Abstract
Chronic spontaneous urticaria (CSU) is one of the most common types of chronic urticaria (CU), with symptoms that recur easily, migrate and are refractory. It is unclear whether association between the differentiation of protein expression levels in the serum of CSU patients and the different duration of wheals exists. In the present study the samples were divided according to the duration of the wheals into group A (wheal duration <2 h) and group B (wheal duration 12–24 h). Differentially expressed proteins in sera of CSU patients with different durations of wheals were identified and validated with isobaric tags for relative and absolute quantitation (iTRAQ) in combination with two-dimensional liquid chromatography/tandem mass spectrometry (2D-LC-MS/MS). Three hundred and seventy CSU serum-related proteins were initially identified. Among these proteins, ~30 had significant differences between the groups. According to the classification of biological functions and upregulated/downregulated values, serum amyloid A (SAA), CFL1, TPM4 and monocyte differentiation antigen (CD14) were chosen and validated by enzyme-linked immunosorbent assay (ELISA). The expression levels of CD14 in sera were not significantly different among the groups. SAA, CFL1 and TPM4 were associated with the wheal duration in CSU patients and therefore could be considered as new potential inflammatory biomarkers associated with CSU.
Keywords: chronic spontaneous urticaria, isobaric tags for relative and absolute quantitation, serum amyloid A protein, cofilin-1
Introduction
Chronic urticaria (CU) is a common distressing skin disorder characterized by recurrent wheals and/or angioedema lasting for >6 weeks. CU can be divided into chronic spontaneous urticaria (CSU), inducible urticaria and other types of urticaria according to different etiology. CSU is a mast cell-driven disease that is defined as the recurrence of wheals, angioedema or both for >6 weeks due to known or unknown causes, excluding chronic inducible urticaria and urticaria vasculitis (1). The incidence of CSU and the etiology is unclear, the prevalence in the general population is ~0.5–1%, and the annual incidence is ~1.4% (1,2). Lapi et al (3) found that the annual prevalence of CSU in Italy increased from 0.02 to 0.38% from 2002 to 2013. CSU is the most common type of CU, accounting for ~2/3 of all cases (4), which is the same as that found in China (5). Due to the lack of specific treatment, the symptoms easily recur and are difficult to cure. CSU seriously affects the patients' quality of life and the stability of the immune environment. Antihistamine is the most commonly used drug for CSU treatment. However, this is far from satisfactory. Due to its complicated disease mechanism, most clinicians diagnose and treat CSU according to their symptoms. The objective and measurable indicators of disease activity are absent, so it is hard to evaluate the degree of disease, treatment efficiency and prognosis correctly. Thus, further investigation is required.
Although a large number of studies have been conducted on the pathogenesis of CSU, many hypotheses on CSU still have no conclusive evidence to confirm its mechanism. The specific allergen can not be found in most CSU patients. So the mechanism can not be well elaborated through IgE-mediated classic type I hypersensitivity. It is also difficult to explain with autoimmune theory, or genetic factors. Several researchers have suggested that blood parameters may indicate disease activity and duration. This would assist monitor treatment and could provide potential prognostic biomarkers of CSU (6–9). Some strong evidence has shown significant differences between patients with CSU and healthy controls in blood levels or values of D-dimer, C-reactive protein (CRP), matrix metalloproteinase-9 (MMP-9), mean platelet volume (MPV), factor VIIa, prothrombin 1+2 (F1+2), tumur necrosis factor, dehydroepiandrosterone sulphate and vitamin D (10). Data have shown that plasma FDP, D-dimer and serum CRP may be well associated with each other and significantly associated with the disease severity of CU, but not with the skin reactions of the autologous serum (11). However, Korean scholars Kim et al (12) have found that the expression level of serum clusterin increases in patients with a positive autologous serum skin test (ASST) than a negative one. Fujii et al (13) further found that, after the urticaria subsided the elevated circulating thrombin-antithrombin (TAT) III complex level recovered to within the normal range, and so did the elevated D-dimer level in all cases except two. This may explain the clinical symptoms of wheals in urticaria patients that vanished gradually by themselves. Researches (11,14) have shown that C3, C4 and CRP are closely related with the severity of CSU in the diagnosis of CSU and/or prognosis. Some authors have studied the inflammatory and cytokines in serum and lesions of CSU patients. This can not clearly explain the exact relationship with the pathogenesis of the disease. Conflicting evidence might also be explained by different patient populations and differences in the analysis of the results (e.g., difference in standardization between the various producers of the assay kits, the existence of various cut-offs and distinct methodological measurements of blood parameters).
