Abstract
Polyamines are small amino-acid derived polycations capable of binding negatively charged macromolecules. Bacterial polyamines are structurally and functionally diverse, and are mainly produced biosynthetically by pyridoxal-5-phosphate-dependent amino acid decarboxylases referred to as Lysine-Arginine-Ornithine decarboxylases (LAOdcs). In a phylogenetically limited group of bacteria, LAOdcs are also induced in response to acid stress. Here, we performed an exhaustive phylogenetic analysis of the AAT-fold LAOdcs which showcased the ancient nature of their short forms in Cyanobacteria and Firmicutes, and emergence of distinct subfamilies of long LAOdcs in Proteobacteria. We identified a novel subfamily of lysine decarboxylases, LdcA, ancestral in Betaproteobacteria and Pseudomonadaceae. We analyzed the expression of LdcA from Pseudomonas aeruginosa, and uncovered its role, intimately linked to cadaverine (Cad) production, in promoting growth and reducing persistence of this multidrug resistant human pathogen during carbenicillin treatment. Finally, we documented a certain redundancy in the function of the three main polyamines—Cad, putrescine (Put), and spermidine (Spd)—in P. aeruginosa by demonstrating the link between their intracellular level, as well as the capacity of Put and Spd to complement the growth phenotype of the ldcA mutant.
Keywords: phylogenetic analysis, bacterial evolution, amino acid decarboxylases, polyamines, Pseudomonas aeruginosa
Introduction
Polyamines are small molecules with two or more amino groups separated by alkyl chains (Tabor and Tabor 1964; Lightfoot and Hall 2014). They perform essential functions in all living organisms by participating in DNA replication, gene expression and protein synthesis, and are generally described as growth factors (Lightfoot and Hall 2014; Michael 2016b). At physiological pH, polyamines behave as polycations and can interact with nucleic acids, membrane phospholipids and proteins (Tabor and Tabor 1964, 1985). They were proposed to bind and structurally modify RNA thereby acting at the level of translation (Igarashi and Kashiwagi 2015, 2006). Strengthening this hypothesis, “polyamine modulons” were identified in Escherichia coli and more recently in eukaryotes (Igarashi and Kashiwagi 2015, 2006). As a possible consequence, bacterial polyamines were shown to participate in expression of proteins essential for growth fitness and viability but also in biofilm formation, antibiotic resistance and virulence (Kwon and Lu 2006; Shah and Swiatlo 2008; Di Martino et al. 2013; Karatan and Michael 2013; Michael 2016a).
The triamine spermidine (Spd) and its diamine precursor putrescine (Put) are ubiquitous in eukaryotes and therefore extensively studied (Michael 2016b). In bacteria, a third polyamine, cadaverine (Cad), is recognized as an important player in enterobacterial acid stress response wherein it decreases porin permeability to protons and alkalinizes the medium (Dela Vega and Delcour 1996; Zhao and Houry 2010). During oxidative stress response, Cad is also capable of scavenging reactive oxygen species (ROS) in Vibrio vulnificus (Kang et al. 2007). In some Negativicutes, Cad was shown to be incorporated in the peptidoglycan and essential for its stability (Kamio et al. 1986; Kamio and Nakamura 1987), whereas in different bacterial species such as Streptomyces coelicolor this polyamine is involved in iron uptake and is required for the synthesis of hydroxamate-type siderophores (Burrell et al. 2012).
Polyamine biosynthesis depends on the activity of basic amino acid decarboxylases using Lysine, Arginine, or Ornithine as specific substrate to produce Cad, Agmatine (Agm), or Put, respectively. These enzymes can therefore be generally referred to as LAOdcs (Lysine-Arginine-Ornithine decarboxylases). The main polyamine biosynthesis pathways are thoroughly described and synthetically summarized in a recent extensive review (Michael 2016a). In most eukaryotes, Put synthesis is carried out by an ornithine decarboxylase (Odc) (Michael 2016a, 2016b). An alternative pathway for Put biosynthesis found in bacteria and plants involves decarboxylation of arginine by arginine decarboxylases (Adc); these enzymes produce Agm which is further converted into Put (Michael 2016a, 2016b). Interestingly, plants (Lee and Cho 2001; Bunsupa et al. 2012) and some bacteria exemplified by Selenomonas ruminantium and V. vulnificus (Takatsuka et al. 2000; Lee et al. 2007) possess a bifunctional Odc/Ldc capable of synthesizing both Put and Cad. Spd is formed by spermidine synthase (SpdSyn) through aminopropylation of Put (Michael 2016a, 2016b). In addition, E. coli SpdSyn transforms Cad into aminopropyl-Cad, a Spd analogue sharing its growth stimulating properties (Kim et al. 2016).
Pyridoxal-5-phosphate (PLP)-dependent LAOdcs responsible for polyamine biosynthesis belong to either the alanine racemase fold (AR-fold) superfamily or the aspartate aminotransferase fold (AAT-fold) superfamily (Eliot and Kirsch 2004). LAOdcs of the AR-fold superfamily are widespread throughout the three domains of life whereas the AAT-fold LAOdcs are found exclusively in Bacteria and a few archaea (Lee et al. 2007; Burrell et al. 2010). Bacterial AAT-fold LAOdcs can be divided in two types according to the presence or absence of a CheY-like response regulator receiver domain, known as the “wing domain”, necessary for the formation of higher-order oligomers (Burrell et al. 2010; Kanjee, Gutsche, Ramachandran, et al. 2011). The short form referred as wing-less LAOdc was found in Firmicutes, Cyanobacteria, and Actinobacteria phyla; a few “wing-less” AAT-fold decarboxylases from Firmicutes and Actinobacteria have an Adc activity, required in particular for biofilm formation in Bacillus subtilis (Burrell et al. 2010). The long, wing domain-containing form likely originated in Proteobacteria (Kanjee, Gutsche, Ramachandran, et al. 2011).
AAT-fold decarboxylases with the wing domain have been intensively studied in Enterobacteria since the early 1940s (Gale and Van Heyningen 1942; Gale and Epps 1944) because of the link between enterobacterial pathogenicity for humans and their capacity to withstand acid stress thanks to the inducible LAOdcs. Consequently, the current understanding of the AAT-fold LAOdc is based on analyses of a very limited number of bacterial species, mostly enterobacteria: E. coli, Salmonella typhimurium, Vibrio cholerae, and V. vulnificus. At a specific acidic pH, expression of the decarboxylase gene is induced by an excess of the target amino acid uptaken by a dedicated inner membrane antiporter (Kanjee and Houry 2013). The enzyme transforms the amino acid substrate into the corresponding polyamine upon consumption of a proton and production of a CO2 molecule. In association with the polyamine excretion by the antiporter, this reaction results in an efficient buffering of the intracellular medium and the extracellular surroundings. These inducible stress response decarboxylases are distinguished from “biosynthetic” enzymes that are involved only in polyamine biosynthesis. Escherichiacoli encodes two biosynthetic decarboxylases (LdcC: Lys->Cad and OdcC/SpeC: Orn->Put) and three acid stress-inducible decarboxylases (LdcI: Lys->Cad, AdcI/AdiA: Arg->Agm, and OdcI/SpeF: Orn->Put) which together constitute a very robust acid stress response system that allows the bacterium to survive upon acid stress as low as pH 2.0 (Zhao and Houry 2010; Kanjee, Gutsche, Alexopoulos, et al. 2011; Kanjee and Houry 2013). The importance of LdcI, often designated as CadA as originally proposed (Tabor et al. 1980), was also demonstrated in S. typhimurium, V. cholerae, and V. vulnificus, where it promotes growth and survival under acidic conditions but also confers protection from oxidative stress insults (Merrell and Camilli 2000; Kang et al. 2007; Viala et al. 2011).
Escherichia coli LdcI but not LdcC interacts with the AAA+ ATPase RavA to assemble into a huge macromolecular cage (Snider et al. 2006; El Bakkouri et al. 2010; Malet et al. 2014; Kandiah et al. 2016). One of the functions of this mysterious complex is to protect LdcI from inhibition by the stringent response alarmone ppGpp, thus enabling the bacterium to efficiently cope with acid and nutrient stresses simultaneously (El Bakkouri et al. 2010; Kanjee, Gutsche, Alexopoulos, et al. 2011; Malet et al. 2014). While investigating why RavA binds only LdcI but not LdcC, we documented numerous inconsistencies in annotation of enterobacterial ldcI and ldcC genes and realized that each of these two families seemed to have a distinct genetic context (Kandiah et al. 2016). Thus, spurred on by the limited nature of the previous studies, here we performed an extensive phylogenetic analysis of prokaryotic AAT-fold LAOdcs to decipher their evolutionary history and their functional evolution. We revealed the ancient nature of the wing-less LAOdcs in Cyanobacteria and Firmicutes, and the complex history of long AAT-fold LAOdcs in Proteobacteria, leading to the emergence of distinct subfamilies. Moreover, we disclosed a novel subfamily of enzymes, widely distributed in Betaproteobacteria and Pseudomonadaceae, and clearly distinct from the well-known LdcI, LdcC, AdcI, OdcI and OdcC families, more related to Ldc and Adc than to Odc. The only previously mentioned LAOdc from these taxa is the lysine decarboxylase LdcA from a major multidrug resistant opportunistic human pathogen Pseudomonas aeruginosa (Chou et al. 2010). Thus, here we went beyond the initial characterization of P. aeruginosa LdcA (Chou et al. 2010), and further analyzed its expression, regulation, and function in the light of the available knowledge in particular on E. coli LdcI and LdcC. This combined phylogenetic and functional study revealed that LdcA belongs to a novel subgroup of the long AAT-fold LAOdcs, and that its function is linked to Cad production and to the general polyamine metabolism rather than to stress response.
