Abstract
Over the course of evolution, mammalian body surfaces have adapted their complex immune system to allow a harmless coexistence with the commensal microbiota. The adaptive immune response, in particular CD4+ T cell-mediated, is crucial to maintain intestinal immune homeostasis by discriminating between harmless (e.g., dietary compounds and intestinal microbes) and harmful stimuli (e.g., pathogens). To tolerate food molecules and microbial components, CD4+ T cells establish a finely tuned crosstalk with the environment whereas breakdown of these mechanisms might lead to chronic disease associated with mucosal barriers and beyond. How commensal-specific immune responses are regulated and how these molecular and cellular mechanisms can be manipulated to treat chronic disorders is yet poorly understood. In this review, we discuss current knowledge of the regulation of commensal bacteria-specific CD4+ T cells. We place particular focus on the key role of commensal-specific CD4+ T cells in maintaining tolerance while efficiently eradicating local and systemic infections, with a focus on factors that trigger their aberrant activation.
Keywords: commensals, microbiota, CD4+ T cells, inflammatory bowel disease (IBD), bacteria–host interaction, immune education, intestinal immunity, autoimmunity
Introduction
The human body is composed of a myriad of microbes that outnumber the whole body human cells (1, 2). The highest density of commensal bacteria is located in the gastrointestinal tract, which is the largest immune organ in the body (3). The vast capabilities of the microbiota have led scientists to call for a “second human genome project” to account for the effect of microbial genes in human health (4). Commensal bacteria serve various functions, from catabolizing certain food molecules to promoting tissue repair. Among other functions, the microbiota controls enteric infections by competing for common resources as well as inducing the production of antimicrobial peptides by gut epithelial cells (5, 6). Importantly, the gut microbiota shapes both local and systemic immune responses. This is most evident in the case of chronic inflammation in the intestines, as this can lead to an unbalance of immune cells as well as dysbiosis of commensals and can result in various diseases, including local inflammatory bowel disease (IBD) and systemic diseases such as diabetes, asthma, and cancer (7). While many immune cells play a role in maintaining homeostasis with the microbiota (8), both effector and regulatory CD4+ T cells are highly enriched in intestinal tissues and represent a major immune cell in mediating homeostatic responses. Moreover, CD4+ T cells are believed to be the main drivers of pathogenicity in IBD and as such are considered a main therapeutic target (9).
Commensals have been shown to play a crucial role in driving physiological CD4+ T cell differentiation at barrier sites. T cells recognize immunological epitopes through engagement of their T cell receptors (TCRs) with cognate antigens presented on major histocompatibility (MHC) molecules on antigen-presenting cells (APCs) (10). Unlike T cells specific for self-antigens expressed by thymic epithelial cells, commensal-specific T cells do not undergo negative selection in the thymus and are present in healthy individuals despite the constant presence of their cognate antigens (11). Importantly, T cell transfer studies have shown the ability of these cells to drive pathogenesis. In this review, we will therefore discuss the multifaceted role of commensal bacteria-specific CD4+ T cells with special emphasis on the regulation and fate of these cells. More in detail, we will illustrate the mechanisms at the basis of commensal-host mutualism and how their disturbance affects local and systemic health. A detailed description of the state-of-the-art models available for the study of microbiota-specific T cell responses will provide major insights into the variety of CD4+ T cell responses that are elicited by selected commensal bacterial species.
Commensal-host mutualism
The relationship between humans and microbes is multifaceted; while several bacterial species can result in pathogenesis upon colonization, the vast majority are innocuous and even beneficial to the host (12). Mutualism between hosts and microbes benefits both parties but relies on a delicate balance between the antimicrobial and tolerogenic effector functions of the immune system as well as the control of pathogenicity of commensals. Commensals aid their hosts by inactivating toxins and catabolizing host-indigestible nutrients, such as some complex carbohydrates, to useful metabolites such as short chain fatty acids (SCFAs) (13–15). The commensal bacteria also control pathogen overgrowth through competition for the same biological niches and release of molecular mediators (16–22). Beyond nutrient breakdown and protection from infections, commensal bacteria also play a role in the development of anatomical structures in the intestines, angiogenesis, maturation of the epithelial layer, and leukocyte imprinting (5, 23, 24).
The gut microbiota has a vast effect on the local immune system. Accordingly, antibiotic-mediated clearance of commensal microbes has been associated with susceptibility to infections, likely due to an unfit immune system (25). Importantly, commensal bacteria can induce both pro- and anti-inflammatory CD4+ T cell responses via different mechanisms. For example, segmented filamentous bacteria (SFB) efficiently induce ileal IL-17 producer T helper (TH17) cell expansion, whereas the production of SCFAs by colonic bacteria promotes the differentiation of colon-resident regulatory T cells (26). The delicate balance between pro-inflammatory and regulatory T helper cells is believed to play a pivotal role for the maintenance of the symbiotic relationship between commensal bacteria and their host.
Regulation of bacteria-host interaction by the mucosal barrier
The maintenance of a symbiotic relationship between commensal microbes and their host relies both on physical segregation of the microbiota to the intestinal lumen and on active sampling of bacteria by the immune system under steady-state conditions. This physical segregation is achieved through a single layer of columnar intestinal epithelial cells (IECs) kept together by tight junctions, that regulate paracellular permeability. In addition, the epithelium is covered by mucous layers of varying thickness and structure along the intestinal tract (27, 28). Disturbance of the inner colonic mucus layer structure, which can be achieved by genetic depletion of Muc2 or a short-term treatment with dextran sulfate sodium (DSS), is sufficient to allow bacteria to relocate to the epithelial layer and cause gut inflammation in mice (29–31). In CA-MLCK mice, which are characterized by constitutive tight junction barrier loss due to transgenic expression of constitutively active myosin light chain kinase, expansion of intestinal CD4+ T cells depends on the presence of commensal bacteria. This response resulted in protection against early Salmonella Typhimurium infection but exacerbated inflammation during chronic infection (32). Similarly, defective mucin production and aberrant expression of epithelial junctional proteins associated with early colorectal neoplastic lesions promoted permeability to commensal bacteria in humans, furthering inflammation and tumorigenesis (33).