With the further development of genomics and proteomics research, as well as the continuous accumulation of protein research data, proteomics research will lead to breakthroughs in the pathogenesis, diagnosis, treatment and new drug development of various skin diseases. Increasing number of proteomic techniques have been applied to skin diseases (15–17), including some allergic skin diseases (18–22). So far, proteomics technology has not been applied in the field of urticaria and is rarely reported. Isobaric tags for relative and absolute quantitation (iTRAQ) technology is a new relative and absolute quantification technique of in vitro isotope labeling of peptides developed by ABI Scientific, Inc. (Sterling, VA, USA) in 2004. Proteomics research based on the iTRAQ labeling method is a powerful tool for discovering protein markers. This technique can quantitatively compare proteins in 8 different samples at the same time. Therefore, it can distinguish the protein changes in quality and quantity between normal and disease samples, pre- and post-treatment, disease progression and cell culture under different conditions.
In this study, iTRAQ labeling combined with two-dimensional liquid chromatography/tandem mass spectrometry (2D-LC-MS/MS) technique was used to study the protein profiling of sera from CSU patients with different durations of wheals and normal subjects. A total of 370 serum proteins associated with CSU were initially identified. Among those identified proteins, ~30 were found to have significant differences between the groups. According to the classification of biological functions and upregulated/downregulated values, SAA, CFL1, TPM4 and monocyte differentiation antigen (CD14) were chosen and validated by enzyme-linked immunosorbent assay (ELISA).
The results showed that in CSU patients with wheal duration of 12–24 h the expression level of SAA increased (P=0.047767), while CFL1 and TPM4 levels decreased (P=0.049229 and P=0.0049, respectively) in comparison with the healthy control group. Compared with the group of patients with wheal duration of <2 h, the expression level of SAA in CSU patients with 12–24 h wheal duration had no significant difference, while the expression levels of CFL1 and TPM4 decreased (P=0.01684 and P=0.0186, respectively). However, there was no significant difference in serum levels of SAA, CFL1 and TPM4 between CSU patients with wheal duration of <2 h and the healthy control group. The expression level of CD14 in serum was no significantly different among the three groups. SAA, CFL1 and TPM4 proteins were associated with wheal duration in CSU patients and might be considered as new potential inflammatory biomarkers associated with CSU.
Materials and methods
Specimens and sample collection
A total of 20 CSU patients were selected and divided into group A and B according to the duration of the wheals. Group A: wheal duration <2 h, 10 cases; group B: wheal duration 12–24 h, 10 cases. All the serum samples were collected before the wheals subsided. All patients conformed to the diagnostic criteria of CSU in EAACI/GA (2) LEN/EDF/WAO guidelines (1).
At the same time, group D, whose outpatient physical examination was positive, served as a healthy control group. The 10 cases comprising the control group underwent blood tests. Liver and kidney function, blood glucose and blood lipids were normal, and there were no previous allergic and systemic diseases.
The study was approved by the Ethics Committee of Pudong New Area People's Hospital (Shanghai, China). Signed informed consents were obtained from the patients or the guardians.
After the differentially expressed CSU serum-associated proteins were identified among the groups, 42 CSU patients were re-selected: 21 patients with wheal duration <2 h (group Y) and 21 patients with wheal duration of 12–24 h (group G), and 21 healthy controls (group J) were selected. The level of differentially expressed serum protein was examined by ELISA test.