Materials and Methods
Phylogeny: Data Set Assembly
Functionally characterized sequences of AAT-fold LAOdcs were retrieved from NCBI: LdcI (NP_418555.1), LdcC (NP_414728.1), Adc (NP_418541.1), OdcC (NP_417440.1), and OdcI (NP_415220.1) from E. coli str. K-12 substr., MG1655 and LdcA (NP_250509.1) from P. aeruginosa PAO1. These sequences were used as seeds to query a local database containing 4,466 complete proteomes of prokaryotes downloaded from the NCBI (ftp://ftp.ncbi.nlm.nih.gov) with the BLASTP 2.2.6 software (Altschul et al. 1997) using default parameters. Sequences with an e-value <10−4 were retrieved and aligned using MAFFT v7 (Katoh and Standley 2013). The resulting multiple alignment was visually inspected with Seaview version 4 (Gouy et al. 2010) and used to infer phylogenetic trees with FastTree 2.1.8 (Price et al. 2009) (-wag -c 4 -gamma) in order to detect atypical/divergent sequences that could correspond to wrongly delimited ORF or nonhomologous sequences. Doubtful sequences were systematically verified using reciprocal BBH. Retained sequences have been realigned and used to build a HMM profile with the HMMbuild program from the HMMER v3.1b1 package (Eddy 2011). This profile was then used to query the local database of complete proteomes with the HMMsearch program. Sequences with e-values lower than 2.2e–13 were retrieved and verified as described above. Finally, the search for potential unannotated sequences was performed using TBLASTN 2.2.6 on genomic sequences corresponding to the 4,466 complete proteomes. This led to the identification of 4,090 homologous sequences, 13 of which were unannotated or annotated as pseudogenes.
Phylogeny: Phylogenetic Inference
To limit taxonomic redundancy for phylogenetic analyses, a sampling of the retrieved sequences by selecting randomly one strain per species has been performed. Multiple alignments were built with MAFFT using the L-INS-i option that allows the construction of accurate multiple alignments and trimmed with BMGE v1.1 with matrix substitution BLOSUM30 (Criscuolo and Gribaldo 2010).
Maximum likelihood (ML) trees were inferred with PhyML 3.1 (Guindon et al. 2010). The best suited evolutionary models were selected using the model test tool implemented in IQ-TREE v1.4.1 according to the Bayesian information criterion (BIC) (Nguyen et al. 2015). The robustness of the inferred trees was assessed using the nonparametric bootstrap procedure implemented in PhyML (100 replicates of the original data sets). All ML trees have also been inferred using IQ-TREE. They are provided in Supplementary Material. Bayesian trees were inferred using MrBayes v3.2.6 (Ronquist and Huelsenbeck 2003). Two runs were launched with four chains for each run (50,000 iterations). The first 25% of the trees were discarded as burn-in and chain convergence has been checked by analysing the evolution of the Ln(L) curve and checking the average standard deviation of split frequencies values. Figures of trees have been generated using EvolView (He et al. 2016) (http://nar.oxfordjournals.org/content/44/W1/W236; last accessed October 23, 2018), Figtree (http://tree.bio.ed.ac.uk/software/figtree/; last accessed October 23, 2018) and iTOL (Letunic and Bork 2016). Genomic context figures have been generated using GeneSpy (https://lbbe.univ-lyon1.fr/GeneSpy/; last accessed October 23, 2018) developed by P.S. Garcia.
Reference phylogenies of Firmicutes, Cyanobacteria, Proteobacteria, and Enterobacteriaceae strains contained in our local database were inferred using ribosomal proteins as suggested elsewhere (Ramulu et al. 2014). The reference tree of Firmicutes has been rooted according to a recent study (Antunes et al. 2016). The reference tree of Cyanobacteria has been rooted by including ribosomal protein sequences from Natranaerobius thermophilus (Firmicutes) and Streptomyces albulus (Actinobacteria). The reference tree of Proteobacteria has been rooted according to Gupta (Gupta 2000). Finally, the reference phylogeny of Enterobacteriaceae has been rooted using with Shewanella baltica (Alteromonadales) and Pasteurella multocida (Pasteurellales) ribosomal protein sequences.
For each analysis, the ribosomal protein sequences were identified using the RiboDB database engine (Jauffrit et al. 2016) and aligned with MAFFT using the L-INS-i option. The resulting multiple alignments were trimmed as described above and combined to build a large supermatrix that was used to build ML phylogenetic trees as described above.
To infer the ancestral presence of a given protein family in a given taxon, we compared the protein family tree with the species tree. To be considered as ancestral in a given taxon, the protein family must be present in the majority of proteomes of the taxon and the phylogeny of the protein family must be consistent with the species phylogeny.
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in supplementary table S1, Supplementary Material online. Bacteria were cultivated aerobically at 37 °C in rich Lysogeny Broth (LB) medium, in Mueller Hinton II (Becton Dickinson) or in minimal medium P (MMP) (30 mM Na2HPO4, 14 mM KH2PO4, 20 mM (NH4)2SO4, 1 mM MgSO4, 4 µM FeSO4, 0.4 µM Pyridoxal-5′-phosphate, pH 7.4; Haas et al. 1977) containing the indicated carbon and nitrogen sources. Pseudomonas aeruginosa was also cultured on Pseudomonas Isolation Agar plates (PIA; Difco). When required, antibiotics were added at the following concentrations (in µg/ml): 100 (ampicillin), 25 (gentamycin), 25 (kanamycin), and 10 (tetracycline) for E. coli, 200 (carbenicillin), 200 (gentamycin), and 200 (tetracycline) for P. aeruginosa.
Genetic Manipulations
To delete ldcA gene, fused upstream and downstream flanking regions of the gene were generated by “Splicing by Overlap Extension” (SOE)-PCR using PAO1 genomic DNA as matrix. The resulting fragment of 819 bp was cloned into pCR-Blunt II-TOPO vector, sequenced and then subcloned into BamHI-HindIII sites of pEXG2. The resulting plasmid pEXG2ΔldcA was mobilized into P. aeruginosa strain by triparental mating, using the conjugative properties of the helper plasmid pRK2013. Co-integration events were selected on PIA plates containing gentamycin. Single colonies were then plated on PIA medium containing 5% (w/v) sucrose to select for the loss of plasmid which carries the counter-selectable sacB marker: The resulting sucrose-resistant strains were checked for antibiotic sensitivity and for ldcA (wild-type or truncated gene) genotype by PCR.
To complement the ldcA mutant, a 2,785 bp-long fragment encompassing the coding sequence and 495 bases upstream the ATG was PCR amplified from PAO1 genomic DNA, cloned into pCR-Blunt II-TOPO and sequenced. The SpeI restriction fragment was subcloned in mini-CTX1, leading to miniCTX-ldcA. To construct the lacZ reporter vector, the 548 bp-long ldcA promoter fragment (–498/+44 relative to translation initiation) was PCR amplified, cloned into pCR-Blunt II-TOPO vector, sequenced, and finally subcloned into the XhoI-EcoRI sites of mini-CTX-lacZ. Both miniCTX-ldcA and mini-CTX-PldcA-lacZ were introduced into P. aeruginosa by triparental conjugation and the transconjugants were selected on PIA plates containing tetracycline. The pFLP2 plasmid was then used to excise the FRT cassette as described (Hoang et al. 1998).
Plasmids and primers used in PCR are listed in supplementary tables S1 and S2, Supplementary Material online respectively.
β-Galactosidase Assays
Bacteria were grown aerobically at 37 °C in 100 ml flasks with agitation (300 rpm). At the indicated OD, β-galactosidase activity was assayed as described (Miller 1972; Thibault et al. 2009).
Time-Dependent Killing Assay
The number of persister cells of PAO1, PAO1ΔldcA and PAO1ΔldcA::ldcA strains were determined as previously described (Manuel et al. 2010) using a challenge with at 500 µg/ml carbenicillin (8× the MIC) of 24 h.
Intracellular Metabolite Analysis
Strains were first isolated from an overnight solid culture at 37 °C on Mueller Hinton II agar 1.5%. Precultures and cultures were performed on a Minitron II rotary shaker at 220 rpm (Infors HT) under aerobic conditions (10% of total volume of Erlenmeyer flask). A few bacterial colonies from the agar plate were precultured overnight at 37 °C and an aliquot was withdrawn and diluted to an OD600 of ∼0.1 in a fresh culture medium for the culture step. Growth curves were determined for each strain and used to determine their respective concentration (CFU/ml) and the OD at which the bacteria should be harvested to correspond to early-, mid-, and late-exponential phases.
The protocol of the intracellular metabolites sampling was adapted from a previously described procedure (Aros-Calt et al. 2015). In brief, a 5 ml aliquot of cell culture broth was taken from the main culture and was filtered in a few seconds using poly(ether sulfone) sterile membrane disc filters (Supor450, 0.45 μm pore size, PALL) mounted on a Millipore filtration device (Darmstadt). The bacteria on the filter were quickly washed with 5 ml of 0.6% NaCl solution maintained at room temperature. The filter was then rapidly transferred to a 50 ml Falcon tube containing 5 ml of cold 60% ethanol (v/v, ≤−20 °C). The Falcon tube was subsequently quickly immersed in liquid nitrogen. Following this quenching step, tubes containing bacteria on filters in the extraction solution were vortexed 10 times on ice to remove the cells from the filter. Then, a 1 ml aliquot of the cell suspension was transferred to 2 ml tubes containing 0.1 mm glass beads (Bertin Technologies). Cell disruption was performed by three cycles in a Precellys 24 homogenizer (Bertin Technologies) for 30 s at 3,800 rpm at ∼4 °C. The glass beads and cell debris were separated from the supernatant by centrifugation for 5 min at 4 °C and 10,000 × g. A 400 μl aliquot of the supernatant was withdrawn and further vacuum-dried using a SpeedVac instrument (Thermo Fisher Scientific) and stored at −80 °C until analysis. Dried extracts were dissolved in an adjusted volume of 95% mobile phase A/5% mobile phase B to obtain the equivalent of 1.25 × 107 CFU in 15 µl before analysis by LC/HR-MS.
To detect intracellular metabolites, LC/HR-MS experiments were performed using a Dionex Ultimate chromatographic system (Thermo Fisher Scientific) coupled to an Exactive (Orbitrap) mass spectrometer from Thermo Fisher Scientific fitted with an electrospray source. The mass spectrometer was externally calibrated before each analysis using the manufacturer’s predefined methods and recommended calibration mixture provided by the manufacturer. Chromatographic separation was performed on a Discovery HS F5 PFPP 5 µm, 2.1 × 250 mm column (Sigma) at 30 °C. The chromatographic system was equipped with an on-line prefilter (Thermo Fisher Scientifics). Mobile phases were 100% water (A) and 100% ACN (B), both of which containing 0.1% formic acid. Chromatographic elution was achieved with a flow rate of 250 µl/min. After injection 15 µl of sample, elution started with an isocratic step of 2 min at 5% phase B, followed by a linear gradient from 5 to 100% of phase B in 18 min. These proportions were kept constant for 4 min before returning to 5% of phase B and letting the system equilibrate for 6 min. The column effluent was directly introduced into the electrospray source of the mass spectrometer, and analyses were performed in the positive ion mode. Source parameters were as follows: Capillary voltage set at 5 kV; capillary temperature at 300 °C; sheath and auxiliary gas (nitrogen) flow rates at 50 and 25 arbitrary units, respectively; mass resolution power of the analyzer set at 50,000 at m/z 200 (full width at half maximum, FWHM) for singly charged ions. The acquisition was achieved from m/z 50 to 250 in the positive ionization mode during the first 12 min of the run. Under these conditions were achieved a good separation and detection (with an average mass accuracy better than 3 ppm) of the targeted molecules (under their [M+H]+ form). These species were readily identified in the extracts through the use of the corresponding commercial molecules obtained from Sigma–Aldrich. Extracted ion chromatograms were generated and resulting peaks integrated using the Xcalibur software (version 2.1, Thermo Fisher Scientific) for Spd ([M+H]+ at theoretical m/z 146.1652, retention time 5.24 min), Put (m/z 89.1073, 3.63 min), and Cad (m/z 103.1230, 3.94 min).
