Abstract
Purpose
The relationship between the pharmacokinetics of irinotecan and outcomes of advanced colorectal cancer is unclear, and few studies have examined individualized irinotecan-based chemotherapy depending on plasma 7-ethyl-10-hydroxy camptothecin (SN-38) levels and dihydropyrimidine dehydrogenase (DPD) activity, particularly for the UGT1A1*6 or UGT1A1*28 heterozygous type.
Methods
This study retrospectively explored the relationship among plasma SN-38 level 1.5 hours after critical enzyme for irinotecan (CPT-11) administration (CSN-38 1.5h), plasma SN-38 level 49 hours after CPT-11 administration (CSN-38 49h), DPD activity, and clinical outcomes for the UGT1A1*6 and UGT1A1*28 heterozygous types.
Results
CSN-38 1.5h and CSN-38 49h of the UGT1A1*6 or UGT1A1*28 heterozygous type were close to those of UGT1A1*6 and UGT1A1*28 wild-types; some of those with relatively high CSN-38 1.5h levels obtained better median progression-free survival (mPFS), whereas others with higher CSN-38 49h concentrations showed a relatively high incidence of adverse reactions possibly because of the decreased activity of DPD.
Conclusion
Increasing the dosage of CPT-11 according to CSN-38 1.5h may improve the efficacy in patients with lower CSN-38 1.5h levels. For cases with comparably low DPD activity, advisable primary and subsequent dose adjustment of 5-fluorouracil based on plasma 5-fluorouracil levels may be a practical strategy for reducing the occurrence of adverse reactions for personalized treatment of the UGT1A1*6 or UGT1A1*28 heterozygous type.
Keywords: irinotecan, pharmacokinetics, enzyme activity, uridine diphosphate glucuronosyl-transferase 1A1, colorectal cancer
Introduction
Single-nucleotide polymorphisms (SNPs) in drug metabolizing enzymes have an considerable effect on drug absorption, metabolism, distribution, and excretion and can lead to completely different efficacies and/or adverse reactions (ADRs).1,2 Uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1), which converts 7-ethyl-10-hydroxy camptothecin (SN-38) to SN-38 glucuronide (SN-38G), is a critical enzyme for irinotecan (CPT-11), which is the first-line drug for treating metastatic colorectal cancer (mCRC). Previous studies demonstrated that the incidence of life-threatening ADRs is often linked to mutant homozygotes in UGT1A1*6 and *28, which reduce or inhibit UGT1A1 activity and increase plasma SN-38 concentrations;3,4 however, the incidence of the homozygous genotype is <10% in Asian population.5,6 Thus, in addition to screening for homozygous genotypes that may cause serious ADRs, the main purpose of CPT-11 individualized therapy for these patients is to elucidate whether the pharmacokinetics of CPT-11 is correlated with clinical outcomes so that the dose can be adjusted within a relatively short period to achieve better results.
There may be a widely variable range of UGT1A1 activity for UGT1A1*6 and/or *28 heterozygous types (including *1/*28-*1/*1, *1/*1-*1/*6, and *1/*28-*1/*6 genotypes). Theoretically, to achieve personalized administration, the best strategy is to relate CPT-11 pharmacokinetics parameters with the UGT1A1*6 and *28 genotypes, rather than relying on one factor.7 However, the relationship between plasma SN-38 levels or concentration–time curve (area under the curve [AUC]) and clinical efficacy remains unclear, which may be related to the different distribution of UGT1A1*28 and UGT1A1*6 genotypes between population and poor clinical operations of calculating SN-38/SN-38G AUC for the following CPT-11 dosage. Thus, the maximum tolerance dose (MTD) is determined only by dose escalation.8,9 Our previous studies showed that plasma SN-38 level 1.5 hours after CPT-11 administration (CSN-38 1.5h) was related to progression-free survival (PFS) for UGT1A1*6 and *28 wild-types and to better clinical efficacy for relatively high CSN-38 1.5h.10,11
In addition, CPT-11 is routinely combined with 5-fluorouracil (5-FU) as a first-line treatment for mCRC, and 80%–85% of 5-FU is metabolized to inactive dihydro fluorouracil (DHFU) by dihydropyrimidine dehydrogenase (DPD) in the liver. Serious ADRs such as neutropenia, diarrhea, and oral mucositis, which are similar to those caused by CPT-11, occur in cases of partial or total deficiency of DPD activity, leading to inhibition of plasma 5-FU clearance;12 accordingly, the identification of CPT-11-associated ADRs may be affected. Therefore, it is important to detect DPD activities before FOLFIRI chemotherapy, which can reduce the probability of 5-FU-related ADRs by decreasing the 5-FU dosage for those with lower DPD activities to improve the effectiveness of CPT-11 individualized medication.
Assessing the SNPs UGT1A1*6 and *28 and DPD activities simultaneously is a feasible strategy for dosage personalization of CPT-11, although few studies have examined this approach.13 Thus, we retrospectively explored the correlation between clinical parameters such as CSN-38 1.5h, plasma SN-38 level 49 hours after CPT-11 administration (CSN-38 49h), DPD activity, and outcomes (efficacy and ADRs) to provide a basis for individualized CPT-11 administration according to plasma SN-38 levels and DPD activity for patients with the UGT1A1*28 or *6 heterozygous genotypes.
Methods
Patient’s eligibility
The SNPs of UGT1A1*6 and *28 were detected in 550 cases before the first chemotherapy treatment from December 2012 to May 2014, and 499 cases met the following inclusion criteria: previously untreated local advanced or mCRC with measurable lesions verified by pathological and imaging data, East Cooperative Oncology Group (ECOG) physical status score of 0–2 points, life expectancy greater than 3 months, no chemotherapy contraindication, written informed consents, serum bilirubin levels, and transaminase levels limited to 1.5-and 5-fold the normal levels, and ability to undergo administration of at least three cycles of FOLFIRI chemotherapy, as well as one assessment. Patients with complete or incomplete intestinal obstruction, chronic enteritis, a history of extensive colectomy, severe allergy to CPT-11 or 5-FU, other malignant tumors and central nervous system metastases, previously treated measurable lesions such as by radiotherapy or local interventional therapy, major organ dysfunction, and poor compliance and pregnancy were ruled out. A total of 234 cases confirmed with UGT1A1*28 and/or *6 heterozygous genotype were analyzed, which include those from the Zhongshan Hospital (54 cases), Cancer Medical Center (43 cases) affiliated with Fudan University Shanghai Medical College, Ruijin Hospital (41 cases), Renji Hospital (36 cases), and General Hospital (30 cases) affiliated with Shanghai Jiaotong University Medical of School and Shanghai Tenth People’s Hospital (20 cases) affiliated with Tongji University (Table 1).
