Abstract
Purpose of review
The aim of this study is to review genetics of congenital heart disease (CHD) with a focus on clinical applications, genetic testing and clinical challenges.
Recent findings
With improved clinical care, there is a rapidly expanding population of adults, especially women, with CHD who have not undergone contemporary genetic assessment and do not understand their risk for having a child with CHD. Many patients have never undergone assessment or had genetic testing. A major barrier is medical geneticist availability, resulting in this burden of care shifting to providers outside of genetics. Even with current understanding, the cause for the majority of cases of CHD is still not known. There are significant gaps in knowledge in the realms of more complex causes such as noncoding variants, multigenic contribution and small structural chromosomal anomalies.
Summary
Standard assessment of patients with CHD, including adult survivors, is indicated. The best first-line genetic assessment for most patients with CHD is a chromosomal microarray, and this will soon evolve to be genomic sequencing with copy number variant analysis. Due to lack of medical geneticists, creative solutions to maximize the number of patients with CHD who undergo assessment with standard protocols and plans for support with result interpretation need to be explored.
Keywords: cardiovascular genetics, congenital heart disease, genetic testing, genetics
INTRODUCTION
As the most common birth defect affecting 1% of live births, congenital heart disease (CHD) is ubiquitous to paediatric practice. Paediatric patients with CHD who undergo surgical intervention in the first year of life made up 51% of the Society for Thoracic Surgeon's Congenital Heart Surgery Database after excluding patent ductus arteriosus ligations [1]. This class of patients present a significant area of complex disease burden. Survival of patients with complex CHDs is continually improving, which has resulted in an increased clinical burden of adults with complex CHD [2]. Adult survival is skewed with more female survivors than male survivors, and the volume of adult CHD survivors has already surpassed the entirety of the paediatric CHD population [3]. This population is creating unique and evolving challenges for clinicians, including evolving extracardiac care guidelines, reinforcing that congenital cardiac lesions result in a lifelong burden of disease that requires specialized care [4]. From a genetics perspective, this group of adult CHD survivors represents a growing population of individuals, particularly women, of reproductive age who have an increased risk of having infants with CHD who have never undergone genetic evaluation. There is limited information on the topic, but available data suggest that fewer than half of adult CHD survivors understand their risk for having a child with CHD [5]. In addition, the number of children born with CHD has increased in recent years [6]. There is a debate regarding if this is attributable to increased survival and reproduction of individuals with higher intrinsic risk to having a child with CHD or increased ability to diagnose minor CHD lesions [6].
Currently, over half of CHD cases have an unknown cause [7▪▪,8▪▪]. The diagnostic rate for patients with CHD is varied. One assessment of infants with CHD less than a year old evaluated in the cardiac ICU by a medical geneticist reported a diagnosis rate of 25%; however, this study only included patients in whom a consult was requested [9]. Our experience has demonstrated a higher diagnosis rate of infants with CHD requiring surgical intervention in the first year of life with an overall diagnostic rate of 36% after instituting a universal genetic testing protocol for these patients [10]. We have created a Clinical Cardiovascular Genetics Program or ‘cardiogenetics’ programme for infants with critical CHD. This involves evaluation by a medical geneticist for all patients with critical CHD not due to Trisomy 21 and our early data showed a diagnostic rate of 39% [10]. Our current unpublished programme diagnosis rate with a larger cohort is 33% (68/205). This suggests that involvement of a dedicated medical geneticist assessing infants with CHD not due to Trisomy 21 (exclusion of Trisomy 21 patients reduces diagnosis rate by ∼10%) increased the overall diagnosis rate for infants with CHD by an additional nearly 7–13%, and is beneficial to patients when resources are available.
Medical geneticist participation in Neurodevelopmental Outcomes Clinic for patients with CHD has also been shown to have a significant effect on diagnosis rate [11]. As CHD postsurgical outcomes are increasingly tied to underlying genetic cause, the importance of routine genetic assessment of patients with CHD is becoming more critical [8▪▪]. Specific early and late surgical outcomes such as length of intubation, transfusion requirement, transplant-free survival, growth and neurodevelopmental outcomes are all known to have genetic risk mediators [8▪▪,12–15].
