Abstract
Background and Objective
Antibodies to human neutrophil antigens (HNAs) have been implicated in transfusion-related acute lung injury and allo- and autoimmune neutropenia. To date, five HNA systems are assigned, and during the last decades enormous efforts have been undertaken to identify the underlying genes and to characterize the antigens. This review of the literature will provide the current genetic, molecular and functional information on HNAs.
Recent Findings
New information on alleles and antigens has been added to nearly each of the five HNA systems. HNA-1d has been added as the antithetical epitope to HNA-1c that is located on the glycoprotein encoded by FCGR3B*02 but not by FCGR3B. FCGR3B*04 and *05 now are included as new alleles. A CD177*787A>T substitution was demonstrated as the main reason for the HNA-2-negative phenotype on neutrophils. The target glycoprotein of HNA-3 antibodies could be identified as choline transporter-like protein 2 (CTL2) encoded by SLC44A2. The conformation sensitive epitope discriminates between arginine and glutamine at position 152 resulting in HNA-3a and HNA-3b. An additional Leu151Phe substitution can impair HNA-3a antibody binding. Recently an alloantibody against HNA-4b which discriminates from HNA-4a by an Arg61His exchange of the glycoprotein encoded by the ITGAM gene was reported in neonatal alloimmune neutropenia. An update of the current HNA nomenclature based on the new findings was provided in 2016 by the ISBT Granulocyte Immunobiology Working Party nomenclature subcommittee.
Conclusions
The molecular basis of each of the five HNA antigen systems has been decoded during the past decades. This enables reliable molecular typing strategies, antibody detection and specification as well as development of new assays based on recombinant antigens. However, research on HNA alleles, antigens, and antibodies is not finally terminated and also in the future will add new findings.
Keywords: Acute lung injury, Alloimmunization, Antibodies, Antigens, Autoimmunity, DNA, Genetic variation, Genotyping, Granulocytes, HNA, Neutrophil, Polymorphism, Single nucleotide, TRALI
Introduction
Human neutrophil antigens (HNAs) are a group of glycoproteins expressed on the surface of human neutrophil granulocytes which play an important role in allo- and autoimmunity. They exhibit an extended polymorphism resulting in a variety of different allo- and isoantigens. The expression on neutrophil granulocytes is essential but some HNAs are also expressed on a variety of other cells and tissues, for example choline transporter-like protein 2 (CTL2), CD11a, and CD11b [1, 2]. Antibodies to HNAs have been shown to be involved in a variety of clinical conditions like transfusion-related acute lung injury (TRALI), neonatal alloimmune neutropenia (NIN), primary and secondary autoimmune neutropenia, febrile transfusion reactions, and refractoriness to granulocyte transfusions [3]. Long before implementing the term HNA in 1998 by the Granulocyte Antigen Working Party (GIWP) of the ISBT [4], a leukocyte-specific antibody was described by Lalezari and colleagues [5] in a case of neonatal neutropenia. Shortly thereafter the former 5b antigen, which is now termed HNA-3a, was demonstrated in a case of TRALI [6]. In the following decades, further neutrophil antigens were described including their molecular organization and the clinical relevance [3], but it took until 2010 to decode the molecular basis of the former 5b antigen [7, 8].
Today, fourteen HNA alleles have been officially assigned to five HNA antigen systems by the ISBT GIWP [9]. HNA alleles are defined as alternative forms of a gene discriminated mainly by single nucleotide exchanges which code for the respective glycoproteins. A HNA glycoprotein can carry one or more HNA epitopes that are specifically recognized by antibodies. The term epitope defines the antigenic determinant or antibody binding region on an immunogenic molecule. This is used in preference to the term ‘antigen’, which is the superordinate term for a molecule inducing immunization, e.g. HNA. Although the coding genes of each of the currently known five HNA systems are published, even today some open questions on the causal connection between serological phenotypes and genotypes remain [10, 11]. The following review will provide an overview of the molecular genetics, structures, and characteristics of the HNAs.
Nomenclature
The nomenclature of the HNA-encoding genes and alleles and of the antigens which are defined by alloantibody reactivity follows the recommendations published by the ISBT GIWP [4, 9]. The allele names comprise the official gene name (according to the Human Genome Nomenclature Committee (www.genenames.org/) followed by an asterisk and a 2-digit allele number. Genetic variations of standard alleles which have been confirmed by the ISBT GIWP are numbered consecutively in the order of publication. Only missense mutations within exons leading to an amino acid exchange are included. The corresponding phenotype on neutrophil granulocytes should be determined by standard granulocyte serology. New mutations without known phenotype and/or clinical data should be described as mutation of the respective standard alleles following the recommendations of the Human Genome Variation Society (HGV; http://varnomen.hgvs.org/). Thus nucleotide positions in this review strictly meet the rule to start with position number 1 at the A of the start-ATG (1st codon). In the case that more than one transcript variant should exist, amino acid numbers refer to the form expressed on neutrophils.
The HNA nomenclature is an epitope-based rather than antigen-based system. The antigen nomenclature is based on serologically defined epitopes on neutrophil glycoproteins. The phenotypic variation within an HNA antigen system is given in lower case letters, e.g. HNA-1a, HNA-1b or HNA-3a.
HNA-1
In humans, the low-affinity FcγRIII exits in two highly homologous forms, the transmembrane Fc gamma receptor IIIa (FcγRIIIa) which is expressed on macrophages and natural killer (NK) cells and the FcγRIIIb which is restricted to neutrophil granulocytes and is anchored to the cell membrane via glycosyl-phospatidylinositol (GPI) [3, 12, 13, 14, 15]. The underlying genes, FCGR3A and FCGR3B, are part of a region with highly homologous Fcγ receptor(FcγR)-encoding genes on the human chromosome 1q23–24 [14, 16]. The alleles of the HNA-1 system are exclusively encoded by the FCGR3B gene. However, the high degree of homology between FCGR3B and FCGR3A complicates the development of genotyping assays. With about 1–2 × 105 copies per cell the low-affinity IgG1/IgG3 FcγRIIIb mediates clearance of immune complexes and phagocytosis of opsonized microorganisms [17, 18, 19]. The membrane proximal domain is critical for IgG Fc binding, whereas the function of the HNA-1 epitope carrying distal domain remains unclear [3]. The HNA-1 protein backbone of 29–33 kDA is highly glycosylated with four N-linked glycosylation sites (50–65 kDa) for the HNA-1a and six (65–80 kDa) for the HNA-1b form [20].
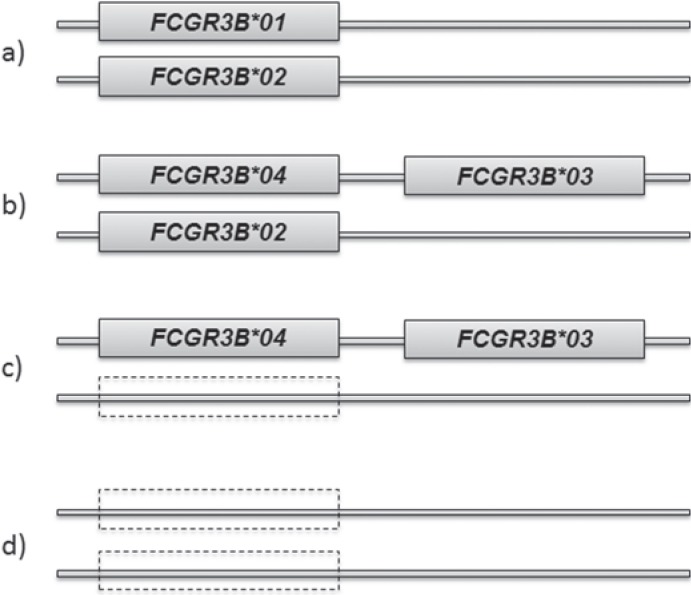
Gene copy number variation seems to be a special feature of FCGR3B. This means that individuals may exhibit between zero and four FCGR3B alleles [21, 22, 23, 24, 25] which can be explained by gene duplication combined with recombination and/or unequal crossing-over during meiosis. This event may result in either two FCGR3B genes in close vicinity on the same chromosome or, conversely, in a FCGR3B gene deficiency [1, 21, 25, 26] (fig. 1). Interestingly, HNA-1 null individuals who completely lack FCGR3B genes on both chromosomes do not suffer from autoimmune or immune complex-mediated diseases or an increased infection rate [23]. The FCGR3B gene copy number variation additionally includes the neighbored FCGR2C gene so that both genes segregate as one haplotype block [27].
Fig. 1.
Some examples of possible FCGR3B allele combinations and copy number variation. From top to bottom a the initial situation with two alleles, here heterozygosity for FCGR3B*01 and FCGR3B*02; b duplication and mutation resulting in FCGR3B*04 and FCGR3B*03 on one chromosome and only one on the 2nd parental chromosome; c allele drop out on one chromosome; d allele drop out on both chromosomes, resulting in an HNA-1null phenotype.