Table 1.
Clinical characteristics between UGT1A1*28 and *6 heterozygous genotypes
| Clinical characteristics | *1/*28-*1/*1 genotype (n=98) | *1/*1-*1/*6 genotype (n=116) | F | P |
|---|---|---|---|---|
| ECOG performance score | 0.10 | 0.75 | ||
| 0 | 52 | 59 | ||
| 1 | 46 | 57 | ||
| Gender | 2.39 | 0.12 | ||
| Male | 61 | 60 | ||
| Female | 37 | 56 | ||
| Median age (years) | 55.55 | 54.90 | 0.03 | 0.87 |
| Primary tumor site | 1.76 | 0.19 | ||
| Colon | 46 | 65 | ||
| Rectum | 52 | 51 | ||
| TMN staging | 0.01 | 0.91 | ||
| IIIb | 8 | 9 | ||
| IV | 90 | 107 | ||
| Metastatic organs | 0.95 | 0.62 | ||
| 1 | 71 | 87 | ||
| 2 | 22 | 26 | ||
| 3 | 5 | 3 | ||
| Chemotherapy cycles | 8.26±2.74 | 9.78±2.32 | 5.09 | 0.03 |
| CPT-11 initial dosage (mg) | 293.60±30.28 | 292.33±28.29 | 0.82 | 0.37 |
| DPD activity (UH2/U) | 4.59±2.17 | 4.83±1.86 | 0.46 | 0.50 |
Abbreviations: CPT-11, irinotecan; DPD, dihydropyrimidine dehydrogenase; ECOG, East Cooperative Oncology Group; UGT1A1, uridine diphosphate glucuronosyltransferase 1A1.
SNPs analysis for UGT1A1
Plasma genomic DNA was collected using a DNA purification kit (Qiagen, Hilden, Germany), and gene fragments containing UGT1A1*6 and *28 polymorphism sites were amplified by PCR (25 µL): 2 µL of 10× PCR buffer (15 mM MgCl2), 2 µL of dNTP (2.5 mM), 0.5 µL of sense and antisense primers (10 µM), 0.2 µL of Taq DNA polymerase (5 U/µL), 1 µL of DNA templates, and 18.8 µL of double-distilled water (ddH2O). The primer pairs for *6 and *28 polymorphism points in the UGT1A1 gene were designed as follows: upstream, 5′-TCCCTGCTACCTTTGTG-GAC-3′; downstream, 5′-AGCAGGCCCAGGACAAGT-3′. The conditions of PCR amplification were as follows: initial denaturation at 94°C for 5 minutes, followed by 40 cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 25 seconds, extension at 72°C for 50 seconds, and then extension at 72°C for 7 minutes. Next, 5 µL of eligible PCR samples showing clear and stable bands in gel electrophoresis were mixed with 2 µL of shrimp ALP (SAP) and then stored at 4°C after incubation at 37°C for 60 minutes and subsequent incubation at 80°C for 15 minutes. Approximately 3 µL of positive PCR enzymatic hydrolysates, 1 µL of sequencing reagent (BigDye® ; Applied Biosystems, Foster City, CA, USA), and 2 µL of sequencing primer containing fragments of UGT1A1*28 and UGT1A1*6 were used for PCR amplification. The primer pairs were designed and synthesized as follows: UGT1A1*28 primer: forward; 5′-CAGCCTCAAGACCCCACA-3′, reverse: 5′-TGCTCCTGCCAGAGGTTC-3′; UGT1A1*6 primer: forward: 5′-TCCCTGCTACCTTTGTGGA-3′, reverse: 5′-AGGAAAGGGTCCGTCAGC-3′. PCR amplification was conducted in 25 cycles of denaturation at 96°C for 10 seconds, annealing at 50°C for 5 seconds, and extension at 60°C after an initial denaturation at 96°C for 1 minute; the temperature was maintained at 4°C after the reaction. Finally, the reaction products were directly sequenced with a DNA sequencer (ABI-373; Thermo Fisher Scientific, Waltham, MA, USA), and the sequencing results were analyzed and displayed by FinchTV® software.
DPD activity (UH2/U) determination
The internal standard, composed of 250 µL of plasma and 50 µL of 5-bromouracil (5-BrU) solution (2 µg/mL), was added to 1.5 mL extraction solution consisting of n-propanol: tert-butyl ether (25:75, v/v), and the mixture was vortexed for 2 minutes and centrifuged at 3,550× g for 5 minutes. The abovementioned steps were repeated after the organic phase was separated. The inorganic phase was dried with nitrogen and redissolved in 18 µL of ddH2O, and then 20 µL of dichlo-romethane was added after vortex mixing for 30 seconds, followed by centrifugation at 2,250× g for 10 minutes. After vortexing for 5 seconds, 10 µL of the supernatant was injected into a high-performance liquid chromatography system. UH2/U detection was performed using the Shimadzu UFLC chromatographic system (Shimadzu Corporation, Kyoto, Japan), which was equipped with two LC-20AD pumps, a model DGU-20A3 degasser unit, an SIL-20A autosampler, a CTO-20AC thermosetted column compartment, and a model RF-10AXL fluorescence detector. Data were processed with Shimadzu LC-Solution chromatography software (version 1.21, SP1). Analytes were separated at room temperature using a Welch Ultimate XB-C18 column (4.6×150 mm, 5 µm). Detection was carried out with 20 µL of injection volume at an excitation wavelength of 385 nm and emission wavelength of 535 nm at a column temperature of 25°C. The mobile phase consisted of acetonitrile:0.05 M Na2HPO4 salt solution:triethylamine (72.5:27.5:0.5, v:v:v) with a flow rate at 1.0 mL/min and was adjusted to pH 5.0 by phosphate.
