Abstract
Vibrio cholerae is a Gram-negative bacterium found in aquatic environments and a human pathogen of global significance. Its transition between host-associated and environmental lifestyles involves the tight regulation of niche-specific phenotypes such as motility, biofilm formation, and virulence. V. cholerae’s transition from the host to environmental dispersal usually involves suppression of virulence and dispersion of biofilm communities. In contrast to this naturally occurring transition, bacterial aggregation by cationic polymers triggers a unique response, which is to suppress virulence gene expression while also triggering biofilm formation by V. cholerae, an artificial combination of traits that is potentially very useful to bind and neutralize the pathogen from contaminated water. Here, we set out to uncover the mechanistic basis of this polymer-triggered bacterial behavior. We found that bacteria–polymer aggregates undergo rapid autoinduction and achieve quorum sensing at bacterial densities far below those required for autoinduction in the absence of polymers. We demonstrate this induction of quorum sensing is due both to a rapid formation of autoinducer gradients and local enhancement of autoinducer concentrations within bacterial clusters as well as the stimulation of CAI-1 and AI-2 production by aggregated bacteria. We further found that polymers cause an induction of the biofilm-specific regulator VpsR and the biofilm structural protein RbmA, bypassing the usual suppression of biofilm during autoinduction. Overall, this study highlights that synthetic materials can be used to cross-wire natural bacterial responses to achieve a combination of phenotypes with potentially useful applications.
Both natural and synthetic cationic macromolecules, such as cationic antimicrobial peptides, cationic polymers, and dendrimers, have been extensively reported as antimicrobial.1−3 Because of their positive charge, these polymers can efficiently bind the negatively charged envelope of Gram-negative and Gram-positive bacteria.4−6 At high concentrations and charge densities, these molecules have the potential to interfere with membrane integrity and decrease bacterial viability.1−3 However, antimicrobial activity is heavily dependent on the length and nature of the polymer and, more importantly, on the nature of the targeted microbe. At low concentrations, cationic polymers are still capable of causing bacterial aggregation by mediating electrostatic interactions but do so without significantly affecting bacterial membrane integrity and growth.7−12
We and others have previously reported that bacteria clustered by subinhibitory concentrations of cationic polymers display interesting phenotypes resembling those of biofilm communities.7−12 For instance, we have recently demonstrated that clustering of the diarrheal pathogen Vibrio cholerae with methacrylamides containing primary or tertiary amines leads to accumulation of biomass and extracellular DNA and represses ToxT-regulated virulence factors including cholera toxin.12 However, what drives these phenotypic changes in response to polymer exposure remains unclear. Similarly, the marine bacterium Vibrio harveyi shows enhanced bioluminescence in response to polymer-mediated clustering.7,8,11 The bacterial components necessary to produce luminescence are encoded by the luxCDABE genes and subject to complex regulatory mechanisms. A major regulatory cascade controlling luminescence is quorum sensing. Because the regulators controlling luminescence are functionally conserved between Vibrio species, expression of V. harveyi luxCDABE genes can be used as a tool to probe quorum sensing in heterologous hosts.13
In V. cholerae, four parallel quorum sensing pathways operate,14 each governed by an autoinducer synthase, which produces a small, freely diffusible molecule that is released in the environment and can be sensed by a corresponding sensor/kinase component that controls the activity of the LuxU/LuxO phosphorelay. Of the four pathways, the LuxS/LuxPQ system, which produces and detects the interspecies autoinducer AI-2 (S-TMHF-borate) and the CqsA/CqsS system, which produces and senses the Vibrio specific autoinducer S-3-hydroxytridecan-4-one (CAI-1), are the best characterized. CqsA and LuxS synthesize the autoinducers, and the hybrid sensor/kinases CqsS and LuxQ sense and respond to their cognate autoinducers by dephosphorylating and inactivating the regulator LuxO. When extracellular autoinducer concentrations are low, LuxO is phosphorylated and activates the transcription of the small RNAs Qrr1–4, which in turn inhibits the expression of HapR, a master regulator controlling diverse cellular functions including luminescence, biofilm formation, and virulence.15 When the extracellular autoinducer concentration increases above a threshold, the sensor/kinases instead act as phosphatases, leading to dephosphorylation and inactivation of LuxO, ultimately allowing the expression of HapR. Consequently, when autoinducer concentration is high, HapR activates luminescence but suppresses virulence and biofilm genes. This regulatory mechanism is thought to mediate the dissipation of host-associated biofilms and enable transmission of V. cholerae from the intestine to the environment.16 Because in V. harveyi exposure to subinhibitory concentrations of cationic polymers leads to enhanced luminescence, we set out here to study whether V. cholerae would also activate quorum sensing in response to polymer-mediated clustering and how quorum sensing could be related to the observed phenotypic changes, including lowered virulence and enhanced biofilm formation.