The mucosal barrier is far from being a passive defense mechanism against microbial translocation. Immunoglobulins A (IgA), the most abundant immunoglobulin class in the body, are produced by B cells and plasma cells that reside in the Peyer's patches and intestinal lamina propria, respectively. Functional importance of this molecule in limiting commensal-specific T cell activation has been demonstrated in studies using the CBir1 TCR transgenic mouse model (Table 1). Activation of adoptively-transferred CBir1 Tg cells in response to orally-administered CBir1 flagellin was specifically blocked in WT mice, while selective impairment of IgA production or mucosal secretion unleashed CBir1 antigen-dependent T cell proliferation (48). Interestingly, IgA-mediated compartmentalization of the mucosal T cell response to the commensal microbiota does not apply to all bacteria, as activation of SFB or Helicobacter hepaticus-specific T cell clones occurs in immunocompetent mice (39, 42) (Figure 1). Furthermore, by the secretion of microbial peptides, epithelial cells actively contribute to the segregation of selected commensal bacteria to the intestinal lumen. These peptides are critical regulators of bacterial immunity and their impaired production is associated with intestinal and systemic inflammation. Mutations in Nod2, which are highly correlated with Crohn's disease, were found to negatively affect the production of a subgroup of intestinal anti-microbial peptides known as cryptidins (56). RegIII lectins are produced by Paneth cells in response to MyD88-dependent recognition of gut microbial patterns (57). Their depletion in RegIIIγ−/− and RegIIIβ−/− mice resulted in increased colonization of the intestinal epithelial surface and bacterial translocation to the liver, respectively, with consequent extra-intestinal inflammation (58, 59). Interestingly, depletion of retinoic acid (RA) receptor alpha specifically in epithelial cells resulted in increased numbers of goblet and Paneth cells and increased RegIIIγ production. In agreement with the role of RegIIIγ in regulating bacterial colonization at the epithelium vicinity, these mice did not have any detectable bacteria as seen by 16S FISH staining (60). Altogether, these data demonstrate that tight regulation of the mucosal barrier is crucial in maintaining microbial homeostasis.
Table 1.
Models to study commensal-specific T cell responses.
| Model | Description | References |
|---|---|---|
| Gnotobiotic mice | • Germ-free (GF) mice, which lack commensal microbes, have immunological defects that extend beyond the intestinal mucosa, with hypoplastic lymphoid tissues lacking B and T-cell compartmentalization | (23, 34) |
| • GF mice colonized with defined bacterial species are useful to study the effects of selected microbes on the development of the immune system | (35) | |
| • GF mice colonized at birth with rat or human microbiota maintain an immature gut immune system compared to specific pathogen-free (SPF) mice. The immune-educating effect of the microbiota is host-specific | (36) | |
| Tetramers | • Soluble MHC/peptide multimers with the capability of binding selected TCRs, useful for the ex vivo study of low-frequency endogenous antigen-specific CD4+ or CD8+ T cell populations | (37, 38) |
| • I-Ab/3340-A6 tetramer allows recognition of segmented filamentous bacteria (SFB)-specific T cells | (39, 40) | |
| • I-Ab-CBir1p tetramer selectively stains cells that recognize CBir1 flagellin, an immunodominant microbiota antigen | (41) | |
| • HH1713172–86 and HH1713230–44 tetramers stain Helicobacter-specific T cells | (42) | |
| TCR transgenic mice | • SFB, a unique member of Clostridiales that inhabits the terminal ileum, has been shown to specifically induce TH17 cell differentiation in vivo • In 7B8 TCR transgenic mice, CD4 T cells recognize SFB-specific antigens • Upon transfer of naïve 7B8 cells to SFB-colonized congenic mice, these cells proliferate and are detected in the small intestinal lamina propria as TH17 cells |
(39, 43, 44) |
| • Commensal Clostridia induce the accumulation of Treg cells in the intestine • CT2 and CT6 mouse lines express microbiota-dependent colonic Treg TCRs • Upon adoptive transfer into young immunocompetent hosts, these naïve TCR Tg cells spontaneously acquired a Treg cell phenotype |
(45–47) | |
| • Increased antibodies against microbial antigens were detected in both Crohn's disease patients and animal models of gut inflammation. The Cbir1 epitope of bacterial flagellin protein was immunodominant in both settings • CBir1 TCR transgenic mice have CD4+ T cells capable of recognizing bacterial flagellin and other commensal microbes • Activation of adoptively transferred naïve Cbir1 TCR cells does not occur under steady-state conditions but can be achieved via disturbance of the intestinal mucosal barrier or lack of immunoregulatory mechanisms |
(48–52) | |
| • HH7-2tg and HH5-1tg mouse strains are transgenic for clonotypic TCRs that recognize Helicobacter hepaticus • HH7-2tg cells acquired a Foxp3+RORγt+ regulatory phenotype under steady state conditions • Intestinal inflammation skews the H. hepaticus-specific response toward a pro-inflammatory TH17/TH1 cell phenotype |
(42, 53) |
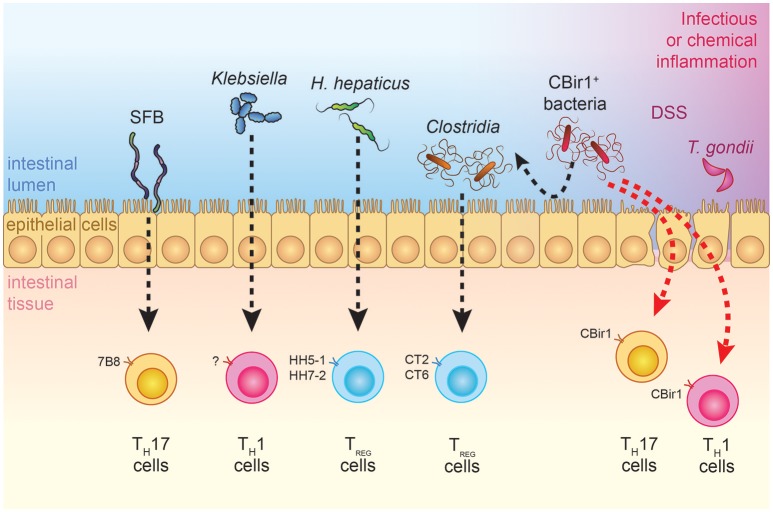
Figure 1.
