
- 52.Miller K, Dunsmore CJ, Fishwick CWG, Chopra I. 2008. Linezolid and tiamulin cross resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother 52:1737–1742. doi: 10.1128/AAC.01015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lannergard J, von Eiff C, Sander G, Cordes T, Seggewiss J, Peters G, Proctor RA, Becker K, Hughes D. 2008. Identification of the genetic basis for clinical menadione auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob Agents Chemother 52:4017–4022. doi: 10.1128/AAC.00668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long KS, Munck C, Andersen TMB, Schaub MA, Hobbie SN, Bottger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross resistance. Antimicrob Agents Chemother 54:4705–4713. doi: 10.1128/AAC.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.