Results
Polymers Enhance Quorum Sensing in V. cholerae
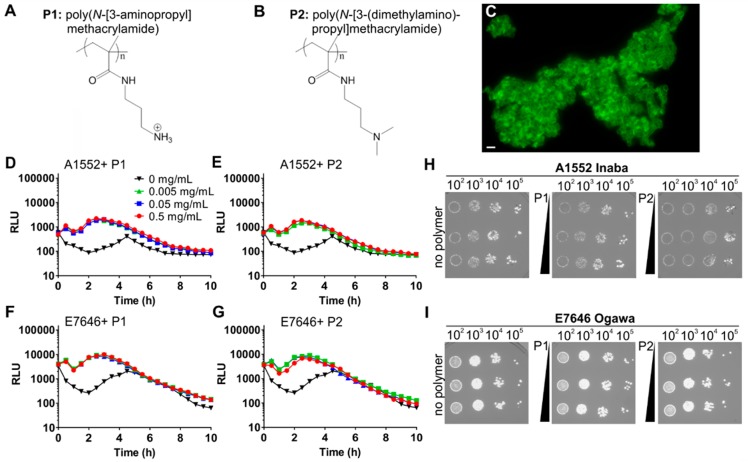
First, we set out to test if clustering by the cationic polymers poly(N-[3-aminopropyl] methacrylamide) (P1, Figure 1A) and poly(N-[3-(dimethylamino)propyl] methacrylamide) (P2, Figure 1B) induced quorum sensing in V. cholerae. As previously shown and detailed here by N-SIM super-resolution microscopy, even low concentrations of polymers induced rapid clustering of bacteria without affecting bacterial viability (Figure 1C). For creating a fast and direct read-out for quorum sensing, the cosmid pBB1, which contains the luxCDABE luminescence genes from V. harveyi,17 was used to transform V. cholerae serogroup O1 biovar El Tor strain A1552 (serotype Inaba). The transformed bacteria were then grown in LB for 20 h, washed with artificial marine water (AMW), and adjusted to an OD of 0.2 in AMW alone or AMW containing polymers at concentrations ranging from 0.005 to 0.5 mg mL–1. In the absence of polymer, luminescence as a read-out of quorum sensing first decreased due to back-dilution of the culture from a high density overnight culture to OD 0.2 and then gradually increased due to accumulation of autoinducers, peaking at 4.5 h (Figure 1D and E, black traces). Interestingly, the quorum induction kinetics were significantly different in the presence of either P1 (Figure 1D) or P2 (Figure 1E) with initial luminescence being sustained and reaching a higher second peak at ∼2.5 h as opposed to 4.5 h in the absence of polymers. The magnitude of induction was higher in polymer-treated cultures (∼5-fold at peak quorum induction) compared to untreated cultures. Results were similar for the Ogawa serotype strain E7646 in that quorum sensing was initially sustained and then further enhanced (∼4-fold at peak quorum induction) by polymer-mediated bacterial aggregation (Figure 1F, G). In both strains, the magnitude of luminescence induction was independent of the polymer concentration used, suggesting a threshold response.
Figure 1.
Polymers enhance quorum sensing in Vibrio cholerae. Chemical structures of P1 (A) and P2 (B). N-SIM super-resolution image of LIVE/DEAD-stained V. cholerae A1552 clustered by P1 (C). V. cholerae El Tor strains A1552 (D, E, H) or E7646 (F, G, I) containing the luminescence reporter pBB1 were adjusted to an OD600 of 0.2 following 16 h of growth and incubated in AMW alone (black) or AMW-containing polymer P1 (D, F) or P2 (E, G) at concentrations of 0.005 (green), 0.05 (blue), or 0.5 (red) mg mL–1. Luminescence was recorded every 30 min for 10 h and plotted as means ± SEM from at least three biological replicates. For the effect of polymers on bacterial viability to be tested, samples were removed after 10 h, serially diluted, and plated on LB (H, I).
For assessing viable bacterial counts at the experimental end point, samples were serially diluted in high-salt buffer to disrupt clusters as previously described12 and plated. Similar numbers of colony-forming units were recovered from untreated or polymer-treated samples (Figure 1H, I), suggesting that the presence of polymers did not affect bacterial viability or proliferation in agreement with our previous data.12 Taken together, our data demonstrate that these cationic polymers that cluster V. cholerae modulate the community behavior to give an accelerated and more robust autoinduction.
Bacterial Density Shapes the Kinetics of Quorum Induction in Response to Polymer
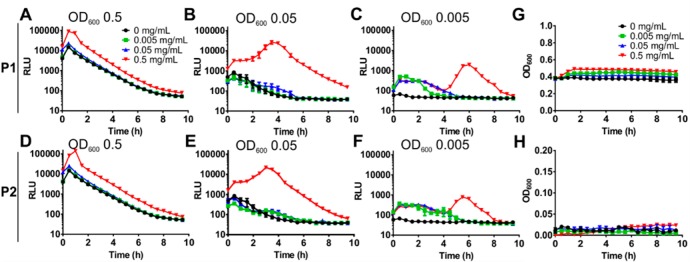
Because quorum sensing is usually tightly linked to bacterial density, we set out to explore how initial culture density affects quorum induction in the presence of polymers. V. cholerae carrying pBB1 as a quorum sensing reporter was adjusted to optical densities of 0.005, 0.05, and 0.5 in AMW alone or AMW containing 0.005–0.5 mg mL–1 P1 or P2, and luminescence was monitored (Figure 2). The onset of autoinduction was not significantly modulated by the addition of polymers P1 or P2 to higher density cultures (OD600 of 0.5), but an approximate 6- (P1) to 9-fold (P2) increase in peak luminescence was observed (Figure 2A, D). At lower culture densities, autoinduction of the untreated cultures was less pronounced (0.05) and eventually ceased (OD 0.005) because not enough autoinducer accumulated to reach the threshold concentration. In the presence of polymers, the initial quorum present in the culture was sustained, even in very dilute cultures, and the enhancement in peak luminescence was much more pronounced compared to that of untreated cultures (Figure 2B–F). Interestingly, clustering of very dilute cultures led to two peaks in luminescence with a gap of ∼4 h (Figure 2C, F). Of note, the luminescence was not a result of bacterial growth, which was negligible under the observed conditions (AMW) and over the time frame described, both at high and low initial densities (Figure 2G, H).
Figure 2.
Bacterial density shapes the kinetics of quorum induction in response to polymer. Cultures of V. cholerae A1552 containing pBB1 were grown for 16 h and diluted into AMW alone (black) or AMW containing 0.005 (green), 0.05 (blue), or 0.5 (red) mg mL–1 of P1 (A–C) or P2 (D–F). Bacterial densities were adjusted to result in OD600 values of 0.5 (A, D), 0.05 (B, E), and 0.005 (C, F), respectively. Luminescence and OD600 were recorded every 30 min for 10 h, and values are means ± SEM from at least three biological replicates. No significant growth was detected over this time frame at either initial density of 0.5 (G) or 0.005 (H).
For the process of bacterial clustering and luminescence induction to be visualized, V. cholerae A1552 containing pBB1 was incubated in the presence of P1, P2, or AMW alone in glass-bottom plates. Bacteria were imaged every 30 min to simultaneously visualize clustering and luminescence induction. Both P1 and P2 bacterial clusters were observed within minutes and remained stable over the duration of the experiment (Figure 3). Luminescence appears to originate from and be restricted to bacterial clusters. Quantification of luminescence based on integrated pixel intensities showed a polymer-mediated enhancement of autoinduction (Figure 3H) in agreement with spectroscopic data.
Figure 3.