Detection of plasma SN-38 levels
SN-38 and internal standard were dissolved in 50% methanol at a concentration of 1.0 mg/mL and stored at −80°C. To draw a calibration curve, an appropriate volume of standard working solution was added to 180 µL aliquots of blank human plasma ranging from 5 to 1,500 ng/mL. All samples were mixed with 100 µL of 7% perchloric acid, vortexed for 3 minutes, and centrifuged at 15,800× g for 10 minutes. Plasma SN-38 levels were detected using the Shimadzu UFLC chromatographic system as described earlier. Data were processed with Shimadzu LC-Solution chromatography software (version 1.21, SP1).
Evaluation and follow-up
The first evaluation was conducted after three cycles of chemotherapy according to evaluation criteria for the curative effect of solid tumor (Response Evaluation Criteria in Solid Tumors, edition 1.1) for all patients. Efficacy reconfirmation was evaluated 4 weeks later for those who achieved complete or partial remission. ADRs were graded under the Common Terminology Criteria for Adverse Events version 3.0 (CTCAE v3.0). Patients could be administered other treatments such as second-line chemotherapy with or without molecular targeted drugs and best support care after progression and were visited every 3 months for survival analysis. The median follow-up time was 15 months (range, 8–22 months).
Statistical methods
The measurement data were presented as the mean±SD, and the enumeration data were expressed as a rate or composition using SPSS® statistic software (version 19.0; IBM Corporation, Armonk, NY, USA). The Student’s t-test was used for the measurement data such as plasma drug level comparison. Chi-squared test and one-way ANOVA were used for intergroup analysis of classification or connectivity evaluation index such as differences of ADRs. Kaplan–Meier test and Log-rank test were used to determine the differences in survival and efficacy between groups. A P-value <0.05 was considered to indicate significant results.
Results
SNPs and proportion for UGT1A1*6 and/or *28 heterozygous genotype
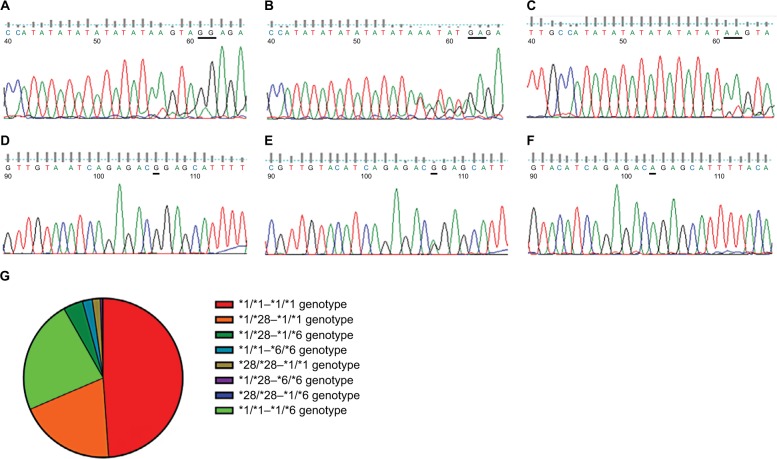
The sequencing results are shown in Figure 1A–F for the 234 cases with the UGT1A1*6 and/or *28 heterozygous genotype (accounting for 46.89%), including 116 cases of the *1/*1-*1/*6 genotype, 98 cases of the *1/*28-*1/*1 genotype, and 20 cases of the *1/*28-*1/*6 genotype. The constituent ratio is shown in Figure 1G.
Figure 1.
Sequencing results of UGT1A1*28 and *6 SNPs and distributive characteristics of different SNP combinations for mCRC patients.
Notes: DNA sequencing for wild-type UGT1A1*28 (A) and UGT1A1*6 (D), heterozygous type UGT1A1*28 (B) and UGT1A1*6 (E), and homozygous type UGT1A1*28 (C) and UGT1A1*6 (E) by FinchTV® software. (G) The pie chart gives the proportion of the different combinations of wild-type (*1/*1-*1/*1 genotype: 244 cases, which accounted for 48.90%), heterozygous type (*1/*28-*1/*1, *1/*1-*1/*6, and *1/*28-*1/*6 genotype: 234 cases, which accounted for 46.89%), and heterozygous type (*28/*28-*1/*1, *1/*1-*6/*6, *1/*28-*6/*6, and *28/*28-*1/*6 genotype: 21 cases, which accounted for only 4.21%).
Abbreviations: mCRC, metastatic colorectal cancer; SNP, single-nucleotide polymorphism; UGT1A1, uridine diphosphate glucuronosyltransferase 1A1.
CSN-38 1.5h and CSN-38 49h between UGT1A1*6 and *28 wild-type and UGT1A1*6 and/or *28 heterozygous genotype
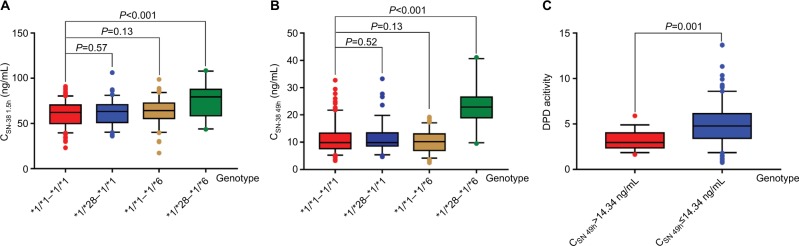
As shown in Figure 2A, the CSN-38 1.5h values of the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes were 61.74±11.49 and 63.10±10.29 ng/mL, respectively, which were not significantly different from that of the *1/*1-*1/*1 genotype (60.84±11.13 ng/mL, P=0.57 and 0.13), but were significantly lower than that of the *1/*28-*1/*6 genotype (75.10±23.16 ng/mL, P<0.001). The same results were observed for CSN-38 1.5h (Figure 2B).