All data are publicly available.
Results
Taxonomic Distribution of AAT-Fold LAOdcs in Prokaryotes
An in-depth survey of 4,466 prokaryotic proteomes representing 1,904 species revealed 4,090 protein sequences belonging to the AAT-fold LAOdcs (supplementary fig. S1A, Supplementary Material online). Representatives of this superfamily are mainly present in Bacteria, especially in Proteobacteria, Firmicutes, Actinobacteria, and Cyanobacteria, whereas only seven sequences were detected in Archaea (supplementary fig. S1A, Supplementary Material online). The corresponding ML tree could be divided in three parts (supplementary fig. S1B, Supplementary Material online). Cluster I encompasses sequences devoid of the wing domain (short AAT-fold LAOdc) (Bootstrap value [BV] = 99%). They are mainly found in Firmicutes, Cyanobacteria, and Actinobacteria. Cluster II gathers nearly all proteobacterial sequences and a few sequences from Firmicutes (BV = 99%), all containing a wing domain. Cluster III sequences branch in-between Cluster I and Cluster II; they correspond to a mix of long and short AAT-fold LAOdc sequences from unrelated taxonomic groups (supplementary fig. S1C, Supplementary Material online). This suggests that punctual horizontal gene transfers (HGT) may be responsible for the observed taxonomic distribution of Cluster III sequences. Because of the low number of available sequences, it is difficult to propose a reliable scenario for the origin of Cluster III itself and to evaluate the possible contribution of hidden paralogy.
Wing-Less AAT-Fold LAOdc Are Ancient in Firmicutes and Cyanobacteria
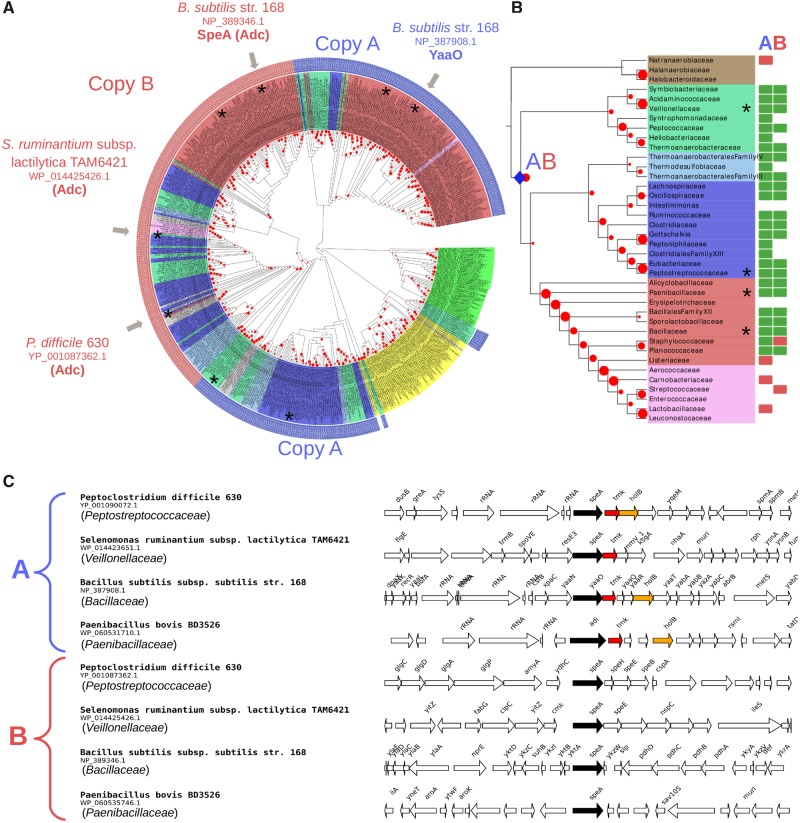
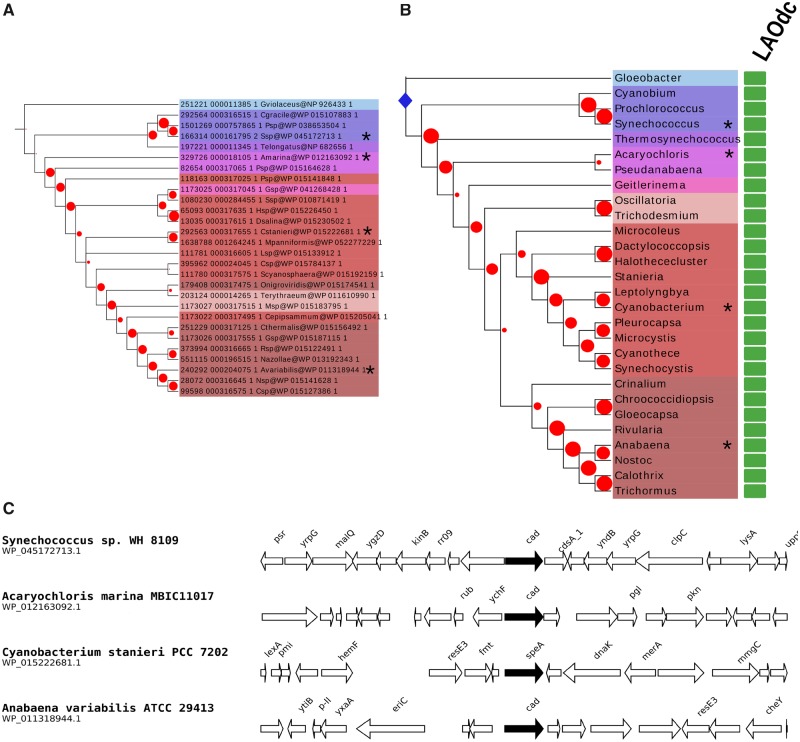
Within Cluster I, all cyanobacterial sequences group together albeit with a weak BV (BV = 17%) (fig. 1A). They are widely distributed in this phylum (fig. 2). The phylogeny inferred with these sequences (fig. 2A) is overall consistent with a reference phylogeny of Cyanobacteria based on ribosomal proteins (fig. 2B), indicating that a gene coding for a wing-less AAT-fold LAOdc was likely present in the ancestor of Cyanobacteria and had been mainly transmitted vertically in Cyanobacteria. The genomic context of LAOdc in Cyanobacteria is not conserved even in relatively close species (fig. 2C).
Fig. 1.
—LAOdcs are ancient in Firmicutes. (A) Unrooted maximum likelihood phylogeny of LAOdc Cluster I (PhyML, LG+I+G4, 504 sequences, 295 amino-acid positions), displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. Leaf colors correspond to taxonomic groups (Firmicutes: Red, pink, blue, light blue, fade green, and brown, Cyanobacteria: Green, Actinobacteria: Yellow, other: Grey). External colored rings correspond to copy A (blue) and B (red). LAOdc sequences discussed in the text or for which functional information is available are indicated with gray arrows. Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Sequences that are displayed in their genomic context are mapped with asterisks. (B) Taxonomic distribution of AAT-fold LAOdc mapped on a ribosomal protein tree of Firmicutes (PhyML, LG+I+G4, 38 sequences, 6,133 amino-acid positions). For clarity the tree is displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Taxa that are represented in (C) are mapped with asterisks. The blue diamonds pinpoint the emergence of copy A and copy B. Rectangles at leaves indicate that at least one genome of the considered taxon encodes one or more AAT-fold LAOdc. A green rectangle indicates that the ancestor of the taxon likely contains one (or more) AAT-fold LAOdc gene, whereas a red rectangle indicates that some members of the taxon acquired secondarily their AAT-fold LAOdc by HGT. (C) Genomic context of LAOdc A and B in a sample of Firmicutes. LAOdc A presents a well-conserved association with thymidylate kinase and DNA polymerase III subunit delta coding genes, while the genomic context of LAOdc B is not conserved. Black arrows: LAOdc coding genes, colored arrows: Conserved neighbor genes.
Fig. 2.
—LAOdcs are ancestral in Cyanobacteria. (A) Maximum likelihood phylogeny of LAOdc of Cyanobacteria (PhyML, LG+I+G4, 28 sequences, 446 amino-acid positions), displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. The tree has been rooted according to the reference phylogeny of Cyanobacteria (see above). Leaf colors correspond to taxonomic groups. Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Sequences that are displayed in their genomic context are mapped with asterisks. (B) Taxonomic distribution of AAT-fold LAOdc mapped on a ribosomal protein tree of Cyanobacteria (PhyML, LG+I+G4, 30 sequences, 6,394 amino acid positions). For clarity the tree is displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Taxa that are represented in (C) are mapped with asterisks. The blue diamond pinpoints the origin of AAT-fold LAOdc in Cyanobacteria. Rectangles at leaves indicate that at least one genome of the considered taxon encodes one or more AAT-fold LAOdc. A green rectangle indicates that the ancestor of the taxon likely contains one (or more) AAT-fold LAOdc gene. (C) Genomic context of LAOdc in a sample of Cyanobacteria. Black arrows: LAOdc genes.
Similarly, sequences of Firmicutes belonging to Cluster I are widely distributed in this phylum (fig. 1). Furthermore, most of the members of Firmicutes harbor two LAOdc copies (referred as A and B, fig. 1B), that can be easily distinguished by their genomic context (fig. 1C). The comparison of the Cluster I phylogeny (fig. 1A) with a reference phylogeny of Firmicutes (fig. 1B) suggests that copies A and B could be ancient in this phylum as they likely originate after the divergence of the Halanaerobiaceae, Halobacteroidaceae, and Natranaerobiaceae orders, the three deepest-branching families of Firmicutes. Worthy of note, a loss of both copies can be inferred in the ancestor of Lactobacillales. Yet, some Streptococcaceae and Carnobacteriaceae have reacquired either copy A or copy B by HGT from different Firmicutes donors (fig. 1A). Finally, Cluster I encompassed a group of sequences from Actinobacteria. Their taxonomic distribution is patchy and their phylogeny is not consistent with the current taxonomy, suggesting that they have been acquired and spread through HGT in Actinobacteria.
