Abstract
Endothelial PAS domain-containing protein 1 (EPAS1) serves a role in angiogenesis, which is important for the development of tumors, including colorectal cancer (CRC). The current study aimed to estimate whether EPAS1 methylation was associated with CRC. A two-stage association study of EPAS1 methylation and CRC was conducted. In the first phase, EPAS1 methylation was evaluated in the tumor and adjacent non-tumor tissue samples from 41 patients with sporadic CRC in Jiangsu province, China. The diagnostic value of methylation of EPAS1 for CRC in the second phase was evaluated in 79 patients with sporadic CRC and 22 normal individuals in Zhejiang province, China. The methylation assay was performed using a quantitative methylation-specific polymerase chain reaction (qMSP) method. The percentage of methylated reference (PMR) was used to quantify the methylation level. The first-stage results indicated that EPAS1 promoter methylation was significantly lower in CRC tumor tissues compared with 5-cm-para-tumor tissues (median PMR, 0.59 vs. 1.22%; P=0.027) and 10-cm-para-tumor tissues (median PMR, 0.59 vs. 1.89%; P=0.001). In addition, the second-stage results indicated that EPAS1 promoter methylation was significantly lower in tumor tissues compared with 5-cm-para-tumor tissues (median PMR, 1.91 vs. 6.25%; P=3×10−7) and normal intestinal tissues from healthy controls (median PMR, 1.91 vs. 28.4%; P=5×10−7). Receiver Operating Characteristic curve analysis of the second-stage data indicated that the highest area under the curve of EPAS1 hypomethylation was 0.851 between Zhejiang CRC tissues and Zhejiang normal intestinal tissues (sensitivity, 95.5%; specificity, 60.8%).
Keywords: colorectal cancer, DNA methylation, endothelial PAS domain-containing protein 1, quantitative methylation-specific polymerase chain reaction
Introduction
In 2015, ~376,300 cases of colorectal cancer (CRC) were diagnosed in China (1). CRC is regarded as one of the most common cancers with high morbidity and mortality (2). Despite the improvement of diagnostic technologies, screening tools and clinical therapy, CRC remains a global challenge for public health due to the absence of a ‘gold standard’ for early diagnosis (2).
The molecular carcinogenic mechanisms of CRC have not been completely elucidated, however, CRC appears to be driven by the accumulation of abnormal genetic and epigenetic alternations in both oncogenes and tumor-suppressor genes (3). Cytosine modification, including DNA methylation, is one of the basic molecular mechanisms involved in the initiation and progression of CRC (2,4–6). Therefore, DNA methylation or epigenetic alterations may be promising markers for the early detection of CRC (4,7).
The endothelium PAS domain protein 1 (EPAS1) gene product is one of the important subunits of oxygen-induced hypoxia inducible factor (HIF) α, which regulates the primary transcriptional response to hypoxic stress (8). Hypoxia is one of the main factors promoting tumor angiogenesis (9,10). In addition, angiogenesis is considered a prerequisite for a range of biological processes, including tumorigenesis and tumor progression (11,12). HIF-2α/EPAS1 serves a role in tumor angiogenesis of different types of cancer, including lung cancer (13,14), renal carcinoma (15,16), liver cancer (17), pheochromocytoma (18–20) and CRC (8,21).
In a previous study including 39 patients with CRC and 43 normal controls, the expression of EPAS1 in the blood of patients with CRC was significantly increased, and was subsequently decreased after surgical resection of the tumor, returning to a normal level (21). Another study revealed significantly increased levels of EPAS1 methylation and significantly lower levels of EPAS1 mRNA expression in 120 primary colon adenocarcinoma tissues compared with paracancerous tissues (8).
In the current study, quantitative methylation-specific polymerase chain reaction (qMSP) was used to measure EPAS1 methylation in 120 Chinese patients with CRC and 22 healthy controls in two-stage experiments to assess whether the methylation of EPAS1 could be used as a biomarker for the diagnosis of CRC.
Patients and methods
Study subjects
The first phase of the association study involved 41 patients with CRC, from whom frozen tumor tissues and adjacent tissues 5 and 10 cm away from the tumor lesions were collected. These patients with CRC were recruited from the Third Affiliated Hospital of Nanjing University of Traditional Chinese Medicine (Nanjing, China) between August 2011 and March 2015 and their average age was 64.03±11.39 years (range, 21–86 years). Of the 41 patients, 28 were male, 12 were female and 1 was missing information. The second phase of the association study was conducted to verify the role of EPAS1 methylation in CRC. The second phase of the association study involved 79 CRC tumor tissues, 79 paired adjacent tissues 5 cm away from the tumor lesions and 22 healthy human intestinal tissues. Patients involved in the second phase of the present study were recruited from the Zhejiang Cancer Hospital (Hangzhou, China) and Shaoxing First People's Hospital (Shaoxing, China) between August 2011 and January 2015. The average age of the 79 patients with CRC in the second phase of the present association study was 60.27±11.74 years. Of the 79 patients, 51 were male and 28 were female. All patients were diagnosed by pathological examination. No radiotherapy or chemotherapy was performed prior to surgery. The age and sex data of healthy controls were not available. All clinical data were extracted between August 2011 and March January 2015 from medical records for subsequent analysis. The Human Research Ethics Committee of Ningbo University (Ningbo, China) granted approval for the present study. Each participant completed the written informed consent form.