Effects of commensal bacteria on T cell differentiation during steady-state and inflammatory conditions. TCR transgenic models that are available for the study of bacteria-specific immune responses are depicted. SFB-specific 7B8 cells are preferentially skewed toward a TH17 cell phenotype, while H. hepaticus-specific TCR transgenic cells and Clostridia-specific CT2-CT6 cells acquire a Treg fate under steady-state conditions (39, 42, 47). Physiological encounter of CBir1 T cells with their cognate antigen does not occur in adult individuals but can be triggered by different infectious or barrier-disrupting events, which shape the type of CD4+ T cell response (48, 49, 54). Ectopic colonization of oral Klebsiella in the intestines has TH1-inducing and pro-inflammatory effects on the gut, although antigen specificity has yet to be investigated (55).
Regulation of CD4+ T cell responses against commensal bacteria
CD4+ T cells orchestrate the immune response through the release of pro- and anti-inflammatory cytokines and expression of co-stimulatory molecules. To this end, they play crucial roles in driving or repressing the response of macrophages, CD8+ T cells, and B cells toward both pathogens and autoimmune antigens [reviewed in (61)]. CD4+ T cells can differentiate into various T helper (TH) subsets with differing effector functions [reviewed in (62, 63)]. The most extensively characterized TH subsets include: TH1 cells, which are characterized by the production of interferon gamma (IFNγ), tumor necrosis factor alpha (TNFα), and expression of the transcription factor T-box expressed in T cells (T-bet); TH2 cells, which produce IL-4 and IL-13 and express the transcription factor GATA-binding protein 3 (GATA-3); and TH17 cells, which express IL-17A/F and IL-22 and the transcription factor RA receptor-related orphan nuclear receptor RORγt. Anti-inflammatory T cell subsets include “natural” CD4+CD25+FoxP3+ regulatory (Treg) cells that develop in the thymus as well as “inducible” regulatory cells, such as FoxP3+ Treg and FoxP3− TR1 cells, which arise in the periphery (64–66). In addition, Bcl6-expressing T follicular helper (TFH) cells reside in germinal centers and coordinate B cells responses through regulation of B cell recruitment, expansion, survival, antibody class-switching, and somatic hypermutation [reviewed in (67)]. Differentiation of T cells into certain TH subsets can be fostered by specific features of the microenvironment. In vitro studies have shown that neutralization of IFNγ reduces the development of TH1 cells, while transforming growth factor beta (TGFβ) promotes the differentiation of TH17 and Treg cells (61, 68).
Adherence of selective microbes to the gut epithelium or intestinal damage can expose commensal bacterial antigens to APCs, which can then initiate commensal-specific T cell responses. Several subsets of APCs inhabit the intestinal lamina propria and have been shown to respond to fluctuations of the commensal microbiota composition (69, 70). For instance, CX3CR1hi mononuclear phagocytes residing in the small intestine were reported to express tight junction proteins that allow them to extend dendrites through the intact intestinal epithelium and sample microbial antigens (71, 72). Moreover, despite being non-migratory under steady state, these APCs were able to migrate to the MLNs and trigger Salmonella-specific T cell responses upon dysbiotic conditions (73). Both IRF4-dependent CD103+CD11b+ dendritic cells (DCs) and CX3CR1+ intestinal macrophages have been shown to play a role in the induction of intestinal TH17 cells in response to epithelium-associated SFB (74, 75). A number of reports demonstrate the ability of CD103+CD11b+ DCs to produce pro-inflammatory cytokines IL-6 and IL-23 in response to detection of microbial patterns, therefore making them good candidates for the induction of TH17 cells (74, 76). Cytokine production by innate immune cells may synergize with or be neutralized by TCR specificity in determining the fate of the TH17 cell response. For instance, joint colonization with SFB and TH1-inducer Listeria monocytogenes does not prevent SFB-specific T cells from acquiring TH17 features (39).
Intestinal innate lymphoid cells (ILCs) represent another innate immune population with a high degree of functional compartmentalization that can be finely shaped by the composition of the commensal microbiota (77). It has been recently demonstrated that some group 3 ILCs (ILC3s) have the capacity to present antigens through MHC class II molecules, and this feature allows them to regulate the commensal-specific T cell response (78). Loss of MHCII expression within this ILC subset resulted in the accumulation and pro-inflammatory activation of commensal microbiota-specific CBir1 Tg T cells in the MLN and colonic lamina propria. In addition, the regulatory function of ILC3 was shown to rely on MHCII- and antigen-dependent induction of CBir1 T cell apoptosis, thus uncovering a novel regulatory mechanism of the commensal-specific T cell response (40).
T cell tolerance toward commensals
Activation of TH cells in the gut-associated lymphoid tissues is subjected to a delicate control, whereby T cells specific for commensal bacteria are either ignorant of their cognate antigens (e.g., CBir1 flagellin-specific cells) or physiologically skewed toward a regulatory function (e.g., CT2 and CT6 commensal-specific T cells) (45, 49) (Figure 1). Understanding the physiological mechanisms that are in place to maintain T cell homeostasis toward commensal antigens is therefore fundamental to understand why inflammation occurs and to potentially revert pathogenic activation of commensal-specific T cells.
The peculiar microenvironment present at mucosal surfaces—largely influenced by microbial metabolic products and soluble factors either digested of produced by IECs—promotes tolerance toward foreign antigens (26, 60, 79). Peripheral tolerance prevents the development of immune-mediated inflammatory diseases (IMIDs) (11, 80) and can be achieved by different mechanisms. The high frequency of Treg cells in the gut, which has been shown to depend at least in part on TCR-mediated recognition of some bacteria residing in the colonic mucosa, namely Clostridia, provides an important mechanism for the maintenance of tolerance to commensals (45). CD4+ regulatory T cells dampen inflammation through the release of cytokines such as interleukin 10 (IL-10) and TGFβ. Several non-CD4+ T cell subsets such as CD8+ T cells and double negative T regulatory cells can also promote tolerance through the expression of anti-inflammatory cytokines or through direct killing of effector cells (81, 82). All of these regulatory cell types can be induced in the context of oral tolerance, which is defined as the active establishment of local and systemic unresponsiveness to antigens acquired via the oral route (83).
