Abstract
Background
Identification of the proteins that are associated with estrogen receptor (ER) status is a first step in selecting drugs against hormone-dependent breast cancer. Recently, the proteins associated with ER status were reported using liquid chromatography and tandem mass spectrometry, and microRNA (miRNA) profiling of breast cancer subtype was demonstrated using real-time-PCR.
Methods
We present herein two cases with differential protein expression and miRNA profiling in ER-positive and -negative breast cancer.
Results
Proteins associated with fatty acid metabolism were uniquely detected in ER-positive breast cancer. The level of miR-181a expression in ER-positive cancer was higher than that in ER-negative cancer, while the expression of miR-27a, miR-107, and miR-195 was lower in ER-positive compared with ER-negative cancer.
Conclusion
These cases suggest that fatty acid synthase (FAS) and FAS-related miRNAs are important in ER-positive breast cancer.
Keywords: breast cancer, estrogen receptor, fatty acid synthase, microRNA
Introduction
It is known that estrogen plays a major role in the development and progression of breast cancer. DNA microarray analysis has demonstrated that estrogen receptor (ER)-positive and -negative breast cancers have unique molecular profiles, has identified several distinct molecular subclasses, and has been used to predict prognosis.1–3 The practical classification of intrinsic subtypes was proposed at the St. Gallen consensus meeting of breast cancer: luminal A type (ER-positive and/or progesterone receptor (PgR)-positive, human epidermal growth factor receptor type 2 (HER2)-negative, and low proliferation), luminal B type (ER-positive and/or PgR-positive, HER2-negative, and high proliferation), HER2-positive type (ER-negative, PgR-negative, and HER2-positive), and triple-negative type (ER-negative, PgR-negative, and HER2-negative).4 Although information exists on the messenger RNA (mRNA) expression signatures of specific breast cancer subtypes, mRNA levels do not necessarily correlate with protein abundance;5,6 thus, comparing the protein expression profiles of ER-positive and -negative breast cancer remains necessary. Protein level information is crucial for functional understanding, and the identification of receptors and intracellular protein kinases will likely assist in the selection of drug targets in breast cancer. MicroRNAs (miRNAs) are small non-coding genes that control gene transcription or protein translation and have been implicated in multiple regulatory roles in various human malignancies. In breast cancer, some miRNAs have been shown to upregulate the functions of oncogenes, and others stimulate tumor supressors.7,8
Previous studies have demonstrated the association of proteins with ER status9 or miRNA signatures in breast cancer subtype,10 while the expression of protein and miRNA in identical breast cancer tissues has not been reported. We present herein two cases with differential protein expression and miRNA profiles in ER-positive and -negative breast cancer tissues.
Methods
Our two patients noted breast tumors on mammography screenings in 2007. One patient was a 47-year-old female with a 1.2 cm mass in her right breast, and the findings of core needle biopsy confirmed invasive breast carcinoma of histological grade 3. Immunohistochemistry for ER and PgR was positive, but that for HER2 was negative. The other patient was a 37-year-old female with a 1.8 cm mass in her left breast, and the findings of core needle biopsy revealed invasive breast carcinoma of histological grade 3. Immunostaining for ER, PgR, and HER2 was negative. The two patients at stage I (T1N0M0) underwent breast conservation surgery followed by adjuvant radiation therapy. The patient with the ER- and PgR-positive tumor was administered toremifene for 5 years after surgery, and the patient with ER-, PgR-, and HER2-negative tumor was given no medication postoperatively. The patients showed no recurrence or metastasis for 10 years after surgery. They provided written informed consent for the examination of tumor characteristics and for this study.
Tumor specimens were fixed in 10% formalin, and samples embedded in paraffin were used for protein assay and miRNA PCR array.
Ethical approval
These case reports and research were approved to be published by the ethics committee of Kameda Medical Center (18-103).
Results
Identification of proteins was performed by nano-flow liquid chromatography and tandem mass spectrometry (LC/MS/MS). Using a microtome, 10 µm slices of formalin-fixed, paraffin-embedded (FFPE) tissue were placed in 1.5 mL microcentrifuge tubes. Proteins from samples for LC/MS/MS were extracted using 10 µL of 8 M urea solution with an ultrasonic homogenizer. Proteins in the samples were digested with trypsin (15–18 units) by overnight incubation at 37°C. After extraction, the samples were stored at –20°C. Peptide samples were loaded into a column (ZORBAX-SB- C18) and an Agilent 1200 HPLC system (Agilent Technologies, Santa Clara, CA, USA) was used for nano-flow liquid chromatography, followed by the use of an Agilent 6520 for mass spectrometry with a Molecular Feature Extractor (Agilent Technologies). Protein identification results for triplicate injections are shown in Figure 1. For ER-positive and -negative tumors, the numbers of proteins were 123 and 177, respectively, and 98 proteins were common to both. Thus, 25 proteins were unique to the ER-positive tumor and 79 were unique to the ER-negative tumor (Figure 1). Among the unique proteins in the ER-positive tumor, seven proteins were associated with cell growth, five with cytoskeletal activities, five with protein catabolism, and five with fatty acid metabolism (Table 1).
