Abstract
Background
The provision of compatible blood products to patients is the most essential task of transfusion medicine. Besides ABO and Rh, a number of additional blood group antigens often have to be considered for the blood supply of immunized or chronically transfused patients. It also applies for platelet antigens (HPA) and neutrophil antigens (HNA) for patients receiving platelet or granulocyte concentrates. Here, we describe the molecular screening for a number of blood group, HPA, and HNA alleles. Based on the screening results we are building up a regional blood donor registry to provide extended matched blood products on demand.
Methods
We developed and validated TaqMan™ PCR and PCR-SSP methods for genetic markers defining 37 clinically relevant blood group antigens (beyond ABO and Rh), 10 HPA, and 11 HNA. Furthermore, we describe a feasible method for fast molecular screening of the HNA-2null phenotype. All data were statistically evaluated regarding genotype distribution. Allele frequencies were compared to ExAC data from non-Finnish Europeans.
Results
Up to now more than 2,000 non-selected regular blood donors in south-west Germany have been screened for blood group, HPA, and HNA alleles. The screening results were confirmed by serology and PCR-SSP methods for selected numbers of samples. The allele frequencies were similar to non-finnish Europeans in the ExAC database except for the alleles encoding the S, HPA-3b and HNA-4b antigens, with significantly lower prevalence in our cohort, as well as the LU14 and the HNA-3b antigens, with a higher frequency compared to the ExAC data. We identified 71 donors with rare blood groups such as Lu(a+b-), Kp(a+b-), Fy(a-b-) and Vel-, and 169 donors with less prevalent HPA or HNA types.
Conclusion
Molecular screening for blood group alleles by using TaqMan™ PCR is an effective and reliable high-throughput method for establishing a rare donor registry.
Keywords: Molecular blood typing, Genotyping, Polymerase chain reaction, High- and low-prevalence antigens, Granulocytes, Platelets, Blood groups
Introduction
The provision of optimally matched blood products with regard to antigen compatibility is an ongoing challenge in transfusion medicine. Patients who repeatedly need red blood cell concentrates (RBCs) due to e.g. hemoglobinopathy or other diseases and also patients who have developed red cell antibodies often have to be transfused with RBCs compatible not only for ABO and Rh but also for several additional blood group antigens [1]. Blood recipients who are negative for high-prevalence antigens such as Vel are at risk for immunization and subsequent hemolytic transfusion reactions when transfused with antigen-positive blood. On the other hand, blood donors positive for low-prevalence antigens can cause antibody formation in antigen-negative patients [2]. Antibodies to low-prevalence antigens are of risk in pregnancy as they can cause a hemolytic disease of the fetus and the newborn (HDFN) and are often not detected in prenatal antibody screening. It also applies for the immunization to human platelet alloantigens (HPA) causing fetal neonatal alloimmune thrombocytopenia (FNAIT) or neutrophil antigens (HNA) causing neonatal immune neutropenia (NIN) [3, 4]. Furthermore, transfusion-associated acute lung injury (TRALI) can be caused by antibodies to HNA antigens.
With the advent of high-throughput screening in molecular blood typing, there is increasing information regarding the prevalence of rare blood groups and other antigens. Several methods have been developed and used for screening larger numbers of blood donors for common and rare antigens [5, 6, 7, 8, 9, 10, 11, 12, 13]. These studies add further information regarding the frequency of blood group alleles and regional differences in the availability of donors with rare blood groups. These approaches have led to the establishment of regional or national rare donor registries [14, 15, 16, 17]. As national legal and logistical considerations can cause problems and critical delay for import and export of rare blood, it might be reasonable to establish such registries on a local level as well. Our institute is located in the Rhein-Neckar metropolitan region in south-west Germany, an area which is home to a population with different ethnic background [18, 19]. This in turn may lead to a higher incidence of diseases such as thalassemia, which require frequent blood transfusions [20]. In addition, those ethnic differences might well translate into a higher incidence of rare blood groups as well [21].
To improve our ability to provide rare blood RBCs on a regional level, it is important to continue the screening activities and to test significant part of our donor population for alleles coding for rare blood groups. We also focused on the genotypes for the most relevant HPAs and HNAs in order to improve the availability of donors for antigen-selected platelet and granulocyte concentrates and for diagnostic purposes.
Material and Methods
DNA Samples of Blood Donors
This study was performed in a urban blood donor cohort from the south-western part of Germany. Donors gave written consent to provide additional 4.5 ml EDTA blood samples, and the use of the blood samples for research purposes was approved by the ethics committee II of Heidelberg University, Medical Faculty Mannheim. DNA was isolated in our laboratory from pseudonymized EDTA-anticoagulated blood samples using a commercial system for 96 well plates (Invisorb Blood Mini HTS 96 Kit; Stratec Biomedical AG, Berlin, Germany).
Genotyping
We developed TaqMan™ PCR methods and PCR with sequence-specific primers (PCR-SSP) for 21 blood group-defining SNPs, 5 SNPs defining HPAs, 3 SNPs defining the HNA-3a/b, HNA-4a/b and HNA-5a/b and the CD177 haplotype causing the HNA-2null phenotype (table 1). The FCGR3B*01, *02, and *03 alleles encoding the HNA-1a/b/c/d antigens were screened by PCR-SSP using a previously published protocol [22] and slightly modified primers (table 1). For each genetic marker, we established PCR-SSP methods according to a standard protocol [23] and with primers listed in table 1. The A4GALT SNP rs2143918 has a 100% correlation with the P1/P2 phenotype in all populations, except for rare individuals of African descent [24]. This SNP was addressed for P genotyping.
Table 1.