The gut-draining mesenteric lymph nodes (MLNs) and CD103+ DCs play a critical role in the induction of antigen-specific Treg cells (84, 85). One mechanism of Treg cell induction involves the ability of CD103+ DCs to metabolize dietary vitamin A to RA (86–90). The difference in TH cell subsets, and in particular percentages of Treg cells, along the intestinal tract could therefore, be ascribed to a differential uptake of vitamin A and other nutrients in selected anatomical locations, as well as to the presence of different consortia of commensal bacteria (45, 46, 91). While the process of tolerance toward food antigens shares some features with tolerance to commensals, the two mechanisms are fundamentally different. Indeed, tolerance to microbes is largely limited to the gut whereas oral tolerance has systemic consequences (92).
Regulatory T cells can be implicated in the maintenance of the host-microbiota commensalism at different levels. First of all, T cells that are specific for selected bacteria, i.e., Clostridium species or H. hepaticus, acquire a Treg phenotype under steady-state conditions (42, 45, 47). For instance, the preferential conversion of H. hepaticus-specific HH7-2tg cells into functional RORγt+Foxp3+ iTreg cells depends on the intrinsic expression of the transcription factor c-MAF, which is known to promote a cellular anti-inflammatory program including production of the cytokine IL-10. Selective depletion of c-MAF in Treg cells resulted in the accumulation of both polyclonal and H. hepaticus-specific TH17 cells in the large intestine (42). Additionally, Treg cells participate indirectly in the restriction of CBir1 flagellin-specific T cell responses by providing help to B cells which, in turn, produce CBir1-specific IgA molecules (48). Thus, Treg cells contribute to tolerance of commensals by various mechanisms.
Commensal-specific T cells in homeostasis
While the general effect of commensals on the immune system is established, it is only of recent that focus has been directed toward the specific T cells that recognize them. Antibiotic treatment has not only been shown to affect Treg cell populations as a whole, but also to alter the TCR repertoire of Treg cells (93). Moreover, enhanced Treg suppressive activity has been observed in the presence of their cognate antigen (93). Indeed, the existence of colonic Treg cells appears to be regulated by TCR specificity and antigen availability, and in vitro stimulation assays of colonic Treg-associated TCRs with host-derived commensals show that many of the local antigens these cells react to are microbiota-derived (45–47). Despite this, the microbiota does not appear to be essential to induce Treg differentiation, since germ-free (GF) mice, which are devoid of microbiota, do not lack this T cell subset. The variable ontogeny of Treg cells inhabiting the intestinal mucosa, either thymic-derived or peripherally induced, might explain the controversial observations regarding the effects of the presence of commensals on Treg cell frequencies [reviewed in (94)]. The effect of the microbiota on TH1 and TH17 induction is more evident, as their numbers are greatly reduced in GF animals (95, 96). In particular, it has been demonstrated that the majority of TH17 cells in the intestinal lumen are specific for commensals (43, 46, 97).
Mouse studies reveal that commensal-specific T cells in the gut mucosa typically develop into either TH17 or Treg cells during steady state and these subsets collectively help maintain homeostasis in these tissues. The fate of commensal-specific T cells is dependent on the microbe they are specific to. For instance, the presence of SFB, a commensal that resides in the ileum, has been shown to drive TH17 differentiation, whereas T cells that are specific to some Clostridia species develop into colonic Treg cells under homeostatic conditions (43, 45, 46, 98). A comprehensive analysis of T cells from human peripheral blood and mucosal samples recently revealed that commensal-specific T cells are similarly present in healthy individuals, mainly have a memory phenotype and are capable to express different cytokines according to their immune specificities (9). In memory phase, commensal-specific CD4+ T cells behave like pathogen-specific CD4+ T cells, persisting after clearance of infection but declining in numbers over time (49, 99). Understanding the mechanisms behind the reduction of commensal-specific T cell numbers despite the persistence of their cognate antigen could shed light on the different capacity of individuals to recover from intestinal challenges (100).
Interestingly, despite the large antigenic load of commensal bacteria present within the intestinal lumen, T cells specific for certain bacterial antigens remain naïve in mice under homeostatic conditions (48). Upon disturbance of the barrier-mediated segregation of bacteria to the intestinal lumen, however, activation of T cells toward such antigens may take place. Whether activation of CD4+ T cell responses toward such antigens is directly contributing to the pathogenesis of inflammation or represents a mere epiphenomenon in the broader context of barrier disruption is still an open question.
Pathogenesis of commensal-specific T cells
Local passage of either pathogenic or normally tolerated bacteria through the mucosal barrier and their systemic translocation can cause disease. As described above, despite the fact that commensal antigens are non-self, their coexistence within the host is finely regulated via multiple immune tolerance mechanisms, spanning from an efficient epithelium and mucus-mediated segregation to the secretion of antimicrobial peptides, and specific antibodies (101–103). Contact between host epithelial cells and commensals within the intestinal lumen is not in itself problematic since some bacterial species have been described to colonize the epithelial surface and induce protective immune responses in an adherence-dependent manner (43, 104).
Mice with genetic defects in the barrier develop spontaneous intestinal inflammation (105, 106) but gut-resident macrophages have been shown to efficiently clear invading pathogens thus preventing their systemic translocation (107). Intestinal immune responses are normally limited to the intestines and associated lymphoid tissue, since DCs that pick up commensal antigens migrate from the gut mucosa to the gut-draining MLNs, but not further into the body (108). However, antibodies against skin- and gut-resident commensals have been identified in humans, suggesting that we are challenged throughout life with commensal antigens and some of these immune responses have systemic effects (109). As a result of a mucosal firewall-breaching pathogen, commensal-specific T cells are activated and expand in the gastrointestinal tract (49, 110, 111) and this may result in the persistent disruption of migratory DC trafficking and impairment of tissue-specific adaptive immunity and tolerance (112). Furthermore, an acute enteric infection model has been shown to result in the differentiation of normally quiescent commensal-specific T cells into pro-inflammatory TH1 cells (49).