Unfortunately, the supply of medical geneticists is limited, with a total of 1583 board-certified medical geneticists in the United States between 1982 and 2017 [16]. The number of medical geneticists actively evaluating patients is significantly lower. For example, as of February 2018, the American Board of Medical Genetics and Genomics listed 20 board-certified medical geneticists in Wisconsin, but the clinical full time effort was 5.5 across eight actively practicing geneticists [17]. This reinforces the need for paediatricians, paediatric cardiologists, paediatric cardiac intensivists and other non-Medical Genetics providers to gain some familiarity with a standardized approach to genetic assessment of patients with CHD.
This review will cover the recommendations for genetic assessment of patients with CHD based on lesion type and provide a framework for evaluation of patients of all ages with CHD.
Box 1.
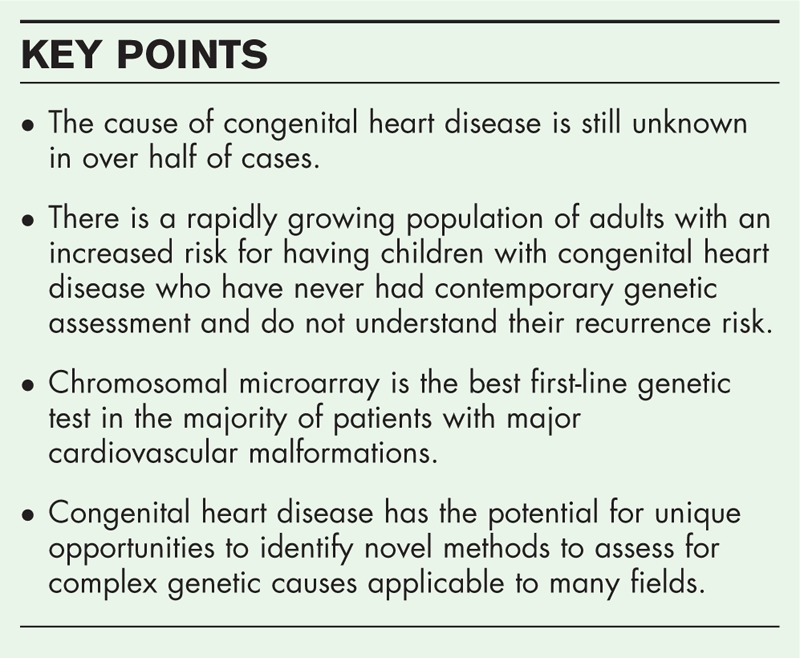
no caption available
CLINICAL ASSESSMENT
There are three main parts to focus on in the clinical assessment of patients with CHD, the physical examination, the pedigree and classification of the cardiac lesion. Patients with CHD require a thorough physical examination to assess for signs of other major and minor anomalies. Describing dysmorphic features is critical to classify minor anomalies. The National Institute of Health's Elements of Morphology series is a valuable resource for those unfamiliar with how to describe dysmorphic features or to find examples of the spectrum of dysmorphisms [18]. We also encourage facial and profile pictures to be part of the patient's medical record to assist when medical geneticists complete a chart review to assist in result interpretation.
Next, a thorough family pedigree should be obtained for all patients, although a first-degree relative with CHD is only reported in nearly 2.2% of cases [19]. A pedigree is also necessary to ensure there are no concerns, such as recurrent miscarriage or consanguinity, which could alter the risk of specific disease causes.
Finally, cardiac lesion classification helps inform physical examination features to evaluate for, genetic testing and recurrence risk. There are multiple classification systems for cardiac anatomy. The most ideal and best system to use from a genetics perspective is the classification system utilized by the National Birth Defects Prevention Study (NBDPS), which was designed specifically to account for underlying genetic and aetiologic drivers in specific cardiac lesions [20]. The largest groupings in this system (Level 3) are divided into eight categories: anomalous pulmonary venous return (APVR), atrioventricular septal defects (AVSDs), complex lesions, conotruncal lesions, heterotaxy, left ventricular outflow tract obstructive (LVOTO) lesions, right ventricular outflow tract obstructive (RVOTO) lesions and septal lesions [20]. It is important to note that over time, the description of the original cardiac lesion can evolve to be more focused on mechanical flow and repair technique versus the true underlying anatomic lesion. For example, a patient with an isolated, severely unbalanced atrioventricular septal defect with small left-sided structures who required single ventricle palliation may evolve into being called ‘hypoplastic left heart’. Although this may not affect the medical management of the patient, as hypoplastic left heart syndrome is highly heritable, it will alter the patient's estimated recurrence risk of having a child with CHD from 1.5–2.5% for atrioventricular septal defects to 21% for hypoplastic left heart syndrome [21].