Alleles
Originally described as a bi-allelic system with the alloantigens NA1 (now HNA-1a) and NA2 (now HNA-1b) [28], the HNA-1 system was later found to include a third (SH, now HNA-1c) [22] and a fourth alloantigen (HNA-1d) [29]. The HNA-1 polymorphism originates in five nucleotide substitutions within exon 3 of the FCGR3B gene. Four missense mutations induce amino acid exchanges at the positions 36, 65, 82, and 106 of the FcγRIIIb glycoprotein [1, 16, 30] (table 1). Currently, the official nomenclature comprises five FCGR3B alleles and one FCGR3B*null allele that indicates FCGR3B deficiency [9]. The single nucleotide polymorphism (SNP) position numbers had been corrected following the recommendations of the HGV, based on FCGR3B transcript variant 2 (NM_000570.4) which corresponds to the amino acid positions of the mature glycoprotein [9]. Thus they now differ from the originally described positions [30]. FCGR3B*01 codes for HNA-1a and differs from FCGR3B*02 which codes for HNA-1b in each of the five nucleotide positions. About 58% of Caucasians and up to 91% of Taiwanese express HNA-1a, and vice versa the HNA-1b phenotype is detected in 88% of Caucasians but only 54% of Taiwanese [31]. In contrast, FCGR3B*03, the basis of HNA-1c distinguishes from FCGR3B*02 only in an additional Ala78Asp substitution [22]. Interestingly, many FCGR3B*03-positive individuals exhibit three HNA-1 antigens with a combination of the HNA-1a and HNA-1c encoding alleles on one chromosome, and, as expected, these individuals present with FcγRIIIb hyperexpression [3, 21, 31, 32]. About 5% of German individuals carry the HNA-1c antigen, whereas it is much more frequent in African populations with 22–38% and missing in Chinese. Because gene duplication can be seen as the counterpart of gene deletion, FCGR3B*null alleles leading to the HNA-1null phenotype are more frequently observed in African populations, too [21, 31] (tables with comprehensive antigen and allele frequencies are available at [1, 3, 31, 32, 33]). The fourth allele, FCGR3B*04 with a 316G>A exchange compared to FCGR3B*01 codes for a glycoprotein with unaltered reactivity with HNA-1a-specific sera and monoclonal antibodies [34, 35] in individuals lacking an additional FCGR3B*01 allele. In a study on German blood donors the FCGR3B*04 allele instead of the FCGR3B*01 allele segregated together with FCGR3B*03 on the same chromosome (own observations A. Reil, abstract at the DGTI Meeting 2011 in Hannover, Germany). The FCGR3B*05-encoded glycoprotein which is characterized by an Asn82Asp exchange compared to a wild type HNA-1b [34, 36] exhibited a reduced reactivity with HNA-1b antibodies in the granulocyte immunofluorescence test (GIFT) [35]. A mother carrying the same variant was immunized by a wild-type HNA-1b of her neonate (own observations A. Reil, abstract at the ESPGI Meeting 2014 in Bad Homburg, Germany). Screening among 67 German blood donors typed FCGR3B*02-positive revealed a second case with the same FCGR3B*244A>G mutation. The neutrophils of this individual were not agglutinated by HNA-1b antibodies and showed a strongly reduced antibody binding in the indirect GIFT [2]. The SNPs contributing to the confirmed FCGR3B alleles are provided in the NCBI SNP database as 108G/C = rs 200688856, 194A/G = rs448740, 233C/A = rs503038, 244G/A = rs147574249, 316G/A = 2290934. In addition to the above listed confirmed alleles, a multitude of genomic variants has been reported [26, 36, 37, 38, 39]. However, varying experimental approaches as well as missing phenotype or clinical data limit the informative value of these observations.
Table 1.
HNA alleles and antigens of the five HNA systemsa
| Allele | Nucleotide positions of the corresponding alleles | Amino acid positions of the corresponding glycoproteins | Epitopes | Glycoprotein | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (141) | (147) | (227) | (266) | (277) | (349) | FcγRIIIb, CD16 | |||||||||
| FCGR3B*01 | 108G* | 114C | 194A | 233C | 244G | 316G | 36Arg | 38Leu | 65Asn | 78Ala | 82Asp | 106Val | HNA-1a | ||
| FCGR3B*02 | 108C | 114T | 194G | 233C | 244A | 316A | 36Ser | 38Leu | 65Ser | 78Ala | 82Asn | 106Ile | HNA-1b | HNA-1d | |
| FCGR3B*03 | 108C | 114T | 194G | 233A | 244A | 316A | 36Ser | 38Leu | 65Ser | 78Asp | 82Asn | 106Ile | HNA-1b | HNA-1c | |
| FCGR3B*04 | 108G | 114C | 194A | 233C | 244G | 316A | 36Arg | 38Leu | 65Asn | 78Ala | 82Asp | 106Ile | HNA-1a | ||
| FCGR3B*05 | 108C | 114T | 194G | 233C | 244G | 316A | 36Ser | 38Leu | 65Ser | 78Ala | 82Asp | 106Ile | HNA-1b** | ||
| FCGR3B*null | no alleles | no glycoprotein (gp) | HNA-1null | no gp | |||||||||||
| CD177 | 787A | 242Lys# | HNA-2# | CD177 | |||||||||||
| 787T | 242X | HNA-2null | no gp | ||||||||||||
| SLC44A2*01 | 451C | 455G | 151Leu§ | 152Arg§ | HNA-3a | CTL2 | |||||||||
| SLC44A2*02 | 451C | 455A | 151Leu | 152Gln | HNA-3b | ||||||||||
| SLC44A2*03 | 451T | 455G | 151Phe | 152Arg | HNA-3a** | ||||||||||
| ITGAM*01 | 230G | Arg61 | HNA-4a | CD11b | |||||||||||
| ITGAM*02 | 230A | His61 | HNA-4b | ||||||||||||
| ITGAL*01 | 2372G | Arg766 | HNA-5a | CD11a | |||||||||||
| ITGAL*02 | 2372C | Thr766 | - | ||||||||||||
All nucleotide and amino acid positions follow the recommendations of the Human Genome Variation Society. Nucleotide numbers start at position 1 with the A of the START codon. The originally published FCGR3B nucleotide positions are provided in brackets.
Numbering according to FCGR3B transcript variant 2 (NM_000570.4) which corresponds to the amino acid positions of the mature neutrophil glycoprotein.
Variation of reactivity with human antisera can be noticed.
Currently, HNA-2 appears to be an isoantigen without epitope variation and therefore should be named HNA-2 (instead of HNA-2a). Amino acid position refers to the mature protein after cleavage of the 21 aa signal peptide. 242X = 242stop.
Numbering according to CTL2 transcript variant 2 (NM_001145056.1), which is expressed on neutrophils.
Epitopes
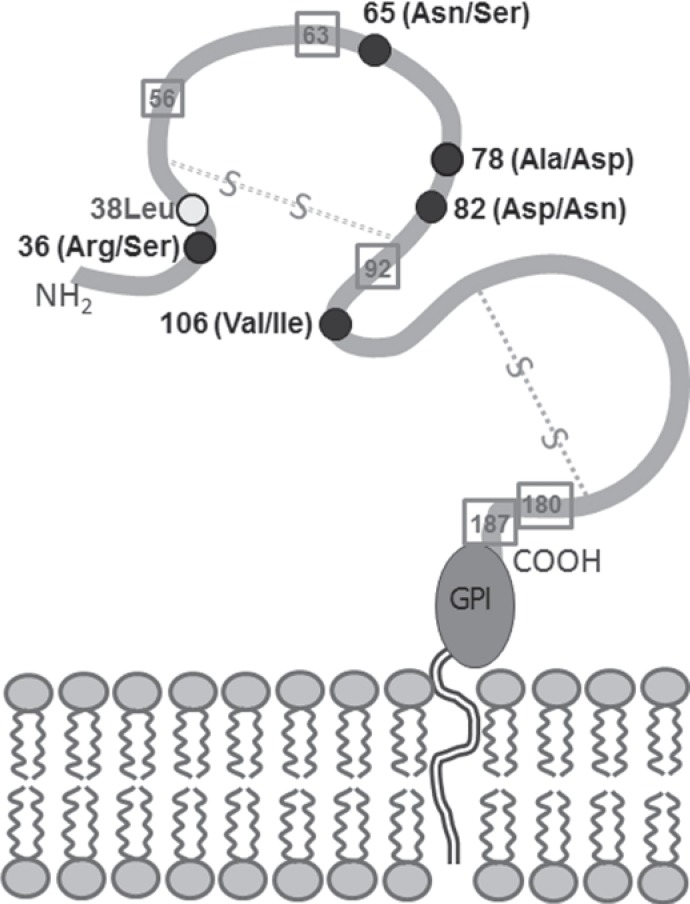
Because the combination of five polymorphic sites within exon 3 of the FCGR3B alleles enable different amino acids at the respective positions within the distal domain of the HNA-1 glycoprotein one molecule can comprise more than one HNA-1 epitope [2]. Both, HNA-1b and HNA-1c are encoded by FCGR3B*03 and HNA-1b and HNA-1d are encoded by FCGR3B*02 (table 1). Vice versa, one epitope can be encoded by more than one allele (e.g. HNA-1a by FCGR3B*01 and FCGR3B*04). Amino acids 65 and 82 seem to be critical for the formation of the HNA-1a and HNA-1b epitopes, but also amino acids 36 and 106 have been described as crucial for the HNA-1a epitope [35, 40] (fig. 2). Because of the high homology with the FcγRIIIa on monocytes and NK cells it is likely that antibodies recognize conformational epitopes rather than linear epitopes covering only few amino acids [3]. The glycoprotein encoded by the FCGR3B*03 allele expresses both the HNA-1c epitope (including 78Asp) and the HNA-1b epitope. In two cases of NIN, maternal antibodies were reactive with HNA-1b on neutrophils from FCGR3B*02-positive, but not FCGR3B*03-positive donors. Both mothers typed as FCGR3B*01+, *02-, *03+. Thus, the antibodies detected the antithetical epitope to HNA-1c covering Ala78 and Asn82 which resides on the same glycoprotein as the HNA-1b epitope on FCGR3B*02-positive neutrophils [29]. The epitope is now assigned to HNA-1d (table 1).