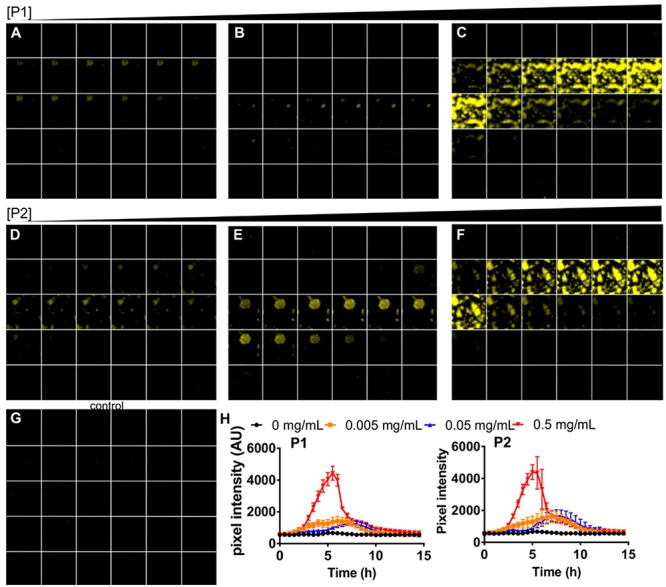
Polymers enhance quorum sensing in V. cholerae. V. cholerae A1552 containing pBB1 was grown for 16 h and then adjusted to an OD600 of 0.2 in the presence of P1 (A–C), P2 (D–F), or AMW alone (G). Polymers were adjusted to final concentrations of 0.005 (A, D), 0.05 (B, E), or 0.5 (C, F) mg mL–1 in AMW. Luminescence of samples was imaged every 30 min for 15 h, and representative images for each time point are shown from 0 h (top left) to 15 h (bottom right) of each panel. Luminescence intensities over time were analyzed by quantifying pixel intensities, and means ± SEM from at least three biological replicates are shown for P1 (left) and P2 (right panel) (H).
Polymer-Mediated Luminescence Is Not Because of Nutrient Starvation within Clusters
Although autoinduction controls HapR and luminescence via the regulator LuxO, other environmental cues, including nutrient availability, have been reported to feed into the LuxO signaling cascade and thus have the potential to affect luminescence. Nutrient sensing and the LuxO signaling pathway converge at the cyclic AMP (cAMP) receptor protein (CRP).18 During limitation of PTS sugars such as glucose, cAMP-CRP is capable of modulating LuxO activation by affecting the expression of autoinducer synthases.19 Because clustering of bacteria by polymers may limit nutrient access and induce starvation, we tested whether a CRP deletion strain would be capable of activating luminescence in the presence of polymers. The V. cholerae Δcrp strain was transformed with pBB1 to monitor luminescence. However, the culture produced very low levels of luminescence both in the presence and absence of polymers (Figure 4A, B), which is in agreement with previous work on V. fischeri CRP.20 Although this suggests that cross-talk between CRP and LuxO signaling is a dominant cue for luminescence induction in V. cholerae as well, this made it unfeasible to determine whether the presence of polymers induced a state of carbon starvation, leading to luminescence induction via CRP. Instead, we repeated the experiment using the pBB1 containing V. cholerae wild-type strain and supplementing the AMW with additional glucose. If clustering would limit nutrient diffusion, increasing the nutrient concentration should be able to overcome this and revert the bacteria to a nonluminescent phenotype. However, polymers still enhanced and sustained luminescence in the presence of 1% glucose to a similar extent as in AMW alone (Figure 4C), suggesting that the effect was not due to nutrient limitation in the clusters.
Figure 4.
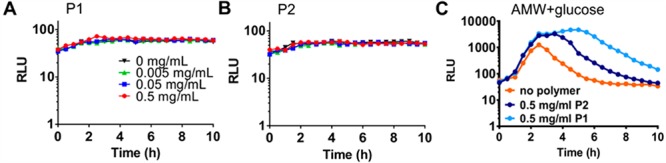
Polymer-mediated luminescence is not due to nutrient starvation within clusters. V. cholerae E7646 Δcrp containing the luminescence reporter pBB1 was grown for 16 h and then diluted to an OD600 of 0.2 in AMW alone (black) or AMW containing P1 (A) or P2 (B) at concentrations of 0.005 (green), 0.05 (blue), or 0.5 (red) mg mL–1. Luminescence was recorded every 30 min for 10 h, and means ± SEM from at least three biological replicates are shown. (C) V. cholerae E7646 wild-type containing pBB1 was grown for 16 h and adjusted to an OD600 of 0.2 in AMW containing 1% glucose alone (orange) or in the presence of 0.5 mg mL–1 of P1 (light blue) or P2 (dark blue). Luminescence was recorded every 30 min for 10 h, and means ± SEM from at least three biological replicates are shown.
Polymer-Mediated Enhancement of Quorum Sensing Is Dominated by CAI-1
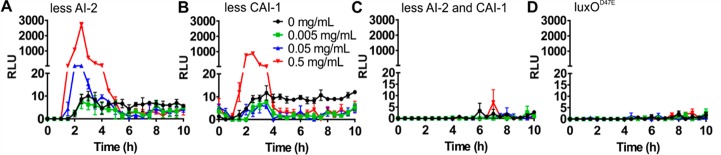
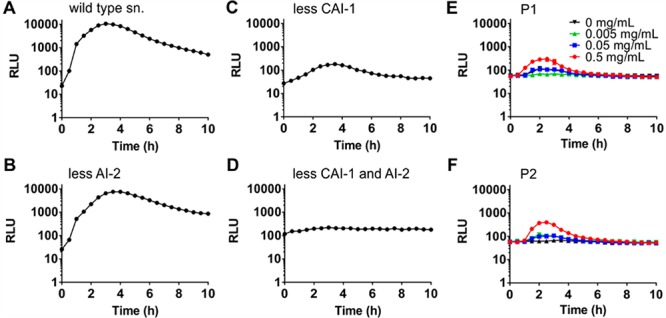
In V. cholerae, at least four parallel quorum sensing pathways converge to control the activity of the quorum sensing regulator LuxO and thus quorum-regulated phenotypes including biofilm formation and virulence.14 We set out to test whether the polymer-mediated effect on quorum sensing was specific for any one pathway. V. cholerae wild-type strain A1552 was mixed with equal numbers of cells of quorum sensing mutants transformed with pBB1 either deficient in the production of AI-2 (DH231) or CAI-1 (WN1103) to give a final OD600 of 0.2. Additionally, the mutants were unable to sense the presence of AI-2 or CAI-1. Cultures producing less AI-2 produced a luminescence profile similar in shape and magnitude to quorum sensing proficient cells in response to polymer P1 addition (Figure 5A). In contrast, cultures producing less CAI-1 still had a comparable response profile upon addition of P1, but the magnitude of the luminescence enhancement was decreased (Figure 5B) compared to that of cultures producing less AI-2 (Figure 5A). In cocultures producing less of both autoinducers (containing strain BH1578), the luminescence response to polymer was abolished (Figure 5C). Similarly, cocultures containing a low-density locked mutant of the downstream quorum regulator LuxO (LuxOD47E) showed no luminescence induction upon addition of polymer P1 (Figure 5D). These data pointed at both autoinducers being involved in the quorum sensing enhancement in response to polymer with CAI-1 being the dominant inducer. Additionally, the luminescence response to the polymer seems to proceed through the canonical LuxO-dependent pathway.