MOLECULAR ASSESSMENT AND ETIOLOGIES OF CONGENITAL HEART DISEASE
To maximize direct clinical relevance, CHD cause is broken into groups that cluster with their ability to be detected by a specific genetic testing modality: karyotype, chromosomal microarray (CMA) and genomic sequencing. There is a summary of associated genetic causes by NBDPS classification in Table 1[20,22–24]. Excellent summaries with tables of extracardiac manifestations of syndromic causes of CHD can be found in reviews by Fahed et al.[22] and Zaidi and Brueckner [7▪▪].
Table 1.
Examples of associated syndromic and isolated genetic anomalies by National Birth Defects Prevention Study lesion classification [20,22–24]
| NBDPS lesion classification | Chromosomal anomalies | Copy number variants | Monogenic disruptions |
| Diagnostic test | Karyotype | Chromosomal microarray | Gene sequencing |
| Anomalous pulmonary venous return | Tetrasomy 22q | 3q22.1q26.1 Del/Dup, 13q14.11 Dup, 15q11.2 Del | ANKRD1, GATA4, GJA1, NODAL, PDGFRA, ZIC3 |
| Atrioventricular septal defects | Trisomy 21 | 3q22.1q26.1 Del/Dup, 8p23.1 Del/Dup, 15q11.2 Del, Xp22.2 Dup | ACVR1, CITED2, CRELD1, GATA4, GATA6, GJA1, NKX2.5, TBX5, TBX20 |
| Complex lesions | Trisomy 13, Trisomy 18 | 11p15.5 Dup, 15q11.2 Del | |
| Conotruncal lesions | Tetrasomy 22q | 1q21.1 Del/Dup, 1p36 Deletion, 3p25.1 Dup, 3q22.1q26.1 Del/Dup, 4p16.3 Del, 4q22.1 Dup, 5p15.2 Del, 8p23.2 Del/Dup, 9q34.3 Del, 13q14.11 Dup, 19p13.3 Del/Dup, 22q11.2 Del, Xp22.2 Dup | ALDH1A2, CHD7, FOXH1, GATA4, GATA6, GDF1, HAND2, JAG1, MED13L, NKX2.5, NKX2.6, NOTCH2, SEMA3E, TBX1, TDGF1, ZFPM2 |
| Heterotaxy | 2p25.1 Dup, 3p24.1 Del, 22q11.2 Del, Xq26.2 Del | ACVR2B, CCDC11, CFC1, CRELD1, DNAH11, FOXH1, GDF1, LEFTY2, NKX2.5, NODAL, ZIC3 | |
| Left ventricular outflow tract obstructive lesions | Monosomy X | 1q21.1 Del, 8p23.1 Del/Dup, 9q34.3 Del, 11p15.5 Dup, 11q23 Del, 13q14.11 Dup, 15q11.2 Del, 16p13.11 Dup | GATA5, GJA1, KDM6A, KMT2D, MYH6, NKX2.5, NOTCH1, TGFB2, TGFBR1, TGFBR2, VEGF |
| Right ventricular outflow tract obstructive lesions | 1q21.1 Del/Dup, 4p16.3 Del | BRAF, CBL, GATA4, GATA6, HRAS, JAG1, KRAS, MAP2K1, MAP2K2, MEK1, NF1, NOTCH2, NRAS, PTPN11, RAF1, SOS1, | |
| Septal lesions | Trisomy 13, Trisomy 18, Trisomy 21 | 1p36 Del, 4p16.3 Del, 5p15.2 Del, 8p23.1 Del/Dup, 11q23 Del, 18q11.1q11.2 Dup | CITED2, CREBBP, GATA4, GATA6, IRX4, NIPBL, NKX2.5, TBX5, TBX20, TDGF1 |
Del, deletion; Del/Dup, deletions and duplications; Dup, duplication; NBDPS, National Birth Defects Prevention Study.