The proteins that flowed through the depletion column were precipitated using ReadyPrep 2-D Cleanup kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer's instructions. After precipitation, protein pellets were re-suspended in dissolution buffer using the iTRAQ experiment kit (AB Sciex, Framingham, MA, USA). The total protein concentration of each sample was determined by Bradford protein assay (Thermo Fisher Scientific, Inc., Waltham, MA, USA), as previously described (7). A total of 100 µg aliquots of each of the 3 samples were then reduced, alkylated, digested with trypsin, and labeled individually with one iTRAQ tag (Applied Biosystems: Thermo Fisher Scientific, Inc., Foster City, CA, USA) according to the manufacturer's instructions. Group A, B and D samples were labeled with 113, 114 and 115 tags, respectively. The labeled samples were then pooled and dried by centrifugal evaporation (Christ Alpha 1–2 and Christ RVC 2–25; Martin Christ Gefriertrocknungsanlagen GmbH, Osterode, Germany).
2D-LC conditions
Chromatographic separation of the pooled samples was performed on an Acquity Ultra Performance LC system (Waters Corp., Milford, MA, USA). Tryptic digested and labeled peptides were first fractionated by a strong cation exchange (SCX) liquid chromatograph using a 0.5×23 mm, 5 µm, 300 Å column (Waters Corp.). Samples were loaded onto the column and eluted stepwise by injecting salt plugs of 10 different molar concentrations of 25, 50, 75, 100, 150, 200, 300, 400, 500, 1,000 mM of NH4Ac. Ten fractions were collected from the SCX column. Each of the fractions was then loaded onto a reverse phase (RP) column, ZORBAX 300SB-C18 column (5 µm, 300 Å, 4.6×50 mm; Agilent Technologies, Inc., Santa Clara, CA, USA). The flow rate used for separation on RP column was 0.4 µl/min. Buffer A was 5% acetonitrile, 95% water, 0.1% formic acid and buffer B was 95% acetonitrile, 5% water, 0.1% formic acid. Elution was performed using a gradient ranging from 5 to 45% buffer B for >90 min.
MS/MS conditions
The LC eluent was subjected to positive ion nanoflow electrospray analysis using a Qstar XL MS/MS system (Applied Biosystems: Thermo Fisher Scientific, Inc.) in an information-dependent acquisition (IDA) mode. In IDA mode, a TOFMS survey scan was acquired (m/z 400–1,800), with up to six most intense multiply charged ions in the survey scan sequentially subjected to product ion analysis. Product ion spectra were accumulated for 2 sec in the mass range m/z 100–2,000 with a modified Enhance All mode Q2 transition setting favoring low mass ions, so that the reporting iTRAQ ion (113, 114, and 115 m/z) intensities were enhanced for quantitation.
Data analysis
All LC MS/MS data were acquired by Analyst QS 3.1 (Applied Biosystems: Thermo Fisher Scientific, Inc.). MS/MS data were analyzed using ProteinPilot v3.0 (Applied Biosystems: Thermo Fisher Scientific, Inc.) which uses the Paragon Algorithm to perform database searching. The search results were further processed by the Pro Group Algorithm to remove redundant hits and comparative quantitation so that the minimal set of justifiable identified proteins could be found. The protein database used for all searches was Swiss-Prot human (downloaded on Sept. 15, 2016). Loading error was normalized by bias correction calculated using ProteinPilot software. All reported data were based on 95% confidence for protein identification as determined by ProteinPilot (Prot score ≥1.3). The relative protein quantitation was calculated as an average ratio. The confidence level of the altered expression of proteins was calculated by ProteinPilot as P-value, which allows the results to be evaluated based on the confidence level of expression change, not just by the magnitude of the change.
ELISA
Blood samples taken from the patients and normal control, were grouped and numbered (5 ml elbow vein blood, low temperature centrifugation, at the speed of 1,500 × g for 15 min at 4°C, supernatant sub-packaging, −80°C in storage) and then recorded. Specific detection was performed according to iTRAQ technology and ELISA kit.