DNA extraction and bisulfite conversion
DNA was extracted from frozen tissues using E.Z.N.A.® Tissue DNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer's protocol. DNA concentrations measurement and bisulfite treatment were performed as previously described (22).
SYBR-Green-based qMSP
qMSP was performed as previously described (23,24). The following thermocycling conditions were used: Initial denaturation at 95°C for 10 min; 45 cycles of 95°C for 20 sec, 58°C for 20 sec and 72°C for 30 sec; melting curve analysis at 95°C for 15 sec, 58°C for 1 min and 60°C for 1 min; and a final cooling stage at 40°C for 10 min. The primer sequences for EPAS1 (95 bp) were forward, 5′-GTTATAGATAGCGTTTGTAGAC-3′ and reverse, 5′-GATTACCACATTCCCGATA-3′; and the primer sequences for ACTB (133 bp) were forward, 5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and reverse, 5′-AACCAATAAAACCTACTCCTCCCTTAA-3′. The percentage of methylated reference (PMR) of EPAS1 for each sample was calculated using the 2−ΔΔCq quantification approach, where ΔΔCq=sample DNA (CtEPAS1- CtACTB control)-fully methylated DNA (CtEPAS1-CtACTB control) (25).
Bioinformatics analysis
The genomic position of the amplified fragment was obtained from University of California Santa Cruz genome browser according to human (GRCh37) assembly (genome.ucsc.edu). To evaluate the association between mRNA expression and EPAS1 methylation, data in the TCGA colorectal adenocarcinoma cohort with 372 samples were downloaded from cBioPortal (www.cbioportal.org).
Statistical analysis
All data were analyzed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA). Due to the skewed distribution of methylation levels, data were presented as the median (interquartile range). Friedman test and Wilcoxon nonparametric test were used to assess the difference in methylation between samples. Mann-Whitney U nonparametric test was used to assess the difference in methylation between groups. Contingency correlation test was used to evaluate the association between EPAS1 methylation and clinical features. Spearman's rank correlation coefficient was used to assess the correlation between EPAS1 methylation and gene expression. Pearson χ2 test or Fisher's exact test were used to assess the difference in clinical features between different sampling locations. The receiver operating characteristic (ROC) analysis was used to evaluate the diagnostic value of EPAS1 promoter methylation for CRC. Two-tailed P<0.05 was considered to indicate a statistically significant difference. All figures were plotted using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Sequencing results were analyzed using Chromas LITE 2.1.1 software (Technelysium Pty, Ltd., Brisbane, Australia).
Results
DNA methylation analysis
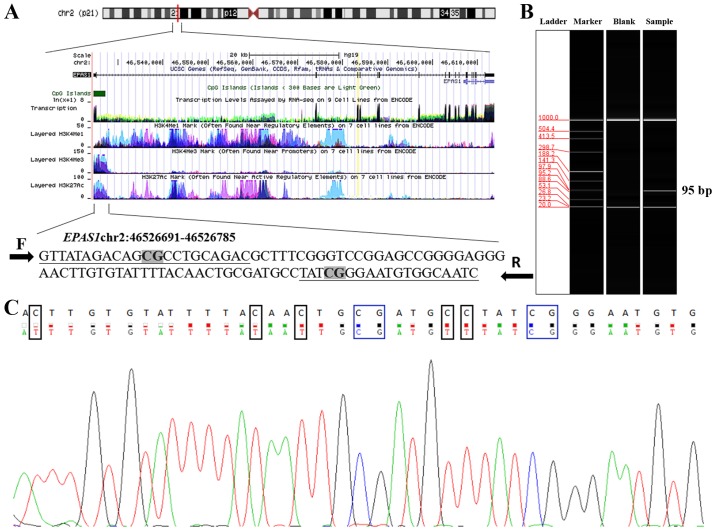
To assess the association between methylation of EPAS1 and CRC, a two-stage association study was conducted. Two cytosine-phosphate-guanine (CpG) sites were identified in the 95 bp fragment of EPAS1 (hg38; chr2:46526691-46526785) (Fig. 1A). The tested EPAS1 fragment was expected to be 95 bp (Fig. 1B). Sequencing results indicated that the amplified fragment matched the target sequence (Fig. 1C).
Figure 1.