Fig. 2.
Predicted structure of the FcγRIIIb molecule which is anchored to the neutrophil membrane by glycosylphosphatidylinositol (GPI). Amino acid positions contributing to the HNA-1 polymorphism are indicated as filled black circles, the first alternative amino acid indicates the situation of HNA-1a. Square symbols indicate potential N-linked glycosylation sites.
HNA-1 alloantibodies have been implicated in NIN and TRALI [3, 41, 42, 43] while especially HNA-1a is the main antibody specificity detected in primary autoimmune neutropenia of infants younger than 3 years [44, 45, 46].
HNA-2
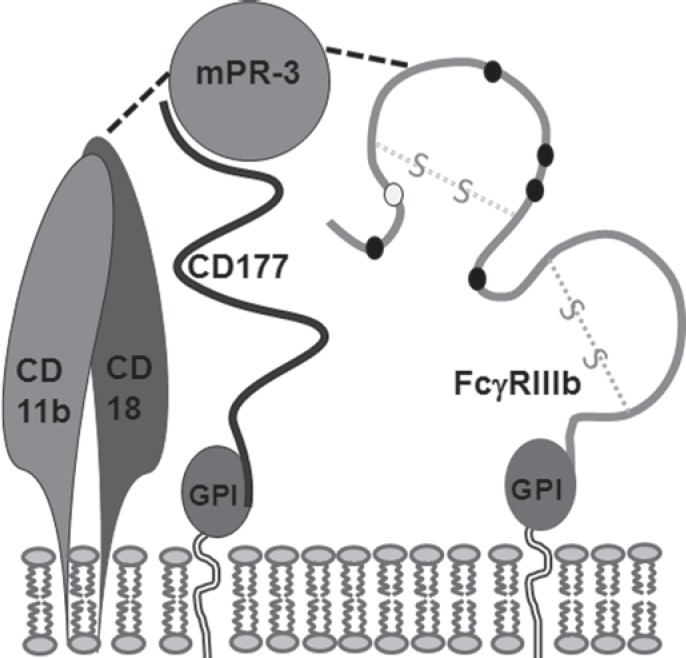
HNA-2 was originally described as neutrophil-specific antigen NB1 in a case of neonatal neutropenia [47]. Later on, the glycoprotein which is anchored to the neutrophil membrane by GPI was identified as CD177 with a molecular mass of 56–64 kDa and three potential N-linked glycosylation sites [48, 49]. HNA-2 and PRV-1, which is overexpressed in neutrophils of polycythemia rubra vera patients, most likely are alleles of the same CD177 gene located on chromosome 19q13.2 [50, 51]. CD177 interacts with proteinase 3 (PR3) leading to the membrane binding of PR3 which usually is stored in intracellular granules and vesicles of neutrophils [52, 53, 54] (fig. 3). The CD177-PR3 interaction aids neutrophil transmigration through the endothelium [55] and enables interaction with CD11b/CD18 and FcγRIIIb in lipid rafts [56]. The function of a pseudo gene (CD177P) located downstream in close vicinity to the 9 exons of CD177 and spanning over exons 4–9 in reverse orientation (19q13.31), remains unclear but it significantly hampers studies to decode the molecular reason of differential gene expression [50]. Between 88% of Japanese and about 97% of Caucasian individuals express the CD177 glycoprotein on their neutrophils while the rest are completely devoid of HNA-2 [1, 57, 58]. Only HNA-2 deficient (HNA-2null) [59] individuals are able to produce HNA-2 isoantibodies that can cause TRALI and immune neutropenia [3, 60]. However, although antibody-mediated clustering of CD177 primes the respiratory burst reaction of neutrophils, HNA-2-negative individuals do not seem to be at increased risk of infection [61]. A differential expression on neutrophil subsets with one or two neutrophil subpopulations expressing the antigen at a high or intermediate level and another subset lacking the glycoprotein is a special feature of HNA-2 [49, 62]. A proportion of HNA-2-positive neutrophils in the blood with either intermediate or high expression between 1 and 100% has been demonstrated in different individuals. This proportion is a lifelong characteristic [10, 59]. However, upon neutrophil activation in severe bacterial infections, in pregnancy and in newborns the expression can be upregulated [57, 63, 64, 65, 66].
Fig. 3.
Potential organization of FcγRIIIb (HNA-1), CD177 (HNA-2), CD11b/CD18 (HNA-4) and membrane-associated proteinase-3 (mPR-3) as lipid raft within the neutrophil membrane.
Molecular Basis of Differential Gene Expression
Neither the reason for the complete HNA-2 deficiency nor for the differential expression on neutrophil subsets is completely understood. The NCBI dbSNP database lists numerous SNPs affecting nearly each of the 437 amino acids (21 of them are part of a signal peptide resulting in a 416 amino acid mature protein). Different SNPs have been attributed to a low or high HNA-2 expression or the presence of one or two antigen-positive subpopulations [67, 68, 69]. A dose effect of the wild-type CD177*787A or heterozygosity for both *787A and *787T has been correlated with the proportion of HNA-2-positive neutrophils [10, 59]. Recently, gene silencing and a novel monoallelic expression pattern caused by disparate CpG and histone methylation have been described as basis for distinct neutrophil subsets thus introducing epigenetic mechanisms [70].
In two individuals the molecular basis for the complete HNA-2 deficiency was demonstrated to result from an incorrect splicing [71]. However, this does not account for each case because a correctly spliced cDNA was found in an immunized HNA-2-deficient women with a HNA-2 isoantibody (own observations, unpublished).
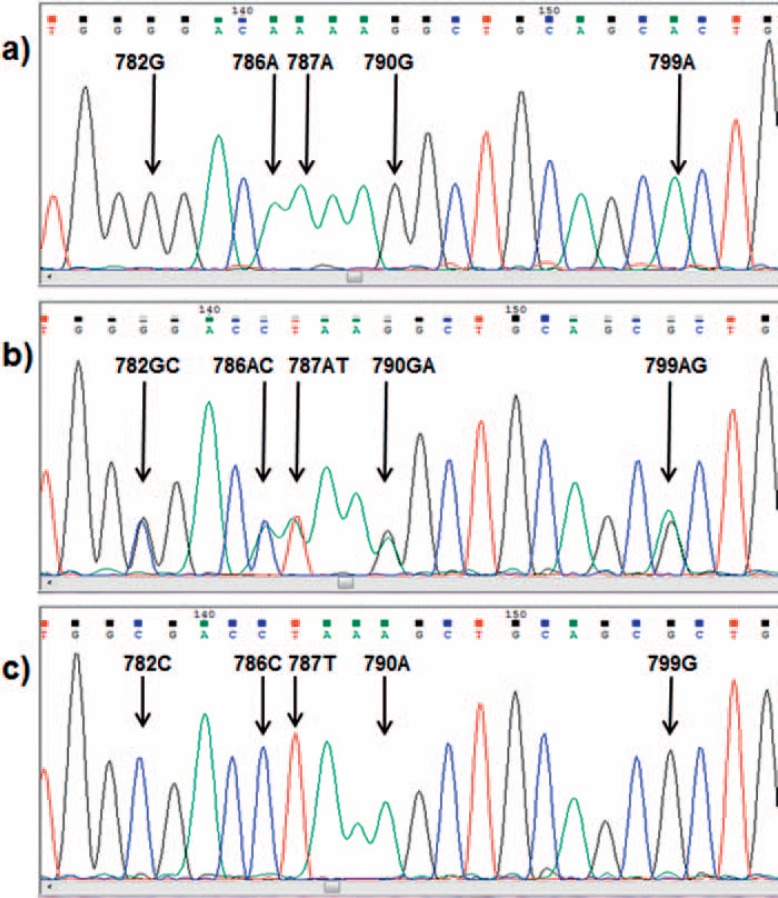
The CD177*787A>T exchange (rs879198465, in [10] reported as rs70950396) leading to a premature stop within exon 7 was identified in a number of HNA-2-deficient individuals, some of them carrying HNA-2 antibodies [10, 11, 59] (table 1). The SNP originally was assigned to position 829 based on a numbering starting at position 1 of the cDNA and not at the initial A of the start codon. The CD177*787A>T substitution was explained by an allelic gene conversion of exon 7 of the CD177P1 pseudo gene rather than a random mutation [59]. The CD177*787A>T exchange is flanked by further nucleotide exchanges [10]. But these do not induce further stop codons (fig. 4). Nine of 9 individuals homozygous for CD177*787T were HNA-2-deficient, and 2 further deficient individuals were heterozygous but carried an additional CD177*955delG mutation leading to a frame shift [10]. Additionally, the percentages of HNA-2-positive neutrophils were significantly lower in heterozygous samples than in those of individuals with a homozygous CD177*787A wild-type allele. The results were largely confirmed by a second group that introduced a typing protocol based on a long-range PCR product covering the whole gene to separate the CD177 gene from the CD177P1 pseudo gene [11]. However, only 3/5 immunized patients and 2/5 HNA-2 antigen-negative probands were homozygous for the 787A>T mutation, whereas it was not detected in any of 10 HNA-2-positive individuals. The 966delG mutation was detected not at all in this study, which leaves a gap of explanation for some HNA-2-deficient individuals. A third study applying deep sequencing also identified the novel stop codon within exon 7 (CD177*787A>T) in HNA-2null individuals [59]. However, the authors did not regard the SNP as a mutation of the CD177 gene itself but explained it by an entire substitution by the pseudo gene (CD177P1) exon 7 as a result from allelic gene conversion. Heterozygosity for ectopic CD177P1 gene conversion correlates with a higher proportion of HNA-2-negative neutrophils, in which both the CD177P1 partially incorporated allele and the paired intact CD177 are transcribed [59].