Figure 1.
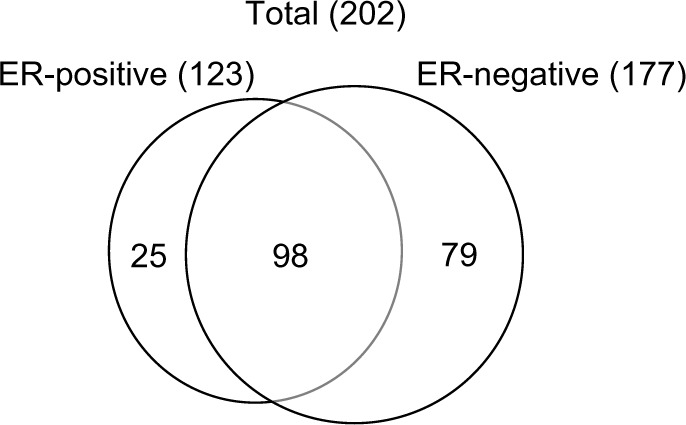
Venn diagram of identified proteins from ER-positive and -negative breast cancer tissues.
Notes: A total of 123 proteins were identified in ER-positive and 177 in ER-negative tumors. About 98 proteins were common to both ER-positive and -negative tumors. The total number of identified proteins was 202.
Abbreviation: ER, estrogen receptor.
Table 1.
The identified unique proteins in ER-positive breast cancer tissues on the biological process
| Biological process | Identified proteins |
|---|---|
|
| |
| Cell growth (n=7) | Anterior gradient protein 2 homolog precursor |
| Activated RNA polymerase transcriptional coactivator p15 | |
| Heterogeneous nuclear ribonucleoprotein F | |
| Thymidine phosphorylase precursor | |
| Thioredoxin | |
| Prolactin-inducible protein precursor | |
| Heterogeneous nuclear ribonucleoprotein U | |
| Cytoskeletal activities (n=5) | Decorin precursor |
| Collagen alpha-1 (XIV) chain precursor | |
| LIM and SH3 domain protein 1 | |
| Glutathione S-transferase | |
| Receptor expression-enhancing protein 6 | |
| Protein catabolism (n=5) | Inter-alpha-trypsin inhibitor heavy chain H1 precursor |
| Lysosome-associated membrane glycoprotein 2 precursor | |
| Serpin B6 | |
| Kelch-like protein 31 | |
| Flavin reductase | |
| Fatty acid metabolism (n =5) | Fatty acid synthase |
| Apolipoprotein A-I precursor | |
| Zic-alpha-2-glycoprotein precursor | |
| Platelet-activating factor acetylhydrolase IB | |
| Acid ceramidase precursor | |
| Others (n=3) | Adenylate kinase isoenzyme 1 |
| Arylamine N-acetyltransferase 1 | |
| Fructose-1,6-bisphosphatase 1 | |
Note: Activated RNA polymerase transcriptional coactivator p15.