Characteristics of the target sequence in EPAS1. (A) Target sequences in EPAS1 promoter region. The genomic position of the amplified fragment from University of California Santa Cruz genome browser according to human (GRCh37) assembly. The primers are underlined and two CpG sites are presented in bold fond with grey highlight. (B) Capillary electrophoresis for the amplified fragment (95 bp). (C) Sanger sequencing results. The top row of the sequence is the original sequence of the fragment. The bottom row of the sequence is the converted sequence; CG dinucleotides that remained unaltered are in blue boxes; and C nucleotides with corresponding converted T nucleotides are in black boxes. F, forward primer; R, reverse primer; EPAS1, endothelial PAS domain-containing protein 1; CpG, cytosine-phosphate-guanine.
Association between EPAS1 hypomethylation in patients with CRC and clinical features
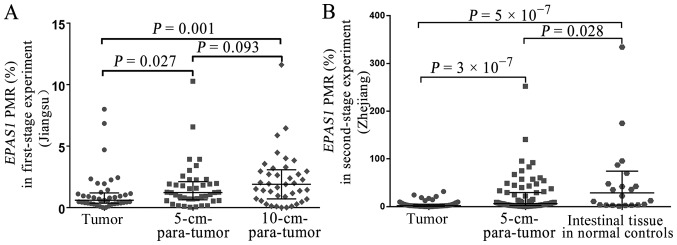
The results of the present study indicated that the EPAS1 methylation was not associated with sex, age, TNM stage, differentiation, tumor size or lymph node metastasis (all P>0.05) (Table I). Methylation levels in the tissues from 41 patients with CRC (Jiangsu, China) and 79 patients with CRC (Zhejiang, China) were examined. The results indicated that EPAS1 promoter methylation levels in CRC tissues (Jiangsu, China) were significantly lower compared with those of the 5-cm-para-tumor tissues (median PMR, 0.59 vs. 1.22%; P=0.027) (Fig. 2A) and 10-cm-para-tumor tissues (median PMR, 0.59 vs. 1.89%; P=0.001) (Fig. 2A). Second-stage experiment was used to further validate the role of EPAS1 methylation in CRC. The results indicated that EPAS1 promoter methylation was significantly lower in CRC tissues (Zhejiang, China) compared with 5-cm-para-tumor tissues (median PMR, 1.91 vs. 6.25%; P=3×10−7) (Fig. 2B) and normal intestinal tissues from healthy controls (median PMR, 1.91 vs. 28.84%; P=5×10−7) (Fig. 2B). Additionally, a significantly lower EPAS1 promoter methylation was found in the paired 5-cm-para-tumor tissues (Zhejiang, China) compared with normal intestinal tissues of healthy controls (median PMR, 6.25 vs. 28.84%; P=0.028) (Fig. 2B). A negative correlation between mRNA expression and EPAS1 methylation was identified in 372 TCGA colorectal adenocarcinoma samples (r=−0.329, P=8×10−11) (Fig. 3).
Table I.
Association between EPAS1 hypermethylation and clinicopathological characteristics of patients with CRC in the two-stage experiment.
| First-stage experiment | Second-stage experiment | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinical characteristic | N | EPAS1 hypomethylation | EPAS1 hypermethylation | P-value | N | EPAS1 hypomethylation | EPAS1 hypermethylation | P-value |
| Total cases | 41 | 31 | 10 | 79 | 57 | 22 | ||
| Sexa | 1.000 | 0.534 | ||||||
| Male | 28 | 21 | 7 | 51 | 38 | 13 | ||
| Female | 12 | 9 | 3 | 28 | 19 | 9 | ||
| Age, yearsa | 0.859 | 0.534 | ||||||
| ≤65 | 21 | 16 | 5 | 57 | 40 | 17 | ||
| >65 | 19 | 14 | 5 | 22 | 17 | 5 | ||
| Differentiationa | NA | 0.440 | ||||||
| Poor | 0 | NA | NA | 14 | 9 | 5 | ||
| Moderate + well | 41 | 31 | 10 | 63 | 47 | 16 | ||
| TNM stageb | NA | 0.945 | ||||||
| I+II | NA | NA | NA | 40 | 29 | 11 | ||
| III+IV | NA | NA | NA | 39 | 28 | 11 | ||
| Tumor size, cma | 0.859 | 0.289 | ||||||
| ≤5 | 21 | 16 | 5 | 40 | 31 | 9 | ||
| 5 | 19 | 14 | 5 | 39 | 26 | 13 | ||
| Lymphatic metastasisa | 1.000 | 0.314 | ||||||
| Yes | 20 | 15 | 5 | 36 | 28 | 8 | ||
| No | 20 | 15 | 5 | 43 | 29 | 14 | ||
Data are presented as the median (interquartile range).
The information for 2 patients was lost.
The TNM stage information in the first-stage experiment was not available. Cutoff value of EPAS1 hypermethylation is set at the mean value. Contingency correlation test was used to evaluate the association between EPAS1 methylation and clinical features. PMR, percentage of methylated reference; N, number; NA, not available; EPAS1, endothelial PAS domain-containing protein 1; CRC, colorectal cancer.