Wing-Domain Containing AAT-Fold LAOdcs Form Four Groups in Proteobacteria
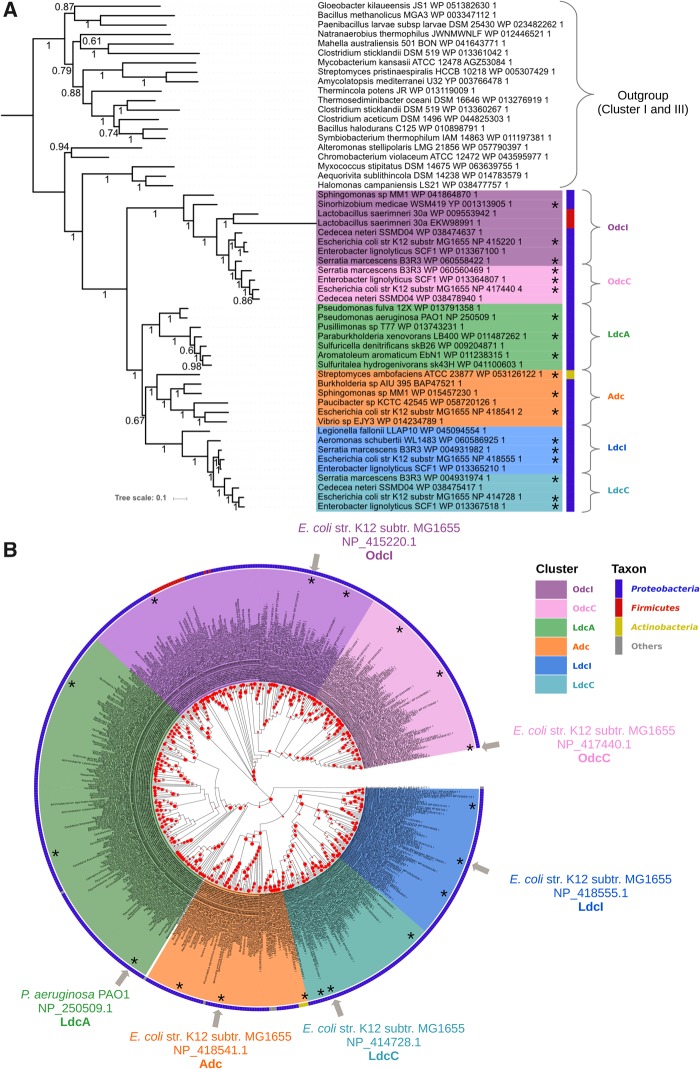
The phylogenetic analysis of proteobacterial AAT-fold LAOdcs composing Cluster II revealed two monophyletic groups, corresponding to Odc (Posterior probabilities [PP] = 1, fig. 3A and BV = 100%, fig. 3B) and LAdc (PP = 1, fig. 3A and BV = 100%, fig. 3B). The LAdc group is further split into LdcI/LdcC (PP = 1, fig. 3A and BV = 100%, fig. 3B), Adc (PP = 1, fig. 3A and BV = 93, fig. 3B), and a hitherto nonidentified subfamily (PP = 1, fig. 3A and BV = 100%, fig. 3B), that will be referred as LdcA in the name of its only previously mentioned member, LdcA from P. aeruginosa (Chou et al. 2010). Whereas the monophyly of OdcI/OdcC, LdcI/LdcC, Adc, and LdcA groups is well supported, the relationships between LdcI/LdcC, Adc, and LdcA are unresolved (PP = 0.67, fig. 3A and BV < 80%, fig. 3B).
Fig. 3.
—Phylogeny of the LAOdc Cluster II. (A) Bayesian phylogeny of cluster II inferred from a sample of representative sequences and rooted with a sample of sequences from clusters I and III (MrBayes, mixed model+G4, 54 sequences, 392 amino acid positions). The scale bar represents the average number of substitutions per site. Numbers at branches correspond to posterior probabilities. The ML tree inferred with the same data set supported the same topology (see Supplementary Material). (B) Maximum likelihood phylogeny of the LAOdc Cluster II (PhyML, LG+I+G4, 551 sequences, 589 amino-acid positions), displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. LAOdc sequences from Escherichia coli and Pseudomonas aeruginosa discussed in the text are indicated with gray arrows. The tree has been rooted according to (A). Colors on the external circle correspond to taxonomic groups: Dark blue: Proteobacteria, red: Firmicutes, yellow: Actinobacteria, gray: Other taxa. Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Sequences that are displayed in their genomic context supplementary figure S2, Supplementary Material online are mapped with asterisks.
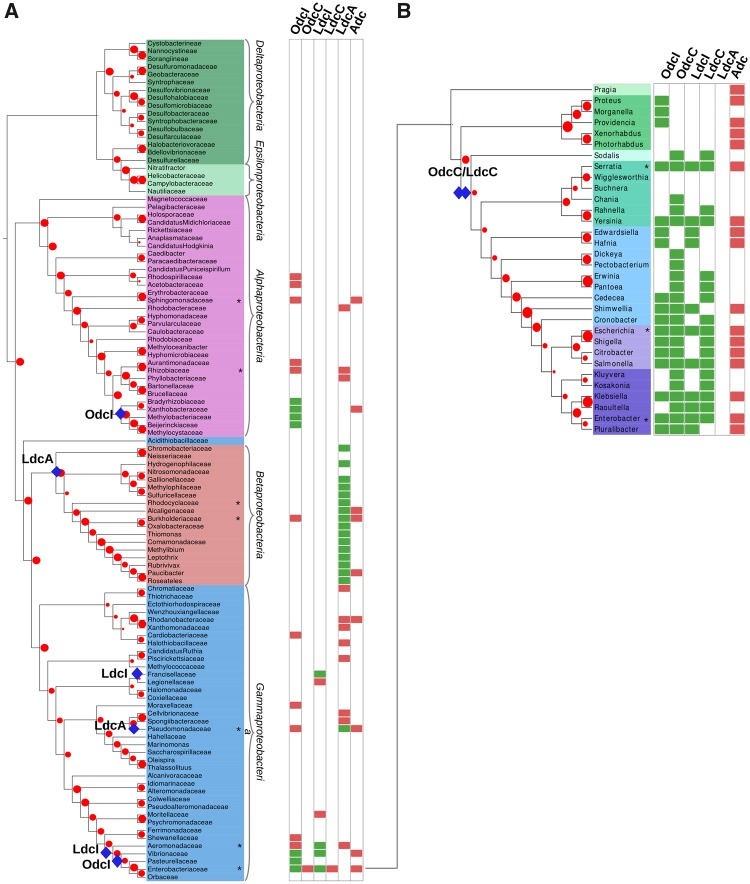
Comparison of the LAOdc phylogeny (fig. 3B) and taxonomic distribution with a reference phylogeny of Proteobacteria (fig. 4A) suggests that odcI could be ancestral in some Alphaproteobacteria, as well as in Vibrionaceae, Pasteurellaceae, and Enterobacteriaceae (all belonging to Gammaproteobacteria). In contrast, odcC seems more recent and appears to result from a gene duplication of odcI that occurred in Enterobacteriaceae, before the divergence of Sodalis (PP = 1, fig. 3A and BV = 75%, fig. 3B, and fig. 4B). Genes coding for OdcI and OdcC are clearly distinguished by their context (supplementary fig. S2, Supplementary Material online). More precisely, odcI is predominantly upstream from a gene (potE) coding for a Put-ornithine antiporter, consistently with the function of OdcI that converts Orn to Put. Interestingly, a few HGT of odcI occurred from Proteobacteria to unrelated firmicutes such as some Lactobacillus.
Fig. 4.
—Taxonomic distribution of LAOdcs in Proteobacteria. (A) Taxonomic distribution of AAT-fold LAOdc mapped on a ribosomal protein tree of Proteobacteria (PhyML, LG+I+G4, 108 sequences, 6,129 amino acid positions). For clarity the tree is displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. Leaf colors correspond to taxonomic groups (Alphaproteobacteria: Pink, Betaproteobacteria: Red, Gammaproteobacteria: Blue, Deltaproteobacteria: Dark green, Epsilonproteobacteria: Light green). Red dots correspond to bootstrap values (BV), ranging from 0% (smallest circles) to 100% (largest circles). Taxa that are represented in supplementary figure S2, Supplementary Material online are mapped with asterisks. A blue diamond indicates the ancestral presence of LAOdc families in the corresponding taxon. Rectangles at leaves indicate that at least one genome of the considered taxon encodes one or more AAT-fold LAOdc. A green rectangle indicates that the ancestor of the taxon likely contains one (or more) AAT-fold LAOdc gene, whereas a red rectangle indicates that some members of the taxon acquired secondarily their AAT-fold LAOdc by HGT. (B) Taxonomic distribution of AAT-fold LAOdc mapped on a ribosomal protein tree of Enterobacteraceae (PhyML, LG+I+G4, 34 sequences, 6,333 amino acid positions). For clarity the tree is displayed as a cladogram. The corresponding phylogram is available at the newick format as supplementary material, Supplementary Material online. Leaf colors correspond to taxonomic groups. Other legend elements are identical to (A).
The ldcI gene, also called cadA, appears to be ancestral in some gammaproteobacterial lineages (Francisellaceae, Aeromonadaceae, Vibrionaceae, and Enterobacteriaceae, fig. 4A). Moreover, similar to odcI/odcC, ldcC apparently derived from ldcI, and more precisely from a gene duplication that occurred, as for odcI, just before the emergence of Sodalis (PP = 1, fig. 3A and BV = 100%, figs. 3B and 4B). The ldcI gene forms the cadBA operon together with the lysine-Cad antiporter encoded by the cadB gene. In Enterobacteriaceae, this operon is regulated by the transcriptional factor CadC integrating three external signals—low pH, high lysine and low Cad levels (Kuper and Jung 2005; Fritz et al. 2009). Our analysis confirms the conserved genomic organization of the cadCBA system (supplementary fig. S2, Supplementary Material online) (Zhao and Houry 2010) allowing Enterobacteria to face acid and oxidative stresses, and shows that the ldcC genomic context is also strongly conserved (supplementary fig. S2, Supplementary Material online).