Primers for PCR-SSP and TaqMan™-PCR assays
| Antigen system | dbSNP No. | Primer sequences for PCR-SSP (5′−3′) | Specificity | TaqMan™ PCR assay* |
|---|---|---|---|---|
| MNS (MN) | rs7687256, | rev: GGCAAGAATTCCTCCATAGTAG | GYPA | (self-designed assay1) |
| rs7658293 | for: CATATCAGCATTAAGTACCACTGGT | c.71G, c.72T | ||
| for: CATATCAGCATTAAGTACCACTGAG | c.71A, c.72G | |||
| MNS (Ss) | rs7683365 | for: caaaatatgattaagaaaaggaaacccg | GYPB | C__34183121_10 |
| rev: cgatggacaagttgtccca | c.143T | |||
| rev: cgatggacaagttgtcccg | c.143C | |||
| P1PK (P1/P2) | rs2143918 | rev: CACAAAGAACCTGGCTTCTCG | A4GALT | C__16072990_20 |
| for: CACATCTTTCCTGGGAAGGAATT | +2857T | |||
| for: CACATCTTTCCTGGGAAGGAATG | +2857G | |||
| LU (Lua/b) | rs28399653 | for: CCAGGGAGACCCATAACAAG | BCAM | C__25764173_10 |
| rev: TCTCAGCCGAGGCTAGGT | c.230A | |||
| rev: TCTCAGCCGAGGCTAGGC | c.230G | |||
| LU (LU8/14) | rs28399656 | rev: GAGGTCAAAGGCCAGCACAG | BCAM | C__32363989_10 |
| for: CTCTCCCAGAGGGCTACAT | c.611T | |||
| for: GATCTCTCCCAGAGGGCTACAA | c.611A | |||
| LU (Aua/b) | rs1135062 | rev: ggttaagctatggtggtcattgc | BCAM | C__1846428_10 |
| for: ccatgtcttccacttcggca | c.1615A | |||
| for: ccatgtcttccacttcggcg | c.1615G | |||
| KEL (K/k) | rs8176058 | for: GGGAGATGGAGATGGAAATGG | KEL | C__1719_20 |
| rev: GACTCATCAGAAGTCTCAGCG | c.578C | |||
| rev: GACTCATCAGAAGTCTCAGCA | c.578T | |||
| KEL (Kpa/b) | rs8176059 | rev: AGGGCACTAGGAGGAAGAAG | KEL | C__25596888_20 |
| for: CTTGTCAATCTCCATCACTTCAC | c.841C | |||
| for: CTTGTCAATCTCCATCACTTCAT | c.841T | |||
| KEL (Jsa/b) | rs8176038 | rev: ggcccttgacacttgcatac | KEL | C__25596899_20 |
| for: tgcctgggggctgcct | c.1790T | |||
| for: tgcctgggggctgccc | c.1790C | |||
| KEL (11/17) | rs61729034 | rev: CCTTAGAGGAGGGACACAAAG | KEL | C__27862879_10 |
| for: GGCAAGCTCTTCCAGATGGT | c.905T | |||
| for: GGCAAGCTCTTCCAGATGGC | c.905C | |||
| FY (Fya/b) | rs12075 | rev: GGCACCACAATGCTGAAGAG | DARC | C__2493442_10 |
| for: CTTCCCAGATGGAGACTATGG | c.125G | |||
| for: CTTCCCAGATGGAGACTATGA | c.125A | |||
| FY (Fy0) | rs2814778 | rev: CAAAGGGAGGGACACAAGAG | DARC | C__15769614_10 |
| for: CCTCATTAGTCCTTGGCTCTTAT | -67T | |||
| for: CCTCATTAGTCCTTGGCTCTTAC | -67C | |||
| JK (Jka/b) | rs1058396 | rev: AGACAGCAAGTGGGCTCAAG | SLC14A1 | C__1727582_10 |
| for: TCTTTCAGCCCCATTTGAGG | c.838G | |||
| for: GTCTTTCAGCCCCATTTGAGA | c.838A | |||
| DI (Dia/b) | rs2285644 | rev: TCCTGCCTGCCCTAGTTCTG | SLC4A1 | C__26654865_10 |
| for: GGGTGGTGAAGTCCACGCT | c.2561T | |||
| for: GGTGGTGAAGTCCACGCC | c.2561C | |||
| DI (Wra/b) | rs75731670 | rev: TGGGAGAATGCCAGGGAAAG | SLC4A1 | C__2259533_10 |
| for: CACTGGGCTTGCGTTCCA | c.1972A | |||
| for: CACTGGGCTTGCGTTCCG | c.1972G | |||
| YT (Yta/b) | rs1799805 | rev: GGGAGGACTTCTGGGACTTC | ACHE | C__8786419_20 |
| for: CATCAACGCGGGAGACTTCC | c.1057C | |||
| for: CATCAACGCGGGAGACTTCA | c.1057A | |||
| SC (SC1/2) | rs56025238 | rev: CCCTTATATTCCGGCATCAGATC | ERMAP | C__32292873_10 |
| for: CTCTCTCCCTCTGGCCCG | c.169G | |||
| for: CTCTCTCCCTCTGGCCCA | c.169A | |||
| DO (Doa/b) | rs11276 | for: CTCCACATCCCTCCTGAAAG | ART4 | C__2687344_20 |
| rev: GTTGACCTCAACTGCAACCAGTT | c.793A | |||
| rev: GTTGACCTCAACTGCAACCAGTC | c.793G | |||
| CO (Coa/b) | rs28362692 | rev: GCACGGAAGATGCTGATCTGG | AQP1 | C__27536484_20 |
| for: GGGGAACAACCAGACGGC | c.134C | |||
| for: GGGGAACAACCAGACGGT | c.134T | |||
| LAN (+/–) | rs149202834 | rev: AGGTGTACCTGGCTCCTTTC | ABCB6 | C_168788417_10 |
| for: TTTAGCCTGTGGGTGCTGC | c.574C | |||
| for: TTTAGCCTGTGGGTGCTGT | c.574T | |||
| VEL (+/–) | rs566629828 | rev: CCAAAGGCTGCGGTTTGCTG | SMIM1 | see [7] |
| for: GCAGCAGGGACGGAGTCA | c.64–80ins | |||
| for: GCAGCAGGGACGGAGTCC | c.64–80del | |||
| HPA-1a/b | rs5918 | rev: gtgcaatcctctggggact | ITGB3 | C__818008_30 |
| for: gacttacaggccctgcctct | ITGB3*176T | |||
| for: acttacaggccctgcctcc | ITGB3*176C | |||
| HPA-2a/b | rs6065 | rev: gccagcgacgaaaatagagg | GP1BA | C__11442703_10 |
| for: cccccagggctcctgac | GP1BA*482C | |||
| for: cccccagggctcctgat | GP1BA*482T | |||
| HPA-3a/b | rs5911 | rev: accagagagcctgctcactac | ITGA2B | C__3017440_10 |
| for: ggactgggggctgcccat | ITGA2B*2621T | |||
| for: ggactgggggctgcccag | ITGA2B*2621G | |||
| HPA-5a/b | rs1801106 | rev: ggcagtacactatacattcaactc | ITGA2 | C__27862812_10 |
| for: aaggaagagtctacctgtttactatcaaag | ITGA2*1600G | |||
| for: aaggaagagtctacctgtttactatcaaaa | ITGA2*1600A | |||
| HPA-15a/b | rs10455097 | for: cagtattatgaccttatgatgacctattc | CD109 | C__3226894_10 |
| rev: gttacttcaaattcttggtaaatcctgg | CD109*2108C | |||
| rev: gttacttcaaattcttggtaaatcctgt | CD109*2108A | |||
| HNA-1a | rs448740 | for: cacagtggtttcacaatgagaa | FCGR3B*01 | none |
| rs368410676 | rev: atggacttctagctgcacc | |||
| HNA-1b/d | rs527909462 | for: gcctcaatggtacagcgtgctt | FCGR3B*02 | none |
| rs5030738 | rev: ctgtcgttgactgtggcag | |||
