LETTER
Vibrio alginolyticus is a Gram-negative halophilic and mesophilic bacterium, particularly associated with severe epidemic vibriosis, which causes high mortality in marine animals, including fish, shellfish, and shrimp (1). The carbapenemase NDM-1 has been found mainly in Enterobacteriaceae, such as Escherichia coli and Klebsiella pneumoniae (2). However, recent reports have described the existence of blaNDM genes in Vibrio parahaemolyticus, Vibrio fluvialis, and Vibrio cholerae from clinical or environmental samples (3, 4). In this study, a Vibrio sp. strain, Vb1394, was isolated from a shrimp sample in a market of Shenzhen, China, in August 2016. The isolate was shown to be resistant to ceftriaxone, cefotaxime, amoxicillin-clavulanate, ampicillin, ciprofloxacin, and trimethoprim-sulfamethoxazole (SXT); intermediate resistant to meropenem and imipenem; and susceptible to tetracycline, amikacin, gentamicin, nalidixic acid, and chloramphenicol (Table 1). The isolate was screened for the known β-lactamase genes by using previously described multiplex PCR assays and was shown to harbor blaNDM-1 (5). A conjugation experiment using azide-resistant Escherichia coli J53 as the recipient strain was performed to determine the transferability of the multidrug resistance property, with results showing that the carbapenem, cephalosporin, and SXT resistance phenotype could be transferred to E. coli J53. Interestingly, the transconjugant, designated TCVb1394, was highly resistant to meropenem and imipenem; however, the parental strain Vb1394 had reduced susceptibility to these two antibiotics in Mueller-Hinton (MH) medium without additional Zn2+, although it carried blaNDM-1, which generally referred to carbapenem resistance (Table 1). A similar observation was also reported from previous studies, in which carbapenems did not appear to offer any substantial activity against these NDM producers, and even a few isolates carrying NDM-1 had low MIC values, as low as 0.125 mg/liter (6); this warrants further investigation. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and hybridization were performed on the donor strain Vb1394 and the transconjugant TCVb1394 using the blaNDM-1 probe, with results confirming that blaNDM-1 was located on a plasmid with a size of ca. 165 kb, which could be transferred by conjugation. To describe the detailed genetic context of this plasmid, pC1394 was extracted from TCVb1394 and sequenced with the Illumina NextSeq 500 platform and the MinION sequencing platform, as the workflow reported previously (7).
TABLE 1.
MICs of different antibiotics against Vibrio alginolyticus strain Vb1394 and its transconjugant TCVb1394 using azide-resistant Escherichia coli J53 as the recipient strain
| Strain | MIC (mg/liter) by antibiotica |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRO | CTX | AMC | AMP | MRP | IMI | TET | AMK | GEN | CIP | NAL | CHL | SXT | |
| J53 | 0.03 | 0.03 | 4/2 | 8 | 0.03 | 0.25 | 2 | 1 | 0.25 | 0.015 | 4 | 4 | 0.25 |
| Vb1394 | >16 | >16 | >64/32 | >64 | 2 | 2 | 2 | 2 | 4 | 16 | 1 | 1 | 8 |
| TCVb1394 | >16 | >16 | >64/32 | >64 | 8 | 16 | 2 | 1 | 0.25 | 0.12 | 8 | 4 | 8 |
CRO, ceftriaxone; CTX, cefotaxime; AMC, amoxicillin-clavulanic acid; Amp, ampicillin; MRP, meropenem; IMI, imipenem; TET, tetracycline; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; NAL, nalidixic acid; CHL, chloramphenicol; SXT, sulfamethoxazole-trimethoprim.