Emerging evidence points toward the intestinal microbiota as a major factor in the establishment, persistence and/or resolution of intestinal, and extra-intestinal immune diseases (Figure 2). Although the definition of a “healthy” microbiota is still under discussion, a balanced microbiota composition is necessary to maintain intestinal immune homeostasis (36). By contrast, dysbiosis has been associated with disease by promoting immune dysregulation and inflammation in the gut and beyond. This unbalance is reflected by modifications of the immune response to physiologically-recognized bacterial antigens and novel recognition of bacteria to which the immune system is normally ignorant. For instance, pro-inflammatory T cell responses are observed toward classical Treg-inducer Clostridium species during gastrointestinal infection (49). Furthermore, immunoglobulins reactive to commensal antigens can be found in patients with active gut inflammation (50).
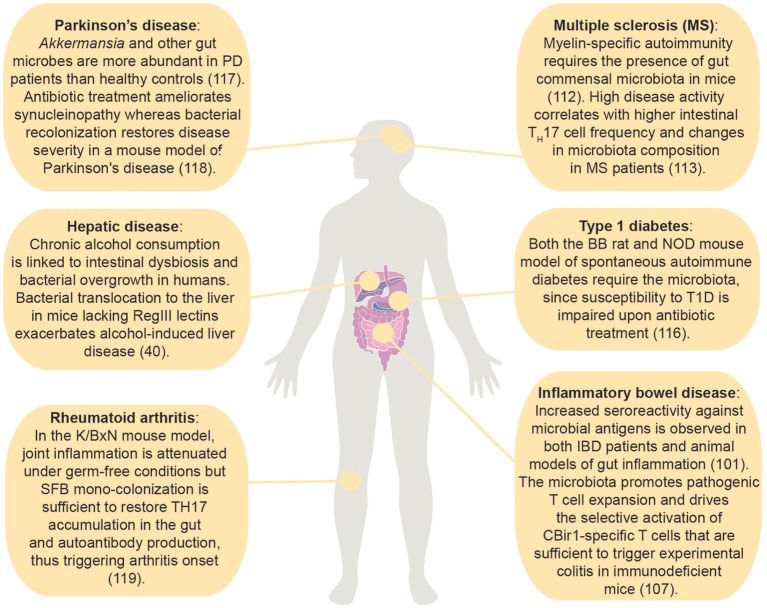
Figure 2.
Effects of the intestinal microbiota on immune mediated diseases within and outside the gastrointestinal tract. (50); (113); (114); (115); (116); (59); (117); (51); (118).
While the microbiota plays an important role in the determination of T cell fate, its effects are buffered by the genetic background of the host. For example, Treg induction following the introduction of altered Schaedler flora (ASF) was shown to be mouse strain-dependent (119). Indeed, while IBD is recognized as a disease caused by an aberrant immune response against commensals in the intestinal mucosa, it is unknown why some people develop chronic inflammation upon barrier damage while others recover (50, 100).
Intestinal disorders
The intestinal mucosa and its immune system, by providing the major interface between gut bacteria and the host, are susceptible to a number of different pathological conditions upon disturbance of their delicate symbiotic relationship. IBD is considered an intestinal disorder characterized by an aberrant immune response against the commensal microbiota in genetically-susceptible hosts, most likely triggered by the environment. Commensal-specific T cells are thought to play a direct role in IBD pathogenesis. For instance, transfer of naïve T cells in lymphopenic mice is sufficient to induce chronic intestinal inflammation in a microbiota-dependent manner (120, 121). Furthermore, increased seroreactivity against a commensal-derived flagellin expressed by a subset of Clostridia (CBir1) was found to induce a systemic adaptive immune response in both Crohn's disease patients and colitic mice, suggesting a microbiota-specific immune response (50, 122, 123). Indeed, the microbiota has been shown to promote pathogenic T cell expansion and drive the selective activation of CBir1-specific T cells that are sufficient to trigger experimental colitis in immunodeficient mice (51). Recent studies suggest that during colitis an effector T cell response is specifically activated against other microbial antigens, e.g., those provided by the mucosal-associated Helicobacter spp. that elicit Treg responses during homeostatic conditions (53, 122). Today, several pathogenic T cells have been identified in mouse and humans, such as the IL-17 and IFNγ double-producer T cells as well as the recently described IL-22BP-producer T cells (124, 125). Whether these T cells are microbiota-specific needs to be demonstrated.
Evidence also suggests that commensal-specific T cells may play a role in the pathogenesis of colorectal cancer (CRC) and dietary conditions. Epidemiological studies reveal that chronic inflammation, such as the one resulting from IBD, increases the risk of CRC (126). The gut microbiota was shown to play a role in both stages of CRC-associated inflammation. Studies in IL-10 knockout mice showed that alteration of the microbiota by introduction of probiotic Lactobacilli can reduce the incidence of both mucosal inflammation and spontaneous CRC development in these mice (127). In addition, commensals have been linked to dietary conditions. For example, through their distinct metabolic patterns, some commensal bacteria are able to modify immunogenicity of dietary proteins, thus triggering gluten-specific T-cell responses and celiac disease (128). Whether bacteria-targeted therapy is effective in treating or preventing these conditions remains to be determined.