Figure 2.
Comparisons of EPAS1 methylation levels between patients and normal controls. (A) Comparisons of methylation levels between tumor tissues, 5-cm-para-tumor tissues and 10-cm-para-tumor tissues (Jiangsu, China). (B) Comparisons of methylation levels between tumor tissues, 5-cm-para-tumor tissues and normal intestinal tissues of healthy controls (Zhejiang, China). EPAS1, endothelial PAS domain-containing protein 1; PMR, percentage of methylated reference.
Figure 3.
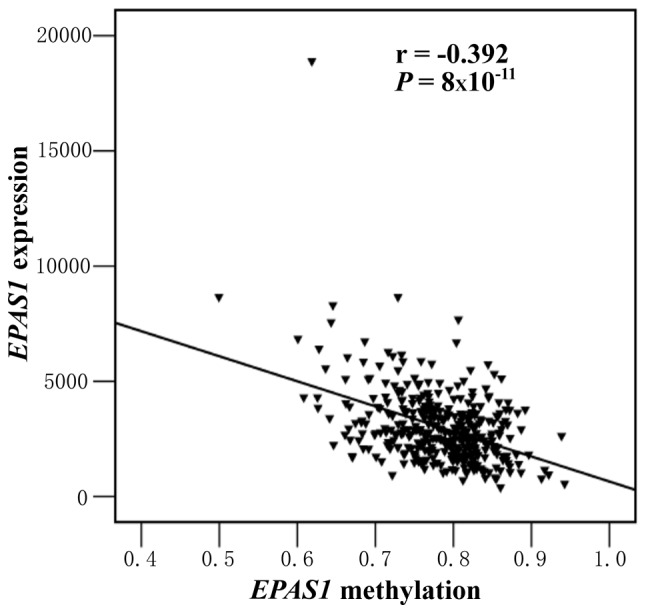
Bioinformatics analysis of the correlation between EPAS1 methylation and expression. An inverse correlation was identified between EPAS1 methylation and mRNA expression in 372 colorectal cancer samples (r=−0.329, P=8×10−11). EPAS1, endothelial PAS domain-containing protein 1.
The present study demonstrated that EPAS1 methylation levels of tumor tissues from patients from Zhejiang province were significantly higher compared with tumor tissues from Jiangsu province (median PMR, 1.91 vs. 0.59%; P=4×10−7) (Table II). Significantly higher EPAS1 methylation levels were observed in the adjacent non-tumor tissues from patients from Zhejiang province compared with patients from Jiangsu province (median PMR, 6.25 vs. 1.22%; P=4×10−7) (Table II). However, the difference in EPAS1 methylation levels between tumor and non-tumor tissues (D=PMRtumor-PMRnon-tumor) was not significant between Jiangsu province and Zhejiang province (P=0.066) (Table III). To clarify the differences in methylation status between the two provinces, the present study further investigated the association between sample locations and the clinical phenotypes of patients with CRC. The results indicated that there was a significant difference in the age at diagnosis between Zhejiang (57/79; 72.2%) compared with Jiangsu (21/41, 51.2%) (χ2=4.541; P=0.033) (Table III). In addition, there was statistically significant difference in differentiation between Jiangsu and Zhejiang province (P=0.002) (Table III). However, there were no significant differences in EPAS1 methylation levels between age subgroups and between differentiation subgroups in both provinces (P>0.05) (Table II).
Table II.
Subgroup analysis by age and differentiation.
| Tumor PMR (%) | Non-tumor PMR (%) | |||||
|---|---|---|---|---|---|---|
| Variable | Jiangsu | Zhejiang | P-value | Jiangsu | Zhejiang | P-value |
| Total | 0.59 (0.35, 1.19) | 1.91 (0.79, 4.90) | 4×10−7 | 1.22 (0.63, 2.11) | 6.25 (2.35, 29.63) | 4×10−7 |
| Age, years | ||||||
| ≤65 | 0.83 (0.38, 1.54) | 1.91 (0.83, 5.22) | 0.008 | 1.06 (0.56, 1.92) | 7.35 (7.24, 30.43) | 3×10−6 |
| >65 | 0.48 (0.28, 1.24) | 1.83 (0.65, 4.02) | 0.009 | 1.53 (0.69, 2.27) | 3.90 (2.33, 24.86) | 2×10−4 |
| P-value | 0.236 | 0.577 | 0.630 | 0.418 | ||
| Differentiation | ||||||
| Poor | NA | 2.39 (1.28, 6.03) | NA | NA | 9.16 (2.32, 39.14) | NA |
| Moderate + well | 0.59(0.35, 1.19) | 1.89 (0.79, 4.72) | 2×10−4 | 1.22 (0.60, 2.18) | 6.19 (2.35, 29.63) | 8×10−9 |
| P-value | NA | 0.235 | NA | 0.530 | ||
Mann-Whitney U nonparametric test were used to assess the difference in methylation between groups. NA, not available; PMR, percentage of methylated reference.