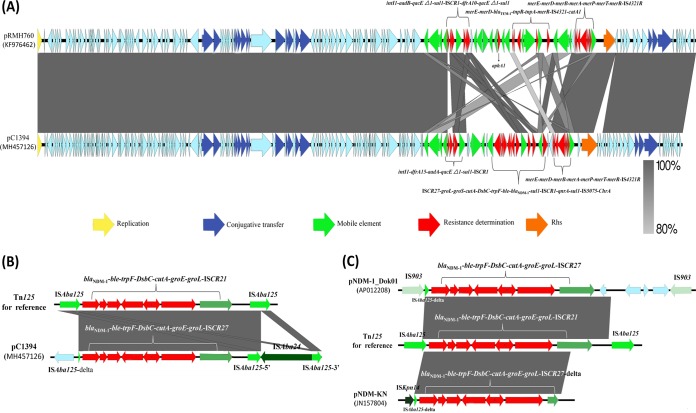
Complete sequence analysis indicated that the blaNDM-1-positive plasmid, designated pC1394 (GenBank accession number MH457126), was found to be a circular IncA/C-type plasmid of 167,140 bp, with 200 predicted coding sequences (CDSs), including a 128-kb backbone (65.5% GC content) and a single 39-kb resistance island (56.1% GC content). Based on the presence or absence of orf1832-orf1847, rhs1-rhs2, i1, and i2, which were the key features distinguishing between type 1 and type 2 A/C2 plasmids (8), pC1394 was considered to be a type 1 A/C2 plasmid. Comparative analysis of pC1394 and pRMH760 showed that the major difference was located in the multiple-resistance region, designated ARI-A (Fig 1A). The ARI-A of pRMH760 contained six resistance genes, aadB, sul1, dfrA10, aphA1, catA1, and blaTEM-1, clustered together in the region of about 45 kb, while that of pC1394 contained a novel complex class 1 integron carrying dfrA15, aadA, sul1, ble, blaNDM-1, and qnrA. According to the previous report, genetic structures surrounding blaNDM-1 were commonly associated with the presence of ISAba125 (9). Notably, a composite transposon named Tn125, composed of two copies of insertion sequence ISAba125 bracketing a ca. 8-kb fragment (blaNDM-1-ble-trpF-tat-dct-groS-groL-ISCR27), has been demonstrated to be responsible for the dissemination of blaNDM-1 in bacteria. A BLAST search of Tn125 of pC1394 has identified a similar region in two other type 1 A/C2 plasmids, pNDM-1_Dok01 (GenBank accession number AP012208) from E. coli and pNDM-KN (GenBank accession number JN157804) from K. pneumoniae. The Tn125 from these three plasmids shared the same core structure of the blaNDM-1 mobile element, while different surrounding insertion sequence (IS) elements suggested their important role in the evolution and mobilization of the blaNDM-1 mobile element (Fig 1B and C). The initial mobilization of blaNDM-1 might occur through a transposition involving ISCR27, and then different mobile elements, such as ISAba125, seem to have moved segments that contain blaNDM-1 into existing multidrug resistant (MDR) plasmids on type 1 A/C2 plasmids. IncA/C plasmids are thought to be vehicles that have an extremely broad bacterial host range and may play an important role in the spread of blaNDM-1 in China (10). All three type 1 A/C2 plasmids studied here harbor segments matching different parts of Tn125, suggesting that different mechanisms appear to be responsible for the independent transfer and further indicating a variation in the ways in which blaNDM-1 has been acquired by the same type plasmids.
FIG 1.
Comparative genetic analysis of pC1394 with other IncA/C2 plasmids. (A) Comparison between pC1394 and pRMH760. Sequence similarity is depicted by the shaded region. Genes coding for proteins of known function are named above and colored according to the figure legend. (B and C) Comparison of ARI-A islands and mobile elements encoding the blaNDM-1 gene from different plasmids. Genes are denoted by arrows. Genes and mobile elements are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity, ≥99%). The GenBank accession numbers for each plasmid are as follows: KF976462 (pRMH760), MH457126 (pC1394), AP012208 (pNDM-1_Dok01), and JN157804 (pNDM-KN).
ACKNOWLEDGMENT
This work was supported by the Health and Medical Research Fund (grants 14130402 and 14130422).
REFERENCES
- 1.Thompson FL, Austin B, Swings J. 2006. The biology of vibrios. ASM Press, Washington, DC. [Google Scholar]
- 2.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury G, Pazhani GP, Sarkar A, Rajendran K, Mukhopadhyay AK, Bhattacharya MK, Ghosh A, Ramamurthy T. 2016. Carbapenem resistance in clonally distinct clinical strains of Vibrio fluvialis isolated from diarrheal samples. Emerg Infect Dis 22:1754–1761. doi: 10.3201/eid2210.151612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 5.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Datta S, Roy S, Ramanan L, Saha A, Viswanathan R, Som T, Basu S. 2016. Carbapenem resistance in Acinetobacter baumannii and other Acinetobacter spp. causing neonatal sepsis: focus on NDM-1 and its linkage to ISAba125. Front Microbiol 7:1126. doi: 10.3389/fmicb.2016.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li R, Xie M, Dong N, Lin D, Yang X, Wong MHY, Chan EW, Chen S. 2018. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Datta S, Mitra S, Chattopadhyay P, Som T, Mukherjee S, Basu S. 2017. Spread and exchange of bla NDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis 36:255–211. doi: 10.1007/s10096-016-2794-6. [DOI] [PubMed] [Google Scholar]
- 10.An J, Guo L, Zhou L, Ma Y, Luo Y, Tao C, Yang J. 2016. NDM-producing Enterobacteriaceae in a Chinese Hospital, 2014–2015: identification of NDM-producing Citrobacter werkmanii and acquisition of blaNDM-1-carrying plasmid in vivo in a clinical Escherichia coli isolate. J Med Microbiol. 65:1253–1259. doi: 10.1099/jmm.0.000357. [DOI] [PubMed] [Google Scholar]