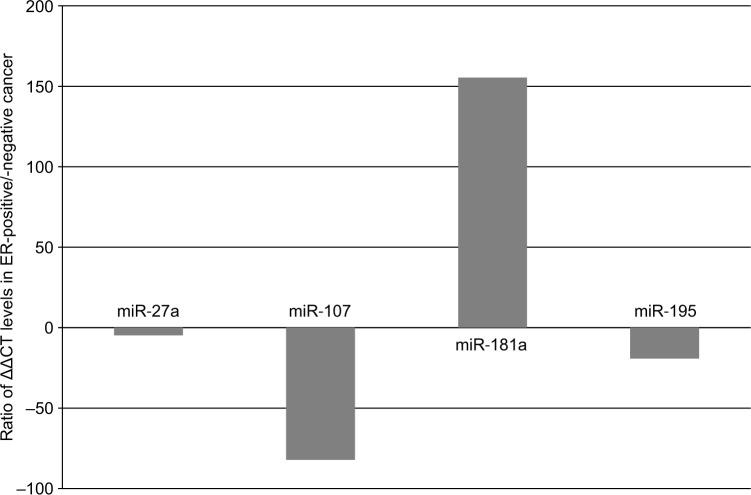
Paraffin in 10 µm slices of FFPE tissue was removed by xylene treatment, and the tissues were washed with ethanol twice to remove xylene. The tissues were then treated with proteinase K at 37°C overnight. Following centrifugation, the supernatant was processed with a silica-based spin column (Toray Industries Inc., Kanagawa, Japan) in order to obtain purified total RNA. The degree of RNA cross-linking and RNA degradation was analyzed by electrophoresis using an Agilent 2100 Bioanalyzer (Agilent Technologies). miRNA was isolated from total RNA using an RT2 qPCR-Grade miRNA Isolation Kit (SABiosciences, Frederick, MD, USA). The miRNA MiScript SYBR Green human breast cancer miRNA PCR kit (Qiagen NV, Venlo, the Netherlands) was used in combination with an ABI PRISM 7000 Sequence System (Thermo Fisher Scientific, Waltham, MA,USA). real-time (RT)-qPCR analyses were performed with regard to 84 miRNAs in the breast cancer panel. The raw RT-qPCR data were deposited in GEO database (GEO accession number: GSE121172). Data were analyzed using the ΔΔCt method (original expression level =2−ΔΔCt).11 Data are expressed as a ratio of ΔΔCt levels in the ER-positive specimen to those in the ER-negative specimen. Measurements were recorded in triplicate. Fatty acid synthase (FAS) in breast cancer was reported to be a predictive factor for poor prognosis.12 We observed that four miRNAs, namely miR-27a, miR-107, miR-181a, and miR-195, were involved in FAS, because only these four miRNAs were available for FAS-related miRNAs in the breast cancer panel which was used in this study. The median expression ratio of miR-181a was 155.42, and those of miR-27a, miR-107, and miR-195 were −4.79,–82.14, and –19.16, respectively (Figure 2). These results indicate that the level of miR-181a expression in the ER-positive tumor was higher than that in the ER-negative tumor, while reduced expression was noted for miR-27a, miR-107, and miR-195 in the ER-positive tumor.
Figure 2.
Changes of microRNA expression for fatty acid synthase in ER-positive and -negative breast cancer tissues.
Notes: The expressions of miR-27a, miR-107, miR-181a, and miR-195 were showed as a ratio of ΔΔCT value (ER-positive/ER-negative breast cancer tissue).
Abbreviation: ER, estrogen receptor.
Discussion
Breast cancer has recently been classified into several intrinsic subtypes.1 These intrinsic subtypes are identified based on a combination of immunohistochemical analyses of ER, PgR, and HER2 expression, and Ki67 labeling index or histological grade in routine clinical practice. The present two cases had similar clinicopathological features (stage I, HER2-negative, and histological grade 3) but different ER status. Identification of the proteins that are associated with ER status is the first step toward better understanding the hormone-dependent nature of breast carcinogenesis. Rezaul et al9 assessed the proteins in breast cancer tissues using LC/MS/MS and revealed that upregulated proteins of ER-positive cancer were over-represented by proteins involved in amino acid metabolism, protea-some, and fatty acid metabolism. FAS is a multi-enzyme complex catalyzing the synthesis of long-chain fatty acids from acetyl-CoA and malonyl-CoA. In most normal human tissues, however, FAS is generally expressed at low levels because cells preferentially use circulating dietary fatty acids for the synthesis of new structural lipids.13 High levels of FAS expression have been found in many human cancers, including breast cancer. Two studies showed that high levels of FAS expression in breast cancer were associated with an increased risk of death when occurring together with a high proliferative index,12 and FASN, which is one of the genes involved in fatty acid metabolism, was reported to be a candidate therapeutic target in breast cancer.14 The fatty acid metabolism pathway was also very significantly enriched in ER-positive breast cancer samples. The degradation of expression of genes involved in fatty acid/lipid metabolism in ER-positive cancer was associated with a relatively good prognosis.15 In our LC/MS/MS results, five proteins associated with fatty acid metabolism, including FAS, were uniquely present in ER-positive cancer. This suggests that FAS is an important pathway in ER-positive breast cancer.