Figure 2.
CSN-38 1.5h and CSN-38 49h between UGT1A1*6 and *28 wild-type and UGT1A1*6 and *28 heterozygous genotype as well as DPD activities between CSN-38 49h>14.34 ng/mL and ≤14.34 ng/mL subgroups.
Notes: The CSN-38 1.5h of *1/*1-*1/*1 genotype was 60.84±11.13 ng/mL, having no significant difference with those of *1/*28-*1/*1 and *1/*1-*1/*6 genotype (61.74±11.49 and 63.10±10.29 ng/mL, P=0.57 and 0.13, respectively), but with statistical difference being found in that of *1/*28-*1/*6 genotype (75.10±23.16 ng/mL, P<0.001, seen in A). Likewise in CSN-38 49h, the CSN-38 49h of *1/*28-*1/*1 and *1/*1-*1/*6 genotype were 11.49±5.06 and 10.29±3.70 ng/mL, respectively, which did not differ obviously from that of *1/*1-*1/*1 genotype (11.13±4.95 ng/mL, P=0.52 and 0.13), while being significantly different from that of *1/*28-*1/*6 genotype (23.16±6.95 ng/mL, P<0.001, shown in B). In C, the DPD activity of CSN-38 49h>14.34 ng/mL subgroup was 3.24±1.02, remarkably lower than that of CSN-38 49h≤14.34 ng/mL subgroup with obvious difference (4.93±2.08, F=11.20, P=0.001).
Abbreviations: CPT-11, irinotecan; CSN-38 1.5h, plasma SN-38 level 1.5 hours after CPT-11 administration; CSN-38 49h, plasma SN-38 level 49 hours after CPT-11 administration; DPD, dihydropyrimidine dehydrogenase; UGT1A1, uridine diphosphate glucuronosyltransferase 1A1.
Moreover, there were no significant differences in gender, age, ECOG performance status, location of the primary tumor, TMN staging, median of chemotherapeutic cycles, initial doses of CPT-11, and DPD activity between the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes (Table 1).
Regression analysis of CSN-38 1.5h and CSN-38 49h with clinical parameters
Stepwise regression analysis was conducted for the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes with CSN-38 1.5h and CSN-38 49h serving as dependent variables, and the initial doses of CPT-11, serum bilirubin levels before and after treatment, chemotherapeutic cycles, short-term response, PFS, overall survival (OS), and ADRs were independent variables. We found that CSN-38 1.5h was related to PFS (t=16.81, P<0.001), whereas CSN-38 49h was related to bone marrow hypocellularity, increased alanine aminotransferase, and diarrhea in the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes (t=8.82, P<0.001; t=5.02, P<0.001; and t=4.84, P<0.001, respectively; Table 2).
Table 2.
Stepwise regression of CSN-38 1.5h, CSN-38 49h, and DPD activity for *1/*28-*1/*1 and *1/*1-*1/*6 genotypes
| Associated clinical parameters | 95% CI | t | P-value | Mean of adjusted predicted value | Standard error of predicted value | ||
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| CSN-38 1.5h (ng/mL) | Constant | 20.21 | 29.36 | 62.51 | 10.69 | ||
| PFS | 5.77 | 7.30 | 16.81 | <0.001 | |||
| CSN-38 49h (ng/mL) | Constant | 6.34 | 7.66 | 20.92 | 10.84 | 3.50 | |
| Bone marrow hypocellularity | 2.14 | 3.37 | 8.82 | <0.001 | |||
| Increased alanine aminotransferase | 1.19 | 2.73 | 5.02 | <0.001 | |||
| Diarrhea | 1.37 | 3.24 | 4.84 | <0.001 | |||
| DPD activity | Constant | 4.89 | 5.75 | 24.20 | 4.66 | 0.53 | |
| Bone marrow hypocellularity | −1.23 | −0.20 | −2.75 | 0.007 | |||
| Increased alanine aminotransferase | −0.81 | −0.17 | −3.03 | 0.003 | |||
Abbreviations: CPT-11, irinotecan; CSN-38 1.5h, plasma SN-38 level 1.5 hours after CPT-11 administration; CSN-38 49h, plasma SN-38 level 49 hours after CPT-11 administration; DPD, dihydropyrimidine dehydrogenase; PFS, progression-free survival.