Fig. 4.
CD177 gene region covering the CD177*787A>T substitution. The electropherogram at the top (a) indicates a homozygous wild type, in the middle (b) a heterozygous sample and at the bottom (c) a homozygously mutated type.
Despite the high number of identified SNPs within the CD177 gene no alloreactivity has been observed. As far as reported, antibodies against HNA-2 do not discriminate between different forms so that the antibody produced by HNA-2null individuals is regarded as an isoantibody [2, 3].
HNA-3
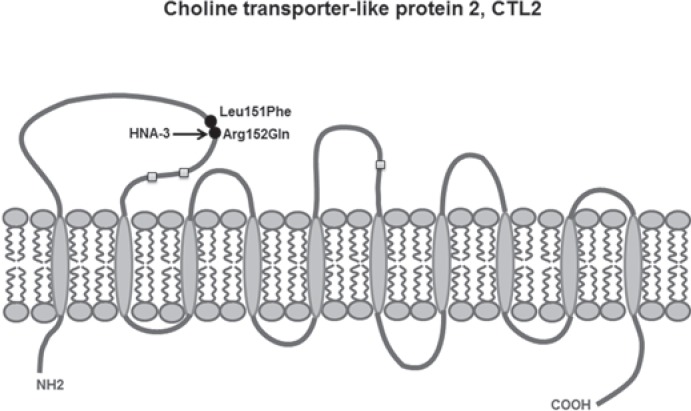
Antibodies to the former neutrophil 5b antigen were described early in 1964 by van Leeuwen et al. [72] as leukocyte agglutinins, but it took until 2010 to localize the antigen to the CTL2 which is encoded by the SLC44A2 gene on chromosome 19p13.1 [7, 8]. The alloantigens HNA-3a (formerly 5b) and HNA-3b (formerly 5a) are expressed on human neutrophils, lymphocytes, platelets, and various tissues such as lung, liver, colon, and the inner ear [7, 73, 74, 75]. Two transcript variants TV1 (also named P2; NM_020428.3) and TV2 (also named P1; NM_001145056.1) differ in the NH2 terminus located within the cytoplasmic compartment, but only the two amino acids longer TV1 enables choline transport through the cell membrane [73]. On human neutrophils, mononuclear cells, platelets, and liver tissue only the shorter TV2 is expressed, whereas lung tissue, especially human microvascular endothelial cells derived from lung blood vessels and human umbilical vein endothelial cells, carry both transcript variants [74, 76]. Differential cell and tissue distribution however does not influence HNA antibody binding as could be demonstrated by transfected HEK-293T cells expressing either TV1 or TV2. CTL2 is a 68–72 kDa transmembrane protein characterized by ten hydrophobic membrane-spanning domains, five extracellular loops, and six intracellular regions with one predicted and two confirmed N-linked glycosylation sites and potential disulfide bonds (fig. 5) [19, 75]. This special conformation aggravates the development of assays for the detection of HNA-3 antibodies based on recombinant CTL2 protein because the posttranslational modifications of the protein and the correct conformation within the neutrophil membrane are required for antibody binding [77, 78, 79, 80].
Fig. 5.
Predicted structure of the CTL2 molecule according to [75]. Amino acid positions critical for the HNA-3 polymorphism are indicated as filled black circles, the first alternative aa indicates the situation of HNA-3a. Square symbols indicate potential N-linked glycosylation sites.
Epitopes
HNA-3a and HNA-3b differ in a SLC44A2*455G>A exchange (rs2288094) leading to an Arg152Gln substitution within the first extracellular loop of CTL2 TV2 [7, 8, 9] (table 1). Different position numbers are reported in the literature because the first reports referred to TV1 where the same amino acid is located at position 154. Since HNA-3 on neutrophils is only expressed as TV2, this was adapted later on. HNA-3a antibody binding strictly requires the presence of Arg152 [7, 79, 81], whereas the HNA-3b epitope includes Gln152 (fig. 5). The HNA-3a encoding allele was demonstrated in Caucasians with a frequency of 0.79, in African Americans with 0.93 and in Chinese with 0.74, and the respective HNA-3b-encoding alleles with 0.21, 0.07 and 0.26 [82, 83, 84]. An additional SLC44A2*451C>T exchange (rs147820753) affecting the amino acid next to Arg152 and inducing a Leu151Phe substitution impairs the HNA-3a epitope in a way that the induction of neutrophil agglutination by some HNA-3a-specific alloantibodies is diminished or even abrogated [85]. Of note is that this additional mutation can influence HNA-3a typing in HNA-3a/3b heterozygous individuals when the HNA-3a allele-specific sense primer binds with reduced affinity so that the typing result is falsely interpreted as HNA-3b homozygous. This methodical problem can be overcome by applying adequate molecular tests [83, 86]. The SLC44A2*451C>T substitution has been demonstrated with a gene frequency of 0.01 and phenotype frequencies of 2.4% in the Caucasian population [80, 85] and of 0.4% in African Americans [83].
Before the introduction of measures to exclude plasma of parous women from therapeutic application without prior testing for HNA and HLA antibodies, HNA-3a antibodies in Germany and other countries represented one of the main causes of fatal TRALI cases [43, 87, 88]. In contrast, immunogenicity of HNA-3b seems to be much lower [82], and also the clinical effect of HNA-3b alloantibodies is less severe.
HNA-4
The HNA-4 system is expressed on the CD11b/αM subunit of the αMβ2-integrin (CD11b/CD18, Mac-1, CR3) [89, 90]. The leukocyte β2 integrin family consists of four members sharing a common β subunit (CD18) which is non-covalently linked to one of four different CD11 subunits (CD11a-d) [91, 92]. CD11b/CD18 is expressed on most leukocytes like neutrophils, monocytes, and NK cells where the complex is functionally inactive as long as the white blood cell is resting. Upon leukocyte activation the β2 integrin also is rapidly activated and enables adhesion of the cell to its counter receptors, transmigration, phagocytosis, and oxidative burst [91]. The CD11b/αM subunit is a transmembrane protein with 19 potential and three confirmed N-linked glycosylation sites [19]. It is encoded by the ITGAM gene on chromosome 16p11.2 and codes for a 1,153 amino acid glycoprotein including a 16 amino acid signal peptide which is cleaved in the mature protein (NM_001145808.1, transcript variant 1) [93, 94].