Table III.
Association between sampling location and clinical characteristics.
| Clinical characteristic | Number | Jiangsu | Zhejiang | P-value |
|---|---|---|---|---|
| Total cases | 120 | 41 | 79 | |
| Sexa | 0.553b | |||
| Male | 79 | 28 | 51 | |
| Female | 40 | 12 | 28 | |
| Age, yearsa | 0.033b | |||
| ≤65 | 78 | 21 | 57 | |
| >65 | 41 | 19 | 22 | |
| Tumor sizea | 0.847b | |||
| <5 cm | 61 | 21 | 40 | |
| ≥5 cm | 58 | 19 | 39 | |
| Differentiationa | 0.002c | |||
| Low and none | 14 | 0 | 14 | |
| High and medium | 103 | 40 | 63 | |
| Lymph node metastasisa | 0.842b | |||
| Negative | 55 | 19 | 36 | |
| Positive | 64 | 21 | 43 | |
| EPAS1 methylation | 0.066b | |||
| Hypomethylation | 91 | 27 | 64 | |
| Hypermethylation | 29 | 14 | 15 |
The information for 1 patient was lost.
Data analyzed using Pearson χ2 test.
Data analyzed using Fisher's exact test. Hypermethylation was determined to be present if PMR detected in the tumor tissue was higher than in the matched normal sample. Hypomethylation was determined to be present in the inverse case. EPAS1, endothelial PAS domain-containing protein 1.
ROC curve analysis
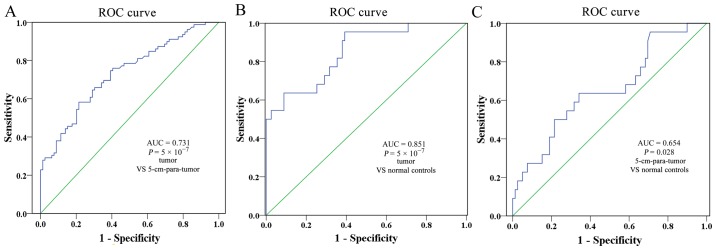
ROC curve analysis was used to measure the diagnostic value of EPAS1 hypomethylation for CRC. The second-stage association results indicated that EPAS1 hypomethylation yielded a significant AUC of 0.731 (95% CI, 0.653–0.808) with a sensitivity of 58.2% and a specificity of 78.5% between cancer tissues and 5-cm-para-tumor tissues (Fig. 4A); a significant AUC of 0.851 (95% CI, 0.760–0.942) with a sensitivity of 95.5% and a specificity of 60.8% between CRC tissues and normal intestinal tissues of healthy controls (Fig. 4B); and a significant AUC of 0.654 (95% CI, 0.522–0.786) with a sensitivity of 63.6% and a specificity of 65.8% between 5-cm-para-tumor tissues and normal intestinal tissues of healthy controls (Fig. 4C). All of the above data supported the hypothesis that hypomethylation of EPAS1 may be a potential diagnostic biomarker for CRC.
Figure 4.
ROC curves for the diagnostic value of EPAS1 hypomethylation in the second-stage experiment. (A) ROC curve for the diagnostic value of EPAS1 hypomethylation between CRC tissues and paired non-cancer tissues (Zhejiang, China). (B) ROC curve for the diagnostic value of EPAS1 hypomethylation between CRC tissues and normal intestinal tissues of healthy controls (Zhejiang, China). (C) ROC curve for the diagnostic value of EPAS1 hypomethylation between paired non-cancer tissues and normal intestinal tissues of healthy controls (Zhejiang, China). ROC, receiver operating characteristic; AUC, area under the curve; EPAS1, endothelial PAS domain-containing protein 1; CRC, colorectal cancer.
Discussion
CRC is regarded as a threat to human health (2), and the study of methylation in the context of CRC is a field of growing interest. In the present study, EPAS1 promoter methylation significantly decreased according to the results of both the first-stage and the second-stage association tests. These results led to a hypothesis that EPAS1 hypomethylation may be associated with the development of CRC. In the subsequent experiment, EPAS1 hypomethylation yielded an AUC of 0.851 (sensitivity, 95.5%; specificity, 60.8%) to distinguish the CRC tumor tissues from normal intestinal tissues, suggesting that EPAS1 hypomethylation could serve as a promising diagnostic biomarker for CRC.