Figure 5.
Polymer-mediated enhancement of quorum sensing is mainly driven by CAI-1. V. cholerae A1552 wild-type (dark) and quorum sensing mutants containing pBB1 were grown for 16 h and then diluted into AMW to give equal cell densities and a total OD600 of 0.2. Strains were grown together in AMW alone (black) or AMW containing P1 at 0.005 (green), 0.05 (blue), or 0.5 (red) mg mL–1. Luminescence was recorded every 30 min for 10 h, and means ± SEM from at least three biological replicates are shown. Mutants grown in coculture with the wild-type were (A) DH231 (ΔluxSΔcqsS) producing no AI-2, (B) WN1103 (ΔluxQΔcqsA) producing no CAI-1, (C) BH1578 (ΔluxSΔcqsA) producing no AI-2 or CAI-1, and (D) BH1651 (luxOD47E).
Enhanced Quorum Sensing Is Driven by Enhanced Production of Autoinducers in Response to Polymers
It has been described that, at least under some conditions, quorum sensing may be subject to positive feedback, where quorum induction leads to increased production of one of the autoinducer synthases.21 Therefore, the enhancement in luminescence in response to polymers could be due to positive feedback as a result of the polymers increasing the local concentration of autoinducers above the threshold. Alternatively, the polymers could have a direct effect on the production of autoinducers. We set out to test this by establishing a reporter assay that allowed us to decouple quorum sensing from the production of autoinducers. For this assay, we used as a luminescence reporter a V. cholerae strain transformed with pBB1 that could sense both CAI-1 and AI-2 but could not produce either molecule (BH1578). This reporter strain was exposed to supernatants from producer strains grown under different conditions to evaluate the effect of the polymer. Initially, we evaluated the assay by growing the reporter in the presence of supernatants harvested from wild-type V. cholerae or strains incapable of producing AI-2, CAI-1, or both autoinducers. Supernatant harvested from the quorum-proficient wild-type strain grown to high cell density triggered the highest level of luminescence in BH1578 (Figure 6A). The luminescence triggered by the AI-2-deficient strain was slightly decreased (Figure 6B), whereas luminescence was significantly decreased in response to supernatant from the CAI-1-deficient strain (Figure 6C) and was abolished in response to the strain deficient in both CAI-1 and AI-2 production (Figure 6D). Hence, the assay was capable of detecting different levels of autoinducers produced by a second strain.
Figure 6.
Enhanced quorum sensing is driven by enhanced production of autoinducers in response to polymers. Cultures of V. cholerae were adjusted to an OD of 0.2 and grown for 16 h in LB medium, and the supernatants were harvested, filtered, and incubated with V. cholerae BH1578 containing pBB1. Strains used to harvest supernatants were (A) wild-type A1552, (B) DH231 (ΔluxSΔcqs), (C) WN1103 (ΔluxQΔcqsA), and (D) BH1578 ((ΔluxSΔcqsA). Luminescence was recorded every 30 min for 10 h, and means ± SEM from at least three biological replicates are shown. V. cholerae wild-type was adjusted to an OD600 of 0.2 in AMW alone or AMW containing 0.005–0.5 mg mL–1 P1 (E) or P2 (F), and the supernatants were harvested and filtered 16 h later. For their autoinducer content to be determined, the supernatants were incubated with the reporter strain V. cholerae BH1578 containing pBB1. Luminescence was recorded every 30 min for 10 h, and means ± SEM from at least three biological replicates are shown.
We took this assay forward and harvested supernatants from wild-type cells exposed to AMW alone or AMW containing 0.005–0.5 mg mL–1 polymers P1 or P2, and exposed the reporter strain to filtered supernatants to test if the levels of autoinducers produced by the wild-type strain in the presence of polymers were different. The reporter strain was not clustered under the assay conditions. Although wild-type cells exposed to AMW alone did not produce a detectable amount of autoinducer and thus no significant luminescence reading in the reporter strain, both polymers P1 and P2 enhanced the production of autoinducers by wild-type V. cholerae, leading to an increase in luminescence upon exposure of the reporter to the supernatants (Figure 6E, F). These data demonstrate that polymer exposure leads to enhanced production of autoinducers by the bacteria.
Polymer-Mediated Quorum Induction Overrides the Canonical Biofilm Dissipation Program in V. cholerae
Contrary to many bacteria that use quorum signaling as a means to induce biofilm formation, in V. cholerae autoinduction promotes repression of biofilm production and dissemination via the regulator HapR.22 However, we had previously observed enhanced accumulation of V. cholerae upon prolonged exposure to cationic polymers, but whether this was accompanied by transcriptional changes at the level of biofilm production was not known. Thus, we grew V. cholerae containing transcriptional fusions to promoters regulating key biofilm components in the presence or absence of P1 and P2 (Figure 7). V. cholerae biofilms contain the structural protein RbmA and require the regulator VpsR, which controls the expression of the vps polysaccharide biosynthesis genes.23−25 Upon exposure to either P1 or P2, vpsR and rbmA were both significantly induced (Figure 7A, B), suggesting that upon polymer-mediated clustering, quorum sensing does not, as usual, suppress genes involved in biofilm production, but instead their transcription is enhanced. AphA, which is another direct target usually induced by VpsR, is suppressed in the presence of polymers (Figure 7C).