The first and for a long time only antigen was the formerly called ‘Marta' (now HNA-4a) [89]. Different non-synonymous mutations of CD11b/αM gene have been reported, but application of recombinant HNA-4 clearly indicated that an arginine at position 61 of the mature protein (position 77 of the precursor protein) is essential for HNA-4a alloantibody binding [92]. An ITGAM*230G>A substitution (rs1143679) is responsible for the translation into either 61Arg or the antithetical form with 61His (table 1) [9]. The HNA-4a allele frequencies in most populations have been determined with about 0.9–1.0 [84, 90, 92]. Only Taiwanese and Brazilians have lower frequencies with 0.65–0.92 [19, 95]. It is not known whether the function of the CD11b/CD18 complex is influenced by the polymorphism [3]. HNA-4a alloantibodies that interfere with CD11b/Mac-1-dependent neutrophil adhesion can induce NIN [96]. Interestingly, antibodies of the same specificity that do not exhibit this functional characteristic were not able to induce NIN [92]. The two types of alloantibodies also differ in their capability to induce the neutrophil respiratory burst, probably by differential epitope recognition at the N-terminal region of CD11b. Recently, alloantibodies directed to the HNA-4 epitope including 61His have been described in two cases of NIN [97, 98]. The antibody has been assigned to HNA-4b. In addition to alloantibodies also autoantibodies against the CD11b/CD18 complex (without clear definition of the epitopes) have been reported not only to induce neutropenia but also to impair neutrophil adhesion [44, 99].
HNA-5
To date the HNA-5 system only comprises one antigen, the HNA-5a, which originally was described as ‘Onda' [89, 100]. HNA-5a alloantibodies recognize an epitope on the αL-subunit (CD11a) of the αLβ2 integrin (CD11a/CD18, LFA-1) which is characterized by an arginine at position 766 of the mature protein (rs2230433). The presence of two mRNA transcripts of the integrin αL gene (ITGAL) complicates assignment of the polymorphism to a definite location. Because only transcript variant 1 (NM_002209.2) is in accordance with the Arg766Thr substitution of the mature protein (table 1), the SNP has been assigned to ITGAL*2372G. HNA-5a gene frequencies vary in different populations between 0.59 and 0.66 with phenotype frequencies between 79 and 88% [101]. So far no alloantibodies against the antithetical form which is characterized by 755Thr (encoded by ITGAL*2372G>A) have been described.
The CD11a/CD18 complex is expressed on all leukocytes and, like the CD11b/CD18 complex, is involved in leukocyte adhesion [3]. It is unknown whether the HNA-5 polymorphism influences integrin function. The first reported HNA-5a antibody was produced by a patient with hypoplastic anemia and thrombocytopenia who did not develop HPA or HLA class I antibodies after transfusion of platelet concentrates of more than 700 random donors. Additionally, he showed a markedly prolonged survival of an experimental skin graft from an HLA-incompatible donor. It was argued that the HNA-5a alloantibody might have blocked leukocyte interactions in the graft [100, 102]. The first case of NIN caused by an HNA-5a alloantibody was described in 2011 [103]. Interestingly, the maternal antibody was only reactive with donor neutrophils and T cells but was negative with B cells and monocytes. Altogether, HNA-5 is the system with the least known facts on the clinical significance of both the antigen and the related HNA-5a antibody.
Conclusions and Future Prospect
During the last two decades the main facts on HNA genetics, molecular basis, and antibody detection have been unraveled. These findings were the basis for further development of in-house and commercial assays for genotyping and antibody detection that facilitate the confirmation of clinically suspected cases of allo- and autoimmune neutropenia and TRALI. Easy availability of these tests may have expedited the awareness of clinical pictures related to HNA antibodies. A higher awareness of NIN and neonatal autoimmune neutropenia could deepen the knowledge on antibodies and antigens involved in these clinical pictures.
However, even if many facts were added to granulocyte immunobiology, some open questions remain. Even if the facts on HNA-1 alleles, antigens and epitopes are well documented, the recent findings on copy number variation and differentially expressed epitopes give a wide scope for further developments. The molecular basis for the enigma of differential HNA-2 expression on neutrophil subsets and for some unresolved cases of HNA-2null phenotypes remains to be illuminated. After decoding the molecular basis of HNA-3 and restrictions to prevent TRALI placed by many national authorities, the HNA-3 system in the meantime disappeared somewhat from the focus. But further studies on functional aspects regarding HNA-3a antibody-induced tissue damage may add additional facts. Less is known on HNA-4 and HNA-5 alloantibodies and their function. Especially data from geographical regions with diverging allele frequencies could add further facts.
However, granulocyte immunobiology is somewhat like a niche discipline where only few researchers and laboratories worldwide have the capability to administer new findings. During the last years many efforts have been made in Eastern Asian Countries in HNA diagnostics. But many regions of the world, especially Africa, Near East, India and Latin America (except for Brazil), to date are scarcely engaged in granulocyte immunobiological diagnostics and research. Including of these regions could add new findings because of possibly differing alleles and frequencies.
Disclosure Statement
The German Red Cross Blood Service West has filed a patent application for the HNA-3a antigen. None of the authors has to declare a conflict of interest relevant to this manuscript.
References
- 1.Bux J. Molecular genetics of granulocyte polymorphisms. Vox Sang. 2000;78((suppl 2)):125–130. [PubMed] [Google Scholar]
- 2.Flesch BK for the International Society of Blood Transfusion (ISBT) HNA nomenclature subcommittee Human neutrophil antigens: a nomenclature update based on new alleles and new antigens. ISBT Sci Se. 2015;10((suppl 1)):243–249. [Google Scholar]
- 3.Bux J. Human neutrophil alloantigens. Vox Sang. 2008;94:277–285. doi: 10.1111/j.1423-0410.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- 4.Bux J. Nomenclature of granulocyte antigens. Transfusion. 1999;39:662–663. doi: 10.1046/j.1537-2995.1999.39060662.x. [DOI] [PubMed] [Google Scholar]
- 5.Lalezari P, Nussbaum M, Gelman S, Spaet TH. Neonatal neutropenia due to maternal isoimmunization. Blood. 1960;15:236–243. [PubMed] [Google Scholar]
- 6.Lalezari P, Bernard GE. Identification of a specific leukocyte antigen: another presumed example of 5b. Transfusion. 1965;5:135–142. doi: 10.1111/j.1537-2995.1965.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 7.Greinacher A, Wesche J, Hammer E, Fürll B, Völker U, Reil A, Bux J. Characterization of the human neutrophil alloantigen-3a. Nat Med. 2010;16:45–48. doi: 10.1038/nm.2070. [DOI] [PubMed] [Google Scholar]
- 8.Curtis BR, Cox NJ, Sullivan MJ, Konkashbaev A, Bowens K, Hansen K, Aster RH. The neutrophil alloantigen HNA-3a (5b) is located on choline transporter like protein 2 (CTL2) and appears to be encoded by an R>Q 154 amino acid substitution. Blood. 2010;115:2073–2076. doi: 10.1182/blood-2009-11-248336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flesch BK, Curtis BR, de Haas M, Lucas G, Sachs UJ. Update on the nomenclature of human neutrophil antigens and alleles. Transfusion. 2016;56:1477–1479. doi: 10.1111/trf.13575. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Mair DC, Schuller RM, Li L, Wu J. Genetic mechanism of human neutrophil antigen 2 deficiency and expression variations. PLoS Genet. 2015;11:e1005255. doi: 10.1371/journal.pgen.1005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayat B, Bein G, Sachs UJ. A sequence-specific polymerase chain reaction method for HNA-2 genotyping: homozygous c.843A>T mutation predicts the absence of CD177. Transfusion. 2016;56:2127–2132. doi: 10.1111/trf.13689. [DOI] [PubMed] [Google Scholar]