Chromosomal anomalies (karyotype)
Large chromosomal anomalies and aneuploidies were the first detectable cause of CHD. A population assessment of children with CHD found 10.8% (480/4430) of patients with a diagnosis of a chromosomal anomaly detectable by karyotype [25]. The majority of the chromosomal anomalies reported were Trisomy 21 (62%, 298/480), Trisomy 18 (15%, 73/480), Trisomy 13 (6.4%, 31/480) and Turner syndrome (3%, 13/480) [25]. In the absence of these classic diagnoses, karyotype yield is low at 1.6% (65/4015) [25].
Clinical implications
For patients with concern for Trisomy 21, Trisomy 18, Trisomy 13 or Turner syndrome karyotype should be completed, but karyotype should not be incorporated into routine assessment without clinical indication or suspicion.
Copy number variation (chromosomal microarray)
Copy number variants (CNVs) are areas of chromosomal deletion or duplication too small to be detected by karyotype but can lead to pathogenically altered gene dosage. The most commonly described pathogenic CNV in CHD is 22q11.2 deletion syndrome. CHD is associated with a constantly expanding, wide range of rare CNVs [26]. The understanding of the underlying pathophysiology of CNVs associated with CHD is still an evolving field due to incomplete penetrance and variable expressivity that complicate quantifying the effect each CNV has on cardiac formation [26]. Fortunately, there are large consortiums of collaborative researchers working to accumulate more information on specific CNVs and their range of effects on cardiac formation [27]. Despite being our most effective diagnostic test currently, CMAs still fail to detect all small pathogenic CNVs due to technical and reporting limitations. Newer evaluation techniques using next-generation genomic sequencing data or higher resolution CMAs hold promise for identifying smaller pathogenic CNVs.
Clinical implications
Given its high diagnostic yield CMA is the first-line evaluation for CHD patients with multiple congenital anomalies, developmental delay or autism [28]. CMA is also recommended as first-line testing for patients with major cardiovascular anomalies [10,29–32].
An example of how routine CMA testing has impacted clinical care is for patients with single ventricle physiology. Early data indicate that in patients with single ventricle physiology, the presence of a rare CNVs larger than 300 kilobases is a risk factor for worse neurodevelopmental outcomes and poor growth, independent of if that CNV is known to cause CHD [12]. Having this information shortly after birth is crucial to help the family and team develop a care plan to monitor for future complications. We routinely assess single ventricle physiology patients with CMA with resolution sufficient to report 300 kilobase CNVs.
Monogenic disruption (gene sequencing)
Many monogenic (single gene mendelian inheritance) causes for CHD are well described, but new discovery is becoming more limited. Assessment of exome data from 2645 CHD patients without detected chromosomal anomalies or CNVs determined rare or de-novo sequence variants in nearly 8% of CHD cases [33]. These data continue to enforce the earlier predictions that monogenic discovery would fail to provide answers for the majority of cases of CHD, and that more complex multigenic causes and epigenetic regulatory mechanism will likely provide more answers [22]. An area of monogenic contribution that is likely quite rare, but underdiagnosed, is mosaic monogenic causes for CHD. Bioinformatics is becoming better at detecting low-level mosaicism from genomic sequencing data allowing increased study of mosaicism. An assessment of exome trio data on 715 patients with CHD found 23 mosaic variants affecting cardiac genes, but the authors ultimately felt only two of these variants were causative of CHD in their cohort [34]. They estimated only nearly 0.5% of CHDs are likely attributable to mosaic pathogenic variants [34].
Clinical implications
As the cost of sequencing decreases, single gene sequencing is becoming uncommon, with larger panels or genomic sequencing being preferred. As CNV detection with genomic sequencing data becomes more routine and costs continue to decrease, it is likely to replace CMA as first-line evaluation for patients with CHD.