Numerous studies demonstrated that EPAS1 served a role in angiogenesis of human cancer (26–28) at the post-transcriptional level (27). Yoshimura et al (11) demonstrated that EPAS1 was overexpressed in aggressive colorectal carcinoma and exhibited a significant direct correlation with tumor microvessel count. Cho et al (16) identified that EPAS1 was bound by TP2399 in the PAS B domain, which diminished its ability to bind to ARNT, causing tumor regression in preclinical kidney cancer models. This means that EPAS1 might be an important factor in the processes of tumorigenesis and cancer progression. In human breast cancer cells, methyl-CpG-binding domain protein 3 can remove the methylation of CpG sites near the promoter of EPAS1, and, therefore, significantly increase the expression of EPAS1 (27). In epithelial cells, DNA (cytosine-5)-methyltransferase 3A (DNMT3A) can silence the expression of EPAS1 (29,30). DNMT3A deficiencies were reported in primary tumors and malignant cells, leading to demethylation of EPAS1 promoter (31) and resulting in the growth of cancer cells under hypoxia (31–34). Re-introducing DNMT3A can restore the silencing of EPAS1, and prevent cell proliferation and viability under hypoxia, inhibiting tumor occurrence (8,31). In addition, our data mining of The Cancer Genome Atlas database discovered that there was an inverse EPAS1 methylation-expression correlation in different cancers. Therefore, the present study further hypothesized that angiogenesis induced by increased expression of EPAS1 could be a reason underlying the occurrence of CRC. Future studies should verify the function of EPAS1 in CRC.
Although several types of biomarkers have been studied in CRC (35,36), a biomarker for early diagnosis remains to be identified. Carcinoembryonic antigen (CEA), a biomarker used globally, did not exhibit the desired diagnostic value according to one study (37). However, Peng et al (38) reported an AUC of 0.690 for CEA in the detection of CRC. In addition, there are two main methods widely used in clinical diagnosis of CRC, including fecal occult blood test (FOBT) and colonoscopy (37). However, FOBT is susceptible to bias resulting from external factors including drugs and diet, and colonoscopy is associated with significant costs and can often cause pain (37). The present qMSP-based study indicated an AUC of 0.851 (sensitivity, 95.5%; specificity, 60.8%) between CRC tissues and normal intestinal tissues of healthy controls.
A previous study indicated that patients with CRC exhibited significantly elevated EPAS1 expression in blood, which decreased significantly to normal levels following surgical resection of the tumor (21). Further analyses of EPAS1 methylation in blood samples of patients with CRC before and after surgery should be conducted to further verify the diagnostic value of EPAS1 hypomethylation.
Benign colorectal disease can gradually progress to advanced adenoma and, subsequently, to invasive adenocarcinoma (39–41). In addition, aberrant methylation patterns exhibit a malignant potential in hyperplastic polyps (39–42). Numerous studies have indicated that there were significant differences in DNA methylation levels between CRC tissues, benign colorectal disease tissues and healthy intestinal tissues (43–45). However, methylation data for benign colorectal disease were not available in the present study. Further investigation is necessary to determine whether EPAS1 methylation serves a role in benign colorectal disease.
Plasma circulating DNA (cell-free DNA) of patients with cancer may originate from circulating tumor cells, which can indicate the occurrence of micrometastasis and invasion of cancer cells (46,47). The present study indicated that the levels of EPAS1 methylation in the tumor tissues were lower compared with the adjacent non-tumor tissues. The detection of EPAS1 methylation in plasma cf-DNA could be performed in the future to avoid variation resulting from differences in tissue sampling sites, surgical techniques and so on.
In conclusion, the present study suggested that EPAS1 hypomethylation may be a diagnostic biomarker for CRC. Further study is necessary to clarify the molecular mechanisms by which EPAS1 hypomethylation may exert its role in carcinogenesis.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- EPAS1
endothelial PAS domain-containing protein 1
- CRC
colorectal cancer
- qMSP
quantitative methylation-specific polymerase chain reaction
- PMR
percent of methylated reference
- CpG
cytosine-phosphate-guanine
- ROC
receiver operating characteristic
Funding
The research was supported in part by K.C. Wong Magna Fund in Ningbo University (to SD) and grant no. U01CA217078 in Northwestern University (to WZ).
Availability of data and materials
Not applicable.