miRNAs are strong post-transcriptional regulators of cell differentiation. The overexpression of miR-181a accelerated the accumulation of lipid droplets, increased the amount of triglycerides, and repressed tumor necrosis factor-alpha protein expression, while miR-181a suppression decreased the expression of fat synthesis-associated genes, includ ing FASN.16 In the present cases, expression of miR-181a increased and FAS was detected as a unique protein in ER-positive cancer. This suggests that miR-181a regulates fatty acid metabolism in the pathogenesis of hormone-dependent breast cancer. Lipid metabolism is reported to be regulated by miR-27a targeting the lipid synthetic transcription factor, and suppression of miR-27a increased the cellular lipid contents of human hepatoma cells.17 miR-195 was shown to be a tumor suppressor during tumorigenesis. Overexpression of miR-195 decreased cell invasion and migration in osteosarcoma cells by targeting FASN.18 Overexpression of miR-107 inhibited FASN levels, while miR-107 inhibitor increased FASN levels in a dose-dependent manner.19 Research on miRNA in breast cancer has reported that various subtypes exhibit different miRNA signatures.10 Expression of miR-107 was shown to be associated with ER-positive subtypes such as luminal A and B; expression of miR-107 decreased in luminal A and increased in luminal B. Overexpression of miR-107 inhibited FASN and reduced FAS levels, while high levels of FAS expression were associated with high cell proliferation. Generally, it is known that cell proliferation in luminal B is higher than that in luminal A. We detected the suppression of miR-107 in ER-positive, HER-2-negative, and histological grade three tumors, such as luminal B type and identified FAS in this sample. Decreases in miR-107 could increase FAS levels in ER-positive breast cancer, and consequently, our ER-positive tumor may have a higher histological grade index.
Conclusion
We presented two cases with different ER status and assessed proteomic findings and miRNA expression signatures. We found that FAS and FAS-related miRNAs indicate a relationship between fatty acid metabolism and pathogenesis in hormone-dependent breast cancer. As our report has a limitation such as small number of patients screened and restricted number of FAS-related miRNAs examined, further studies are necessary to support this hypothesis.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Gruvberger S, Ringnér M, Chen Y, et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61(16):5979–5984. [PubMed] [Google Scholar]
- 3.West M, Blanchette C, Dressman H, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98(20):11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coates AS, Winer EP, Goldhirsch A, et al. Panel Members Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26(8):1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. N Engl J Med. 2002;346(5):340–352. doi: 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- 6.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18(3-4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 7.Serpico D, Molino L, Di Cosimo S. microRNAs in breast cancer development and treatment. Cancer Treat Rev. 2014;40(5):595–604. doi: 10.1016/j.ctrv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dys-regulation in breast cancer. Cancer Res. 2013;73(22):6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 9.Rezaul K, Thumar JK, Lundgren DH, et al. Differential protein expression profiles in estrogen receptor-positive and -negative breast cancer tissues using label-free quantitative proteomics. Genes Cancer. 2010;1(3):251–271. doi: 10.1177/1947601910365896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Hori-guchi J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Hum Genet. 2017;62(1):15–24. doi: 10.1038/jhg.2016.89. [DOI] [PubMed] [Google Scholar]
- 11.Hu C, Shen SQ, Cui ZH, Chen ZB, Li W. Effect of microRNA-1 on hepatocellular carcinoma tumor endothelial cells. World J Gastroenterol. 2015;21(19):5884–5892. doi: 10.3748/wjg.v21.i19.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alò PL, Visca P, Trombetta G, et al. Fatty acid synthase (FAS) predictive strength in poorly differentiated early breast carcinomas. Tumori. 1999;85(1):35–40. doi: 10.1177/030089169908500108. [DOI] [PubMed] [Google Scholar]
- 13.Lupu R, Menendez JA. Targeting fatty acid synthase in breast and endo-metrial cancer: An alternative to selective estrogen receptor modulators? Endocrinology. 2006;147(9):4056–4066. doi: 10.1210/en.2006-0486. [DOI] [PubMed] [Google Scholar]
- 14.Menendez JA, Lupu R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin Ther Targets. 2017;21(11):1001–1016. doi: 10.1080/14728222.2017.1381087. [DOI] [PubMed] [Google Scholar]
- 15.Sørlie T, Wang Y, Xiao C, et al. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127–141. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Chen X, Guan L, et al. MiRNA-181a regulates adipogenesis by targeting tumor necrosis factor-α (TNF-α) in the porcine model. PLoS One. 2013;8(10):e71568. doi: 10.1371/journal.pone.0071568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirasaki T, Honda M, Shimakami T, et al. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87(9):5270–5286. doi: 10.1128/JVI.03022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao JH, Zhou RP, Peng AF, et al. microRNA-195 suppresses osteosar-coma cell invasion and migration in vitro by targeting FASN. Oncol Lett. 2012;4(5):1125–1129. doi: 10.3892/ol.2012.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatia H, Verma G, Datta M. miR-107 orchestrates ER stress induction and lipid accumulation by post-transcriptional regulation of fatty acid synthase in hepatocytes. Biochim Biophys Acta. 2014;1839(4):334–343. doi: 10.1016/j.bbagrm.2014.02.009. [DOI] [PubMed] [Google Scholar]