An example of how decreasing cost of sequencing has affected our practice is our initial diagnostic approach for all patients with heterotaxy. Patients with heterotaxy have complex disease, including a high association of ciliary dysfunction. For patients with heterotaxy, we molecularly assess all patients for primary ciliary dyskinesia with a large sequencing panel. A diagnosis of primary ciliary dyskinesia alters care for the patient and increases their morbidity and mortality, making universal testing of heterotaxy patients reasonable [13,35,36]. In another specific group, we complete sequencing in patients with hypoplastic left heart syndrome. We sequence MYH6 in all patients with hypoplastic left heart syndrome. Patients with a pathogenic MYH6 disruption have decreased transplant free survival so knowing their status can help with risk stratification [14].
Variants of uncertain significance
As genomic sequencing becomes more standard, an increasing number of patients will be found to have variants of uncertain significance (VUSs), or sequence variants with unknown clinical impact. VUS in clearly actionable genes can result in care challenges, with complex clinical discussions regarding how to manage a patient when it is unclear if they have a clinically actionable genetic disease. This is especially challenging as much of the burden of ordering and interpreting genetic testing is falling to providers outside of Medical Genetics.
Having a plan in place for how to address abnormal genetic testing results, including VUS, and resources to provide support for result interpretation should be carefully considered as genetic testing protocols are designed. For example, setting up telemedicine contracts with medical geneticists and genetic counsellors in academic institutions to support result interpretation and disclosure may be a reasonable solution for some centres. In some cases, genetic testing laboratories provide support for result interpretation relating to testing they have performed as an intrinsic part of their services. This may be an extremely beneficial service to providers who have limited access to a medical geneticist.
Emerging molecular and environmental causes
Much of the genetic etiologic understanding for CHD is based on large cohort or familial large CNV analysis or exonic gene sequencing with an overall focus on mendelian inheritance patterns. This results in a large gap of knowledge regarding how more complex genetic changes (such as noncoding variants, multigenic nonmendelian contributions and small structural chromosomal anomalies, which may result in CNVs, inversions, chromothripsis or translocations with fusion protein expression) contribute to CHD [7▪▪]. Overall, these are poorly understood areas of genetics with technical limitations to detection, and standard bioinformatics assays for next-generation sequencing specifically exclude data of this type [37]. At the present time, assessments of these types of genetic anomalies are highly investigational, making this an area of active exploration [38]. Epigenetic anomalies resulting in pathogenically altered gene expression and regulation is an area wherein there is known aetiologic contribution to CHD. Continued debates of what tissue and what time in development is optimal for testing gene expression make teasing apart these effects to a point of understanding extremely difficult [7▪▪,39].
Methods to assess complex genetic causes, prioritizing clinical application, should be a priority for investigators of genetic disease in general, and CHD is a uniquely suited area for these types of investigation with its known multigenic roots in ciliopathies and robust, well characterized cohorts [7▪▪,32,40–43].
There are also many well described, but poorly understood, nonheritable contributions to CHD. Exposure to teratogens such as alcohol and glucose are known to increase risk for a child with CHD [44,45]. There continue to be conflicting data for the role of folate supplementation and folate levels in the risk for CHD with the data seeming to favour that periconceptional folate supplementation may decrease the risk of CHD, outside of Norway at least [46–49]. Being a twin increases your risk for CHD independent of the increased risk for CHD associated with artificial reproductive technologies [50].
CONCLUSION
The use of genetic information in direct care of patient with CHD is an area of clear clinical consequence, as well as an area with the potential for improvement. Current limitations with integration of genetic information to care is, at least partially, due to limited access to medical geneticists to support clinical CHD programmes. Designing protocols for genetic testing is becoming easier as advancements in molecular testing allow us to utilize a narrow spectrum of testing while expanding the number of diagnoses detected. The use of genetic testing protocols has been shown to be beneficial for infants with CHD [10]. Utilization of genetic testing protocols that includes consideration of older CHD survivors and can be universally implemented would be beneficial for these patients. A proposed genetic testing protocol is illustrated in Table 2. Over the next few years, the first-line assessment for the majority of CHD patients will switch from CMA to genomic sequencing with CNV analysis. As such, implemented protocols need to have periodic reassessment and mechanisms to institute change for evolving technology. Utilizing CHD testing protocols from clinical CHD programmes with medical geneticist support will help an increased number of CHD patients access genetic testing but will create challenges in the realms of result interpretation.