| HNA-1b/c | rs527909462 | for: gcctcaatggtacagcgtgctt | FCGR3B*03 | none |
| rs5030738 | rev: ctgtcgttgactgtggcaT | |||
| HNA-2 (+/–) | rs1164364335 | for: catggagaaggtgacctttgag | CD177 | (self-designed assay2) |
| rev: CAACAGTGCTGCAGCCTTT | CD177*787A | |||
| rev: CAACAGCGCTGCAGCTTTA | CD177*787T | |||
| HNA-3a/b | rs2288904 | rev: ctgcatggagcagaggatgg | SLC44A2 | C__11789692_10 |
| for: GAGTGGCTGAGGTGCTTCG | SLC44A2*461G | |||
| for: GGAGTGGCTGAGGTGCTTCA | SLC44A2*461A | |||
| HNA-4a/b | rs1143679 | rev: aaggaggtctgacggtgaag | ITGAM | C__2847895_1_ |
| for: ctcatgcgagcccatccg | ITGAM*230G | |||
| for: ctcatgcgagcccatcca | ITGAM*230A | |||
| HNA-5a/b | rs2230433 | rev: tccaccttgcggaaggagagtc | ITGAL | C___25754090_10 |
| for: atcatcccccacagatccag | ITGAL*2372G | |||
| for: catcccccacagatccac | ITGAL*2372C | |||
TaqMan™-PCR assays were commercially available (Life Technologies) under the given assay number
self-designed TaqMan™-PCR assay for MN genotyping with forward primer CTCAGTCACCTCGTTCTTAATC, reverse primer GGCAAGAATTCCTCCATAGTAG, FAM-probe CACTGGTGTGGCAA (M), VIC-probe CACTGAGGTGGCAA (N)
self-designed TaqMan™-PCR assay for HNA-2 genotyping with forward primer CCCTCAGGACTCACATCAAC, reverse primer CTGAGTGGATGGTGGTCTTC, FAM-probe CAGCCTTTTGTCCC (CD177 wild type), VIC-probe CAGCTTTAGGTCGC (CD177 null variant).
For screening by TaqMan™ PCR we used commercial assays (Life Technologies, Darmstadt, Germany), except for the alleles encoding the M/N, VEL, and HNA-2 antigens. All TaqMan™ PCR analyses were performed by endpoint fluorescence detection on a ABI 7000 real-time PCR cycler using the standard program for genotyping (Applied Biosystems, Darmstadt, Germany). The method for VEL screening was described before [12]. For the M/N antigens we developed primers specific for GYPA and probes specific for the corresponding GYPA*01 and GYPA*02 alleles (table 1).
Homozygosity for a nonsense mutation c.787A>T in CD177 was identified as the most frequent cause of the HNA-2null phenotype [25]. But genotyping is hampered by the CD177P1 pseudogene with identical DNA sequence in the corresponding region. The 787A>T mutation can also occur in the CD177P1 gene but does not affect the HNA-2 phenotype. In order to differentiate between the CD177*c.787A>T and the CD177P1*787A>T genotype, a long-range PCR for the specific amplification of the CD177 gene followed by PCR-SSP was described [26]. However, this method is not appropriate for screening of samples in larger scales. We developed a TaqMan™ PCR method with primers and probes listed in table 1 by which individuals homozygous for the 787A>T mutation in both genes CD177 and CD177P1 can be identified.
Validation
The results from TaqMan™ PCR screening were verified by PCR-SSP for significant numbers of samples representing the different genotypes and for all samples with rare genotypes. Donors homozygous for the CD177*c.787A>T mutation were re-analyzed using the long-range and PCR-SSP method described recently [26] with slightly modified primers for the PCR-SSP (table 1). As far as available genotyping data were confirmed by serology for some of the antigens such as MNS, P, K/k, Aua/b, Vel, HPA-1, and HPA-5. In all validation samples the serologically determined phenotype matched the genotype. In addition, 8 donors homozygous for the CD177*c.787A>T mutation identified by the TaqMan™-PCR method were phenotyped for the HNA-2 antigen (flow cytometry using the monoclonal anti-CD177 MEM-166) and the HNA-2null phenotype was confirmed in all samples.
Statistics
The typing data for each genetic marker was reviewed with regard to deviation from the Hardy-Weinberg equilibrium. The minor allele frequencies (MAF) in our donor cohort were compared to the allele frequencies of non-Finnish Europeans in the ExAC database [27]. Significance of differences was calculated by using appropriate tests in the SPSS software package (SPSS Vers. 12.0; IBM, Armonk, NY, USA).
Results
Blood donors of our institute have been enrolled in the genotyping study since June 2016 irrespective of age, gender or ABO and Rh blood group (table 2). Using TaqMan™ PCR, the donors were screened for alleles defining 37 blood group antigens, 10 HPAs, and 11 HNAs including the HNA-2null phenotype. In total, 2,084 donors were typed for all antigens (table 3). Some antigens have already been typed in higher numbers of donors.
Table 2.
Demographic characteristics of the screened donor population
| % of all donors | |
|---|---|
| Age, years | |
| <20 | 6.7 |
| 20–29 | 37.8 |
| 30–39 | 15.9 |
| 40–49 | 17.2 |
| 50–59 | 14.5 |
| >60 | 7.9 |
| Sex | |
| Male | 60.4 |
| Female | 39.6 |
| Blood groups | |
| O pos | 31.0 |
| O neg | 11.4 |
| A pos | 30.8 |
| A neg | 8.9 |
| B pos | 9.1 |
| B neg | 2.7 |
| AB pos | 4.8 |
| AB neg | 1.3 |
Table 3.