In sharp contrast with OdcI/OdcC and LdcI/LdcC, the Adc group presents a patchy taxonomic distribution and the relationships among Adc sequences are at odds with current systematics, with sequences from different classes of Proteobacteria being intermixed on the tree (figs. 3B and 4). This indicates that the Adc subfamily was heavily impacted by HGT and possibly by hidden paralogy. The genomic context of adc is not conserved (supplementary fig. S2, Supplementary Material online), precluding identification of potential functional partners.
Phylogenetic Analyses of LAOdc Disclose a Fourth Group of LAOdcs in Proteobacteria
As specified above, beside OdcI/C, LdcI/C, and Adc, a fourth group of LAOdcs is present in the tree. This group contains LdcA from P. aeruginosa, a highly versatile bacterium that efficiently grows on arginine but not on lysine (Fothergill and Guest 1977; Rahman and Clarke 1980). In this organism exhibiting tightly interconnected lysine and arginine catabolism networks (Chou et al. 2010; Madhuri Indurthi et al. 2016), the PA1818 gene was identified as a part of the ArgR regulon upon growth on excess arginine, but was surprisingly found to code for a lysine decarboxylase, and not for an arginine decarboxylase as one would have logically supposed, and was therefore called ldcA (Chou et al. 2010; Madhuri Indurthi et al. 2016). Our analysis identifies homologues of LdcA in other Pseudomonadaceae and in Betaproteobacteria. Both groups of sequences are well separated on the tree and widely distributed in these two taxa, with relationships globally in agreement with the current taxonomy. This suggests that a LdcA homologue was present in the ancestor of Pseudomonadaceae and in the ancestor of Betaproteobacteria and has been globally well conserved along their diversification. The genetic environment of ldcA is not conserved (supplementary fig. S2, Supplementary Material online).
Pseudomonas aeruginosa is a major cause of gram-negative infections, especially in patients with compromised and weakened immune system. This opportunistic pathogen is also a well-identified threat for patients suffering from Cystic Fibrosis (CF), because the chronic respiratory infections associated to host inflammatory responses lead to pulmonary tissue destruction and lung failure (Bodey et al. 1983; Gellatly and Hancock 2013). The occurrence and persistence of P. aeruginosa in the CF patients’ lungs, whose secretions were shown to be acidified and to become oxidative (Pezzulo et al. 2012), hints to a possible role of LdcA in promoting bacterial fitness. Therefore, in the following sections, we chose to deepen the present knowledge on expression, regulation and functional characterization of P. aeruginosa LdcA.
ldcA Expression Is Growth-Phase Dependent
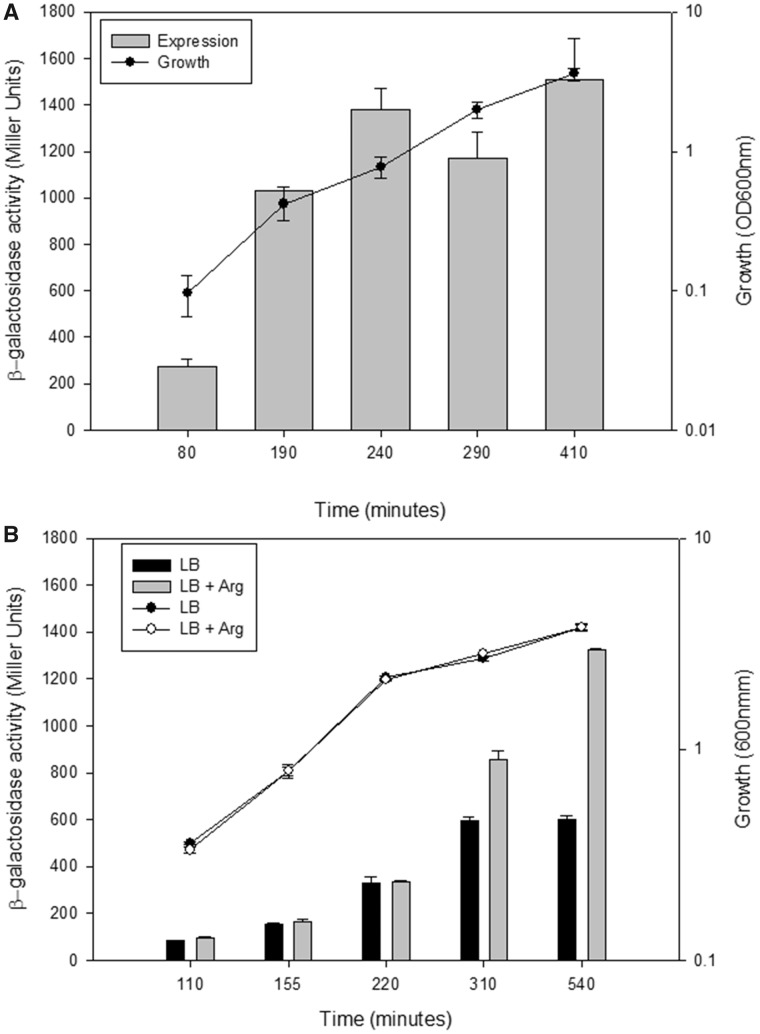
To determine conditions of the ldcA expression in P. aeruginosa, we created a transcriptional fusion between the ldcA promoter (PldcA) and the reporter lacZ gene, integrated it into the chromosome of the PAO1 reference strain, and measured the β-galactosidase activity of the PAO1::PldcA-lacZ strain grown in MMP medium containing glutamate as a carbon source and lysine or arginine as additives. Unlike lysine, arginine was able to induce the expression of ldcA in PAO1::PldcA-lacZ, in agreement with published data identifying ArgR as a positive regulator of ldcA expression in the PAO1 strain (Lu et al. 2004; Chou et al. 2010). The pattern of ldcA expression was followed during the growth both in minimal and rich media. In MMP medium supplemented with glutamate and arginine (MMP-GR), the β-galactosidase activity increased slightly but continuously along the growth and reached a maximum in the stationary phase (fig. 5A). Expression of ldcA in LB rich medium followed the same pattern of expression (fig. 5B), paralleled by an increase in LdcA amount assessed by western blot (not shown). Expression in LB was two-fold lower than in MMP-GR. Addition of 20 mM arginine to LB did not change the growth rate of the bacteria but the ldcA promoter activity increased during the transition to the stationary phase to reach a level similar to the one measured in MMP-GR (fig. 5B), indicating a probable limiting arginine concentration in LB at late growth.
Fig. 5.
—Factors influencing ldcA expression. Activity of ldcA promoter fused to lacZ reporter gene was assessed during growth in different media and genetic backgrounds. Measurements of the β-galactosidase activity of PAO1::PldcA-lacZ strain grown either in minimal medium P (MMP) containing 20 mM l-glutamate and 20 mM arginine (A), or in LB containing or not 20 mM arginine (B) were performed at times indicated. β-Galactosidase activity is expressed in Miller Units (left Y-axis) and presented in the bar graphs. Growth was performed in 125 ml flasks, followed by measure of OD600 (right Y-axis) and plotted on lines. Results are the average of values from three independent experiments ± standard deviation (SD).
ldcA Expression Differs from That of ldcC and ldcI
The ldcA genomic context being different from that of ldcI and ldcC genes (supplementary fig. S2, Supplementary Material online), we wondered if the regulation of ldcA was also different. Whereas ldcC expression is induced in stationary phase in LB medium by RpoS, the sigma factor of stationary phase (Kikuchi et al. 1998), we observed that the ldcA expression was not affected in a rpoS mutant background (data not shown), in agreement with transcriptomic analyses that do not list ldcA as a part of the RpoS regulon (Schuster et al. 2004). To compare with ldcI, despite the absence of a CadC homologue in P. aeruginosa, we assessed if acid stress could activate the expression of ldcA in a CadC-independent manner. Hence, we induced acid stress by decreasing the pH of the medium to 5 during the exponential phase of growth and documented an absence of effect of this treatment on ldcA expression (supplementary fig. S3A, Supplementary Material online). In addition, ldcI from V. vulnificus was reported to be induced by SoxR upon H2O2 stress (Kim et al. 2006). In P. aeruginosa, SoxR is not a key player in the oxidative stress response, but H2O2 activates the global regulator OxyR that orchestrates the defense against ROS (Ochsner et al. 2000). Yet, addition of 1 mM of H2O2 in the culture did not affect ldcA expression (supplementary fig. S3A, Supplementary Material online). To conclude, P. aeruginosa does not overexpress ldcA to respond to low pH or oxidative stress conditions.
Low pH Survival Is Not Affected in Absence of LdcA
To clarify the function of LdcA in P. aeruginosa, a mutant deleted of the ldcA gene, as well as a complemented strain in which one copy of ldcA driven by its own promoter was inserted in the chromosome, were engineered. Using Biolog system, a Phenotype MicroArray analysis was carried out to assess a potential role of LdcA in detoxifying P. aeruginosa and protecting it against acid, alkaline and oxidative stress, antibiotic treatment and toxic molecules causing DNA damage, nitrosative stress, and membrane destabilization. Analysis of the growth fitness of wild-type, mutant and complemented strains indicated that P. aeruginosa could grow optimally in a pH range from 5 to 10 without considerable effect on the metabolism and biomass growth. At pH 4–5, the bacterial growth was strongly inhibited and the strains were unable to grow below pH 4 (supplementary fig. S3B, Supplementary Material online). This set of experiments did not show any significant difference between the mutant and the wild-type and complemented strains, pointing to a noninvolvement of LdcA in survival at low pH. Similarly, no significant effect of ldcA absence on resistance against antibiotics, oxidative and toxic agents could be detected at their concentrations used in Biolog plates (not shown). Hence, under these conditions LdcA seems not to be important for stress response.