Figure 7.
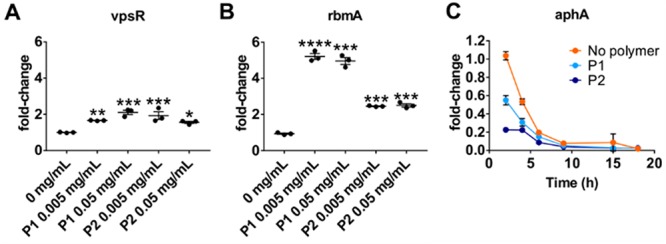
Polymer-mediated quorum induction overrides the canonical biofilm dissipation program in V. cholerae. V. cholerae wild-type A1552 containing pRW50T lacZ reporters for the promoters of rbmA (A) or vpsR (B) were grown for 16 h and then diluted into AMW alone or containing 0.005 or 0.05 mg mL–1 P1 or P2 as indicated to give an OD600 of 0.2. Following 16 h of incubation, clustered bacteria were removed and either processed for β-galactosidase assays or treated with high-salt PBS to disperse the cultures for OD600 measurements. Transcriptional activities were calculated and normalized to untreated cultures. Shown are means ± SEM and individual measurements for three biological replicates. Statistical significance was determined by ANOVA and a Dunnett’s multiple comparison test and is depicted as ****p-values ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05, and ns for not significant (p ≥ 0.05). (C) V. cholerae wild-type A1552 containing pRW50T lacZ reporter for the aphA promoter was grown in AMW alone (orange) or containing P1 (light blue) at 0.05 mg mL–1 or P2 (dark blue) at 0.5 mg mL–1 for 18 h. Clustered bacteria were removed at indicated times and either processed for β-galactosidase assays or treated with high-salt PBS to disperse the cultures for OD600 measurements. Transcriptional activities were calculated and normalized to the activities of untreated cultures at 2 h. Shown are means ± SEM and individual measurements for three biological replicates.
Discussion
Traditionally, work on cationic polymers has been carried out with the development of antimicrobial materials as a main goal.1−3 However, recent work by our groups and others has demonstrated that such cationic polymers can be titrated against bacteria to achieve a charge balance that allows for the rapid and efficient clustering of bacteria but avoids membrane disruption and bacterial cell death.7−12
The use of cationic polymers to induce rapid bacterial clustering in this way has proven as an interesting path to study effects of cell aggregation and crowding on bacterial physiology. Although such behaviors are often studied in batch cultures, by incubating bacterial cultures over a prolonged time, this means aggregation is accompanied by bacterial growth and eventually nutrient limitation, which makes it difficult to establish the primary cause of the observed phenotypes. In contrast, cationic polymers induce cell aggregation rapidly, within minutes, which allows us to study these phenomena independent of cellular proliferation and nutrient limitation.
We and others have previously observed that cationic polymers and dendrimers can, under certain conditions, trigger bioluminescence in the marine bacterium V. harveyi, suggesting they may induce or enhance quorum sensing.7,8,10,11 In a more recent study where we extended this work to the human pathogen V. cholerae, we observed that polymer-mediated clustering led to enhanced deposition of biomass and extracellular DNA, whereas it interfered with the induction of virulence genes in an infection model.12 Because virulence and biofilm production are both regulated by quorum sensing but are usually both regulated concurrently, the goal of this study was to test whether cationic polymers would trigger quorum sensing in V. cholerae and how this would affect downstream transcription of biofilm genes.
We used V. cholerae strains heterologously expressing the luxCDABE luminescence genes (on cosmid pBB1) from V. harveyi to be able to use luminescence as a direct readout for autoinduction. Over 16 h, V. cholerae would grow to high cell densities and as a result was strongly luminescent. Upon dilution into artificial marine water, cell density and autoinducer concentration would rapidly decrease, resulting in a decline in luminescence. After several hours, cells would eventually accumulate sufficient autoinducer to reach the quorum threshold and induce luminescence again. This behavior was observed in AMW alone (Figure 1) and is in agreement with commonly observed results from such experiments.14,21 In contrast, when cells were diluted into media containing polymers, they would undergo extensive clustering almost instantaneously, and luminescence readouts never dropped but instead further increased immediately (Figure 1), suggesting that clustering not only countered the dilution effect but further increased autoinducer concentration within the clusters. Interestingly, this behavior was observed over a broad space of cell densities (at least 2 orders of magnitude), including in dilute cultures that did not by themselves experience autoinduction (Figure 2C, F), suggesting that during clustering polymers create pockets containing strongly increased concentrations of autoinducers around bacterial aggregates.
We further demonstrated that both CAI-1- and AI-2-dependent quorum sensing cascades are activated in response to polymers and that clustering leads to an enhanced production of both autoinducers (Figures 5 and 6). The effect of the Vibrio-specific autoinducer CAI-1 dominated the clustering-driven luminescence phenotype (Figure 5) in line with previous results obtained for batch cultures of V. cholerae in rich medium.21
Some studies have hypothesized that luminescence could be a result of limited diffusion of nutrients in the polymer-mediated bacterial aggregates.10 Catabolite repression of luminescence has been reported for V. fischeri, where cAMP-CRP stimulates luxCDABE expression.26 However, this effect is alleviated by high concentrations of autoinducer.20 In our hands, CRP was essential for luminescence, both triggered by high cell density in the absence of polymers, in line with previous findings for an E. coli Δcrp mutant,20 as well as in response to polymer-induced clustering. Additionally, supplementation of the media with excess glucose did not quench luminescence even in the absence of autoinduction. This suggests that nutrient limitation within the clusters is not a major cue for luminescence induction but further underpins that cross-talk between nutrient sensing and quorum sensing pathways exists.