Median PFS (mPFS) of corresponding CSN-38 1.5h and CSN-38 49h subgroups in *1/*28-*1/*1 and *1/*1-*1/*6 genotypes
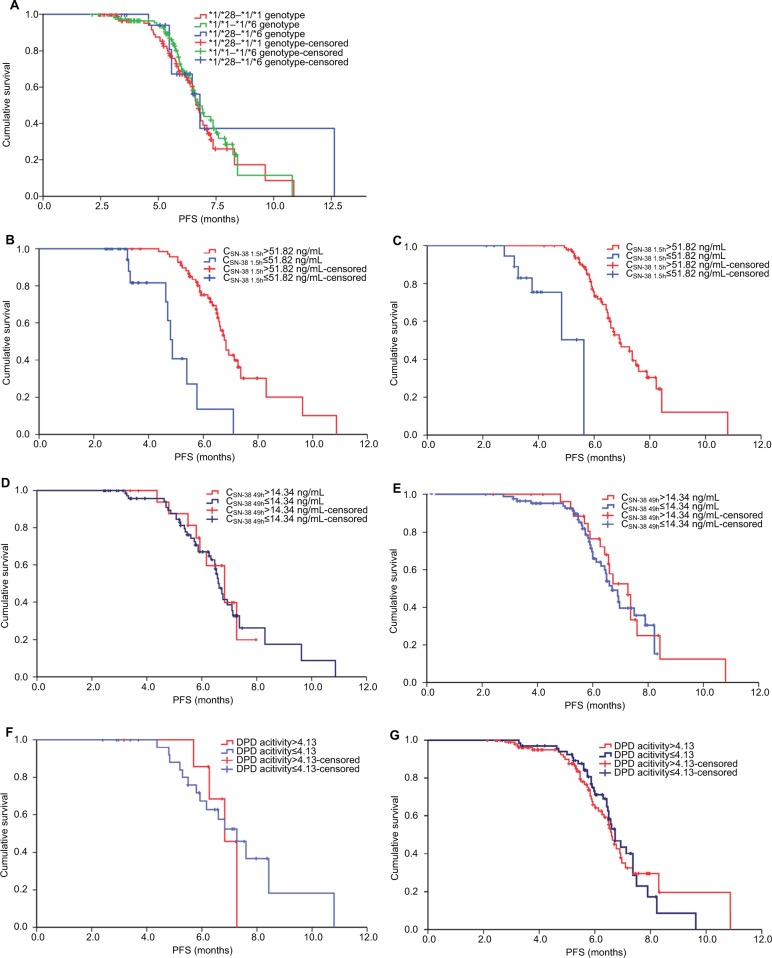
As shown in Figure 3, the mPFS of the *1/*28-*1/*1, *1/*1-*1/*6, and *1/*28-*1/*6 genotypes were 6.73±0.13, 6.73±0.18, and 6.80±0.32 months, respectively, with no significant difference between groups (χ2=1.11, P=0.57). However, a comparison of the CSN-38 1.5h>51.82 ng/mL and CSN-38 49h>14.34 ng/mL subsets with the ≤51.82 and≤14.34 ng/mL subsets, respectively, grouped according to the adjusted predictive values and stand errors of the plasma SN-38 levels in the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes revealed that the mPFS of the CSN-38 1.5h>51.82 ng/mL subgroup was significantly longer than that of the ≤51.82 ng/mL subgroup (6.83±0.17 vs 4.87±0.13 months, 6.93±0.34 vs 5.63±0.31 months; P<0.001, P<0.001), but no significant difference was observed in mPFS between the CSN-38 49h>14.34 ng/mL subgroup and ≤14.34 ng/mL subgroup (6.83±0.48 vs 6.63±0.13 months, 7.27±0.35 vs 6.70±0.21 months, P=0.80 and P=0.59).
Figure 3.
mPFSs of the corresponding CSN-38 1.5h and CSN-38 49h subgroups in *1/*28-*1/*1 and *1/*1-*1/*6 genotype and mPFS of DPD activities between >4.13 and ≤4.13 subsets in CSN-38 49h >14.34 ng/mL and ≤14.34 ng/mL subgroups, respectively, accordingly.
Notes: No statistical difference was observed about the mPFS among *1/*28-*1/*1,*1/*1-*1/*6, and *1/*28-*1/*6 genotypes (A) (6.73±0.13 months vs 6.73±0.18 months vs 6.80±0.32 months, χ2=1.11, P=0.57), but differences were displayed clearly between the mPFS of CSN-38 1.5h>51.82 ng/mL and that of ≤51.82 ng/mL subgroup in *1/*28-*1/*1 (B) and *1/*1-*1/*6 genotypes (C) (6.83±0.17 vs 4.87±0.13 months, P<0.001; 6.93±0.34 vs 5.63±0.31 months, P<0.001), which were divided by the adjusted predictive values and stand errors of CSN-38 1.5h, while the mPFS did not differ between CSN-38 49h>14.34 ng/mL and ≤14.34 ng/mL subgroups grouped by the same way in *1/*28-*1/*1 (D) and *1/*1-*1/*6 genotypes (E) (6.83±0.48 vs 6.63±0.13 months, P=0.80; 7.27±0.35 vs 6.70±0.21 months, P=0.59). The mPFS of DPD activities >4.13 and ≤4.13 subset divided based on the adjusted predictive values and stand errors did not differ obviously in CSN-38 49h>14.34 ng/mL and ≤14.34 ng/mL subgroups of *1/*28-*1/*1 (F) and *1/*1-*1/*6 genotypes (G) (6.83±0.33 vs 7.27±0.53 months, χ2=0.04, P=0.85; 6.60±0.12 vs 6.73±0.22 months, χ2=0.07, P=0.79).
Abbreviations: CPT-11, irinotecan; CSN-38 1.5h, plasma SN-38 level 1.5 hours after CPT-11 administration; CSN-38 49h, plasma SN-38 level 49 hours after CPT-11 administration; DPD, dihydropyrimidine dehydrogenase; mPFS, median PFS; PFS, progression-free survival.
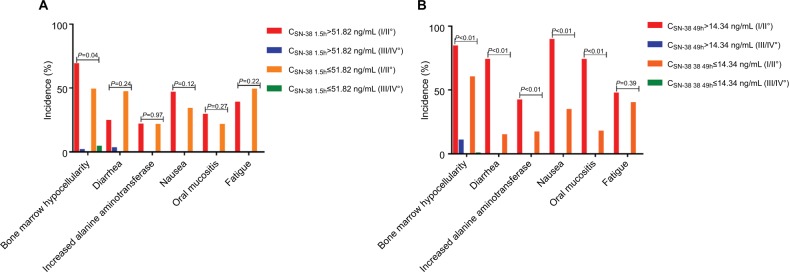
ADRs between corresponding CSN-38 1.5h and CSN-38 49h subgroups in *1/*28-*1/*1 and *1/*1-*1/*6 genotypes
Given the relationship between CSN-38 49h and bone marrow hypocellularity in the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes, the incidence of ADRs was compared between the CSN-38 49h>14.34 and≤14.34 ng/mL subgroups; the results showed that the incidence of bone marrow hypocellularity, diarrhea, increased alanine aminotransferase, nausea, and oral mucositis in the CSN-38 49h>14.34 ng/mL subgroup was significantly higher than that in the ≤14.34 ng/mL subgroup (P<0.001, P<0.001, P<0.001, P<0.001, and P<0.001); however, the difference between the CSN-38 1.5h>51.82 and ≤51.82 ng/mL subgroups was not significant (P=0.04, P=0.24, P=0.97, P=0.12, and P=0.27; A, B).