Results from genotype screening of blood donors
| Antigen system | dbSNP No. | Allele | Genotype | Phenotype1 | Number of donors | % of donors |
|---|---|---|---|---|---|---|
| Blood groups | ||||||
| MNS | rs7687256, | GYPA*01 | c.71G, c.72T | M+N– | 589 | 28.26 |
| (002) | rs7658293 | GYPA*02 | c.71G/A, c.72T/G | M+N+ | 1,078 | 51.58 |
| c.71A, c.72G | M–N+ | 420 | 20.15 | |||
| MNS | rs7683365 | GYPB*03 | c.143T | S+s– | 212 | 9.33 |
| (002) | GYPB*04 | c.143T/C | S+s+ | 995 | 43.77 | |
| c.143C | S–s+ | 1,066 | 46.90 | |||
| P1PK | rs21439182 | A4GALT*P1.01 | +2857T | P1 | 555 | 26.58 |
| (003) | A4GALT*P2.01 | +2857T/G | P1 | 1,017 | 48.56 | |
| +2857G | P2 | 518 | 24.86 | |||
| LU | rs28399653 | LU*01 | c.230A | Lu(a+b–) | 4 | 0.19 |
| (005) | LU*02 | c.230A/G | Lu(a+b+) | 153 | 7.34 | |
| c.230G | Lu(a–b+) | 1,927 | 92.47 | |||
| LU | rs28399656 | LU*02 | c.611T | LU:8,–14 | 1,987 | 95.35 |
| (005) | LU*02.14 | c.611T/A | LU:8,14 | 96 | 4.61 | |
| c.611A | LU:–8,14 | 1 | 0.05 | |||
| LU | rs1135062 | LU*02 | c.1615A | Au(a+b–) | 1,000 | 47.98 |
| (005) | LU*02.19 | c.1615A/G | Au(a+b+) | 933 | 44.77 | |
| c.1615G | Au(a–b+) | 151 | 7.25 | |||
| KEL | rs8176058 | KEL*01 | c.578C | K+k– | 7 | 0.25 |
| (006) | KEL*02 | c.578C/T | K+k+ | 222 | 8.08 | |
| c.578T | K–k+ | 2,518 | 91.67 | |||
| KEL | rs8176059 | KEL*02 | c.841C | Kp(a–b+c–) | 2,047 | 98.22 |
| (006) | KEL*02.03 | c.841C/T | Kp(a+b+c–) | 36 | 1.73 | |
| c.841T | Kp(a+b–c–) | 1 | 0.05 | |||
| KEL | rs8176038 | KEL*02 | c.1790T | Js(a–b+) | 2,083 | 99.95 |
| (006) | KEL*02.06 | c.1790T/C | Js(a+b+) | 1 | 0.05 | |
| c.1790C | Js(a+b–) | 0 | 0.00 | |||
| KEL | rs61729034 | KEL*02 | c.905T | KEL:11,–17 | 2,078 | 99.71 |
| (006) | KEL*02.17 | c.905T/C | KEL:11,17 | 6 | 0.29 | |
| c.905C | KEL:–11,17 | 0 | 0.00 | |||
| FY | rs12075 | FY*01 | c.125G | Fy(a+b–) | 402 | 19.29 |
| (008) | FY*02 | c.125G/A | Fy(a+b+) | 998 | 47.89 | |
| c.125A | Fy(a–b+) | 684 | 32.82 | |||
| FY | rs2814778 | FY*01N.01 or | -67T | 2,032 | 97.50 | |
| (008) | FY02N.01 | -67T/C | 46 | 2.21 | ||
| -67C | Fy(a–b–) | 6 | 0.29 | |||
| JK | rs1058396 | JK*01 | c.838G | Jk(a+b–) | 564 | 27.06 |
| (009) | JK*02 | c.838G/A | Jk(a+b+) | 999 | 47.94 | |
| c.838A | Jk(a–b+) | 521 | 25.00 | |||
| DI | rs2285644 | DI*01 | c.2561T | Di(a+b–) | 0 | 0.00 |
| (010) | DI*02 | c.2561T/C | Di(a+b+) | 3 | 0.11 | |
| c.2561C | Di(a–b+) | 2,746 | 99.89 | |||
| DI | rs75731670 | DI*02 | c.1972A | Wr(a+b–) | 0 | 0.00 |
| (010) | DI*02.03 | c.1972A/G | Wr(a+b+) | 3 | 0.14 | |
| c.1972G | Wr(a–b+) | 2,081 | 99.86 | |||
| YT | rs1799805 | YT*01 | c.1057C | Yt(a+b–) | 2,427 | 88.64 |
| (011) | YT*02 | c.1057C/A | Yt(a+b+) | 295 | 10.77 | |
| c.1057A | Yt(a–b+) | 16 | 0.58 | |||
| SC | rs56025238 | SC*01 | c.169G | SC:1,–2 | 2,727 | 99.27 |
| (013) | SC*02 | c.169G/A | SC:1,2 | 20 | 0.73 | |
| c.169A | SC:–1,2 | 0 | 0.00 | |||
| DO | rs11276 | DO*01 | c.793A | Do(a+b–) | 319 | 15.31 |
| (014) | DO*02 | c.793A/G | Do(a+b+) | 1,029 | 49.38 | |
| c.793G | Do(a−b+) | 736 | 35.32 | |||
| CO | rs28362692 | CO*01 | c.134C | Co(a+b−) | 1,931 | 92.66 |
| (015) | CO*02 | c.134C/T | Co(a+b+) | 151 | 7.25 | |
| c.134T | Co(a–b+) | 2 | 0.10 | |||
| LAN | rs149202834 | ABCB6*01 | c.574C | Lan+ | 2,064 | 99.04 |
| (033) | ABCB6*01N.13 | c.574C/T | Lan+ | 20 | 0.96 | |
| c.574T | Lan– | 0 | 0.00 | |||
| VEL | rs566629828 | VEL*01 | c.64–80ins | Vel+ | 2,653 | 96.54 |
| (034) | VEL*-01 | c.64–80ins/del | Vel+ | 94 | 3.42 | |
| c.64–80del | Vel– | 1 | 0.04 | |||
| Platelet antigens (HPA) | ||||||
| HPA-1 | rs5918 | ITGB3*176T | c.176T | HPA-1(a+b–) | 1,764 | 72.03 |
| ITGB3*176C | c.176T/C | HPA-1(a+b+) | 636 | 25.97 | ||
| c.176C | HPA-1(a–b+) | 49 | 2.00 | |||
| HPA-2 | rs6065 | GP1BA*482C | c.482C | HPA-2(a+b–) | 2,040 | 82.93 |
| GP1BA*482T | c.482C/T | HPA-2(a+b+) | 395 | 16.06 | ||
| c.482T | HPA-2(a–b+) | 25 | 1.02 | |||
| HPA-3 | rs5911 | ITGA2B*2621T | c.2621T | HPA-3(a+b–) | 909 | 38.47 |
| ITGA2B*2621G | c.2621T/G | HPA-3(a+b+) | 1,116 | 47.23 | ||
| c.2621G | HPA-3(a–b+) | 338 | 14.30 | |||
| HPA-5 | rs1801106 | ITGA2*1600G | c.1600G | HPA-5(a+b–) | 1,925 | 81.33 |
| ITGA2*1600A | c.1600G/A | HPA-5(a+b+) | 422 | 17.83 | ||
| c.1600A | HPA-5(a–b+) | 20 | 0.84 | |||
| HPA-15 | rs10455097 | CD109*2108C | c.2108C | HPA-15(a+b–) | 598 | 24.28 |
| CD109*2108A | c.2108C/A | HPA-15(a+b+) | 1,227 | 49.82 | ||
| c.2108A | HPA-15(a–b+) | 638 | 25.90 | |||
| Neutrophil antigens (HNA) | ||||||
| HNA-1 | rs448740 | FCGR3B*01 | c.227A | HNA-1(a+b–c–d–) | 349 | 12.71 |
| rs368410676 | FCGR3B*02 | c.227A/c.147T,266C | HNA-1(a+b+c–d+) | 1,174 | 42.75 | |
| rs527909462 | FCGR3B*03 | c.147T,266C | HNA-1(a–b+c–d+) | 1,100 | 40.06 | |