Finally, we followed up on our earlier observation that exposure to cationic polymers causes deposition of V. cholerae on inorganic surfaces and release of extracellular DNA, both hallmarks of biofilm formation.12 Here, we showed that this phenotype is the result of transcriptional activation of genes involved in biofilm production in response to polymer exposure. Biofilm induction may explain the enhanced resistance toward antimicrobials of bacteria that have been exposed to cationic polymers, as previously described by others.10 The expression of the biofilm regulator VpsR and the biofilm structural protein RbmA were both induced upon exposure to the polymers (Figure 7). This upregulation is in contrast to the canonical biofilm regulation where biofilm genes are repressed during autoinduction. VpsR is a master regulator of biofilm formation and a two component system response regulator. Although no cognate histidine kinase has been identified, VpsR is epistatic to the intracellular hybrid sensor histidine kinase VpsS.27 Induction of vpsR likely leads to the downstream induction of rbmA we observed here because rbmA is a direct target of VpsR regulation.28 However, vpsR induction in the presence of polymers seems to happen despite autoinduction, which should normally lead to suppression of vpsR. What is also different from a regular biofilm response is that VpsR in the presence of polymer fails to upregulate one of its other direct targets, aphA. We showed that, in contrast to this canonical response, aphA is strongly suppressed by the presence of polymers (Figure 7C). When Shikuma et al. identified VpsS as a regulator of VpsR, they established the existence of a pathway that proceeds from VpsS through the quorum regulators LuxU and LuxO and results in the VpsR-dependent activation of biofilm production independent of HapR.27 It may be that, in the presence of polymers, this pathway is active and dominates the effects of the CAI-1 and AI-2 pathways on biofilm. Unfortunately, the cognate signal activating VpsS is as yet unidentified.
Conclusions
We showed here that clustering of V. cholerae in response to cationic polymers leads to autoinduction due to a rapid increase of local autoinducer concentration in the vicinity of aggregated bacteria. Moreover, we demonstrate that stimulation of further autoinducer synthesis is also observed and involves at least two of the four known quorum sensing systems, CAI-1 and AI-2. We speculate that the third quorum sensing pathway, which proceeds through the intracellular hybrid sensor kinase VpsS,14,27 is also activated and leads to the production of biofilm in response to polymer-driven aggregation. Our previous work together with the data presented here rules out membrane disruption and nutrient limitation within clusters, respectively, as cues leading to the phenotypes observed here. Our future work will aim to further dissect the pathway(s) triggered in response to polymer exposure to clarify whether VpsS is indeed involved and activated in response to polymers. Polymeric materials that inhibit bacterial dissemination, both mechanically and transcriptionally, may be useful for applications to enhance wastewater treatment for the decontamination of water from V. cholerae.
Materials and Methods
Bacterial Strains and Culture Conditions
V. cholerae El Tor strains used in this study (Table 1) were derived from A155229 and E7946.30 The E. coli K12 strains JCB38731 DH5α32 and SM10 λpir33 were used for general cloning and conjugation procedures. Strains were propagated at 37 °C in lysogeny broth (LB) supplemented with 10 μg/μL of tetracycline or 30 μg/μL of kanamycin for selection when required. Plasmids were introduced into V. cholerae strains by triparental mating with E. coli DH5α carrying the desired plasmid (donor) and E. coli SM10 (helper strain) carrying the conjugative machinery on pRK2013. Cultures were mixed at a volumetric ratio of 1:2:2 of recipient:helper:donor in 250 μL and spotted onto brain-heart infusion (BHI) agar for incubation overnight at 37 °C. Spots of bacteria were dislodged after an overnight incubation and resuspended in 3 mL of sterile PBS. Then, 100 μL of serial dilutions were plated onto TCBS plates containing 10 μg/μL of tetracycline. The resulting colonies were checked by PCR in the case of pRW50T constructs, and pBB1 transconjugants were screened for luminescence (Table 2).
Table 1. Bacterial Strains Used in This Study.
| strain | description or genotype | source or ref |
|---|---|---|
| Vibrio cholerae | ||
| A1552 | wild-type; O1 biovar El Tor serotype Inaba | (29) |
| E7956 | wild-type; O1 biovar El Tor serotype Ogawa | (30) |
| BH1651 | luxOD47E | (15) |
| BH1578 | ΔluxSΔcqsA | (36) |
| DH231 | ΔluxSΔcqsS | (37) |
| WN1103 | ΔluxQΔcqsA | (37) |
| E7956 Δcrp | Δcrp KanR | gift from D. Grainger |
| NP5005 | A1552 pRW50T containing upstream region of aphA promoter; TetR | (12) |
| Escherichia coli | ||
| DH5α | donor and maintenance of pBB1 | (32) |
| JCB387 | donor and maintenance of pRW50T | (31) |
| SM10 | helper strain; λpir pRK2013; KanR | (33) |
Table 2. Plasmids Used in This Study.
| plasmid | description | source or ref |
|---|---|---|
| pRW50T | pRW50 derivative with a oriT sequence from pRK2; TetR | gift from D. Grainger |
| pRW50T-rbmA | pRW50T containing 273 bp of the upstream region of rbmA, cloned between EcoRI and HindIII restriction sites; TetR | this study |
| pRW50T-vpsR | pRW50T containing 195 bp of the upstream region of vpsR, cloned between EcoRI and HindIII restriction sites; TetR | this study |
| pRW50T-aphA | pRW50T containing the upstream region of aphA, cloned between EcoRI and HindIII sites; TetR | (12) |
| pBB1 | luxCDABE cosmid; TetR | (35) |
β-Galactosidase Assays
pRW50T derivative construction was described before.12 Regions encoding aphA, rbmA, or vpsR promoters were amplified by PCR and cloned into pRW50T using EcoRI and HindIII sites. The insertion was checked by PCR using external primers. Measurement of β-galactosidase activity as a readout for transcriptional activity was done as previously described34 with some modifications to accommodate testing of aggregated bacteria. Small cultures of reporter strains were grown in the absence or presence of polymers P1 and P2 and incubated overnight at 37 °C with shaking. Clustered bacteria were split in two and either used for transcriptional assays or washed with high salt PBS (200 mM NaCl) to disrupt aggregation and enable OD600 measurements.