Extra-intestinal disorders
Remarkably, gut commensal-specific T cells circulate systemically (9) and the gut microbiota affects the balance between pro-inflammatory and anti-inflammatory T cell responses also in extra-intestinal autoimmune diseases. For example, myelin-specific autoimmunity requires the presence of gut commensal microbiota, as confirmed by lack of spontaneous EAE in GF compared to SPF susceptible mice (113). Similarly, in multiple sclerosis patients, high disease activity correlated with higher intestinal TH17 cell frequency, and changes in microbiota composition (114). Furthermore, recent evidence unveiled the importance of the microbiota for the development of spontaneous RA (129, 130). The association between the intestinal environment and type 1 diabetes (T1D) relies on a constantly increasing number of studies both in T1D patients and mouse models of the disease. The requirement for the gut microbiota in the pathogenesis of T1D has been proven in both the BB rat and NOD mouse model of spontaneous autoimmune diabetes, whose susceptibility to the disease is impaired upon antibiotic treatment (117). Unlike in MS and RA models, however, the effect of the microbiota on the induction of pathogenic or protective immune cell responses is not unanimously accepted in T1D. However, since the antigen-specificity of microbiota-induced T cells has been poorly characterized in these disease settings, further studies are required to understand the role of commensal-specific T cells in tissue-specific autoimmune diseases. A functional link between the microbiota and Parkinson's disease (PD), a neurodegenerative disorder, was also recently delineated. Akkermansia and other gut microbes were more abundant in PD patients than healthy controls (115). Antibiotic treatment was capable of ameliorating synucleinopathy (motor dysfunctions derived from the aggregation of the protein α-synuclein) in a mouse model of Parkinson's disease, and recolonization of adult mice with commensal bacteria derived from PD patients or administration of selected bacterial metabolites, namely SCFAs, was sufficient to restore disease penetrance (116). Whether or not reestablishment of a normal microbiota composition can revert PD and other neurogenerative conditions remains to be determined.
Concluding remarks
The use of experimental mouse models has provided key insights into the complex regulation of intestinal immune homeostasis by the microbiota. Descriptive studies comparing IBD patients and healthy individuals have implicated CD4+ T cell-mediated immune responses against commensal bacteria as a potential key factor in the progression of the disease. Moreover, emerging evidence points toward far-reaching consequences of altered microbiota-mediated T cell responses originated in the gut. For instance, aberrant T cell differentiation in the gut have shown effects in skin tumor growth, the onset of T1D as well as MS. Hence, it is broadly accepted that, besides its gut-centric effects, the microbiota has also extra-intestinal repercussions on its host. The role of commensal-specific T cells in various disease settings is yet to be determined and only with deeper understanding of the molecular and cellular mechanism driving commensal specific T cell differentiation and function will the goal of targeting T cell responses to cure IBD become feasible. Toward this goal, the advances in microbiota analysis, use of gnotobiotic mice and TCR transgenic experimental models provides the potential to discern such mechanisms. Moreover, progress in available tools to track commensal specific T cells in humans are urgently needed. Accomplishing these goals will require tight collaboration between researchers and the clinics.
Author contributions
CS, RFC, and EJV outlined and wrote the manuscript. CS and EJV designed and made the figures. NG revised the manuscript. All authors approved the manuscript for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by funding from the Swedish Research Council VR grants K2015-68X-22765-01-6 and 2017-02932, the Wallenberg Academy Fellow (WAF) program and the European Union's Horizon 2020 research and innovation programme (grant agreement No 746628).
References
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124:837–48. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. (2006) 7:688–93. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. (2009) 28:623–67. 10.1146/annurev-immunol-030409-101330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol. (2001) 9:206–8. 10.1016/S0966-842X(01)02041-8 [DOI] [PubMed] [Google Scholar]
- Sekirov I, Finlay BB. The role of the intestinal microbiota in enteric infection. J Physiol. (2009) 587:4159–67. 10.1113/jphysiol.2009.172742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. (2013) 14:685–90. 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. (2010) 8:171–84. 10.1038/nrmicro2297 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. (2010) 10:159–69. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, et al. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology (2017) 153:1320–37.e16. 10.1053/j.gastro.2017.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. (2006) 24:419–66. 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol. (2014) 14:377–91. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature (2007) 449:811–8. 10.1038/nature06245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ. Polysaccharide breakdown by anaerobic microorganisms inhabiting the mammalian gut. Adv Appl Microbiol. (2004) 56:89–120. 10.1016/S0065-2164(04)56003-3 [DOI] [PubMed] [Google Scholar]
- LeBlanc JG, Chain F, Martin R, Bermudez-Humaran LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact (2017) 16:79. 10.1186/s12934-017-0691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool-Zobel B, Veeriah S, Böhmer F. Modulation of xenobiotic metabolising enzymes by anticarcinogens—focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mut Res. (2005) 591:74–92. 10.1016/j.mrfmmm.2005.04.020 [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. (2007) 9:1101–11. 10.1111/j.1462-2920.2007.01281.x [DOI] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature (2011) 469:543–7. 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe (2009) 6:187–96. 10.1016/j.chom.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Bruce AW. Probiotics to prevent urinary tract infections: the rationale and evidence. World J Urol. (2006) 24:28–32. 10.1007/s00345-005-0043-1 [DOI] [PubMed] [Google Scholar]
- Brook I. The role of bacterial interference in otitis, sinusitis and tonsillitis. Otolaryngol Head Neck Surg. (2005) 133:139–46. 10.1016/j.otohns.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. (2004) 28:405–40. 10.1016/j.femsre.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Tilman D. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA. (2004) 101:10854–61. 10.1073/pnas.0403458101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. (1963) 42:471–83. [PMC free article] [PubMed] [Google Scholar]
- Esterhazy D, Canesso MC, Muller PA, Lockhart A, Mesin L, Faria AM, et al. Compartmentalized lymph node drainage dictates intestinal adaptive immune responses. BioRxiv [Preprint] (2018). 10.1101/299628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. (2008) 76:4726–36. 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature (2009) 461:1282–6. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunlist M, Van Landeghem L, Mahe MM, Derkinderen P, des Varannes SB, Rolli-Derkinderen M. The digestive neuronal-glial-epithelial unit: a new actor in gut health and disease. Nat Rev Gastroenterol Hepatol. (2013) 10:90–100. 10.1038/nrgastro.2012.221 [DOI] [PubMed] [Google Scholar]