Preparation of the reagents, samples and standards: 100 µl of sample were added, standard or blank to each well, and incubated for 90 min at 37°C. A total of 100 µl of 1X biotinylated detection anti-human SAA (EA8001-1; AssayPro, St. Charles, MO, USA), CD14 (227920; Biomol GmbH, Hamburg, Germany), CFL1 (LS-F20976; LifeSpan BioSciences, Inc., Seattle, WA, USA) and TPM4 (EKC35896; Biomatik Corp., Cambridge, ON, Canada) polyclonal antibodies (1:300) were aspirated and added, and then incubated for 1 h at 37°C. Then aspirated and washed 3 times. 1X HRP conjugate (100 µl) was added and incubated for 30 min at 37°C. Aspirated and washed 5 times. TMB substrate solution (90 µl) was added and incubated for 15 min at 37°C. Finally, 50 µl of stop solution were added and reading was performed immediately at 450 nm.
Serum measurement: ELISA assays for IGFBP2 and LCAT (Cloud-Clone Corp., Wuhan, China), SHBG (R&D Systems Europe, Ltd., Abingdon, UK), GRP78 (Enzo Life Sciences, Inc., Exeter, UK) and calprotectin (BioLegend, Inc., San Diego, CA, USA) were performed in duplicate using commercial kits following the manufacturer's instructions. The mean coefficients of variance for duplicate analysis for each assay were as follows: IGFBP2, 8.1%; LCAT, 8.4%; SHBG, 7.4%; GRP78, 3.1%; and calprotectin, 4.5%.
Statistical analysis
All reported data were based on 95% confidence for protein identification as determined by ProteinPilot (Prot score ≥1.3). The relative protein quantitation was calculated as an average ratio. The confidence level of the altered expression of proteins was calculated by ProteinPilot as P-value, which allows the results to be evaluated based on the confidence level of expression change, not just by the magnitude of the change. P<0.05 was considered as a statistically significant difference.
Results
Proteomic and biological functional analysis in serum
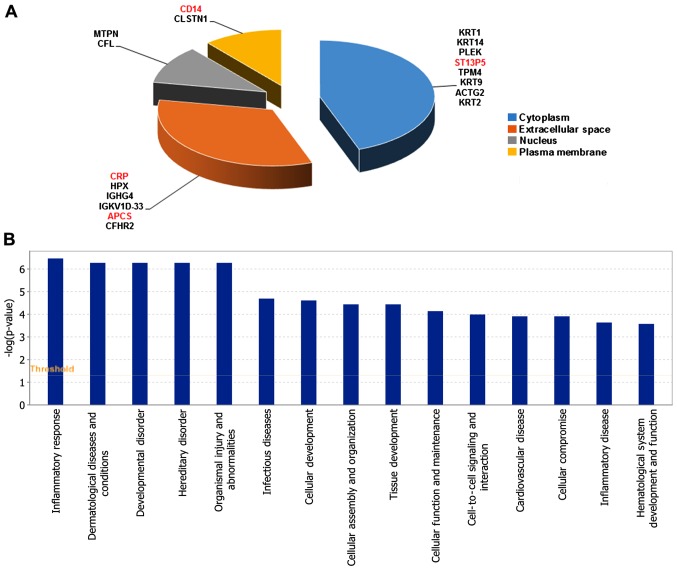
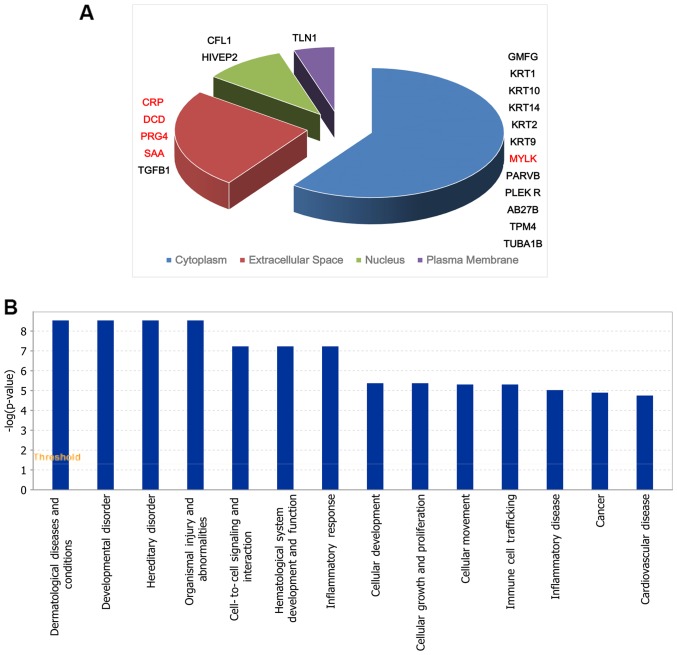
There were in total 370 proteins identified in sera of groups A, B and D using iTRAQ labeling combined with 2D-LC-MS/MS. There were 18 significant differentially expressed proteins between group B and A. Among them, 4 proteins were upregulated and 14 were downregulated (Table I). The cell location and functional distribution of the 18 differentially expressed proteins are displayed in Fig. 1A and B, respectively. There were 20 significantly different proteins in group D compared with group B, 5 proteins were upregulated and 15 were downregulated (Table II). Fig. 2A and B, shows the 20 differentially expressed proteins in cell location and functional distribution. These differentially expressed proteins were classified in cell components by GO. They were both mainly distributed in cytoplasm, outer matrix components, plasma membrane and the nucleus.