Authors' contributions
SD and RP conceived and designed the experiments. JY, DW, CZ, XY, JZ, HY, JD, BW, YM and YZ performed the experiments. RP, CZ and HY analyzed the data. RP, WZ and SD contributed to the completion of figures and tables, and writing of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149:1177–1190.e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: Emerging biomarkers. Gastroenterology. 2015;149:1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D, Tang W, Su G, Cai M, An HX, Zhang Y. PCDH18 is frequently inactivated by promoter methylation in colorectal cancer. Sci Rep. 2017;7:2819. doi: 10.1038/s41598-017-03133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezvani N, Alibakhshi R, Vaisi-Raygani A, Bashiri H, Saidijam M. Detection of SPG20 gene promoter-methylated DNA, as a novel epigenetic biomarker, in plasma for colorectal cancer diagnosis using the MethyLight method. Oncol Lett. 2017;13:3277–3284. doi: 10.3892/ol.2017.5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Zhang X, Lu X, You L, Song Y, Luo Z, Zhang J, Nie J, Zheng W, Xu D, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawłuszko-Wieczorek AA, Horbacka K, Krokowicz P, Misztal M, Jagodziński PP. Prognostic potential of DNA methylation and transcript levels of HIF1A and EPAS1 in colorectal cancer. Mol Cancer Res. 2014;12:1112–1127. doi: 10.1158/1541-7786.MCR-14-0054. [DOI] [PubMed] [Google Scholar]
- 9.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 10.Robins JC, Akeno N, Mukherjee A, Dalal RR, Aronow BJ, Koopman P, Clemens TL. Hypoxia induces chondrocyte-specific gene expression in mesenchymal cells in association with transcriptional activation of Sox9. Bone. 2005;37:313–322. doi: 10.1016/j.bone.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura H, Dhar DK, Kohno H, Kubota H, Fujii T, Ueda S, Kinugasa S, Tachibana M, Nagasue N. Prognostic impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal cancer patients: Correlation with tumor angiogenesis and cyclooxygenase-2 expression. Clin Cancer Res. 2004;10:8554–8560. doi: 10.1158/1078-0432.CCR-0946-03. [DOI] [PubMed] [Google Scholar]
- 12.Tanigawa N, Amaya H, Matsumura M, Lu C, Kitaoka A, Matsuyama K, Muraoka R. Tumor angiogenesis and mode of metastasis in patients with colorectal cancer. Cancer Res. 1997;57:1043–1046. [PubMed] [Google Scholar]
- 13.Putra AC, Eguchi H, Lee KL, Yamane Y, Gustine E, Isobe T, Nishiyama M, Hiyama K, Poellinger L, Tanimoto K. The A Allele at rs13419896 of EPAS1 is associated with enhanced expression and poor prognosis for non-small cell lung cancer. PLoS One. 2015;10:e0134496. doi: 10.1371/journal.pone.0134496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwamoto S, Tanimoto K, Nishio Y, Putra AC, Fuchita H, Ohe M, Sutani A, Kuraki T, Hiyama K, Murakami I, et al. Association of EPAS1 gene rs4953354 polymorphism with susceptibility to lung adenocarcinoma in female Japanese non-smokers. J Thorac Oncol. 2014;9:1709–1713. doi: 10.1097/JTO.0000000000000309. [DOI] [PubMed] [Google Scholar]
- 15.Xia G, Kageyama Y, Hayashi T, Kawakami S, Yoshida M, Kihara K. Regulation of vascular endothelial growth factor transcription by endothelial PAS domain protein 1 (EPAS1) and possible involvement of EPAS1 in the angiogenesis of renal cell carcinoma. Cancer. 2001;91:1429–1436. doi: 10.1002/1097-0142(20010415)91:8<1429::AID-CNCR1149>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, Carvo I, Signoretti S, Bruick RK, Josey JA, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sena JA, Wang L, Heasley LE, Hu CJ. Hypoxia regulates alternative splicing of HIF and non-HIF target genes. Mol Cancer Res. 2014;12:1233–1243. doi: 10.1158/1541-7786.MCR-14-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welander J, Andreasson A, Brauckhoff M, Bäckdahl M, Larsson C, Gimm O, Söderkvist P. Frequent EPAS1/HIF2α exons 9 and 12 mutations in non-familial pheochromocytoma. Endocr Relat Cancer. 2014;21:495–504. doi: 10.1530/ERC-13-0384. [DOI] [PubMed] [Google Scholar]
- 19.Baba Y, Nosho K, Shima K, Irahara N, Chan AT, Meyerhardt JA, Chung DC, Giovannucci EL, Fuchs CS, Ogino S. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol. 2010;176:2292–2301. doi: 10.2353/ajpath.2010.090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favier J, Plouin PF, Corvol P, Gasc JM. Angiogenesis and vascular architecture in pheochromocytomas: Distinctive traits in malignant tumors. Am J Pathol. 2002;161:1235–1246. doi: 10.1016/S0002-9440(10)64400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collado M, Garcia V, Garcia JM, Alonso I, Lombardia L, Diaz-Uriarte R, Fernández LA, Zaballos A, Bonilla F, Serrano M. Genomic profiling of circulating plasma RNA for the analysis of cancer. Clin Chem. 2007;53:1860–1863. doi: 10.1373/clinchem.2007.089201. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Yang Y, Liu J, Li B, Xu Y, Li C, Xu Q, Liu G, Chen Y, Ying J, Duan S. NDRG4 hypermethylation is a potential biomarker for diagnosis and prognosis of gastric cancer in Chinese population. Oncotarget. 2017;8:8105–8119. doi: 10.18632/oncotarget.