DPD activities between CSN-38 49h>14.34 and ≤14.34 ng/mL subgroups
Comparison of the DPD activities between the CSN-38 49h>14.34 and ≤14.34 ng/mL subgroups showed that the enzyme activities of the former were clearly lower than those of the latter (3.24±1.02 vs 4.93±2.08, F=11.20, P=0.001; Figure 2C).
mPFS of DPD activities between >4.13 and ≤4.13 subsets in CSN-38 49h>14.34 and ≤14.34 ng/mL subgroups
By setting DPD activities as dependent variables and clinical parameters such as short-term response, PFS, OS, and ADRs as independent variables, stepwise regression indicated that DPD activities were related to the bone marrow hypocellularity and increased alanine aminotransferase (t=–3.03 and t=–2.75, P=0.003 and P=0.007; Table 2), and the mPFS of DPD activities of the >4.13 and≤4.13 subsets divided based on the adjusted predictive values and stand errors did not greatly differ in the CSN-38 49h>14.34 ng/mL subgroup (6.83±0.33 vs 7.27±0.53 months, χ2=0.07, P=0.85; Figure 3).
Discussion
Dosage individualization of chemotherapeutic drugs is an important factor in personalized cancer treatment, and it has been widely acknowledged in mCRC that CPT-11-associated life-threatening ADRs can be avoided by screening out the UGT1A1 homozygous genotype before administration of CPT-11-based chemotherapy;14,15 however, meta-analysis and studies did not confirm the relationship between the UGT1A1*6 and *28 genotypes and clinical outcomes,3,16–19 but individual dose adjustment is difficult based only on the UGT1A1 genotype. Moreover, most Asian populations have wild-type UGT1A1 or a heterozygous genotype, and the risks of CPT-11-associated serious ADRs are much lower than those for the homozygous genotype according to some meta-analyses, as SN-38 glucuronidation of the former two has been completely saturated.20 Therefore, the main purpose of personalized CPT-11 administration is to improve the therapeutic effect by dosage adjustment based on SN-38 pharmacokinetics. The MTD restricts dose escalation because of the factors such as the dose increase extent, escalation intervals, and patients’ compliance, leading to different subclinical doses administered to patients and distress in judging the outcomes of CPT-11. Accordingly, it is necessary to take SN-38 pharmacokinetics into account when the correlation between different genotypes and outcomes are evaluated, particularly for the heterozygous genotype, which accounts for a large proportion of patients and shows variable UGT1A1 activities.5,21
Previous studies of pharmacokinetics showed that the plasma CPT-11 levels reached a peak at 1.5 hours and decreased to minimum levels at 25.5 hours after intravenous CPT-11 infusion,22 and thus, the plasma SN-38 levels at 1.5 and 49 hours after CPT-11 infusion were evaluated to reflect the metabolism of CPT-11 in this study to examine CPT-11 dose individualization over a relatively short period. It is difficult to determine MTD by computing the AUC8,23 because of factors such as repeated blood sampling, high cost of the examination, long submission cycle, difficult promotion, and poor compliance of patients. Our results showed that the CSN-38 1.5h and CSN-38 49h of the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes were close to that of the *1/*1-*1/*1 genotype, but significantly lower than that of the *1/*28-*1/*6 genotype (as shown in Figure 2), indicating that the UGT1A1 activities of the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes were similar to that of the *1/*1-*1/*1 genotype. Thus, we selected the *1/*28-*1/*1 and *1/*1-*1/*6 genotypes for retrospective analysis because of the relatively low risk of ADRs for dose personalized adjustment of CPT-11. Table 1 shows that the clinical characteristics of the two genotypes were comparable in combined analysis, and stepwise regression analysis revealed that CSN-38 1.5h was relevant to PFS and CSN-38 49h was associated with ADRs such as bone marrow hypocellularity. Further analysis indicated that the mPFS of the CSN-38 1.5h>51.82 ng/mL subgroup was significantly longer than that of the ≤51.82 ng/mL subgroup, whereas no difference of mPFS was observed between the CSN-38 49h>14.34 and≤14.34 ng/mL subgroups, suggesting that the efficacy can be improved by increasing the CPT-11 dose while monitoring CSN-38 1.5h in the CSN-38 1.5h≤51.82 ng/mL subgroup.
For ADRs, we found no significant difference between the CSN-38 1.5h>51.82 and≤51.82 ng/mL subgroups; however, the ADRS incidence in the CSN-38 49h>14.34 ng/mL subgroup was significantly higher than that in the ≤14.34 ng/mL subgroup. Although a previous study indicated that 5-FU did not change the metabolic process of CPT-11 within a regular dose range,24 the determination of CPT-11-associated ADRs can be affected because similar 5-FU-correlated ADRs are caused by decreased or inhibited DPD activities. To evaluate the high incidence of ADRs in the CSN-38 49h>14.34 ng/mL subgroup, stepwise regression was conducted, which showed that when DPD activities were correlated with bone marrow hypocellularity and increased alanine aminotransferase, the DPD activities of CSN-38 49h>14.34 ng/mL subset were remarkably lower than that of the ≤14.34 ng/mL subset; however, there was no significant difference in mPFS between the DPD activities >4.13 subset and ≤4.13 subset (Figure 2C), indicating that 5-FU-associated ADRs due to decreased activities of DPD were misclassified as CPT-11-related ADRs caused by reduced UGT1A1 activities. This leads to mistaken down-regulation of the CPT-11 dose, and thus, the activity or SNPs of DPD must be determined before adjusting the 5-FU dosage before CPT-11-based chemotherapy. In addition, reducing the doses of 5-FU did not affect the outcomes.
Subsequent dose individualization of CPT-11 and its effect on outcomes require further analysis via plasma 5-FU levels monitoring, improving the stability and repeatability of the method to detect the plasma SN-38 levels, and conducting prospective randomized controlled studies with larger samples. In addition, other biomarkers such as members of the ATP-Binding Cassette Subfamily C (ABCC),25–27 organic anion-transporting polypeptide 1B1,28,29 and other factors including obesity30 and human organ function31,32 require further analysis to identify better biomarkers or combinations of biomarkers for predicting the efficacy and/or ADRs of CPT-11-based chemotherapy.