The Cad Pool Generated by LdcA Impacts Persistence Phenotype and Polyamine Content
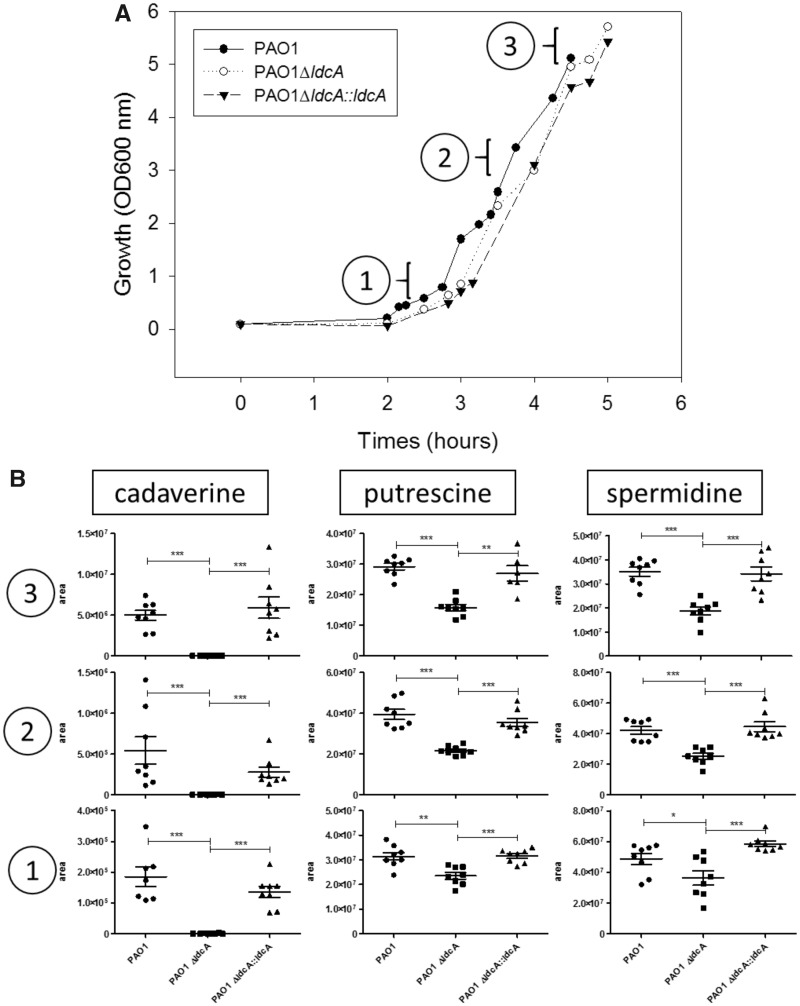
Cad production was shown to lead to a reduction of the dormant P. aeruginosa cells that form an antibiotic-tolerant subpopulation in MH medium (Manuel et al. 2010). Therefore, to assess the role of LdcA in persistence, we first evaluated the impact of ldcA mutation on Cad production in this rich MH medium. To do so, we quantified the intracellular Cad amounts in the bacterial strains by liquid chromatography coupled to high resolution mass spectrometry (LC/HRMS) during early-, mid- and late-exponential growth phases. Whereas Cad was absent in the ldcA mutant, complementation with a wild-type ldcA copy restored its original level indicating that, in PAO1, this polyamine is produced exclusively through the enzymatic activity of LdcA under this growth condition (fig. 6). Moreover, the wild-type strain showed an impressive increase (around 25-fold) of Cad concentration during the growth, probably reflecting the ldcA expression pattern in rich medium (increase during exponential growth, fig. 5B).
Fig. 6.
—Intracellular Cad in rich medium is produced by the lysine decarboxylase LdcA. (A) Growth curves of the wild-type strain, the ldcA mutant and the complemented strain in the rich Mueller Hinton medium. Data show a representative experiment from four independent biological replicates. (B) Intracellular concentrations, expressed in area of chromatogram peak, of the three indicated polyamines at 1) early-, 2) mid-, and 3) late-exponential growth phases as indicated, measured in duplicate in four biological replicates.
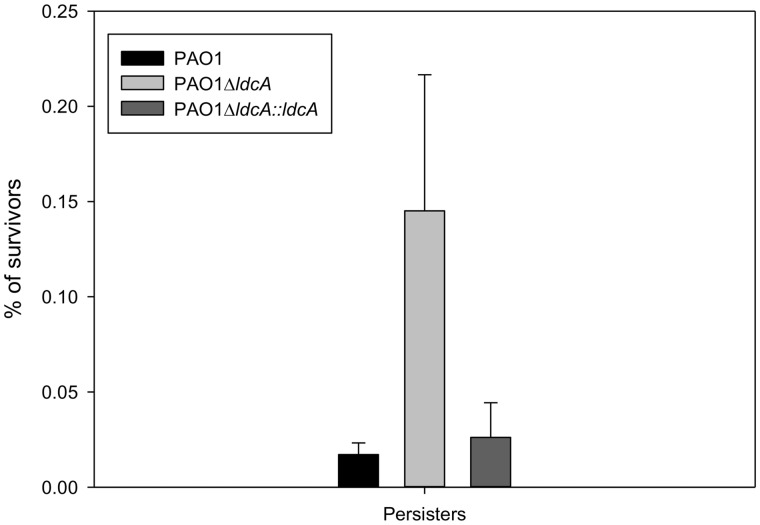
Then, the impact of ldcA on the number of persisters during carbenicillin treatment was addressed. As anticipated, ldcA mutant showed a number of persisters significantly higher compared with the wild-type PAO1 and complemented strains (fig. 7), revealing the importance of LdcA activity in the persistence phenotype.
Fig. 7.
—LdcA function affects carbenicillin persistence. Percentage of survivors in rich medium (cation-adjusted Mueller Hinton Broth) after 24 h of carbenicillin treatment. Growth was performed in Erlenmeyer flasks at 37 °C with agitation (300 rpm). Percentage of survivors was calculated from CFU counting after 24 h of antibiotic treatment at 500 µg/ml (8× MIC). The results are the mean values of experiments performed four times and the error bars indicate the standard deviations.
In parallel to Cad, we also quantified the intracellular concentrations of two other polyamines, Put and Spd, in the wild-type strain and ldcA mutant (fig. 6). While in the wild-type PAO1, the amount of Cad is clearly growth-phase dependent, the amounts of Put and Spd were found to be abundant and constant in the MH medium. Remarkably, inactivation of ldcA affected not only the production of Cad but also the amount of intracellular Put and Spd which were reduced by around two-fold in the ldcA mutant at late exponential growth. This could either indicate a higher turnover of polyamine metabolism in the ldcA mutant or a reduced production of Put and Spd as a compensation for the absence of Cad.
Polyamines Are Important for Growth Fitness
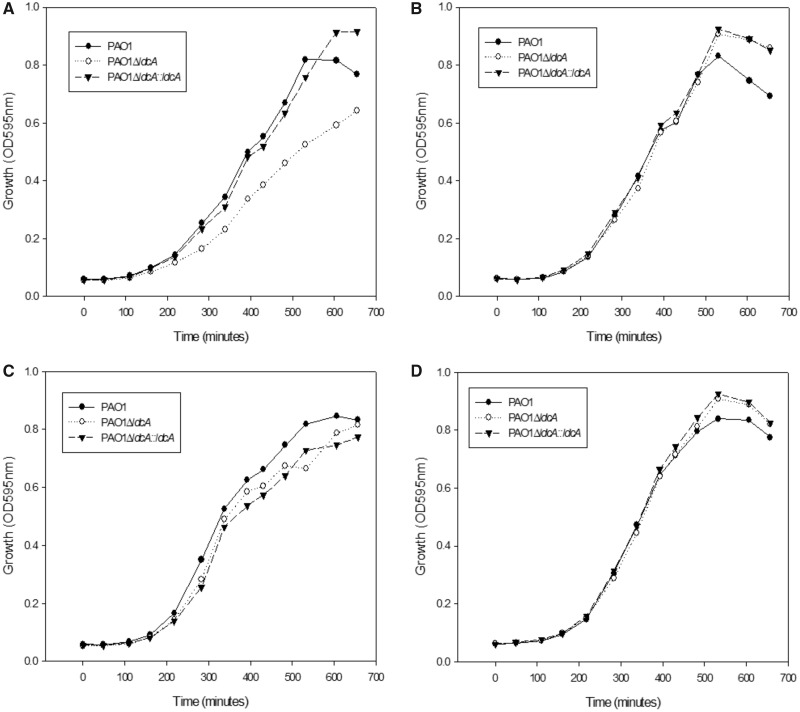
As mentioned above, LdcA synthesis is induced in the minimal MMP medium in the presence of arginine. Therefore, we monitored the growth of the ldcA mutant in this condition in the presence of either lysine or Cad. The observed clear reduction of growth of the ldcA mutant highlighted the metabolic role of the enzyme, whereas wild-type growth was restored in the complemented strain (fig. 8A). Addition of exogenous Cad was sufficient to restore a normal growth in the mutant indicating that the limiting factor was indeed the LdcA enzymatic reaction product Cad (fig. 8B). Strikingly, growth was also restored when Put (fig. 8C) or Spd (fig. 8D) were added to the growth medium at the same concentrations, indicating that these polyamines can substitute for Cad as a growth factor.
Fig. 8.
—Polyamines are important for growth fitness in minimal medium. Growth of the wild-type PAO1 strain, the ldcA mutant and the complemented strain in minimal MMP medium supplemented with 20 mM glutamate, 1 mM arginine and 5 mM of (A) lysine, (B) Cad, (C) Put, or (D) Spd in 96-well plates. The experiments are representative of two experiments.
Discussion
The present exhaustive phylogenetic analysis of the AAT-fold LAOdcs superfamily confirms earlier reports suggesting the ancient nature of the wing-less LAOdcs in Firmicutes (Sekowska et al. 1998; Burrell et al. 2010), and highlights also their ancestral presence in Cyanobacteria. Yet, no LAOdc activity of a cyanobacterial AAT-fold enzyme has yet been documented. We show that wing domain-containing AAT-fold LAOdc emerged during the diversification of Proteobacteria, suggesting that short AAT-fold LAOdc could be more ancient and that the acquisition of the wing domain occurred likely secondarily, as previously proposed (Burrell et al. 2010; Kanjee, Gutsche, Alexopoulos, et al. 2011). The absence of AR-fold decarboxylases in Firmicutes (Burrell et al. 2010; Michael 2016a) implies that their AAT-fold decarboxylases would be the only route for polyamine biosynthesis and thus emphasizes the importance of these enzymes in this phylum. The Firmicutes AAT-fold LAOdc correspond to two copies, likely paralogues (A and B), present in most species and easily distinguishable by their genomic context. Copy A encompasses the B. subtilis yaaO (Sekowska et al. 1998), whereas copy B contains a B. subtilis speA gene coding for an arginine decarboxylase (fig. 1A; Burrell et al. 2010). Two other Firmicutes proteins corresponding to copy B sequences (fig. 1A) were also shown to possess an arginine decarboxylase activity (Liao et al. 2008; Burrell et al. 2010), which led to a hypothesis that all copy B AAT-fold decarboxylases would be Adcs (Burrell et al. 2010). The function of copy A awaits further investigation and to our knowledge, the only study focused on the yaaO gene (copy A) concluded that in B. subtilis this protein had no effect on polyamine production (Sekowska et al. 1998).