Table I.
Differentially expressed proteins of group A and B.
| Accession no. | Entry name | Protein names | Ratio B/A |
|---|---|---|---|
| P02743 | SAMP_HUMAN | SAP-component | 1.719164386 |
| P67936 | TPM4_HUMAN | Tropomyosin α-4 chain (TM30p1) | 0.55495 |
| P02741 | CRP_HUMAN | CRP | 2.707280979 |
| P35527 | K1C9_HUMAN | CK-9 | 0.378568685 |
| P04264 | K2C1_HUMAN | CK-1 | 0.458821195 |
| P01861 | IGHG4_HUMAN | Ig γ-4 chain C region | 0.654559002 |
| P08567 | PLEK_HUMAN | Pleckstrin | 0.605110697 |
| P35908 | K22E_HUMAN | CK-2e | 0.490515564 |
| P23528 | COF1_HUMAN | Cofilin-1 | 0.283736309 |
| P36980 | FHR2_HUMAN | Complement factor H-related protein 2 | 0.56605 |
| P02790 | HEMO_HUMAN | Hemopexin | 0.6199 |
| P02533 | K1C14_HUMAN | K14 | 0.664 |
| P08571 | CD14_HUMAN | Monocyte differentiation antigen CD14 | 1.66208565 |
| Q8NFI4 | F10A5_HUMAN | Putative protein FAM10A5 | 1.5919 |
| P58546 | MTPN_HUMAN | Myotrophin (protein V-1) | 0.5265 |
| O94985 | CSTN1_HUMAN | Calsyntenin-1 | 0.5115 |
| P01608 | KVD33_HUMAN | Immunoglobulin κ variable 1D-33 | 0.6313 |
| P63267 | ACTH_HUMAN | Actin | 0.66 |
Group A: wheal duration <2 h, 10 cases; group B: wheal duration 12–24 h, 10 cases. SAP, serum amyloid P; CRP, C-reactive protein; CK, cytokeratin; K14, keratin-14.
Figure 1.
(A and B) The subcellular location and functional distribution of differentially expressed proteins of group A and B. Group A: wheal duration <2 h, 10 cases; group B: wheal duration 12–24 h, 10 cases. Red, upregulated; black, downregulated. CRP, C-reactive protein.
Table II.
Differentially expressed proteins of group B and D.