Conclusion
According to our analyses, a dose increase of CPT-11 based on CSN-38 1.5h may improve the efficacy in patients with lower CSN-38 1.5h levels. For cases with relatively low DPD activity, advisable primary and subsequent dose adjustment of 5-FU based on the plasma 5-FU levels may be a practical strategy for reducing the incidence of 5-FU-associated ADRs for individualized administration of CPT-11 to those with the UGT1A1*6 or *28 heterozygous type.
Ethics approval and consent to participate
The plan of the research has taken full consideration in the principles of safety and fairness and would be risk free to the patients. This article does not contain any studies with human participants or animals performed by any of the authors. The investigator would protect the patients’ rights and privacy to the maximum extent and make sure that there were no conflicts of interest between the contents and the results of the research. Although no formal consent is required for this type of retrospective research, to ensure the implementation of the project, the patients were admitted to the study providing that they come across this principle of “Ethics, consent and permissions”. Before the plasma specimen being collected, the patients were fully informed as follows: the purposes and methods of the study, the plasma specimen as part of the context, the project would not increase the extra medical costs and pain of patients, and the materials and results of the study were used for the purposes of scientific research without conflict of interest. Any report and publication of the individual patient data (in the form of images, videos, voice recordings, etc) needed the approval of the patients enrolled in the study.
Figure 4.
ADRs between corresponding CSN-38 1.5h and CSN-38 49h subgroups in *1/*28-*1/*1 and *1/*1-*1/*6 genotypes.
Notes: It was not significantly distinguished between CSN-38 1.5h>51.82 ng/mL and ≤51.82 ng/mL subgroups (A) (F=6.58, P=0.04; F=2.86, P=0.24; F=0.002, P=0.97; F=2.39, P=0.12; and F=1.20, P=0.27). However, the incidence of bone marrow hypocellularity, diarrhea, increased alanine aminotransferase, nausea, and oral mucositis in CSN-38 49h>14.34 ng/mL subgroup was higher than that in ≤14.34 ng/mL subgroup with statistical difference (B) (F=26.09, P<0.001; F=57.92, P<0.001; F=11.63, P<0.001; F=38.04, P<0.001; and F=49.20, P<0.01). ADRs were graded by CTCAE v 4.03.
Abbreviations: ADR, adverse reaction; CPT-11, irinotecan; CTCAE, Common Terminology Criteria for Adverse Events; CSN-38 1.5h, plasma SN-38 level 1.5 hours after CPT-11 administration; CSN-38 49h, plasma SN-38 level 49 hours after CPT-11 administration.
Acknowledgments
The authors thank pharmacists Guanhua Zhu, Shengying Gu, and Yankun Guo from the Department of Clinical Pharmacy of Shanghai General Hospital for the detection of polymorphisms of UGT1A1 *28 and *6, plasma SN-38 levels, and DPD activities; and Doctor Mei Kang (statistician) from the Clinical Trial Institution of Shanghai General Hospital for statistical work.
Footnotes
Author contributions
This work was completed with the cooperation of all authors. Xun Cai and Rongyuan Zhuang defined the research objective. Xun Cai, Chuan Tian, and Haifeng Ying designed methods and experiments, carried out the laboratory experiments, analyzed the data, interpreted the results, and wrote the manuscript. Rongyuan Zhuang, Haifeng Ying, Xiaowei Zhang, Hongmin Lu, Hui Wang, Qi Li, and Chungang Wang worked together on patient screening and associated data collection, and Shuowen Wang provided guidance on the pharmacokinetic tests. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Deeken JF, Figg WD, Bates SE, Sparreboom A. Toward individualized treatment: prediction of anticancer drug disposition and toxicity with pharmacogenetics. Anticancer Drugs. 2007;18(2):111–126. doi: 10.1097/CAD.0b013e3280109411. [DOI] [PubMed] [Google Scholar]
- 2.Lee W, Lockhart AC, Kim RB, Rothenberg ML. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist. 2005;10(2):104–111. doi: 10.1634/theoncologist.10-2-104. [DOI] [PubMed] [Google Scholar]
- 3.Onoue M, Terada T, Kobayashi M, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Cancer Chemother Pharmacol. 2014;14(2):136–142. doi: 10.1007/s10147-008-0821-z. [DOI] [PubMed] [Google Scholar]
- 4.Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16(15):3832–3842. doi: 10.1158/1078-0432.CCR-10-1122. [DOI] [PubMed] [Google Scholar]
- 5.Jada SR, Lim R, Wong CI, et al. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98(9):1461–1467. doi: 10.1111/j.1349-7006.2007.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Xu JM, Shen L, et al. Polymorphisms of UGT1A gene and irinotecan toxicity in Chinese colorectal cancer patients. Zhonghua Zhong Liu Za Zhi. 2007;29(12):913–916. Chinese. [PubMed] [Google Scholar]
- 7.Patel JN, Papachristos A. Personalizing chemotherapy dosing using pharmacological methods. Cancer Chemother Pharmacol. 2015;76(5):879–896. doi: 10.1007/s00280-015-2849-x. [DOI] [PubMed] [Google Scholar]