| rs5030738 | c.227A/c.147T,266A | HNA-1(a+b+c+d–) | 41 | 1.49 | ||
| c.227A/c.147T,266C/A | HNA-1(a+b+c+d+) | 64 | 2.33 | |||
| c.147T,266C/A | HNA-1(a–b+c+d+) | 18 | 0.66 | |||
| HNA-2 | rs1164364335 | CD177/CD177P1 | c.787A, c.787A/T | HNA-2(+/–) | 3,315 | 97.53 |
| c.787T | HNA-2(–) | 84 | 2.47 | |||
| HNA-3 | rs2288904 | SLC44A2*461G | c.461G | HNA-3(a+b–) | 1,329 | 58.42 |
| SLC44A2*461A | c.461G/A | HNA-3(a+b+) | 819 | 36.00 | ||
| c.461A | HNA-3(a–b+) | 126 | 5.58 | |||
| HNA-4 | rs1143679 | ITGAM*230G | c.230G | HNA-4(a+b–) | 1,797 | 79.06 |
| ITGAM*230A | c.230G/A | HNA-4(a+b+) | 454 | 19.97 | ||
| c.230A | HNA-4(a–b+) | 22 | 0.97 | |||
| HNA-5 | rs2230433 | ITGAL*2372G | c.2372G | HNA-5(a+b–) | 1,151 | 50.66 |
| ITGAL*2372C | c.2372G/C | HNA-5(a+b+) | 934 | 41.11 | ||
| c.2372C | HNA-5(a–b+) | 187 | 8.23 | |||
Phenotype deduced from the genotype
rs2143918 has a 100 % correlation with the P1/P2 phenotype in all populations except for rare individuals of African descent [21]. It was therefore used to determine the P1/P2 genotype even though it is not the causative mutation.
The most frequent cause of the HNA-2null phenotype is the CD177 haplotype carrying the nonsense mutation c.787A>T (linked to additional 4 SNPs) in exon 7 [25]. However, the CD177P1 pseudogene with almost identical DNA sequence in the exon 4 to 9 region can also harbor this haplotype making CD177-specific genotyping difficult. Pre-amplification of the CD177 gene using long-range PCR followed by PCR-SSP typing of the c.787A>T mutation was described as a genotyping method [26]. Due to the long-range PCR this method is, however, time-consuming and cost-intensive. With our fast (70 min) and easy TaqMan™ PCR method we were able to screen 3,399 donors within a short period of time and identified 84 donors (2.47%) homozygous for the null haplotype in both genes CD177 and CD177P1 (fig. 1). Because this screening method does not distinguish between CD177 and CD177P1, individuals homozygous for the c.787A>T mutation in the CD177 gene, but with one or two wild-type alleles in the CD177P1 gene, are not identified as HNA-2null. For all 84 donors we could confirm the CD177*c.787T homozygosity using the CD177-specific long-range PCR and PCR-SSP. For 8 of the donors we obtained fresh blood samples and confirmed the HNA-2null phenotype by flow cytometry (data not shown). Thus, we describe here a suitable method for fast molecular screening of the HNA-2null phenotype.
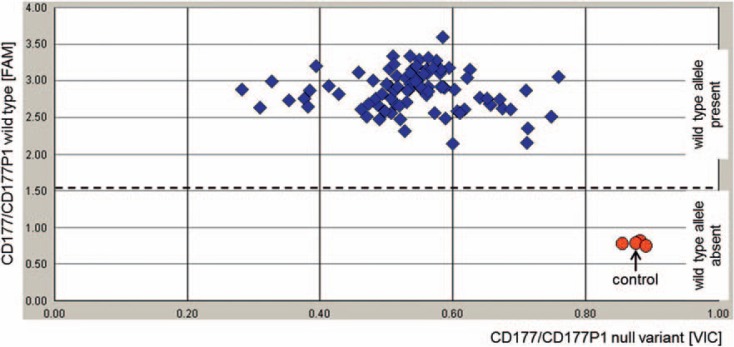
Fig. 1.
Representative result of TaqMan™ PCR-based typing of 96 samples for the c.787A>T mutation in CD177 and CD177P1. Primers were designed to amplify a 88 bp fragment from both genes CD177 and CD177P1. The FAM-labeled probe detects the wild-type allele and the VIC-labeled probe is specific for the null haplotype including the c.787A>T mutation. FAM signals below the threshold (dashed line) indicate absence of the wild type allele in both genes and, therefore, indicate the HNA-2null phenotype. Control: donor sample with confirmed homozygosity for the c.787A>T mutation by CD177 genomic sequencing and confirmed HNA-2null phenotype by flow cytometry. In addition to the control three donors revealed absence of the wild type allele, i.e. homozygosity for the c.787A>T mutation. As expected, the HNA-2null phenotype was confirmed for these samples.
The prevalence of the different genotypes was within the expected range, with a few exceptions. For each measured distribution, basic probability testing was performed using a chi-square test comparing the observed distribution of genotypes to the expected distribution based on observed allele frequencies and the Hardy-Weinberg equilibrium. All but two results (Aua/b and Fynull) were in line with the expected genotype frequencies. For the Aua/b-encoding SNP we identified a trend (p = 0.054) for more heterozygotes (933 vs. 869) and less Aub homozygotes (151 vs. 183) as expected. Re-typing of 144 selected samples using PCR-SSP confirmed the result from TaqMan™ PCR. In addition, phenotyping of 11 samples for the Aua/b antigens by means of serology also confirmed the genotyping results. The overrepresentation of Aua/b donors in our cohort could be a characteristic of our donor population or is just a coincidental finding. The promoter mutation FY*-67T>C was screened for identification of the Fynull phenotype. Based on the allele frequency of 0.0139 for the mutation, we could expect one homozygote in 5,164 donors. We found 6 homozygotes in 2,084 typed donors leading to a significant deviation (p < 0.0001) from the Hardy-Weinberg equilibrium. Most likely, the Fynull donors are of African ancestry because the mutation is very common in this population.