- 12.Selvaraj P, Rosse WF, Silber R, Springer TA. The major Fc receptor in blood has a phosphatidylinositol anchor and is deficient in paroxysmal nocturnal haemoglobinuria. Nature. 1988;333:565–567. doi: 10.1038/333565a0. [DOI] [PubMed] [Google Scholar]
- 13.Flesch BK, Neppert J. Functions of the Fc receptors for immunoglobulin G. J Clin Lab Anal. 2000;14:141–156. doi: 10.1002/1098-2825(2000)14:4<141::AID-JCLA3>3.0.CO;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 15.Qiu WQ, de Bruin D, Brownstein BH, Pearse R, Ravetch JV. Organization of the human and mouse low-affinity Fc gamma R genes: duplication and recombination. Science. 1990;248:732–735. doi: 10.1126/science.2139735. [DOI] [PubMed] [Google Scholar]
- 16.Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med. 1989;170:481–497. doi: 10.1084/jem.170.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bux J, Sohn M, Hachmann R, Mueller-Eckhardt C. Quantitation of granulocyte antibodies in sera and determination of their binding sites. Br J Haematol. 1992;82:20–25. doi: 10.1111/j.1365-2141.1992.tb04588.x. [DOI] [PubMed] [Google Scholar]
- 18.Fleit HB, Wright SD, Unkeless JC. Human neutrophil Fc gamma receptor distribution and structure. Proc Natl Acad Sci U S A. 1982;79:3275–3279. doi: 10.1073/pnas.79.10.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muschter S, Berthold T, Greinacher A. Developments in the definition and clinical impact of human neutrophil antigens. Curr Opin Hematol. 2011;18:452–460. doi: 10.1097/MOH.0b013e32834babdd. [DOI] [PubMed] [Google Scholar]
- 20.Ory PA, Goldstein IM, Kwoh EE, Clarkson SB. Characterization of polymorphic forms of Fc receptor III on human neutrophils. J Clin Invest. 1989;83:1676–1681. doi: 10.1172/JCI114067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flesch BK, Doose S, Siebert R, Ntambi E, Neppert J. FCGR3 variants and expression of human neutrophil antigen-1a, -1b, and -1c in the populations of northern Germany and Uganda. Transfusion. 2002;42:469–475. doi: 10.1046/j.1525-1438.2002.00087.x. [DOI] [PubMed] [Google Scholar]
- 22.Bux J, Stein EL, Bierling P, Fromont P, Clay M, Stroncek D, Santoso S. Characterization of a new alloantigen (SH) on the human neutrophil Fc gamma receptor IIIb. Blood. 1997;89:1027–1034. [PubMed] [Google Scholar]
- 23.de Haas M, Kleijer M, van Zwieten R, Roos D, von dem Borne AE. Neutrophil Fc gamma RIIIb deficiency, nature, and clinical consequences: a study of 21 individuals from 14 families. Blood. 1995;86:2403–2413. [PubMed] [Google Scholar]
- 24.Flesch BK, Achtert G, Bauer F, Neppert J. The NA'null' phenotype of a young man is caused by an Fc gammaRIIIB gene deficiency while the products of the neighboring Fc gammaRIIA and Fc gammaRIIIA genes are present. Ann Hematol. 1998;76:215–220. doi: 10.1007/s002770050392. [DOI] [PubMed] [Google Scholar]
- 25.Koene HR, Kleijer M, Roos D, de HM von dem Borne AE. Fc gamma RIIIB gene duplication: evidence for presence and expression of three distinct Fc gamma RIIIB genes in NA(1+,2+)SH(+) individuals. Blood. 1998;91:673–679. [PubMed] [Google Scholar]
- 26.Blum KS, Tong Y, Siebert R, Marget M, Humpe A, Neppert J, Flesch BK. Evidence for gene recombination in FCGR3 gene variants. Vox Sang. 2009;97:69–76. doi: 10.1111/j.1423-0410.2009.01178.x. [DOI] [PubMed] [Google Scholar]
- 27.Mueller M, Barros P, Witherden AS, Roberts AL, Zhang Z, Schaschl H, Yu CY, Hurles ME, Schaffner C, Floto RA, Game L, Steinberg KM, Wilson RK, Graves TA, Eichler EE, Cook HT, Vyse TJ, Aitman TJ. Genomic pathology of SLE-associated copy-number variation at the FCGR2C/FCGR3B/FCGR2B locus. Am J Hum Genet. 2013;92:28–40. doi: 10.1016/j.ajhg.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huizinga TW, Kleijer M, Tetteroo PA, Roos D, von dem Borne AE. Biallelic neutrophil Na-antigen system is associated with a polymorphism on the phospho-inositol-linked Fc gamma receptor III (CD16) Blood. 1990;75:213–217. [PubMed] [Google Scholar]
- 29.Reil A, Sachs UJ, Siahanidou T, Flesch BK, Bux J. HNA-1d: a new human neutrophil antigen located on Fcgamma receptor IIIb associated with neonatal immune neutropenia. Transfusion. 2013;53:2145–2151. doi: 10.1111/trf.12086. [DOI] [PubMed] [Google Scholar]
- 30.Ory PA, Clark MR, Kwoh EE, Clarkson SB, Goldstein IM. Sequences of complementary DNAs that encode the NA1 and NA2 forms of Fc receptor III on human neutrophils. J Clin Invest. 1989;84:1688–1691. doi: 10.1172/JCI114350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kissel K, Hofmann C, Gittinger FS, Daniels G, Bux J. HNA-1a, HNA-1b, and HNA-1c (NA1, NA2, SH) frequencies in African and American Blacks and in Chinese. Tissue Antigens. 2000;56:143–148. doi: 10.1034/j.1399-0039.2000.560205.x. [DOI] [PubMed] [Google Scholar]
- 32.Hessner MJ, Curtis BR, Endean DJ, Aster RH. Determination of neutrophil antigen gene frequencies in five ethnic groups by polymerase chain reaction with sequence-specific primers. Transfusion. 1996;36:895–899. doi: 10.1046/j.1537-2995.1996.361097017176.x. [DOI] [PubMed] [Google Scholar]
- 33.Hessner MJ, Shivaram SM, Dinauer DM, Curtis BR, Endean DJ, Aster RH. Neutrophil antigen (FcgRIIIB) SH gene frequencies in six racial groups. Blood. 1999;93:1115–1116. [PubMed] [Google Scholar]
- 34.Reil A, Flesch B, Bux J. FCGR3B*04 - a novel allele of the human Fc gamma receptor IIIb gene. Transfus Med Hemother. 2011;38((suppl 1)):69. [Google Scholar]
- 35.Matsuo K, Procter JL, Chanock SJ, Stroncek DF. The expression of NA antigens in people with unusual Fcg receptor III genotypes. Transfusion. 2001;41:775–782. doi: 10.1046/j.1537-2995.2001.41060775.x. [DOI] [PubMed] [Google Scholar]
- 36.Matsuo K, Procter J, Stroncek D. Variations in genes encoding neutrophil antigens NA1 and NA2. Transfusion. 2000;40:645–653. doi: 10.1046/j.1537-2995.2000.40060645.x. [DOI] [PubMed] [Google Scholar]
- 37.Covas DT, Kashima S, Guerreiro JF, dos Sontos SEB, Zago MA. Variation in the FcgR3B gene among distinct Brazilian populations. Tissue Antigens. 2005;65:178–182. doi: 10.1111/j.1399-0039.2005.00352.x. [DOI] [PubMed] [Google Scholar]
- 38.Terzian CC, Chiba AK, Santos VC, Silva NP, Bordin JO. FCGR3B*03 allele inheritance pattern in Brazilian families and some new variants of gene FCGR3B. Transfusion. 2011;52:629–634. doi: 10.1111/j.1537-2995.2011.03326.x. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen KR, Koelbaek MD, Varming K, Baech J, Steffensen R. Frequencies of HNA-1, HNA-3, HNA-4, and HNA-5 in the Danish and Zambian populations determined using a novel TaqMan real time polymerase chain reaction method. Tissue Antigens. 2012;80:249–253. doi: 10.1111/j.1399-0039.2012.01912.x. [DOI] [PubMed] [Google Scholar]
- 40.Bauer C, de Haas M, Bein G, Santoso S, Sachs U. A detailed mapping of HNA-1a and HNA-1b epitopes of FcgammarIIIb. Vox Sang. 2010;99((suppl 2)):35. [Google Scholar]
- 41.Popovsky MA, Moore SB. Diagnostic and pathogenetic considerations in transfusion-related acute lung injury. Transfusion. 1985;25:573–577. doi: 10.1046/j.1537-2995.1985.25686071434.x. [DOI] [PubMed] [Google Scholar]
- 42.Goldman M, Webert KE, Arnold DM, Freedman J, Hannon J, Blajchman MA. Proceedings of a consensus conference: towards an understanding of TRALI. Transfus Med Rev. 2005;19:2–31. doi: 10.1016/j.tmrv.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 43.Keller-Stanislawski B, Reil A, Günay S, Funk MB. Frequency and severity of transfusion-related acute lung injury - German haemovigilance data (2006–2007) Vox Sang. 2010;98:70–77. doi: 10.1111/j.1423-0410.2009.01232.x. [DOI] [PubMed] [Google Scholar]
- 44.Bux J, Behrens G, Jaeger G, Welte K. Diagnosis and clinical course of autoimmune neutropenia in infancy: analysis of 240 cases. Blood. 1998;91:181–186. [PubMed] [Google Scholar]