- 8.Innocenti F, Schilsky RL, Ramírez J, et al. Dose-finding and pharmacokinetic study to optimize the dosing of irinotecan according to the UGT1A1 genotype of patients with cancer. J Clin Oncol. 2014;32(22):2328–2334. doi: 10.1200/JCO.2014.55.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toffoli G, Sharma MR, Marangon E, et al. Genotype-guided dosing study of FOLFIRI plus bevacizumab in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23(4):918–924. doi: 10.1158/1078-0432.CCR-16-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Cao W, Ding H, et al. Analysis of UGT1A1*28 genotype and SN-38 pharmacokinetics for irinotecan-based chemotherapy in patients with advanced colorectal cancer: results from a multicenter, retrospective study in Shanghai. J Cancer Res Clin Oncol. 2013;139(9):1579–1589. doi: 10.1007/s00432-013-1480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai X, Tian C, Wang L, et al. Correlative analysis of plasma SN-38 levels and DPD activity with outcomes of FOLFIRI regimen for metastatic colorectal cancer with UGT1A1 *28 and *6 wild type and its implication for individualized chemotherapy. Cancer Biol Ther. 2017;18(3):186–193. doi: 10.1080/15384047.2017.1294286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maring JG, van Kuilenburg AB, Haasjes J, et al. Reduced 5-FU clearance in a patient with low DPD activity due to heterozygosity for a mutant allele of the DPYD gene. Br J Cancer. 2002;86(7):1028–1033. doi: 10.1038/sj.bjc.6600208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falvella FS, Cheli S, Martinetti A, et al. DPD and UGT1A1 deficiency in colorectal cancer patients receiving triplet chemotherapy with fluoropyrimidines, oxaliplatin and irinotecan. Br J Clin Pharmacol. 2015;80(3):581–588. doi: 10.1111/bcp.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516. doi: 10.1093/annonc/mds236. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T, Itabashi M, Shimada Y, et al. Japanese Society for Cancer of the Colon and Rectum Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20(2):207–239. doi: 10.1007/s10147-015-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satoh T, Ura T, Yamada Y, et al. Genotype-directed, dose-finding study of irinotecan in cancer patients with UGT1A1*28 and/or UGT1A1*6 polymorphisms. Cancer Sci. 2011;102(10):1868–1873. doi: 10.1111/j.1349-7006.2011.02030.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association between UGT1A1*28 polymorphisms and clinical outcomes of irinotecan-based chemotherapies in colorectal cancer: a meta-analysis in Caucasians. PLoS One. 2013;8(3):e58489. doi: 10.1371/journal.pone.0058489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias MM, Pignon JP, Karapetis CS, et al. The effect of the UGT1A1*28 allele on survival after irinotecan-based chemotherapy: a collaborative meta-analysis. Pharmacogenomics J. 2014;14(5):424–431. doi: 10.1038/tpj.2014.16. [DOI] [PubMed] [Google Scholar]
- 19.Takano M, Sugiyama T2. UGT1A1 polymorphisms in cancer: impact on irinotecan treatment. Pharmgenomics Pers Med. 2017;10:61–68. doi: 10.2147/PGPM.S108656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ichikawa W, Araki K, Fujita K, et al. Re: UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2008;100(3):224–225. doi: 10.1093/jnci/djm302. author reply 225. [DOI] [PubMed] [Google Scholar]
- 21.Zhang A, Xing Q, Qin S, et al. Intra-ethnic differences in genetic variants of the UGT-glucuronosyltransferase 1A1 gene in Chinese populations. Pharmacogenomics J. 2007;7(5):333–338. doi: 10.1038/sj.tpj.6500424. [DOI] [PubMed] [Google Scholar]
- 22.Sumiyoshi H, Fujiwara Y, Ohune T, Yamaoka N, Tamura K, Yamakido M. High-performance liquid chromatographic determination of irinotecan (CPT-11) and its active metabolite (SN-38) in human plasma. J Chromatogr B Biomed Appl. 1995;670(2):309–316. doi: 10.1016/0378-4347(95)00130-1. [DOI] [PubMed] [Google Scholar]
- 23.Kim KP, Hong YS, Lee JL, et al. A phase I study of UGT1A1 *28/*6 genotype-directed dosing of irinotecan (CPT-11) in Korean patients with metastatic colorectal cancer receiving FOLFIRI. Oncology. 2015;88(3):164–172. doi: 10.1159/000368674. [DOI] [PubMed] [Google Scholar]
- 24.Ducreux M, Ychou M, Seitz JF, et al. Irinotecan combined with bolus fluorouracil, continuous infusion fluorouracil, and high-dose leucovorin every two weeks (LV5FU2 regimen): a clinical dose-finding and pharmacokinetic study in patients with pretreated metastatic colorectal cancer. J Clin Oncol. 1999;17(9):2901–2908. doi: 10.1200/JCO.1999.17.9.2901. [DOI] [PubMed] [Google Scholar]
- 25.Mathijssen RH, Marsh S, Karlsson MO, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9(9):3246–3253. [PubMed] [Google Scholar]
- 26.Sai K, Kaniwa N, Itoda M, et al. Haplotype analysis of ABCB1/MDR1 blocks in a Japanese population reveals genotype-dependent renal clearance of irinotecan. Pharmacogenetics. 2003;13(12):741–757. doi: 10.1097/00008571-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura Y, Kusuhara H, Sugiyama Y. Functional characterization of multidrug resistance-associated protein 3 (mrp3/abcc3) in the basolateral efflux of glucuronide conjugates in the mouse small intestine. J Pharmacol Exp Ther. 2010;332(2):659–666. doi: 10.1124/jpet.109.156943. [DOI] [PubMed] [Google Scholar]
- 28.Iusuf D, van de Steeg E, Schinkel AH. Hepatocyte hopping of OATP1B substrates contributes to efficient hepatic detoxification. Clin Pharmacol Ther. 2012;92(5):559–562. doi: 10.1038/clpt.2012.143. [DOI] [PubMed] [Google Scholar]
- 29.Teft WA, Welch S, Lenehan J, et al. OATP1B1 and tumour OATP1B3 modulate exposure, toxicity, and survival after irinotecan-based chemotherapy. Br J Cancer. 2015;112(5):857–865. doi: 10.1038/bjc.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelps MA, Sparreboom A. Irinotecan pharmacogenetics: a finished puzzle? J Clin Oncol. 2014;32(22):2287–2289. doi: 10.1200/JCO.2014.56.3387. [DOI] [PubMed] [Google Scholar]
- 31.Schaaf LJ, Hammond LA, Tipping SJ, et al. Phase 1 and pharmacokinetic study of intravenous irinotecan in refractory solid tumor patients with hepatic dysfunction. Clin Cancer Res. 2006;12(12):3782–3791. doi: 10.1158/1078-0432.CCR-05-2152. [DOI] [PubMed] [Google Scholar]
- 32.Fujita K, Sunakawa Y, Miwa K, et al. Delayed elimination of SN-38 in cancer patients with severe renal failure. Drug Metab Dispos. 2011;39(2):161–164. doi: 10.1124/dmd.110.035451. [DOI] [PubMed] [Google Scholar]