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Chen X, Jiang Y, Yang Y, Zhong J, Zhou C, Hu H, Duan S. CCL2 promoter hypomethylation is associated with gout risk in Chinese Han male population. Immunol Lett. 2017;190:15–19. doi: 10.1016/j.imlet.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Chen X, Hu H, Jiang Y, Yu H, Dai J, Mao Y, Duan S. Elevated UMOD methylation level in peripheral blood is associated with gout risk. Sci Rep. 2017;7:11196. doi: 10.1038/s41598-017-11627-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive melting analysis after real time-methylation specific PCR (SMART-MSP): High-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Lou Y, Zhang J, Fu Q, Wei T, Sun X, Chen Q, Yang J, Bai X, Liang T. Hypoxia-inducible factor-2α promotes tumor progression and has crosstalk with Wnt/β-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16:119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J, Duan B, Zhao X, Chen Y, Sun S, Deng W, Zhang Y, Du J, Chen Y, Gu L. MBD3 mediates epigenetic regulation on EPAS1 promoter in cancer. Tumour Biol. 2016;37:13455–13467. doi: 10.1007/s13277-016-5237-1. [DOI] [PubMed] [Google Scholar]
- 28.Zhu DM, Li DC, Zhang ZX, Zhang XY. Effect of endothelial PAS domain protein 1 and hypoxia inducible factor 1alpha on vascular endothelial growth factor expression in human pancreatic carcinoma. Chin Med J (Engl) 2008;121:2258–2264. [PubMed] [Google Scholar]
- 29.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/S0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CM, Vocke C, Torres-Cabala C, Yang Y, Schmidt L, Walther M, Linehan WM. Expression of hypoxia inducible factor-1alpha and 2alpha in genetically distinct early renal cortical tumors. J Urol. 2006;175:1908–1914. doi: 10.1016/S0022-5347(05)00890-6. [DOI] [PubMed] [Google Scholar]
- 31.Lachance G, Uniacke J, Audas TE, Holterman CE, Franovic A, Payette J, Lee S. DNMT3a epigenetic program regulates the HIF-2α oxygen-sensing pathway and the cellular response to hypoxia. Proc Natl Acad Sci USA. 2014;111:7783–7788. doi: 10.1073/pnas.1322909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, Jackson AL, Nikolinakos P, Ospina B, Naumov G, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–2170. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2alpha oncogenic axis. Proc Natl Acad Sci USA. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravegnini G, Zolezzi Moraga JM, Maffei F, Musti M, Zenesini C, Simeon V, Sammarini G, Festi D, Hrelia P, Angelini S. Simultaneous analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate-related gene polymorphisms: The potential for a novel blood-based colorectal cancer biomarker. Int J Mol Sci. 2015;16:28486–28497. doi: 10.3390/ijms161226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Tan ZR, Yu J, Li H, Lv QL, Shao YY, Zhou HH. DNA hypermethylated status and gene expression of PAX1/SOX1 in patients with colorectal carcinoma. OncoTargets Ther. 2017;10:4739–4751. doi: 10.2147/OTT.S143389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Song L, Gong Y, He B. Detection of colorectal cancer by DNA methylation biomarker SEPT9: Past, present and future. Biomark Med. 2014;8:755–769. doi: 10.2217/bmm.14.8. [DOI] [PubMed] [Google Scholar]
- 38.Peng HX, Yang L, He BS, Pan YQ, Ying HQ, Sun HL, Lin K, Hu XX, Xu T, Wang SK. Combination of preoperative NLR PLR and CEA could increase the diagnostic efficacy for I-III stage CRC. J Clin Lab Anal. 2017;31 doi: 10.1002/jcla.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 40.Stella S, Bruzzese A, Chiarini S. Biological bases of the colorectal adenoma-carcinoma sequence. G Chir. 1996;17:473–476. (In Italian) [PubMed] [Google Scholar]
- 41.Takami K, Yana I, Kurahashi H, Nishisho I. Multistep carcinogenesis in colorectal cancers. Southeast Asian J Trop Med Public Health. 1995;26(Suppl 1):S190–S196. [PubMed] [Google Scholar]
- 42.Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang HL, Liu P, Zhou PY, Zhang Y. Promoter methylation of the RASSF1A gene may contribute to colorectal cancer susceptibility: A meta-analysis of cohort studies. Ann Hum Genet. 2014;78:208–216. doi: 10.1111/ahg.12059. [DOI] [PubMed] [Google Scholar]
- 44.Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, Zhu RM, Li GL, Xia XY, Wei XW, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol. 2008;14:3074–3080. doi: 10.3748/wjg.14.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YW, Kong FM, Zhou JP, Dong M. Aberrant promoter methylation of the vimentin gene may contribute to colorectal carcinogenesis: A meta-analysis. Tumour Biol. 2014;35:6783–6790. doi: 10.1007/s13277-014-1905-1. [DOI] [PubMed] [Google Scholar]
- 46.Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanché H, Girard N, Trédaniel J, Guilleminault L, Gervais R, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res. 2014;20:4613–4624. doi: 10.1158/1078-0432.CCR-13-3063. [DOI] [PubMed] [Google Scholar]
- 47.Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker-a critical appraisal of the literature. Clin Chim Acta. 2010;411:1611–1624. doi: 10.1016/j.cca.2010.07.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.