| Accession no. | Entry name | Protein names | Ratio B/D |
|---|---|---|---|
| P0DJI8 | SAA1_HUMAN | SAA-1 protein | 3.118620690 |
| P67936 | TPM4_HUMAN | Tropomyosin α-4 chain | 0.225594016 |
| Q9Y490 | TLN1_HUMAN | Talin-1 | 0.425138687 |
| P02741 | CRP_HUMAN | CRP | 4.628658177 |
| P35527 | K1C9_HUMAN | CK-9 | 0.377987984 |
| P13645 | K1C10_HUMAN | CK-10 | 0.424446977 |
| P04264 | K2C1_HUMAN | CK-1 | 0.393632417 |
| Q92954 | PRG4_HUMAN | Proteoglycan 4 | 1.678792288 |
| P08567 | PLEK_HUMAN | Pleckstrin | 0.180288202 |
| Q15746 | MYLK_HUMAN | Myosin light chain kinase | 1.783790265 |
| P81605 | DCD_HUMAN | Dermcidin | 2.342048293 |
| P35908 | K22E_HUMAN | CK-2e | 0.226904558 |
| P31629 | ZEP2_HUMAN | Transcription factor HIVEP2 | 0.666550825 |
| P01137 | TGFB1_HUMAN | Transforming growth factor β-1 | 0.629120347 |
| Q9HBI1 | PARVB_HUMAN | β-parvin (affixin) | 0.182152123 |
| P23528 | COF1_HUMAN | Cofilin-1 | 0.226166454 |
| P02533 | K1C14_HUMAN | CK-14 | 0.431996357 |
| O00194 | RB27B_HUMAN | Ras-related protein Rab-27B | 0.405605322 |
| P68363 | TBA1B_HUMAN | Tubulin α-1B chain | 0.609584125 |
| O60234 | GMFG_HUMAN | Glia maturation factor γ | 0.447163743 |
Group B: wheal duration 12–24 h, 10 cases; group D: healthy control group, 10 cases. SAA, serum amyloid A; CRP, C-reactive protein; CK, cytokeratin.
Figure 2.
(A and B) The subcellular location and functional distribution of differentially expressed proteins between groups B and D. Group B: wheal duration 12–24 h, 10 cases; group D: healthy control group, 10 cases. Red, upregulated; black, downregulated. CRP, C-reactive protein.
There were 31 significantly different proteins between group A and D, 12 proteins were upregulated and 19 were downregulated (Table III). These 31 differentially expressed proteins were classified in cell components by GO, and were mainly distributed in extracellular matrix components, cytoplasm, plasma membrane and nucleus (Fig. 3A). For IPA bio function, the first three ranks are cell-to-cell signaling and interaction, hematological system development and function and inflammatory response (Fig. 3B).
Table III.
Differentially expressed proteins of group A and D.
| Accession no. | Entry name | Protein names | Ratio A/D |
|---|---|---|---|
| P01008 | ANT3_HUMAN | ATIII | 0.522774597 |
| P0DJI8 | SAA1_HUMAN | SAA-1 protein | 2.838068966 |
| Q96RU2 | UBP28_HUMAN | Ubiquitin carboxyl-terminal hydrolase 28 | 0.662010526 |
| P67936 | TPM4_HUMAN | Tropomyosin-4 | 0.406512327 |
| Q9Y490 | TLN1_HUMAN | Talin-1 | 0.595211961 |
| P02741 | CRP_HUMAN | CRP | 1.709707346 |
| Q15485 | FCN2_HUMAN | Ficolin-2 | 0.553688195 |
| P13645 | K1C10_HUMAN | CK-10 | 0.554630198 |
| O43866 | CD5L_HUMAN | CD5 antigen-like | 0.662852444 |
| P02753 | RET4_HUMAN | Retinol-binding protein 4 | 1.523612657 |
| Q92954 | PRG4_HUMAN | Proteoglycan 4 | 1.818843216 |
| P08567 | PLEK_HUMAN | Pleckstrin | 0.297942514 |
| Q15746 | MYLK_HUMAN | MLCK, smooth muscle | 1.598593238 |
| P02675 | FIBB_HUMAN | Fibrinogen β chain | 0.648407879 |
| P51587 | BRCA2_HUMAN | Breast cancer type 2 susceptibility protein | 0.413992962 |
| P55056 | APOC4_HUMAN | Apolipoprotein C-IV | 2.046185998 |
| P81605 | DCD_HUMAN | Dermcidin | 1.578517902 |
| P35749 | MYH11_HUMAN | Myosin-11 | 1.744439599 |
| P35908 | K22E_HUMAN | CK-2e | 0.46258381 |
| P31629 | ZEP2_HUMAN | Transcription factor HIVEP2 | 0.579206487 |
| P01705 | LV223_HUMAN | Immunoglobulin λ variable 2–23 | 1.68477803 |
| Q9HBI1 | PARVB_HUMAN | β-parvin (affixin) | 0.273112113 |
| P01019 | ANGT_HUMAN | Angiotensinogen (serpin A8) | 0.644745326 |
| P00748 | FA12_HUMAN | Coagulation factor XII | 1.885902876 |
| P02790 | HEMO_HUMAN | Hemopexin (β-1B-glycoprotein) | 1.76584849 |
| P02533 | K1C14_HUMAN | CK-14 | 0.650596923 |
| P08571 | CD14_HUMAN | Monocyte differentiation antigen CD14 | 0.537576093 |
| O94985 | CSTN1_HUMAN | Calsyntenin-1 | 2.147996993 |
| O00194 | RB27B_HUMAN | Ras-related protein Rab-27B (C25KG) | 0.480318932 |
| O00602 | FCN1_HUMAN | Ficolin-1 | 0.644163875 |
| O60234 | GMFG_HUMAN | Glia maturation factor γ (GMF-γ) | 0.584795322 |
Group A: wheal duration <2 h, 10 cases; group D: healthy control group, 10 cases. ATIII, antithrombin-III; SAA, serum amyloid A; CRP, C-reactive protein; CK, cytokeratin; MLCK, myosin light chain kinase.