- Pelaseyed T, Bergstrom JH, Gustafsson JK, Ermund A, Birchenough GM, Schutte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. (2014) 260:8–20. 10.1111/imr.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science (2002) 295:1726–9. 10.1126/science.1069094 [DOI] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. (2008) 105:15064–9. 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut (2014) 63:281–91. 10.1136/gutjnl-2012-303207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelblum KL, Sharon G, Singh G, Odenwald MA, Sailer A, Cao S, et al. The microbiome activates CD4 T-cell-mediated immunity to compensate for increased intestinal permeability. Cell Mol Gastroenterol Hepatol. (2017) 4:285–97. 10.1016/j.jcmgh.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature (2012) 491:254–8. 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Watkins KC, Popper H. Immunologic competence and phagocytosis in germfree animals with and without stress. JAMA (1964) 187:715–8. 10.1001/jama.1964.03060230043011 [DOI] [PubMed] [Google Scholar]
- Martin R, Bermudez-Humaran LG, Langella P. Gnotobiotic rodents: an in vivo model for the study of microbe-microbe interactions. Front Microbiol. (2016) 7:409. 10.3389/fmicb.2016.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell (2012) 149:1578–93. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science (1996) 274:94–6. 10.1126/science.274.5284.94 [DOI] [PubMed] [Google Scholar]
- Sims S, Willberg C, Klenerman P. MHC–peptide tetramers for the analysis of antigen-specific T cells. Expert Rev Vaccines (2010) 9:765–74. 10.1586/erv.10.66 [DOI] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature (2014) 510:152–6. 10.1038/nature13279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science (2015) 348:1031–5. 10.1126/science.aaa4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Cao AT, Weaver CT, Elson CO, Cong Y. Interleukin-12 converts Foxp3+ regulatory T cells to interferon-gamma-producing Foxp3+ T cells that inhibit colitis. Gastroenterol. (2011) 140:2031–43. 10.1053/j.gastro.2011.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature (2018) 554:373–7. 10.1038/nature25500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139:485–98. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe (2011) 10:260–72. 10.1016/j.chom.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature (2011) 478:250–4. 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (2011) 331:337–41. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutsch K, Chai JN, Ai TL, Russler-Germain E, Feehley T, Nagler CR, et al. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep. (2016) 17:206–20. 10.1016/j.celrep.2016.08.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. (2009) 106:19256–61. 10.1073/pnas.0812681106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science (2012) 337:1553–6. 10.1126/science.1220961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. (2004) 113:1296–306. 10.1172/JCI200420295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. (2010) 207:1321–32. 10.1084/jem.20092253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaranunt P, Tometich JT, Ji J, Hand TW. T cell proliferation and colitis are initiated by defined intestinal microbes. J Immunol. (2018) 201:243–50. 10.4049/jimmunol.1800236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. (2017) 2:eaal5068. 10.1126/sciimmunol.aal5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol. (2017) 2:eaao1314. 10.1126/sciimmunol.aao1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science (2017) 358:359–65. 10.1126/science.aan4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science (2005) 307:731–4. 10.1126/science.1104911 [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. (2008) 105:20858–63. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science (2011) 334:255–8. 10.1126/science.1209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe (2016) 19:227–39. 10.1016/j.chom.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijon HB, Suarez-Lopez L, Diaz OE, Das S, De Calisto J, Yaffe MB, et al. Intestinal epithelial cell-specific RARalpha depletion results in aberrant epithelial cell homeostasis and underdeveloped immune system. Mucosal Immunol. (2018) 11:703–15. 10.1038/mi.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. (2010) 28:445–89. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K, Nakayama T. CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm. Int Immunol. (2016) 28:163–71. 10.1093/intimm/dxw006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askar M. T helper subsets & regulatory T cells: rethinking the paradigm in the clinical context of solid organ transplantation. Int J Immunogenet. (2014) 41:185–94. 10.1111/iji.12106 [DOI] [PubMed] [Google Scholar]
- Peterson RA. Regulatory T-cells: diverse phenotypes integral to immune homeostasis and suppression. Toxicol Pathol. (2012) 40:186–204. 10.1177/0192623311430693 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell (2008) 133:775–87. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Gagliani N, Magnani CF, Huber S, Gianolini ME, Pala M, Licona-Limon P, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. (2013) 19:739–46. 10.1038/nm.3179 [DOI] [PubMed] [Google Scholar]
- Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. (2015) 15:185–9. 10.1038/nri3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Li MO. TGF-beta: guardian of T cell function. J Immunol. (2013) 191:3973–9. 10.4049/jimmunol.1301843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol. (2014) 15:98–108. 10.1038/ni.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeris T, Muller-Luda K, Agace WW, Mowat AM. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol. (2017) 10:845–64. 10.1038/mi.2017.22 [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. (2001) 2:361–7. 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. (2006) 203:2841–52. 10.1084/jem.20061884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature (2013) 494:116–20. 10.1038/nature11809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity (2013) 38:958–69. 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B, et al. Intestinal monocyte-derived macrophages control commensal-specific Th17 responses. Cell Rep. (2015) 12:1314–24. 10.1016/j.celrep.2015.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity (2012) 36:276–87. 10.1016/j.immuni.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell (2016) 166:1231–46.e13. 10.1016/j.cell.2016.07.043 [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature (2013) 498:113–7. 10.1038/nature12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner HL, da Cunha AP, Quintana F, Wu H. Oral tolerance. Immunol Rev. (2011) 241:241–59. 10.1111/j.1600-065X.2011.01017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux FP, Lim JB, Cauley AW, Dikiy S, Ertelt J, Mariani TJ, et al. CD4+ T cell tolerance to tissue-restricted self antigens is mediated by antigen-specific regulatory T cells rather than deletion. Immunity (2015) 43:896–908. 10.1016/j.immuni.2015.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligocki AJ, Niederkorn JY. Advances on non-CD4 + Foxp3+ T regulatory cells: CD8+, type 1, and double negative T regulatory cells in organ transplantation. Transplantation (2015) 99:1553–9. 