Regarding the evolutionary history of the long LAOdcs, our analysis reveals that proteobacterial LAOdcs form two monophyletic groups, Odc and LAdc. Thus, although the relationships among the two Ldc families (LdcI/C and LdcA) and the Adc family are not resolved (PP < 0.5), lysine and arginine decarboxylases appear more closely related to each other than to Odcs (fig. 3A). Furthermore, we showed that the proteobacterial biosynthetic OdcC and LdcC emerged from inducible Odc and Ldc (i.e., OdcI and LcdI, respectively) through two independent gene duplication events that occurred in Enterobacteriaceae, after the divergence of Sodalis. Given that both duplication events seem to occur in the same branch of the species tree, it is tempting to hypothesize that they are linked, and that a functional connection between both biosynthetic subgroups may have existed. The emergence of biosynthetic enzymes from inducible ones may appear contra-intuitive at the first glance, but may reflect an expansion and a diversification of polyamine functions in these lineages. This would also explain why Enterobacteriaceae possess also a constitutive pathway of polyamine synthesis through an AR-fold Adc.
The exhaustive phylogenetic analysis of AAT-fold decarboxylases discloses multiple cases of HGT (e.g., within Cluster III, from Firmicutes to Firmicutes and to Actinobacteria within Cluster I, but also from Proteobacteria to Firmicutes within cluster II). Interestingly, the two long AAT-fold LAOdc coding genes found in L. saerimneri 30a were proposed to result from a HGT of an acid stress inducible Odc from Enterobacteriaceae, followed by a gene duplication event: One of the two resulting paralogues is thought to have kept the original function, whereas the other acquired the capacity to use lysine as substrate (Romano et al. 2013, 2014). Instead, our analysis points to a different scenario (supplementary fig. S4, Supplementary Material online). Remarkably, these two enzymes rely on the same antiporter capable to exchange both ornithine/Put and lysine/Cad pairs, resulting in a unique three-component decarboxylation system involved in acid stress response (Romano et al. 2013). This case of substrate specificity shift may not be an exception, because Burkholderia sp. AIU 395 AAT-fold Ldc (Sugawara et al. 2014) branches within the Adc cluster in phylogenetic trees (fig. 3). Beside substrate shift, existence of dual specificity has been documented in the case of AR-fold LOdcs, exemplified by bifunctional enzymes of S. ruminantium (Takatsuka et al. 2000) and V. vulnificus (Lee et al. 2007). In particular, crystal structures of the V. vulnificus enzyme in complex with either Put or Cad revealed that the dual substrate specificity is based on a bridging water molecule necessary for the binding of a shorter Put ligand in addition to the longer Cad. A similar dual substrate specificity mechanism may also exist in the case of the AAT-fold LAOdc enzymes.
Altogether, our data indicate that functional changes affecting gene regulation, substrate fixation, and cellular function occurred several times during the evolution of AAT-fold enzymes. In the light of these observations, one may wonder if the so-called Adc, LdcC, LdcI, OdcC, and OdcI clusters defined according to phylogenetic criteria correspond indeed to homogeneous functional groups and thus if phylogenetic criteria/sequence similarity-based measures are good predictors of the function of the AAT-fold enzymes. The very restricted number of experimentally characterized enzymes calls for caution and for the urgent need for additional experimental data.
One of the main results of the presented phylogenetic analysis is the identification of a novel large family of decarboxylases, called LdcA, ancestral in Betaproteobacteria and in Pseudomonadaceae. The ldcA gene belongs to the core genome of P. aeruginosa, comprising at least 4,000 conserved genes (Hilker et al. 2015; Valot et al. 2015). Similarly to ldcI, in P. aeruginosa, Pseudomonas resinovorans, Pseudomonas denitrificans, and Pseudomonas knackmusii, ldcA is organized in a gene cluster with a gene coding for a homologue of the CadB antiporter, although cadB (PA1819 in P. aeruginosa) appears downstream, and not upstream, of the lysine decarboxylase-encoding gene. The presence of a dedicated Lys/Cad transporter could be important from a physiological standpoint because CadB is involved not only in substrate/product exchange but also in the generation of proton motive force (Soksawatmaekhin et al. 2004). Remarkably, the proximity of cadB and ldcA is an exception in the ldcA subfamily (supplementary fig. S2, Supplementary Material online), which does not rule out a hypothesis that LdcA could have another function in other taxa. The whole of our data suggests that LdcI and P. aeruginosa LdcA are different in terms of regulation and function. Indeed, we clearly show that LdcA is not involved in acid or oxidative stress response, and its expression is triggered by neither of these stresses; instead it is controlled by ArgR. More similar to LdcC, P. aeruginosa LdcA, and certainly the other members of this novel lysine decarboxylase subfamily, are biosynthetic enzymes responsible for the Cad production by the bacterium.
Importantly, LdcA is the only Cad producing enzyme in P. aeruginosa PAO1, as demonstrated by the measurement of the intracellular contents of Cad (fig. 6). This conclusion was unexpected considering that the product of another gene, PA4115, was previously reported to be responsible for 25% of Cad production in overnight cultures grown in the same medium as the one used in our assays, and therefore proposed to be an Ldc (Manuel et al. 2010). After carboxypenicillin treatment, the PA4115 mutant exhibited an increased number of persisters that was significantly reduced upon addition of exogenous Cad, further supporting the hypothesis of PA4115 being an Ldc (Manuel et al. 2010). Yet, our careful inspection of its sequence revealed that PA4115 belongs to the family of Lonely Guy (LOG) proteins because it contains a highly conserved PGGxGTxxE motif and a nucleotide-binding Rossmann fold. Remarkably, LOG proteins were shown to be often misannotated as lysine decarboxylases, whereas they actually possess a cytokinin-specific phosphoribohydrolase activity (Dzurova et al. 2015; Seo and Kim 2017) or a pyrimidine/purine nucleotide 5′-monophosphate nucleosidase activity (Sevin et al. 2017). Recently, PPnN (or YghD) from E. coli, a close homologue sharing 56% identity with PA4115, has been shown to catalyze the hydrolysis of N-glycosidic bond of AMP, GMP, IMP, CMP, dTMP, and UMP to form ribose 5-phosphate and the corresponding free base. Hence, it is quite likely that PA4115 catalyzes the same reaction and plays a role in maintaining the nucleotide pool homeostasis by degrading excess nucleotides in P. aeruginosa (Sevin et al. 2017). Therefore, the reduced production of Cad in PA4115 mutant needs to be reevaluated because the relationship between PA4115 and LdcA activity is not clear and could involve indirect causes.
The persistence phenotype observed in ldcA mutants is consistent with the beneficial effect of LdcA on growth fitness. Indeed, recent research on bacterial persistence uncovered intracellular ATP concentrations as one of the major factors affecting the amount of persister cells, and demonstrated that ATP levels are sufficient to predict bacterial tolerance to antibiotics (Conlon et al. 2016; Shan et al. 2017). Considering that Cad produced by LdcA is metabolized and used up by the Krebs cycle to create ATP and that the activity of the lysine/Cad antiporter generates proton motive force essential for ATP synthesis (Soksawatmaekhin et al. 2004), one would expect that in P. aeruginosa the ldcA mutation may lead to a decrease in ATP levels, which in turn would result in an increase of persisters’ population. This hypothesis could be challenged by blocking the Cad degradation pathway or the CadB antiporter activity.
The capacity of Put and Spd to complement the growth phenotype of the ldcA mutant and boost P. aeruginosa cultures in minimal medium suggests that in P. aeruginosa the three major polyamines share certain properties. In the same lines, Cad was shown to substitute for Put and Spd as growth factors in E. coli cells depleted of these two polyamines (Igarashi et al. 1986). The growth phenotype described in our work is observed under specific growth conditions. It reveals the importance of Cad when growing P. aeruginosa in minimal medium and highlights a certain redundancy in the function of polyamines. It remains to be investigated whether the phenotype is linked to a regulatory effect of Cad or to its anabolism. Recent literature about Eikenella corrodens has pointed out the importance of an AAT-fold LdcI as a virulence factor against eukaryotic cells that acts through depletion of essential lysine (Lohinai et al. 2015). Therefore, the potential role of LdcA in the virulence of P. aeruginosa warrants further investigation. In the present study, we observed that the absence of the enzyme did not affect T3SS-dependent cytotoxicity or the mobility relying on flagellum and Type IV pili (not shown). However, it would also be relevant to probe the importance of LdcA during mouse infection, where the proper functioning of P. aeruginosa metabolism is primordial for virulence.
Our study reveals that the ldcA gene is relatively ancient in Proteobacteria, being ancestral in Betaproteobacteria and in Pseudomonadaceae, two taxa that cover a wide range of ecological niches. Information about regulation and function of an enzyme of the previously unknown LdcA subfamily enables a step further towards understanding of the evolution and the importance of Cad metabolism in bacteria.
Supplementary Material
Acknowledgments
We thank Rémi Peyraud and Ina Attrée for helpful discussions. This work was supported by Grenoble Alliance for Integrated Structural Cell Biology [ANR-10-LABX-49-01]; the European Union’s Horizon 2020 research and innovation programme [grant agreement No. 647784]; the Agence Nationale de la Recherche [ANR-16-CE02-0005]; by CEA; the French Ministry of Research; and National Research Agency as part of the French metabolomics and fluxomics infrastructure [MetaboHUB, ANR-11-INBS-0010 grant]. Laboratory of excellence Grenoble Alliance for Integrated Structural Cell Biology LABEX GRAL and Association pour la Recherche sur le Cancer ARC Santé from the Region Auvergne Rhône-Alpes provided the PhD grant to [D.C.] and [P.S.G.], respectively.