Figure 3.
(A and B) The subcellular location and function distribution of differentially expressed proteins between group A and D. Group A: wheal duration <2 h, 10 cases; group D: healthy control group, 10 cases. Red, upregulated; black, downregulated. CRP, C-reactive protein.
The differentially expressed proteins identified among these three groups were also compared. SAA, CD14, CFL1 and TPM4 proteins had significant change. CRP, as a non-specific inflammatory marker, has been studied and reported in literature. Further 4 proteins, SAA, TPM4, CFL1 and CD14, based on the biological and functional information have close relationship with dermatological diseases and therefore were chosen to be validated as potential disease biomarkers by ELISA.
ELISA
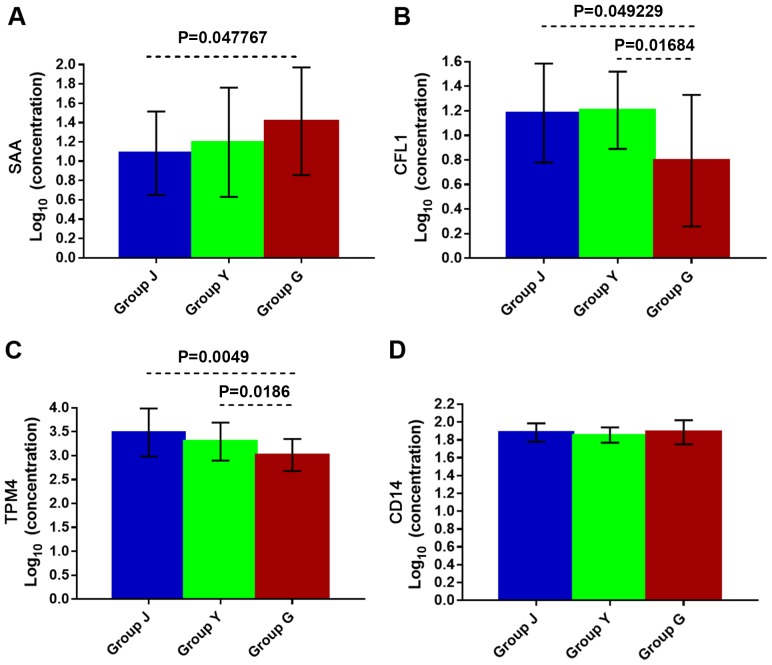
The expression level of SAA in the sera of group G (wheal duration of 12–24 h) was higher than that of healthy control; while there was no significant difference in the expression of SAA between group Y (wheal duration <2 h) and G, and between group Y and J (healthy group) (Fig. 4A) indicating that serum SAA levels are significantly elevated in CSU patients with longer duration of wheals. In other words, the severity of symptoms in CSU patients is directly proportional to the serum level of SAA.
Figure 4.