Table 2.
Genetic testing protocol for patients with congenital heart disease
| Patient features | Testing recommendations |
| Features suggestive of Trisomy 13 or Trisomy 18 | STAT FISH for aneuploidy (13, 18, 21, X and Y) |
| Features suggestive of Monosomy X or Trisomy 21 | Karyotype with reflex to chromosomal microarray if normal |
| Heterotaxy | Chromosomal microarray and heterotaxy panel that includes primary ciliary dyskinesia genes |
| Hypoplastic left heart syndrome | Chromosomal microarray and MYH6 sequencing |
| Other significant cardiovascular malformations | Chromosomal microarray |
When possible involvement of a medical geneticist is encouraged for all patients. For patients with possible Trisomy 13 or Trisomy 18, we recommend ordering FISH for aneuploidy with a rapid turnaround, as these diagnoses critically alter care options. For patients with Trisomy 21 or possible Turner Syndrome, we recommend a karyotype with reflexive addition of chromosomal microarray testing in the event the karyotype is nondiagnostic. The majority of patients should have first-line testing with chromosomal microarray. If a prenatal microarray has been completed, the resolution of that delay should be noted in that chart and repeated if not adequate. Patients with heterotaxy should undergo molecular assessment for primary ciliary dyskinesia. The primary ciliary dyskinesia genes we currently assess in patients with heterotaxy include ARMC4, C21orf59, CCDC103, CCDC114, CCDC151, CCDC39, CCDC40, CCDC65, CCNO, DNAAF1, DNAAF2, DNAAF3, DNAAF5, DNAH1, DNAH11, DNAH5, DNAH8, DNAI1, DNAI2, DNAL1, DRC1, DYX1C1, GAS8, LRRC6, MCIDAS, NME8, RSPH1, RSPH3, RSPH4A, RSPH9, SPAG1 and ZMYND10. Patients with hypoplastic left heart syndrome should undergo MYH6 sequencing.
FISH, fluorescence in-situ hybridization.
Acknowledgements
We would like to thank Drs. Donald Basel, Peter Frommelt, Jeanne James, Steven Kindel, Aaron Kinney and Robert Lane for their support of the development and continuation of the Herma Heart Institute's Clinical Cardiovascular Genetics Program.
Financial support and sponsorship
The Herma Heart Institute's Clinical Cardiovascular Program is supported by the Medical College of Wisconsin's Department of Pediatrics.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Husain SA, Pasquali SK, Jacobs JP, et al. Congenital heart operations performed in the first year of life: does geographic variation exist? Ann Thorac Surg 2014; 98:912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thakkar AN, Chinnadurai P, Lin CH. Adult congenital heart disease: magnitude of the problem. Curr Opin Cardiol 2017; 32:467–474. [DOI] [PubMed] [Google Scholar]
- 3.Gilboa SM, Devine OJ, Kucik JE, et al. Congenital heart defects in the United States: estimating the magnitude of the affected population in 2010. Circulation 2016; 134:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lui GK, Saidi A, Bhatt AB, et al. Diagnosis and management of noncardiac complications in adults with congenital heart disease: a scientific statement from the American Heart Association. Circulation 2017; 136:e348–e392. [DOI] [PubMed] [Google Scholar]
- 5.van Engelen K, Baars MJ, van Rongen LT, et al. Adults with congenital heart disease: patients’ knowledge and concerns about inheritance. Am J Med Genet A 2011; 155A:1661–1667. [DOI] [PubMed] [Google Scholar]
- 6.Bregman S, Frishman WH. Impact of improved survival in congenital heart disease on incidence of disease. Cardiol Rev 2018; 26:82–85. [DOI] [PubMed] [Google Scholar]
- 7▪▪.Zaidi S, Brueckner M. Genetics and genomics of congenital heart disease. Circ Res 2017; 120:923–940. [DOI] [PMC free article] [PubMed] [Google Scholar]; This summary of current knowledge regarding the cause of CHD is comprehensive and well organized to guide readers through the different levels of complexity of the genetics of CHD.