Literature Cited
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LC, et al. 2016. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. Elife 5 doi: 10.7554/eLife.14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aros-Calt S, et al. 2015. Annotation of the Staphylococcus aureus metabolome using liquid chromatography coupled to high-resolution mass spectrometry and application to the study of methicillin resistance. J Proteome Res. 14(11):4863–4875. [DOI] [PubMed] [Google Scholar]
- Bodey GP, Bolivar R, Fainstein V, Jadeja L.. 1983. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 5(2):279–313. [DOI] [PubMed] [Google Scholar]
- Bunsupa S, et al. 2012. Lysine decarboxylase catalyzes the first step of quinolizidine alkaloid biosynthesis and coevolved with alkaloid production in leguminosae. Plant Cell. 24(3):1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell M, Hanfrey CC, Kinch LN, Elliott KA, Michael AJ.. 2012. Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol Microbiol. 86(2):485–499. [DOI] [PubMed] [Google Scholar]
- Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ.. 2010. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem. 285(50):39224–39238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HT, Hegazy M, Lu CD.. 2010. L-lysine catabolism is controlled by l-arginine and ArgR in Pseudomonas aeruginosa PAO1. J Bacteriol. 192(22):5874–5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, et al. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 1:16051. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Vega AL, Delcour AH.. 1996. Polyamines decrease Escherichia coli outer membrane permeability. J Bacteriol. 178(13):3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino ML, et al. 2013. Polyamines: emerging players in bacteria-host interactions. Int J Med Microbiol. 303(8):484–491. [DOI] [PubMed] [Google Scholar]
- Dzurova L, et al. 2015. The three-dimensional structure of “Lonely Guy” from Claviceps purpurea provides insights into the phosphoribohydrolase function of Rossmann fold-containing lysine decarboxylase-like proteins. Proteins 83:1539–1546. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bakkouri M, et al. 2010. Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc Natl Acad Sci USA. 107(52):22499–22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF.. 2004. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 73(1):383–415. [DOI] [PubMed] [Google Scholar]
- Fothergill JC, Guest JR.. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J Gen Microbiol. 99(1):139–155. [DOI] [PubMed] [Google Scholar]
- Fritz G, et al. 2009. Induction kinetics of a conditional pH stress response system in Escherichia coli. J Mol Biol. 393(2):272–286. [DOI] [PubMed] [Google Scholar]
- Gale EF, Epps HM.. 1944. Studies on bacterial amino-acid decarboxylases: 1. l(+)-lysine decarboxylase. Biochem J. 38(3):232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale EF, Van Heyningen WE.. 1942. The effect of the pH and the presence of glucose during growth on the production of alpha and theta toxins and hyaluronidase by Clostridium welchii. Biochem J. 36(7–9):624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly SL, Hancock RE.. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 67(3):159–173. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27(2):221–224. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Gupta RS. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 24(4):367–402. [DOI] [PubMed] [Google Scholar]
- Haas D, Holloway BW, Schambock A, Leisinger T.. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 154(1):7–22. [DOI] [PubMed] [Google Scholar]
- He Z, et al. 2016. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44(W1):W236–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilker R, et al. 2015. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol. 17(1):29–46. [DOI] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP.. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212(1):77–86. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K.. 2015. Modulation of protein synthesis by polyamines. IUBMB Life. 67(3):160–169. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K.. 2006. Polyamine Modulon in Escherichia coli: genes involved in the stimulation of cell growth by polyamines. J Biochem. 139(1):11–16. [DOI] [PubMed] [Google Scholar]
- Igarashi K, et al. 1986. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J Bacteriol. 166(1):128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauffrit F, et al. 2016. RiboDB database: a comprehensive resource for prokaryotic systematics. Mol Biol Evol. 33(8):2170–2172. [DOI] [PubMed] [Google Scholar]
- Kamio Y, Nakamura K.. 1987. Putrescine and cadaverine are constituents of peptidoglycan in Veillonella alcalescens and Veillonella parvula. J Bacteriol. 169(6):2881–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y, Poso H, Terawaki Y, Paulin L.. 1986. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J Biol Chem. 261(14):6585–6589. [PubMed] [Google Scholar]
- Kandiah E, et al. 2016. Structural insights into the Escherichia coli lysine decarboxylases and molecular determinants of interaction with the AAA+ ATPase RavA. Sci Rep. 6:24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang IH, Kim JS, Kim EJ, Lee JK.. 2007. Cadaverine protects Vibrio vulnificus from superoxide stress. J Microbiol Biotechnol. 17(1):176–179. [PubMed] [Google Scholar]
- Kanjee U, Gutsche I, Alexopoulos E, et al. 2011. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30(5):931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee U, Gutsche I, Ramachandran S, Houry WA.. 2011. The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry 50(43):9388–9398. [DOI] [PubMed] [Google Scholar]
- Kanjee U, Houry WA.. 2013. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol. 67(1):65–81. [DOI] [PubMed] [Google Scholar]
- Karatan E, Michael AJ.. 2013. A wider role for polyamines in biofilm formation. Biotechnol Lett. 35(11):1715–1717. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Kurahashi O, Nagano T, Kamio Y.. 1998. RpoS-dependent expression of the second lysine decarboxylase gene in Escherichia coli. Biosci Biotechnol Biochem. 62(6):1267–1270. [DOI] [PubMed] [Google Scholar]
- Kim JS, Choi SH, Lee JK.. 2006. Lysine decarboxylase expression by Vibrio vulnificus is induced by SoxR in response to superoxide stress. J Bacteriol. 188(24):8586–8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, et al. 2016. The essential role of spermidine in growth of agrobacterium tumefaciens is determined by the 1, 3-diaminopropane moiety. ACS Chem Biol. 11(2):491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper C, Jung K.. 2005. CadC-mediated activation of the cadBA promoter in Escherichia coli. J Mol Microbiol Biotechnol. 10(1):26–39. [DOI] [PubMed] [Google Scholar]
- Kwon DH, Lu CD.. 2006. Polyamines induce resistance to cationic peptide, aminoglycoside, and quinolone antibiotics in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother. 50(5):1615–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Michael AJ, Martynowski D, Goldsmith EJ, Phillips MA.. 2007. Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases. J Biol Chem. 282(37):27115–27125. [DOI] [PubMed] [Google Scholar]
- Lee YS, Cho YD.. 2001. Identification of essential active-site residues in ornithine decarboxylase of Nicotiana glutinosa decarboxylating both l-ornithine and l-lysine. Biochem J. 360(Pt 3):657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, et al. 2008. Occurrence of agmatine pathway for putrescine synthesis in Selenomonas ruminatium. Biosci Biotechnol Biochem. 72(2):445–455. [DOI] [PubMed] [Google Scholar]
- Lightfoot HL, Hall J.. 2014. Endogenous polyamine function – the RNA perspective. Nucleic Acids Res. 42(18):11275–11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohinai Z, et al. 2015. Biofilm lysine decarboxylase, a new therapeutic target for periodontal inflammation. J Periodontol. 86(10):1176–1184. [DOI] [PubMed] [Google Scholar]
- Lu CD, Yang Z, Li W.. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol. 186(12):3855–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhuri Indurthi S, Chou HT, Lu CD.. 2016. Molecular characterization of lysR-lysXE, gcdR-gcdHG and amaR-amaAB operons for lysine export and catabolism: a comprehensive lysine catabolic network in Pseudomonas aeruginosa PAO1. Microbiology 162(5):876–888. [DOI] [PubMed] [Google Scholar]
- Malet H, et al. 2014. Assembly principles of a unique cage formed by hexameric and decameric E. coli proteins. Elife 3:e03653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel J, Zhanel GG, de Kievit T.. 2010. Cadaverine suppresses persistence to carboxypenicillins in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother. 54(12):5173–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Camilli A.. 2000. Regulation of vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol. 182(19):5342–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AJ. 2016a. Biosynthesis of polyamines and polyamine-containing molecules. Biochem J. 473(15):2315–2329. [DOI] [PubMed] [Google Scholar]
- Michael AJ. 2016b. Polyamines in eukaryotes, bacteria, and archaea. J Biol Chem. 291(29):14896–14903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. 1972. Experiments in molecular genetics In: Press CSHL, editor. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ.. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: oxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 182(16):4533–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo AA, et al. 2012. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 487(7405):109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Clarke PH.. 1980. Genes and enzymes of lysine catabolism in Pseudomonas aeruginosa. J Gen Microbiol. 116(2):357–369. [DOI] [PubMed] [Google Scholar]
- Ramulu HG, et al. 2014. Ribosomal proteins: toward a next generation standard for prokaryotic systematics? Mol Phylogenet Evol. 75:103–117. [DOI] [PubMed] [Google Scholar]
- Romano A, Ladero V, Alvarez MA, Lucas PM.. 2014. Putrescine production via the ornithine decarboxylation pathway improves the acid stress survival of Lactobacillus brevis and is part of a horizontally transferred acid resistance locus. Int J Food Microbiol. 175:14–19. [DOI] [PubMed] [Google Scholar]
- Romano A, Trip H, Lolkema JS, Lucas PM.. 2013. Three-component lysine/ornithine decarboxylation system in Lactobacillus saerimneri 30a. J Bacteriol. 195(6):1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Schuster M, Hawkins AC, Harwood CS, Greenberg EP.. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 51(4):973–985. [DOI] [PubMed] [Google Scholar]
- Sekowska A, Bertin P, Danchin A.. 1998. Characterization of polyamine synthesis pathway in Bacillus subtilis 168. Mol Microbiol. 29(3):851–858. [DOI] [PubMed] [Google Scholar]
- Seo H, Kim KJ.. 2017. Structural basis for a novel type of cytokinin-activating protein. Sci Rep. 7:45985.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin DC, Fuhrer T, Zamboni N, Sauer U.. 2017. Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli. Nat Methods. 14(2):187–194. [DOI] [PubMed] [Google Scholar]
- Shah P, Swiatlo E.. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol. 68(1):4–16. [DOI] [PubMed] [Google Scholar]
- Shan Y, et al. 2017. ATP-dependent persister formation in Escherichia coli. MBio 8 doi: 10.1128/mBio.02267-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J, et al. 2006. Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J Biol Chem. 281(3):1532–1546. [DOI] [PubMed] [Google Scholar]
- Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K.. 2004. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol. 51(5):1401–1412. [DOI] [PubMed] [Google Scholar]
- Sugawara A, et al. 2014. Characterization of a pyridoxal-5'-phosphate-dependent l-lysine decarboxylase/oxidase from Burkholderia sp. AIU 395. J Biosci Bioeng. 118(5):496–501. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H.. 1985. Polyamines in microorganisms. Microbiol Rev. 49:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H, Hafner EW, Tabor CW.. 1980. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 144:952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H, Tabor CW.. 1964. Spermidine, spermine, and related amines. Pharmacol Rev. 16:245–300. [PubMed] [Google Scholar]
- Takatsuka Y, Yamaguchi Y, Ono M, Kamio Y.. 2000. Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylases from eukaryotes. J Bacteriol. 182(23):6732–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J, Faudry E, Ebel C, Attree I, Elsen S.. 2009. Anti-activator ExsD forms a 1:1 complex with ExsA to inhibit transcription of type III secretion operons. J Biol Chem. 284(23):15762–15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valot B, et al. 2015. What it takes to be a Pseudomonas aeruginosa? The core genome of the opportunistic pathogen updated. PLoS One 10(5):e0126468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala JP, et al. 2011. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS One 6(7):e22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Houry WA.. 2010. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem Cell Biol. 88(2):301–314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.