The MAF of blood group antigens, HPAs, and HNAs in our study cohort were compared to the data of non-Finnish Europeans in the ExAC exome sequencing study [27]. The S antigen encoding GYPB*c.143T allele, the HPA-3b antigen encoding ITGA2B*c.2621G allele, and the HNA-4b antigen encoding ITGAM*c.230A allele was significantly less prevalent in our cohort (table 4). The LU*c.611T allele encoding the LU14 antigen and the SLC44A2*c.461A allele encoding the HNA-3b antigen showed a higher frequency compared to the ExAC data.
Table 4.
Minor allele frequencies in our donor population compared to the non-Finnish European population in the ExAC database
| Antigen | dbSNP No. | Minor allele | MAF in our cohort | MAF in ExAC | Significance* |
|---|---|---|---|---|---|
| Blood groups | |||||
| S | rs7683365 | GYPB*c.143T | 0.3121 | 0.3802 | <0.001 |
| Lu(a) | rs28399653 | LU*c.230A | 0.0390 | 0.0382 | 0.935 |
| LU14 | rs28399656 | LU*c.611T | 0.0234 | 0.0146 | 0.048 |
| Au(b) | rs1135062 | LU*c.1615G | 0.2962 | 0.2993 | 0.809 |
| K | rs8176058 | KEL*c.578T | 0.0430 | 0.0412 | 0.753 |
| Kp(a) | rs8176059 | KEL*c.841T | 0.0091 | 0.0105 | 0.634 |
| Js(a) | rs8176038 | KEL*c.1790C | 0.0002 | 0.0005 | 0.548 |
| KEL17 | rs61729034 | KEL*c.905C | 0.0014 | 0.0018 | 0.726 |
| Fy(a) | rs12075 | FY*c.125G | 0.4325 | 0.4244 | 0.632 |
| Jk(b) | rs1058396 | SLC14A1*c.838A | 0.4897 | 0.4912 | 0.950 |
| Di(a) | rs2285644 | SLC4A1*c.2561T | 0.0005 | 0.0004 | 0.823 |
| Wr(a) | rs75731670 | SLC4A1*c.1972A | 0.0007 | 0.0002 | 0.114 |
| Yt(b) | rs1799805 | ACHE*c.1057A | 0.0597 | 0.0502 | 0.211 |
| Sc2 | rs56025238 | ERMAP*c.169A | 0.0036 | 0.0050 | 0.628 |
| Do(a) | rs11276 | ART4*c.793A | 0.4000 | 0.3849 | 0.348 |
| Co(b) | rs28362692 | AQP1*c.134T | 0.0372 | 0.0398 | 0.623 |
| LAN- | rs149202834 | ABCB6*c.574T | 0.0048 | 0.0030 | 0.454 |
| Platelet antigens (HPA) | |||||
| HPA-1b | rs5918 | ITGB3*c.176C | 0.1499 | 0.1553 | 0.629 |
| HPA-2b | rs6065 | GP1BA*c.482T | 0.0904 | 0.0865 | 0.697 |
| HPA-3b | rs5911 | ITGA2B*c.2621G | 0.3792 | 0.4443 | <0.001 |
| HPA-5b | rs1801106 | ITGA2*c.1600A | 0.0976 | 0.0918 | 0.553 |
| HPA-15a | rs10455097 | CD109*c.2108C | 0.4919 | 0.4975 | 0,752 |
| Neutrophil antigens (HNA) | |||||
| HNA-3b | rs2288904 | SLC44A2*c.461A | 0.2358 | 0.2094 | 0.048 |
| HNA-4b | rs1143679 | ITGAM*c.230A | 0.1095 | 0.1769 | <0.001 |
| HNA-5b | rs2230433 | ITGAL*c.2372C | 0.2879 | 0.2856 | 0.889 |
Bold numbers indicate significant differences (p < 0.05) of MAF between both populations.
By screening of more than 2,000 donors we found a number of rare blood types, such as Lu(a+b-), Kp(a+b-), Fynull, Vel-, HPA-1(a-b+), and HNA-2null (table 5).
Table 5.
Rare blood types identified in the screened donor cohort
| Phenotype* | Number of donors |
|---|---|
| Negative for HP blood groups | |
| Lu(a+b–) | 4 |
| LU:–8,14 | 1 |
| K+k– | 7 |
| Kp(a+b–) | 1 |
| Fy(a–b–) | 6 |
| Yt(a–b+) | 16 |
| Co(a–b+) | 2 |
| Vel– | 1 |
| Positive for LP blood groups | |
| Js(a+) | 1 |
| KEL:17 | 6 |
| Di(a+) | 3 |
| Wr(a+) | 3 |
| SC:2 | 20 |
| HPA / HNA | |
| HPA-1(a–b+) | 49 |
| HPA-2(a–b+) | 25 |
| HPA-5(a–b+) | 20 |
| HNA-2null | 84 |
| HNA-4(a–b+) | 22 |
Negative for high-prevalence (HP) antigens; positive for low-prevalence (LP) antigens.
Discussion
Molecular screening of the local blood donor population is an approach to enable a compatible blood supply on demand. We describe a TaqMan™ PCR-based typing of the corresponding genetic markers (mostly SNPs). This method is feasible, as it is associated with limited workload and can be easily adapted with regard to higher throughput of samples and additional alleles.
Up to now we obtained 72,337 genotypes from 33 genetic markers in more than 2,000 donors, and it is an ongoing screening program including all new donors at our institute. Based on the genotyping data we were able to deduce the phenotypes for 37 blood group antigens, 10 HPAs, and 11 HNAs including the HNA-2null phenotype. The complete antigen profile could be achieved for 2,084 donors. Some of the antigens are already typed in higher numbers of donors such as Vel (2,748 donors), Yta/b (2,738), HPA-1 (2,449), HNA-1 (2,746), HNA-2 (3,399) and others (table 3).
In this study we also describe a feasible method for fast and easy molecular screening to identify HNA-2null individuals. The method is based on TaqMan™ PCR with endpoint fluorescence detection and can be easily adapted to medium or high throughput. Using the 96-well format in standard PCR cyclers more than 1,000 donors can be typed per day. CAVE: only individuals homozygous for the null mutation c.787A>T in both genes CD177 and CD177P1 are typed as HNA-2null. If the mutation is homozygous in CD177 but heterozygous or absent in CD177P1, these individuals are also HNA-2null, but are not recognized by the TaqMan™ PCR method. Other null variants of CD177 are also not detected.