- 8▪▪.Russell MW, Chung WK, Kaltman JR, Miller TA. Advances in the understanding of the genetic determinants of congenital heart disease and their impact on clinical outcomes. J Am Heart Assoc 2018; 7: [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides an overview of interactions between genetics and clinical outcomes and is organized both by genetic cause and clinical outcome making the information accessible.
- 9.Ahrens-Nicklas RC, Khan S, Garbarini J, et al. Utility of genetic evaluation in infants with congenital heart defects admitted to the cardiac intensive care unit. Am J Med Genet A 2016; 170:3090–3097. [DOI] [PubMed] [Google Scholar]
- 10.Geddes GC, Basel D, Frommelt P, et al. Genetic testing protocol reduces costs and increases rate of genetic diagnosis in infants with congenital heart disease. Pediatr Cardiol 2017; 38:1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldenberg PC, Adler BJ, Parrott A, et al. High burden of genetic conditions diagnosed in a cardiac neurodevelopmental clinic. Cardiol Young 2017; 27:459–466. [DOI] [PubMed] [Google Scholar]
- 12.Carey AS, Liang L, Edwards J, et al. Effect of copy number variants on outcomes for infants with single ventricle heart defects. Circ Cardiovasc Genet 2013; 6:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swisher M, Jonas R, Tian X, et al. Increased postoperative and respiratory complications in patients with congenital heart disease associated with heterotaxy. J Thorac Cardiovasc Surg 2011; 141:637–644. 644.e631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomita-Mitchell A, Stamm KD, Mahnke DK, et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiol Genomics 2016; 48:912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner MK, Clarke S, Mahnke DK, et al. Effect of 22q11.2 deletion on bleeding and transfusion utilization in children with congenital heart disease undergoing cardiac surgery. Pediatr Res 2016; 79:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. [[Accessed 29 June 2018]]. American Board of Medical Genetics and Genomics. Number of certified specialist in genetics by specialty. 2017. http://www.abmgg.org/pdf/Statistics%20for%20Webpage.pdf. [Google Scholar]
- 17. [[Accessed 29 June 2018]]. American Board of Medical Genetics and Genomics. Number of certified specialists in genetics by state. 2018. http://www.abmgg.org/pdf/SpecialistsByState%20February%202018.pdf. [Google Scholar]
- 18.Allanson JE, Biesecker LG, Carey JC, Hennekam RC. Elements of morphology: introduction. Am J Med Genet A 2009; 149A:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Øyen N, Poulsen G, Boyd HA, et al. Recurrence of congenital heart defects in families. Circulation 2009; 120:295–301. [DOI] [PubMed] [Google Scholar]
- 20.Botto LD, Lin AE, Riehle-Colarusso T, et al. Seeking causes: classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res A Clin Mol Teratol 2007; 79:714–727. [DOI] [PubMed] [Google Scholar]
- 21.Cowan JR, Ware SM. Genetics and genetic testing in congenital heart disease. Clin Perinatol 2015; 42:373–393. ix. [DOI] [PubMed] [Google Scholar]
- 22.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res 2013; 112:707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azhar M, Ware SM. Genetic and developmental basis of cardiovascular malformations. Clin Perinatol 2016; 43:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowan JR, Tariq M, Shaw C, et al. Copy number variation as a genetic basis for heterotaxy and heterotaxy-spectrum congenital heart defects. Philos Trans R Soc Lond B Biol Sci 2016; 371:pii: 20150406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartman RJ, Rasmussen SA, Botto LD, et al. The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatr Cardiol 2011; 32:1147–1157. [DOI] [PubMed] [Google Scholar]
- 26.Costain G, Silversides CK, Bassett AS. The importance of copy number variation in congenital heart disease. NPJ Genom Med 2016; 1:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton RB, McBride KL, Bleyl SB, et al. Rationale for the cytogenomics of cardiovascular malformations consortium: a phenotype intensive registry based approach. J Cardiovasc Dev Dis 2015; 2:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86:749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geddes GC, Stamm K, Mitchell M. Ciliopathy variant burden and developmental delay in children with hypoplastic left heart syndrome. Genet Med 2017; 19:711–714. [DOI] [PubMed] [Google Scholar]