- 45.Bruin M, Dassen A, Pajkrt D, Buddelmeyer L, Kuijpers T, de HM. Primary autoimmune neutropenia in children: a study of neutrophil antibodies and clinical course. Vox Sang. 2005;88:52–59. doi: 10.1111/j.1423-0410.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- 46.Bux J, Kissel K, Nowak K, Spengel U, Mueller-Eckhardt C. Autoimmune neutropenia: clinical and laboratory studies in 143 patients. Ann Hematol. 1991;63:249–252. doi: 10.1007/BF01698373. [DOI] [PubMed] [Google Scholar]
- 47.Lalezari P, Murphy GB, Allen FH., Jr NB1, a new neutrophil-specific antigen involved in the pathogenesis of neonatal neutropenia. J Clin Invest. 1971;50:1108–1115. doi: 10.1172/JCI106582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stroncek DF, Skubitz KM, McCullough JJ. Biochemical characterization of the neutrophil-specific antigen NB1. Blood. 1990;75:744–755. [PubMed] [Google Scholar]
- 49.Goldschmeding R, van Dalen CM, Faber N, Calafat J, Huizinga TW, Van Der Schoot CE, Clement LT, von dem Borne AE. Further characterization of the NB 1 antigen as a variably expressed 56–62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br J Haematol. 1992;81:336–345. doi: 10.1111/j.1365-2141.1992.tb08237.x. [DOI] [PubMed] [Google Scholar]
- 50.Bettinotti MP, Olsen A, Stroncek D. The use of bioinformatics to identify the genomic structure of the gene that encodes neutrophil antigen NB1, CD177. Clin Immunol. 2002;102:138–144. doi: 10.1006/clim.2001.5154. [DOI] [PubMed] [Google Scholar]
- 51.Caruccio L, Bettinotti M, Director-Myska AE, Arthur DC, Stroncek D. The gene overexpressed in polycythemia rubra vera, PRV-1, and the gene encoding a neutrophil alloantigen, NB1, are alleles of a single gene, CD177, in chromosome band 19q13.31. Transfusion. 2006;46:441–447. doi: 10.1111/j.1537-2995.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 52.Korkmaz B, Kuhl A, Bayat B, Santoso S, Jenne DE. A hydrophobic patch on proteinase 3, the target of autoantibodies in Wegener granulomatosis, mediates membrane binding via NB1 receptors. J Biol Chem. 2008;283:35976–35982. doi: 10.1074/jbc.M806754200. [DOI] [PubMed] [Google Scholar]
- 53.Abdgawad M, Gunnarsson L, Bengtsson AA, Geborek P, Nilsson L, Segelmark M, Hellmark T. Elevated neutrophil membrane expression of proteinase 3 is dependent upon CD177 expression. Clin Exp Immunol. 2010;161:89–97. doi: 10.1111/j.1365-2249.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu N, Westra J, Huitema MG, Bijl M, Brouwer E, Stegeman CA, Heeringa P, Limburg PC, Kallenberg CG. Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase 3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum. 2009;60:1548–1557. doi: 10.1002/art.24442. [DOI] [PubMed] [Google Scholar]
- 55.Kuckleburg CJ, Tilkens SB, Santoso S, Newman PJ. Proteinase 3 contributes to transendothelial migration of NB1-positive neutrophils. J Immunol. 2012;188:2419–2426. doi: 10.4049/jimmunol.1102540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jerke U, Rolle S, Dittmar G, Bayat B, Santoso S, Sporbert A, Luft F, Kettritz R. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J Biol Chem. 2011;286:7070–7081. doi: 10.1074/jbc.M110.171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kissel K, Santoso S, Hofmann C, Stroncek D, Bux J. Molecular basis of the neutrophil glycoprotein NB1 (CD177) involved in the pathogenesis of immune neutropenias and transfusion reactions. Eur J Immunol. 2001;31:1301–1309. doi: 10.1002/1521-4141(200105)31:5<1301::AID-IMMU1301>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 58.Bierling P, Poulet E, Fromont P, Seror T, Bracq C, Duedari N. Neutrophil-specific antigen and gene frequencies in the French population. Transfusion. 2001;30:848–849. doi: 10.1046/j.1537-2995.1990.30991048794.x. [DOI] [PubMed] [Google Scholar]
- 59.Wu Z, Liang R, Ohnesorg T, Cho V, Lam W, Abhayaratna WP, Gatenby PA, Perera C, Zhang Y, Whittle B, Sinclair A, Goodnow CC, Field M, Andrews TD, Cook MC. Heterogeneity of human neutrophil CD177 expression results from CD177P1 pseudogene conversion. PLoS Genet. 2016;12:e1006067. doi: 10.1371/journal.pgen.1006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bux J, Becker F, Seeger W, Kilpatrick D, Chapman J, Waters A. Transfusion-related acute lung injury due to HLA-A2-specific antibodies in recipient and NB1-specific antibodies in donor blood. Br J Haematol. 1996;93:707–713. doi: 10.1046/j.1365-2141.1996.d01-1703.x. [DOI] [PubMed] [Google Scholar]
- 61.Sachs UJH, Hattar K, Weissmann N, Bohle RM, Weiss T, Sibelius U, Bux J. Antibody-induced neutrophil activation as a trigger for transfusion-related acute lung injury in an ex vivo rat lung model. Blood. 2006;107:1217–1219. doi: 10.1182/blood-2005-04-1744. [DOI] [PubMed] [Google Scholar]
- 62.Stroncek DF, Plachta LB, Herr GP, Dalmasso AP. Analysis of the expression of neutrophil-specific antigen NB1:characterization of neutrophils that react with but are not agglutinated by anti-NB1. Transfusion. 1993;33:656–660. doi: 10.1046/j.1537-2995.1993.33893342747.x. [DOI] [PubMed] [Google Scholar]
- 63.Caruccio L, Bettinotti M, Matsuo K, Sharon V, Stroncek D. Expression of human neutrophil antigen-2a (NB1) is increased in pregnancy. Transfusion. 2003;43:357–363. doi: 10.1046/j.1537-2995.2003.00320.x. [DOI] [PubMed] [Google Scholar]
- 64.Taniguchi K, Nagata H, Katsuki T, Nakashima C, Onodera R, Hiraoka A, Takata N, Kobayashi M, Kambe M. Significance of human neutrophil antigen-2a (NB1) expression and neutrophil number in pregnancy. Transfusion. 2004;44:581–585. doi: 10.1111/j.1537-2995.2004.03298.x. [DOI] [PubMed] [Google Scholar]
- 65.Wolff JC, Goehring K, Heckmann M, Bux J. Sex-dependent up regulation of CD 177-specific mRNA expression in cord blood due to different stimuli. Transfusion. 2006;46:132–136. doi: 10.1111/j.1537-2995.2005.00676.x. [DOI] [PubMed] [Google Scholar]
- 66.Göhring K, Wolff J, Doppl W, Schmidt KL, Fenchel K, Pralle H, Sibelius U, Bux J. Neutrophil CD177 (NB1 gp, HNA-2a) expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol. 2004;126:252–254. doi: 10.1111/j.1365-2141.2004.05027.x. [DOI] [PubMed] [Google Scholar]
- 67.Moritz E, Chiba AK, Kimura EY, Albuquerque D, Guirao FP, Yamamoto M, Costa FF, Bordin JO. Molecular studies reveal that A134T, G156A and G1333A SNPs in the CD177 gene are associated with atypical expression of human neutrophil antigen-2. Vox Sang. 2010;98:160–166. doi: 10.1111/j.1423-0410.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- 68.Wolff J, Brendel C, Fink L, Bohle RM, Kissel K, Bux. J. Lack of NB1 GP (CD177/HNA-2a) gene transcription in NB1 GP- neutrophils from NB1 GP-expressing individuals and association of low expression with NB1 gene polymorphisms. Blood. 2003;102:731–733. doi: 10.1182/blood-2002-09-2831. [DOI] [PubMed] [Google Scholar]
- 69.Caruccio L, Walkovich K, Bettinotti M, Schuller R, Stroncek D. CD177 polymorphisms: correlation between high-frequency single nucleotide polymorphisms and neutrophil surface protein expression. Transfusion. 2004;44:77–82. doi: 10.1046/j.0041-1132.2004.00606.x. [DOI] [PubMed] [Google Scholar]
- 70.Eulenberg-Gustavus C, Bahring S, Maass PG, Luft FC, Kettritz R. Gene silencing and a novel monoallelic expression pattern in distinct CD177 neutrophil subsets. J Exp Med. 2017;214:2089–2101. doi: 10.1084/jem.20161093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kissel K, Scheffler S, Kerowgan M, Bux J. Molecular basis of NB1 (HNA-2a, CD177) deficiency. Blood. 2002;99:4231–4233. doi: 10.1182/blood.v99.11.4231. [DOI] [PubMed] [Google Scholar]
- 72.van Leeuwen A, Eernisse JG, van Rood JJ. A new leucocyte group with two alleles: leucocyte group five. Vox Sang. 1964;9:431–446. doi: 10.1111/j.1423-0410.1964.tb03311.x. [DOI] [PubMed] [Google Scholar]
- 73.Kommareddi PK, Nair TS, Thang LV, Galano MM, Babu E, Ganapathy V, Kanazawa T, McHugh JB, Carey TE. Isoforms, expression, glycosylation, and tissue distribution of CTL2/SLC44A2. Protein J. 2010;29:417–426. doi: 10.1007/s10930-010-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flesch BK, Wesche J, Berthold T, Goldmann T, Hundt M, Greinacher A, Bux J. Expression of the CTL2 transcript variants in human peripheral blood cells and human tissues. Transfusion. 2013;53:3217–3223. doi: 10.1111/trf.12160. [DOI] [PubMed] [Google Scholar]