10.1097/TP.0000000000000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. (2012) 30:531–64. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commins SP. Mechanisms of oral tolerance. Pediatr Clin North Am. (2015) 62:1523–9. 10.1016/j.pcl.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. (2005) 202:1063–73. 10.1084/jem.20051100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, et al. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. (2006) 203:519–27. 10.1084/jem.20052016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. (2007) 204:1775–85. 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. (2008) 205:2139–49. 10.1084/jem.20080414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani B, Villablanca EJ, De Calisto J, Wang S, Mora JR. Vitamin A and immune regulation: role of retinoic acid in gut-associated dendritic cell education, immune protection and tolerance. Mol Aspects Med. (2012) 33:63–76. 10.1016/j.mam.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity (2011) 34:237–46. 10.1016/j.immuni.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Villablanca EJ. Retinoic acid-producing DCs and gut-tropic FOXP3(+) regulatory T cells in the induction of oral tolerance. Oncoimmunology (2013) 2:e22987. 10.4161/onci.22987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature (2013) 500:232–6. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. (2012) 5:232–9. 10.1038/mi.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature (2013) 497:258–62. 10.1038/nature12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai TL, Solomon BD, Hsieh C. T-cell selection and intestinal homeostasis. Immunol Rev. (2014) 259:60–74. 10.1111/imr.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Leithäuser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. (2008) 180:559–68. 10.4049/jimmunol.180.1.559 [DOI] [PubMed] [Google Scholar]
- Asseman C, Read S, Powrie F. Colitogenic Th1 cells are present in the antigen-experienced T cell pool in normal mice: control by CD4 regulatory T cells and IL-10. J Immunol. (2003) 171:971–8. 10.4049/jimmunol.171.2.971 [DOI] [PubMed] [Google Scholar]
- Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. (2015) 6:639. 10.3389/fimmu.2015.00639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4:337–49. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8 but declining CD4 T-cell memory. Nat Med. (2001) 7:913–9. 10.1038/90950 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho M. Immunological pathogenesis of inflammatory bowel disease. Intestinal Res. (2018) 16:26–42. 10.5217/ir.2018.16.1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. (2009) 227:221–33. 10.1111/j.1600-065X.2008.00731.x [DOI] [PubMed] [Google Scholar]
- Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut (2008) 57:764–71. 10.1136/gut.2007.141481 [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 Cell induction by adhesion of microbes to intestinal epithelial cells. Cell (2015) 163:367–80. 10.1016/j.cell.2015.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science (1995) 270:1203–7. 10.1126/science.270.5239.1203 [DOI] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. (2008) 5:e54. 10.1371/journal.pmed.0050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk M, Mueller C. Adaptations of intestinal macrophages to an antigen-rich environment. Semin Immunol. (2007) 19:84–93. 10.1016/j.smim.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann NY Acad Sci. (2004) 1029:36–43. 10.1196/annals.1309.005 [DOI] [PubMed] [Google Scholar]
- Zimmermann K, Haas A, Oxenius A. Systemic antibody responses to gut microbes in health and disease. Gut Microbes (2012) 3:42–7. 10.4161/gmic.19344 [DOI] [PubMed] [Google Scholar]
- Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. (2006) 177:8785–95. 10.4049/jimmunol.177.12.8785 [DOI] [PubMed] [Google Scholar]
- Meinzer U, Barreau F, Esmiol-Welterlin S, Jung C, Villard C, Léger T, et al. Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates caspase-1 to induce intestinal barrier dysfunction. Cell Host Microbe (2012) 11:337–51. 10.1016/j.chom.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Hand TW, Han SJ, Gerner MY, Glatman Zaretsky A, Byrd AL, et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell (2015) 163:354–66. 10.1016/j.cell.2015.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature (2011) 479:538–41. 10.1038/nature10554 [DOI] [PubMed] [Google Scholar]
- Cosorich I, Dalla-Costa G, Sorini C, Ferrarese R, Messina MJ, Dolpady J, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. (2017) 3:e1700492. 10.1126/sciadv.1700492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz-Buschart A, Pandey U, Wicke T, Sixel-Doring F, Janzen A, Sittig-Wiegand E, et al. The nasal and gut microbiome in Parkinson's disease and idiopathic rapid eye movement sleep behavior disorder. Mov Disord. (2018) 33:88–98. 10.1002/mds.27105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's Disease. Cell (2016) 167:1469.e12–80.e12. 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugman S, Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, et al. Antibiotic treatment partially protects against type 1 diabetes in the bio-breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia (2006) 49:2105–8. 10.1007/s00125-006-0334-0 [DOI] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity (2010) 32:815–27. 10.1016/j.immuni.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity (2011) 34:794–806. 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Powrie F, Correa-Oliveira R, Mauze S, Coffman RL. Regulatory interactions between CD45RBhigh and CD45RBlow CD4+ T cells are important for the balance between protective and pathogenic cell-mediated immunity. J Exp Med. (1994) 179:589–600. 10.1084/jem.179.2.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. (2007) 13:1202–11. 10.1002/ibd.20221 [DOI] [PubMed] [Google Scholar]
- Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, et al. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. (2007) 13:1191–201. 10.1002/ibd.20237 [DOI] [PubMed] [Google Scholar]
- Wu W, Liu HP, Chen F, Liu H, Cao AT, Yao S, et al. Commensal A4 bacteria inhibit intestinal Th2-cell responses through induction of dendritic cell TGF-beta production. Eur J Immunol. (2016) 46:1162–7. 10.1002/eji.201546160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. (2007) 204:1849–61. 10.1084/jem.20070663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature (2012) 491:259–63. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature (2008) 454:436–44. 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- O'Mahony L, Feeney M, O'Halloran S, Murphy L, Kiely B, Fitzgibbon J, et al. Probiotic impact on microbial flora, inflammation and tumour development in IL-10 knockout mice. Aliment Pharmacol Ther. (2001) 15:1219–25. 10.1046/j.1365-2036.2001.01027.x [DOI] [PubMed] [Google Scholar]
- Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK, et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology (2016) 151:670–83. 10.1053/j.gastro.2016.06.041 [DOI] [PubMed] [Google Scholar]
- Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. (1994) 180:2359–64. 10.1084/jem.180.6.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, et al. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. (2005) 201:949–60. 10.1084/jem.20041758 [DOI] [PMC free article] [PubMed] [Google Scholar]