Luminescence Assays
Luminescence assays were done using V. cholerae pBB1 transconjugants. The pBB1 cosmid35 was introduced into V. cholerae strains by triparental mating in the same conditions as for pRW50T. Overnight cultures of V. cholerae pBB1 were adjusted to OD600 of 0.5, 0.1, and 0.01 in artificial marine water with 10 μg/μL of tetracycline. Polymers P1 and P2 were added at concentrations of 0.005, 0.05, and 0.5 mg mL–1 in DMEM or AMW in 200 μL final volume using a dark-wall clear-bottom 96-well plate. The plate was incubated up to 15 h at 37 °C with shaking a 200 rpm, whereas luminescence and OD600 were recorded every 30 min using a FLUOstar Omega plate reader. The following assays were done with bacterial cultures with OD600 adjusted to 0.2. Cells were recovered after the assay and washed with high-salt PBS containing 200 mM NaCl to disrupt charge-based aggregation and plated onto LB with tetracycline to determine the viability. Plates were imaged using a BioRad Gel Doc XR System, and the images were processed with ImageJ.
Luminescence Time-Lapse Imaging
Overnight culture of V. cholerae A1552 pBB1 was diluted to an OD600 of 0.2 in artificial marine water or clear DMEM with 10 μg/μL of tetracycline and polymers at concentrations of 0.005, 0.05, and 0.5 mg mL–1. Samples were prepared in 200 μL using a glass-bottom 96-well plate and incubated at 37 °C with 5% CO2 in a microscope imaging chamber. Images were taken every 30 min with 10 s of exposure at 40× magnification using an Evolve 512 EMCCD camera mounted on a Nikon-Eclipse TE2000-U microscope. Image acquisition was done using Nikon NIS-Elements software, and final images were processed with ImageJ. Pixel intensity was determined from several clusters within frame using ImageJ.
Super-Resolution Microscopy of Bacterial Clusters
V. cholerae A1552 was incubated with 0.05 mg mL–1 P1 in PBS for 1 h. For membrane integrity to be visualized, the sample was stained using the LIVE/DEAD BacLight Bacterial Viability Kit (Life Technologies) for 10 min at RT. The sample was mounted with ProLong Gold Antifade Mountant and covered with a coverslip. Images were taken on a Nikon N-SIM super-resolution microscope fitted with SR Apo TIRF 100× lens at 100 ms exposure. Deconvolution was carried out using the Nikon NIS elements software.
Luminescence Assays Using V. cholerae BH1578 pBB1 as a Reporter
V. cholerae BH1578 pBB1 was used to determine the effect of polymers on the production of autoinducers. V. cholerae strains at an OD600 of 0.2 were clustered with polymers at concentrations of 0.005, 0.05, and 0.5 mg mL–1 in artificial marine water. Supernatants were recovered by centrifugation and used to resuspend V. cholerae BH1578 pBB1 previously adjusted to an OD600 of 0.2 in 200 μL. Luminescence was recorded at 37 °C using a FLUOStar Omega plate reader. Similarly, V. cholerae strains and V. cholerae BH1578 pBB1 were cocultured in 200 μL final volume, and polymers were added at concentrations of 0.005, 0.05, and 0.5 mg mL–1 in artificial marine water. Both strains were adjusted to a final OD600 of 0.1 each (0.2 total density). Incubation was done at 37 °C with shaking at 200 rpm, and luminescence and OD600 were measured every 30 min using a FLUOStar Omega plate reader.
Acknowledgments
We thank B. Bassler for the generous gift of quorum sensing mutants BH1578, DH23, and WN1103 and the cosmid pBB1. We thank H. Kaplan and the Krachler and Fernandez-Trillo laboratories for critical reading of the manuscript and for suggestions on how to improve this study. This work was supported by University of Birmingham Fellowships (to A.M.K. and F.F.-T.), Wellcome Trust grant 177ISSFPP (to A.M.K. and F.F.-T), BBSRC grants BB/M021513/1 (to A.M.K.) and BB/L007916/1 (to A.M.K.), a CONICYT fellowship (to N.P.-S.), BBSRC MIBTP scholarship BB/M01116X/1 (to O.C.), and a UT Systems Science and Technology Acquisition and Retention Award (to A.M.K.).
The authors declare no competing financial interest.
References
- Hartlieb M.; Williams E. G. L.; Kuroki A.; Perrier S.; Locock K. E. S. (2017) Antimicrobial Polymers: Mimicking Amino Acid Functionality, Sequence Control and Three-dimensional Structure of Host-defense Peptides. Curr. Med. Chem. 24, 2115–2140. 10.2174/0929867324666170116122322. [DOI] [PubMed] [Google Scholar]
- Palermo E. F.; Kuroda K. (2009) Chemical structure of cationic groups in amphiphilic polymethacrylates modulates the antimicrobial and hemolytic activities. Biomacromolecules 10, 1416–1428. 10.1021/bm900044x. [DOI] [PubMed] [Google Scholar]
- Tew G. N.; Liu D.; Chen B.; Doerksen R. J.; Kaplan J.; Carroll P. J.; Klein M. L.; DeGrado W. F. (2002) De novo design of biomimetic antimicrobial polymers. Proc. Natl. Acad. Sci. U. S. A. 99, 5110–5114. 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevc G. (1990) Membrane electrostatics. Biochim. Biophys. Acta, Rev. Biomembr. 1031, 311–382. 10.1016/0304-4157(90)90015-5. [DOI] [PubMed] [Google Scholar]
- Sonohara R.; Muramatsu N.; Ohshima H.; Kondo T. (1995) Difference in surface properties between Escherichia coli and Staphylococcus aureus as revealed by electrophoretic mobility measurements. Biophys. Chem. 55, 273–277. 10.1016/0301-4622(95)00004-H. [DOI] [PubMed] [Google Scholar]
- Devi U. V.; Puri P.; Sharma N. N.; Ananthasubramanian M. (2014) Electrokinetics of Cells in Dielectrophoretic Separation: A Biological Perspective. BioNanoScience 4, 276–287. 10.1007/s12668-014-0140-y. [DOI] [Google Scholar]
- Xue X.; Pasparakis G.; Halliday N.; Winzer K.; Howdle S. M.; Cramphorn C. J.; Cameron N. R.; Gardner P. M.; Davis B. G.; Fernandez-Trillo F.; Alexander C. (2011) Synthetic polymers for simultaneous bacterial sequestration and quorum sense interference. Angew. Chem., Int. Ed. 50, 9852–9856. 10.1002/anie.201103130. [DOI] [PubMed] [Google Scholar]