- 75.Nair TS, Kozma KE, Hoefling NL, Kommareddi PK, Ueda Y, Gong T-W, Lomay MI, Lansford CD, Telian SA, Satar B, Arts A, El-Kashlan HK, Berryhill WE, Raphel Y, Carey TE. Identification and characterization of choline transporter-like protein 2, an inner ear glycoprotein of 68 and 72 kDa that is the target of antibody-induced hearing loss. J Neurosci. 2004;24:1772–1779. doi: 10.1523/JNEUROSCI.5063-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bayat B, Tjahjono Y, Sydykov A, Werth S, Hippenstiel S, Weissmann N, Sachs UJ, Santoso S. Anti-human neutrophil antigen-3a induced transfusion-related acute lung injury in mice by direct disturbance of lung endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:2538–2548. doi: 10.1161/ATVBAHA.113.301206. [DOI] [PubMed] [Google Scholar]
- 77.Kanack AJ, Peterson JA, Sullivan MJ, Bougie DW, Curtis BR, Aster RH. Full-length recombinant choline transporter-like protein 2 containing arginine 154 reconstitutes the epitope recognized by HNA-3a antibodies. Transfusion. 2011;52:1112–1116. doi: 10.1111/j.1537-2995.2011.03411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wozniak MJ, Bowring C, Lucas G, Ridgwell K. Detection of HNA-3a and -3b antibodies using transfected cell lines and recombinant proteins. Transfusion. 2011;52:1458–1467. doi: 10.1111/j.1537-2995.2011.03490.x. [DOI] [PubMed] [Google Scholar]
- 79.Berthold T, Wesche J, Kuhnert K, Fürll B, Hippe H, Hoppen J, Reil A, Muschter S, Bux J, Greinacher A. Epitope mapping of antibodies directed against the human neutrophil alloantigen 3a. Transfusion. 2011;51:2160–2167. doi: 10.1111/j.1537-2995.2011.03115.x. [DOI] [PubMed] [Google Scholar]
- 80.Bayat B, Tjahjono Y, Werth S, Berghofer H, Reil A, Kroll H, Sachs UJ, Santoso S. Implication of transfected cell lines for the detection of alloantibodies against human neutrophil antigen-3. Transfusion. 2012;52:613–621. doi: 10.1111/j.1537-2995.2011.03303.x. [DOI] [PubMed] [Google Scholar]
- 81.Curtis BR, Sullivan MJ, Holyst MT, Szabo A, Bougie DW, Aster RH. HNA-3a-specific antibodies recognize choline transporter-like protein-2 peptides containing arginine, but not glutamine at Position 154. Transfusion. 2011;51:2168–2174. doi: 10.1111/j.1537-2995.2011.03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reil A, Wesche J, Greinacher A, Bux J. Geno- and phenotyping and immunogenicity of HNA-3. Transfusion. 2011;51:18–24. doi: 10.1111/j.1537-2995.2010.02751.x. [DOI] [PubMed] [Google Scholar]
- 83.Huvard MJ, Schmid P, Stroncek DF, Flegel WA. Frequencies of SLC44A2 alleles encoding human neutrophil antigen-3 variants in the African American population. Transfusion. 2012;52:1106–1111. doi: 10.1111/j.1537-2995.2011.03396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xia W, Bayat B, Sachs U, Chen Y, Shao Y, Xu X, Deng J, Ding H, Fu Y, Ye X, Santoso S. The frequencies of human neutrophil alloantigens in the Chinese Han population of Guangzhou. Transfusion. 2010;51:1271–1277. doi: 10.1111/j.1537-2995.2010.02979.x. [DOI] [PubMed] [Google Scholar]
- 85.Flesch BK, Reil A, Bux J. Genetic variation of the HNA-3a encoding gene. Transfusion. 2011;51:2391–2397. doi: 10.1111/j.1537-2995.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 86.Lopes LB, Baleotti W, Suzuki RB, Fabron A, Chiba AK, Vieira-Filho JP, de Souza Castro B, Midori Kunioshi A, Bordin JO. HNA-3 gene frequencies in Brazilians and a new polymerase chain reaction-restriction fragment length polymorphism method for HNA-3a/3b genotyping. Transfusion. 2014;54:1619–1621. doi: 10.1111/trf.12493. [DOI] [PubMed] [Google Scholar]
- 87.Reil A, Keller-Stanislawski B, Günay S, Bux J. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang. 2008;95:313–317. doi: 10.1111/j.1423-0410.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 88.Davoren A, Curtis BR, Shulman IA, Mohrbacher AF, Bux J, Kwiatkowska BJ, McFarland JG, Aster RH. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion. 2003;43:641–645. doi: 10.1046/j.1537-2995.2003.00374.x. [DOI] [PubMed] [Google Scholar]
- 89.Simsek S, Van Der Schoot CE, Daams M, Huiskes E, Clay M, McCullough J, van DC, Stroncek D, von dem Borne AE. Molecular characterization of antigenic polymorphisms (Ond(a) and Mart(a)) of the beta 2 family recognized by human leukocyte alloantisera. Blood. 1996;88:1350–1358. [PubMed] [Google Scholar]
- 90.Kline WE, Press C, Clay M, Keashen-Schnell M, Hackel E, McCullough J. Three sera defining a new granulocyte-monocyte-T-lymphocyte antigen. Vox Sang. 1986;50:181–186. doi: 10.1111/j.1423-0410.1986.tb04875.x. [DOI] [PubMed] [Google Scholar]
- 91.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 92.Sachs UJ, Chavakis T, Fung L, Lohrenz A, Bux J, Reil A, Ruf A, Santoso S. Human alloantibody anti-Mart interferes with Mac-1-dependent leukocyte adhesion. Blood. 2004;104:727–734. doi: 10.1182/blood-2003-11-3809. [DOI] [PubMed] [Google Scholar]
- 93.Corbi AL, Kishimoto TK, Miller LJ, Springer TA. The human leukocyte adhesion glycoprotein Mac-1 (complement receptor type 3, CD11b) alpha subunit. Cloning, primary structure, and relation to the integrins, von Willebrand factor and factor B. J Biol Chem. 1988;263:12403–12411. [PubMed] [Google Scholar]
- 94.Hickstein DD, Hickey MJ, Ozols J, Baker DM, Back AL, Roth GJ. cDNA sequence for the alpha M subunit of the human neutrophil adherence receptor indicates homology to integrin alpha subunits. Proc Natl Acad Sci U S A. 1989;86:257–261. doi: 10.1073/pnas.86.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cardone JD, Bordin JO, Chiba AK, Norcia AM, Vieira-Filho JP. Gene frequencies of the HNA-4a and -5a neutrophil antigens in Brazilian persons and a new polymerase chain reaction-restriction fragment length polymorphism method for HNA-5a genotyping. Transfusion. 2006;46:1515–1520. doi: 10.1111/j.1537-2995.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 96.Fung YL, Pitcher LA, Willett JE, Reed C, Mison L, Bux J, Eiber G, Minchinton RM. Alloimmune neonatal neutropenia linked to anti-HNA-4a. Transfus Med. 2003;13:49–52. doi: 10.1046/j.1365-3148.2003.00413.x. [DOI] [PubMed] [Google Scholar]
- 97.Curtis BR, Roman AS, Sullivan MJ, Raven CS, Larison J, Weitekamp LA. Two cases of maternal alloimmunization against human neutrophil alloantigen-4b, one causing severe alloimmune neonatal neutropenia. Transfusion. 2016;56:101–106. doi: 10.1111/trf.13287. [DOI] [PubMed] [Google Scholar]
- 98.Mraz GA, Crighton GL, Christie DJ. Antibodies to human neutrophil antigen HNA-4b implicated in a case of neonatal alloimmune neutropenia. Transfusion. 2016;56:1161–1165. doi: 10.1111/trf.13463. [DOI] [PubMed] [Google Scholar]
- 99.Bux J, Hartmann C, Mueller-Eckhardt C. Alloimmune neonatal neutropenia resulting from immunization to a high-frequency antigen on the granulocyte Fc gamma receptor III. Transfusion. 1994;34:608–611. doi: 10.1046/j.1537-2995.1994.34794330016.x. [DOI] [PubMed] [Google Scholar]
- 100.Decary F, Verheugt FW, van Helden-Henningheim L, von RE, Schreuder-van GR, von dem Borne AE, Engelfriet CP. Recognition of a non-HLA-ABC antigen present on B and T lymphocytes and monocytes only detectable with the indirect immunofluorescence test. Vox Sang. 1979;36:150–158. doi: 10.1111/j.1423-0410.1979.tb04416.x. [DOI] [PubMed] [Google Scholar]
- 101.Sachs UJ, Reil A, Bauer C, Bux J, Bein G, Santoso S. Genotyping of human neutrophil antigen-5a (Ond) Transfus Med. 2005;15:115–117. doi: 10.1111/j.0958-7578.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 102.Pegels JG, Bruynes EC, Korthals Altes HR, Engelfriet CP, von dem Borne AE. Immune unresponsiveness to platelets. A case study. Vox Sang. 1982;42:211–216. [PubMed] [Google Scholar]
- 103.Porcelijn L, Abbink F, Terraneo L, Onderwater-vd HL, Huiskes E, de HM. Neonatal alloimmune neutropenia due to immunoglobulin G antibodies against human neutrophil antigen-5a. Transfusion. 2011;51:574–577. doi: 10.1111/j.1537-2995.2010.02858.x. [DOI] [PubMed] [Google Scholar]