- 30.Wu XL, Li R, Fu F, et al. Chromosome microarray analysis in the investigation of children with congenital heart disease. BMC Pediatr 2017; 17:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng J, Picker J, Zheng Z, et al. Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genomics 2014; 15:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landis BJ, Ware SM. The current landscape of genetic testing in cardiovascular malformations: opportunities and challenges. Front Cardiovasc Med 2016; 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2871 congenital heart disease probands. Nat Genet 2017; 49:1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manheimer KB, Richter F, Edelmann LJ, et al. Robust identification of mosaic variants in congenital heart disease. Hum Genet 2018; 137:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrod AS, Zahid M, Tian X, et al. Airway ciliary dysfunction and sinopulmonary symptoms in patients with congenital heart disease. Ann Am Thorac Soc 2014; 11:1426–1432. [DOI] [PubMed] [Google Scholar]
- 36.Nakhleh N, Francis R, Giese RA, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation 2012; 125:2232–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abou Tayoun AN, Krock B, Spinner NB. Sequencing-based diagnostics for pediatric genetic diseases: progress and potential. Expert Rev Mol Diagn 2016; 16:987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins RL, Brand H, Redin CE, et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 2017; 18:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomford NE, Dzobo K, Yao NA, et al. Genomics and epigenomics of congenital heart defects: expert review and lessons learned in Africa. OMICS 2018; 22:301–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klena NT, Gibbs BC, Lo CW. Cilia and ciliopathies in congenital heart disease. Cold Spring Harb Perspect Biol 2017; 9:pii: a028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart E, Adams PS, Tian X, et al. Airway ciliary dysfunction: association with adverse postoperative outcomes in nonheterotaxy congenital heart disease patients. J Thorac Cardiovasc Surg 2018; 155:755–763.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geddes GC, Stamm K, Mitchell M, et al. Ciliopathy variant burden and developmental delay in children with hypoplastic left heart syndrome. Genet Med 2017; 19:711–714. [DOI] [PubMed] [Google Scholar]
- 43.Hoang TT, Goldmuntz E, Roberts AE, et al. The Congenital Heart Disease Genetic Network Study: cohort description. PLoS One 2018; 13:e0191319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helle EIT, Biegley P, Knowles JW, et al. First trimester plasma glucose values in women without diabetes are associated with risk for congenital heart disease in offspring. J Pediatr 2018; 195:275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, Zhao W, Pan B, et al. Alcohol exposure causes overexpression of heart development-related genes by affecting the histone H3 acetylation via BMP signaling pathway in cardiomyoblast cells. Alcohol Clin Exp Res 2017; 41:87–95. [DOI] [PubMed] [Google Scholar]
- 46.Elizabeth KE, Praveen SL, Preethi NR, et al. Folate, vitamin B12, homocysteine and polymorphisms in folate metabolizing genes in children with congenital heart disease and their mothers. Eur J Clin Nutr 2017; 71:1437–1441. [DOI] [PubMed] [Google Scholar]
- 47.Leirgul E, Gildestad T, Nilsen RM, et al. Periconceptional folic acid supplementation and infant risk of congenital heart defects in Norway 1999–2009. Paediatr Perinat Epidemiol 2015; 29:391–400. [DOI] [PubMed] [Google Scholar]
- 48.Arjmandnia M, Besharati M, Rezvan S. Studying the determinant factors leading to congenital heart disease in newborns. J Educ Health Promot 2018; 7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abqari S, Gupta A, Shahab T, et al. Profile and risk factors for congenital heart defects: a study in a tertiary care hospital. Ann Pediatr Cardiol 2016; 9:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panagiotopoulou O, Fouzas S, Sinopidis X, et al. Congenital heart disease in twins: the contribution of type of conception and chorionicity. Int J Cardiol 2016; 218:144–149. [DOI] [PubMed] [Google Scholar]