- Lui L. T.; Xue X.; Sui C.; Brown A.; Pritchard D. I.; Halliday N.; Winzer K.; Howdle S. M.; Fernandez-Trillo F.; Krasnogor N.; Alexander C. (2013) Bacteria clustering by polymers induces the expression of quorum-sensing-controlled phenotypes. Nat. Chem. 5, 1058–1065. 10.1038/nchem.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louzao I.; Sui C.; Winzer K.; Fernandez-Trillo F.; Alexander C. (2015) Cationic polymer mediated bacterial clustering: Cell-adhesive properties of homo- and copolymers. Eur. J. Pharm. Biopharm. 95, 47–62. 10.1016/j.ejpb.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Lu H.; Chen H.; Zhang J.; Liu L.; Lv F.; Wang S. (2016) Cationic Conjugated Polymers-Induced Quorum Sensing of Bacteria Cells. Anal. Chem. 88, 2985–2988. 10.1021/acs.analchem.5b03920. [DOI] [PubMed] [Google Scholar]
- Leire E.; Amaral S. P.; Louzao I.; Winzer K.; Alexander C.; Fernandez-Megia E.; Fernandez-Trillo F. (2016) Dendrimer mediated clustering of bacteria: improved aggregation and evaluation of bacterial response and viability. Biomater. Sci. 4, 998–1006. 10.1039/C6BM00079G. [DOI] [PubMed] [Google Scholar]
- Perez-Soto N.; Moule L.; Crisan D. N.; Insua I.; Taylor-Smith L. M.; Voelz K.; Fernandez-Trillo F.; Krachler A. M. (2017) Engineering microbial physiology with synthetic polymers: cationic polymers induce biofilm formation in Vibrio cholerae and downregulate the expression of virulence genes. Chem. Sci. 8, 5291–5298. 10.1039/C7SC00615B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B.; Skorupski K.; Lenz D. H.; Taylor R. K.; Bassler B. L. (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Jung S. A.; Chapman C. A.; Ng W. L. (2015) Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog. 11, e1004837 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen S. L.; Waters C. M.; Bassler B. L. (2008) A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode. Genes Dev. 22, 226–238. 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschler J. K.; Zamorano-Sanchez D.; Utada A. S.; Warner C. J.; Wong G. C.; Linington R. G.; Yildiz F. H. (2015) Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 13, 255–268. 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J.; Silverman M. (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 81, 4154–4158. 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J.; Benitez J. A. (2004) Transcriptional regulation of Vibrio cholerae hemagglutinin/protease by the cyclic AMP receptor protein and RpoS. J. Bact. 186, 6374–6382. 10.1128/JB.186.19.6374-6382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.; Pascual-Montano A.; Silva A. J.; Benitez J. A. (2007) The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153, 2964–2975. 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- Dunlap P. V.; Greenberg E. P. (1988) Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-luxR protein regulatory circuit. J. Bacteriol. 170, 4040–4046. 10.1128/jb.170.9.4040-4046.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley A.; Bassler B. L. (2017) Asymmetric regulation of quorum-sensing receptors drives autoinducer-specific gene expression programs in Vibrio cholerae. PLoS Genet. 13, e1006826 10.1371/journal.pgen.1006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Miller M. B.; Vance R. E.; Dziejman M.; Bassler B. L.; Mekalanos J. J. (2002) Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99, 3129–3134. 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H.; Dolganov N. A.; Schoolnik G. K. (2001) VpsR, a Member of the Response Regulators of the Two-Component Regulatory Systems, Is Required for Expression of vps Biosynthesis Genes and EPS(ETr)-Associated Phenotypes in Vibrio cholerae O1 El Tor. J. Bact. 183, 1716–1726. 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong J. C.; Karplus K.; Schoolnik G. K.; Yildiz F. H. (2006) Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bact. 188, 1049–1059. 10.1128/JB.188.3.1049-1059.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong J. C.; Yildiz F. H. (2007) The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bact. 189, 2319–2330. 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich W. F.; Greenberg E. P. (1983) Glucose repression of luminescence and luciferase in Vibrio fischeri. Arch. Microbiol. 134, 87–91. 10.1007/BF00407937. [DOI] [Google Scholar]
- Shikuma N. J.; Fong J. C.; Odell L. S.; Perchuk B. S.; Laub M. T.; Yildiz F. H. (2009) Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bact. 191, 5147–5158. 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamorano-Sanchez D.; Fong J. C.; Kilic S.; Erill I.; Yildiz F. H. (2015) Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J. Bacteriol. 197, 1221–1235. 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H.; Schoolnik G. K. (1998) Role of rpoS in stress survival and virulence of Vibrio cholerae. J. Bact. 180, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L.; Dirita V. J.; Mekalanos J. J. (1989) Identification of Toxs, a Regulatory Gene Whose Product Enhances Toxr-Mediated Activation of the Cholera-Toxin Promoter. J. Bacteriol. 171, 1288–1293. 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L.; Griffiths L.; Cole J. A. (1990) Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch. Microbiol. 154, 349–354. 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- Taylor R. G.; Walker D. C.; McInnes R. R. (1993) E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678. 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R.; Priefer U.; Puhler A. (1983) A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio/Technology 1, 784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- Bell A. I.; Gaston K. L.; Cole J. A.; Busby S. J. (1989) Cloning of binding sequences for the Escherichia coli transcription activators, FNR and CRP: location of bases involved in discrimination between FNR and CRP. Nucleic Acids Res. 17, 3865–3874. 10.1093/nar/17.10.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L.; Wright M.; Showalter R. E.; Silverman M. R. (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9, 773–786. 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Hammer B. K.; Bassler B. L. (2007) Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104, 11145–11149. 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L.; Wei Y.; Perez L. J.; Cong J.; Long T.; Koch M.; Semmelhack M. F.; Wingreen N. S.; Bassler B. L. (2010) Probing bacterial transmembrane histidine kinase receptor-ligand interactions with natural and synthetic molecules. Proc. Natl. Acad. Sci. U. S. A. 107, 5575–5580. 10.1073/pnas.1001392107. [DOI] [PMC free article] [PubMed] [Google Scholar]