Most of the allele frequencies observed in our cohort were similar to the frequencies reported for non-Finnish Europeans in the ExAC database [27]. Only 5 of the 25 alleles with clear information in the database revealed a significant difference. This probably results from the demographic characteristics of the urban area in which our institute is located. As of 2016, the population of Mannheim includes 44.7% immigrants, of which at least 70% belong to countries with a predominantly Caucasian population [18]. About 12% have Asian ancestry and 5% are African. However, we do not know the portion of immigrants in our donor population. At least for the Fynull phenotype that is most prevalent in Africans and which we found in 6 of our donors (0.3%), we can assume a significant number of donors with African ancestry. Another example could be the SLC44A2*c.461A allele encoding the HNA-3b antigen. According to ExAC data it is more prevalent in East Asians (0.3091) than in non-Finnish Europeans (0.2094), and the MAF was 0.2358 in our cohort. Thus, the differences in allele frequencies in our donor population compared to the general European population could be attributed to a certain number of donors with African or Asian ancestry.
Immigration to Central Europe in general may increase the number of patients with sickle cell anemia or other hemoglobinopathies. The transfusion therapy of such patients is challenging because of a frequent blood demand and because of antigens with different prevalence in other populations. The concept of extended matching in transfusion therapy of such patients is appropriate to prevent alloimmunization events [28, 29, 30]. Especially, patients who were already immunized by past transfusion could benefit from extensive antigen matching beyond C/c, E/e, and K [31, 32]. As recently reported in a large cohort of transfused patients, the most immunogenic antigens were in order K, E, CW, e, Jka, c, and Fya[33]. Among the 2,084 donors typed for these antigens, we could identify 22 donors negative for K, E, Jka, c, and Fya as well as 22 donors negative for D, K, E, Jka, and Fya. Three donors were negative for a number of clinically relevant antigens: A, B, C, D, E, K, Jka, Fya, M, and S.
Based on our experiences we attempt to establish a regional registry of extended typed blood donors to provide fresh RBC units with special blood types on demand. Similar concepts for the extended matched blood supply are established in different countries worldwide [34, 35, 36, 37].
Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Franchini M, Forni GL, Liumbruno GM. Is there a standard-of-care for transfusion therapy in thalassemia? Curr Opin Hematol. 2017;24:558–564. doi: 10.1097/MOH.0000000000000373. [DOI] [PubMed] [Google Scholar]
- 2.Avent ND. Large-scale blood group genotyping: clinical implications. Br J Haematol. 2009;144:3–13. doi: 10.1111/j.1365-2141.2008.07285.x. [DOI] [PubMed] [Google Scholar]
- 3.Moncharmont P. Platelet component transfusion and alloimmunization: where do we stand? Transfus Clin Biol. 2018 doi: 10.1016/j.tracli.2018.01.003. DOI: 10.1016/j.tracli.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Flesch B, Reil A. Molecular genetics of human neutrophil antigens. Transfus Med Hemother. 2018;45((5)) doi: 10.1159/000491031. DOI: 10.1159/000491031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashmi G, Shariff T, Zhang Y, Cristobal J, Chau C, Seul M, Vissavajjhala P, Baldwin C, Hue-Roye K, Charles-Pierre D, Lomas-Francis C, Reid ME. Determination of 24 minor red blood cell antigens for more than 2000 blood donors by high-throughput DNA analysis. Transfusion. 2007;47:736–747. doi: 10.1111/j.1537-2995.2007.01178.x. [DOI] [PubMed] [Google Scholar]
- 6.Wagner FF, Bittner R, Petershofen EK, Doescher A, Müller TH. Cost-efficient sequence-specific priming-polymerase chain reaction screening for blood donors with rare phenotypes. Transfusion. 2008;48:1169–1173. doi: 10.1111/j.1537-2995.2008.01682.x. [DOI] [PubMed] [Google Scholar]
- 7.Jungbauer C, Hobel CM, Schwartz DW, Mayr WR. High-throughput multiplex PCR genotyping for 35 red blood cell antigens in blood donors. Vox Sang. 2012;102:234–242. doi: 10.1111/j.1423-0410.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- 8.Haer-Wigman L, Ait Soussan A, Ligthart P, de Haas M, van der Schoot CE. Molecular analysis of immunized Jr(a-) or Lan- patients and validation of a high-throughput genotyping assay to screen blood donors for Jr(a-) and Lan- phenotypes. Transfusion. 2014;54:1836–1846. doi: 10.1111/trf.12544. [DOI] [PubMed] [Google Scholar]
- 9.Veldhuisen B, Porcelijn L, van der Schoot CE, de Haas M Molecular typing of human platelet and neutrophil antigens (HPA and HNA) Transfus Apher Sci. 2014;50:189–199. doi: 10.1016/j.transci.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Meyer S, Vollmert C, Trost N, Brönnimann C, Gottschalk J, Buser A, Frey BM, Gassner C. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014;54:3198–3207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 11.Meyer S, Trost N, Frey BM, Gassner C. Parallel donor genotyping for 46 selected blood group and 4 human platelet antigens using high-throughput MALDI-TOF mass spectrometry. Methods Mol Biol. 2015;1310:51–70. doi: 10.1007/978-1-4939-2690-9_5. [DOI] [PubMed] [Google Scholar]
- 12.Wieckhusen C, Rink G, Scharberg EA, Rothenberger S, Komurcu N, Bugert P. Molecular screening for Vel- blood donors in southwestern Germany. Transfus Med Hemother. 2015;42:356–360. doi: 10.1159/000440791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner FF, Doescher A, Bittner R, Müller TH. Extended donor typing by pooled capillary electrophoresis: impact in a routine setting. Transfus Med Hemother. 2018;45:225–237. doi: 10.1159/000490155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crottet SL, Henny C, Meyer S, Still F, Stolz M, Gottschalk J, Neuenschwander K, Taleghani BM, Gowland P, Frey BM, Fontana S, Hustinx H, Niederhauser C, Gassner C. Implementation of a mandatory donor RHD screening in Switzerland. Transfus Apher Sci. 2014;50:169–174. doi: 10.1016/j.transci.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Flegel WA, Gottschall JL, Denomme GA. Implementing mass-scale red cell genotyping at a blood center. Transfusion. 2015;55:2610–2615. doi: 10.1111/trf.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong YJ, Chung Y, Hwang SM, Park JS, Kwon JR, Choi YS, Kim JN, Lee DH, Kwon SY, Cho NS, Song EY, Park KU, Song J, Han KS. Genotyping of 22 blood group antigen polymorphisms and establishing a national recipient registry in the Korean population. Ann Hematol. 2016;95:985–991. doi: 10.1007/s00277-016-2645-7. [DOI] [PubMed] [Google Scholar]
- 17.Gassner C, Degenhardt F, Meyer S, Vollmert C, Trost N, Neuenschwander K, Merki Y, Portmann C, Sigurdardottir S, Zorbas A, Engström C, Gottschalk J, Amar el Dusouqui S, Waldvogel-Abramovski S, Rigal E, Tissot J, Tinguely C, Mauvais SM, Sarraj A, Bessero D, Stalder M, Infanti L, Buser A, Sigle J, Weingand T, Castelli D, Braisch MC, Thierbach J, Heer S, Schulzki T, Krawczak M, Franke A, Frey BM. Low-frequency blood group antigens in Switzerland. Transfus Med Hemother. 2018;45:239–250. doi: 10.1159/000490714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einwohner mit Migrationshintergrund in kleinräumiger Gliederung Statistische Daten Mannheim N° 3/2017. Mannheim, Stadt Mannheim. 2017:p. 27. https://www.mannheim.de/sites/default/files/2018-06/d201703_migrationshintergrund__2016.pdf (last accessed September 10, 2018) [Google Scholar]
- 19.Bevölkerung und Erwerbstätigkeit Bevölkerung mit Migrationshintergrund - Ergebnisse des Mikrozensus 2016 Statistisches Bundesamt (Destatis) 2016:p 8. [Google Scholar]
- 20.Rachmilewitz EA, Giardina PJ. How I treat thalassemia. Blood. 2011;118:3479–3488. doi: 10.1182/blood-2010-08-300335. [DOI] [PubMed] [Google Scholar]
- 21.Nance ST. How to find, recruit and maintain rare blood donors. Curr Opin Hematol. 2009;16:503–508. doi: 10.1097/MOH.0b013e3283316bed. [DOI] [PubMed] [Google Scholar]
- 22.Reil A, Bux J. Geno- and phenotyping of human neutrophil antigens. Methods Mol Biol. 2015;1310:193–203. doi: 10.1007/978-1-4939-2690-9_16. [DOI] [PubMed] [Google Scholar]
- 23.Rink G, Scharberg EA, Bugert P. PCR with sequence-specific primers for typing of diallelic blood groups. Methods Mol Biol. 2015;1310:71–81. doi: 10.1007/978-1-4939-2690-9_6. [DOI] [PubMed] [Google Scholar]
- 24.Lai YJ, Wu WY, Yang CM, Yang LR, Chu CC, Chan YS, Lin M, Yu LC. A systematic study of single-nucleotide polymorphisms in the A4GALT gene suggests a molecular genetic basis for the P1/P2 blood groups. Transfusion. 2014;54:3222–3231. doi: 10.1111/trf.12771. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Mair DC, Schuller RM, Li L, Wu J. Genetic mechanism of human neutrophil antigen 2 deficiency and expression variations. PLoS Genet. 2015;11:e1005255. doi: 10.1371/journal.pgen.1005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayat B, Bein G, Sachs UJ. A sequence-specific polymerase chain reaction method for HNA-2 genotyping: homozygous c.843A>T mutation predicts the absence of CD177. Transfusion. 2016;56:2127–2132. doi: 10.1111/trf.13689. [DOI] [PubMed] [Google Scholar]
- 27.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasalle-Williams M, Nuss R, Le T, Cole L, Hassell K, Murphy JR, Ambruso DR. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center (CME) Transfusion. 2011;51:1732–1739. doi: 10.1111/j.1537-2995.2010.03045.x. [DOI] [PubMed] [Google Scholar]
- 29.Casas J, Friedman DF, Jackson T, Vege S, Westhoff CM, Chou ST. Changing practice: red blood cell typing by molecular methods for patients with sickle cell disease. Transfusion. 2015;55:1388–1393. doi: 10.1111/trf.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putzulu R, Piccirillo N, Orlando N, Massini G, Maresca M, Scavone F, Ricerca BM, Zini G. The role of molecular typing and perfect match transfusion in sickle cell disease and thalassaemia: an innovative transfusion strategy. Transfus Apher Sci. 2017;56:234–237. doi: 10.1016/j.transci.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Sins JW, Biemond BJ, van den Bersselaar SM, Heijboer H, Rijneveld AW, Cnossen MH, Kerkhoffs JL, van Meurs AH, von Ronnen FB, Zalpuri S, de Rijke YB, van der Schoot CE, de Haas M, van der Bom JG, Fijnvandraat K. Early occurrence of red blood cell alloimmunization in patients with sickle cell disease. Am J Hematol. 2016;91:763–769. doi: 10.1002/ajh.24397. [DOI] [PubMed] [Google Scholar]
- 32.Yee MEM, Josephson CD, Winkler AM, Webb J, Luban NLC, Leong T, Stowell SR, Fasano RM. Red blood cell minor antigen mismatches during chronic transfusion therapy for sickle cell anemia. Transfusion. 2017;57:2738–2746. doi: 10.1111/trf.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evers D, Middelburg RA, de Haas M, Zalpuri S, de Vooght KM, van de Kerkhof D, Visser O, Péquériaux NC, Hudig F, Schonewille H, Zwaginga JJ, van der Bom JG. Red-blood-cell alloimmunisation in relation to antigens' exposure and their immunogenicity: a cohort study. Lancet Haematol. 2016;3:e284–292. doi: 10.1016/S2352-3026(16)30019-9. [DOI] [PubMed] [Google Scholar]
- 34.Shao CP, Zhao CJ, Wu CL, Xu H, Wang XD, Wu XY, Yi P, Dang XT. Rh-matched transfusion through molecular typing for β-thalassemia patients is required and feasible in Chinese. Transfus Med Hemother. 2018;45:252–257. doi: 10.1159/000489471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castilho L, Dinardo CL. Optimized antigen-matched in sickle cell disease patients: chances and challenges in molecular times - the Brazilian way. Transfus Med Hemother. 2018;45:258–262. doi: 10.1159/000490713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Floch A, Tournamille C, Chami B, Pirenne F. Genotyping in sickle cell disease patients: the French strategy. Transfus Med Hemother. 2018;45:264–270. doi: 10.1159/000490858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan J, Delaney M. Transfusion support of minority patients: extended antigen donor typing and recruitment of minority blood donors. Transfus Med Hemother. 2018;45:271–276. doi: 10.1159/000491883. [DOI] [PMC free article] [PubMed] [Google Scholar]