(A) The SAA, (B) the CFL1, (C) the TPM4, and (D) the CD14 levels in serum of CSU patients of different groups by ELISA. Group Y: wheal duration <2 h, 21 cases; group G: wheal duration 12–24 h, 21 cases; group J: healthy control group, 21 cases. Statistical significance, P<0.05. SAA, serum amyloid A; CSU, chronic spontaneous urticaria; ELISA, enzyme-linked immunosorbent assay.
The expression levels of CFL1 and TPM4 were decreased in group G compared to group Y and J (Fig. 4B and C). The expression levels of CFL1 and TPM4 were significantly decreased in the serum of CSU patients with longer wheal duration compared to shorter wheal duration and healthy control as shown by ELISA. The expression levels of CFL1 and TPM4 had no significant change in the serum of CSU patients with shorter wheal duration and healthy control.
The results showed that in CSU patients with wheal duration of 12–24 h the expression level of SAA increased (P=0.047767), but CFL1 and TPM4 levels decreased (P=0.049229 and P=0.0049, respectively) in comparison with healthy controls. Compared with the group of wheal duration of <2 h, the expression level of SAA in CSU patients with 12–24 h wheal duration had no significantly difference, while the expression level of CFL1 and TPM4 decreased (P=0.01684 and P=0.0186, respectively). However, there was no significant difference in sera levels of SAA, CFL1 and TPM4 between CSU patients with wheal duration of <2 h group and healthy control. There was no statistically significant change in the expression level of CD14 of the three groups (Fig. 4D).
Discussion
CSU is an inflammatory disease, characterized by acute phase response (APR) (23,24). The impact of APR on the body is a change in plasma protein concentration accompanied by a series of physiological and biochemical changes. Among them, the plasma protein concentration increased by >25%, is known as positive acute phase protein (APP), such as CRP, SAA and fibrinogen (FIB). CSU might be caused by an interactive combination of immune, genetic, and environmental factors, including infections (25–27). In addition, the altered function of the neuroendocrine-immune system has been recognized in CSU pathogenesis (28,29). Relevant research between coagulation and inflammation markers and the severity of acute exacerbation of the level of indicators and CU severity is also one of the hot spots in recent years; D-dimer and CRP levels are closely related to the activation of CU (11,30,31).
In the present study, the expression level of SAA in serum of CSU patients with wheal duration of 12–24 h increased significantly compared with the healthy control group as resulted by ELISA test. While there was no difference between healthy group and wheal duration of <2 h group. Serum amyloid A (SAA) is one of the most dominent positive acute-phase proteins, which increases sharply in serum due to inflammation, infection, neoplasia and tissue injury (32–34). The high expression level of SAA in the serum of CSU patients plays an important role in the long duration of the wheals. SAA is highly induced during the inflammatory response and is involved in the systemic regulation of the innate and adaptive immune responses. SAA possesses a number of cytokine-like properties reported in recent research. It can also stimulate the release of mature IL-1β from neutrophils, mast cells, macrophages and fibroblasts (33,35,36). IL-1β is a potent pro-inflammatory cytokine involved in many inflammatory diseases, including autoimmune and allergic diseases (37). The expression level of SAA was also significantly increased in allergic rhinitis and asthma patients (38).
Interestingly, in our experiment the expression levels of CFL1 and TPM4 were significantly downregulated in sera of CSU patients with longer duration of wheals compared to healthy and CSU patients with shorter duration of wheals. There was no difference between the serum in CSU patients with shorter duration of wheals and healthy groups. The role of CFL1 and TPM4 in the disease mechanism of CSU is still unclear. CFL1 is a key member of the actin-binding protein family that cleaves microfilaments and binds to actin monomers, to accelerate actin conversion rate in vivo.
Eosinophils are rich in actin, and the cytoskeleton is dynamic and very sensitive to changes in the cell's environment. This desirable property may benefit a motile cell such as the eosinophil to migrate into an inflammatory site. The dual function of cofilin, namely depolymerization and severing, serve as a key moducule controlling the actin dynamics. Thus, cofilin might be a possible candidate which connects early signal transduction events with the functional role of the cytoskeleton upon eosinophil activation (39). The number of eosinophils is increased in patients with CSU peripheral blood (40) and skin tissue (39). It has also been reported that the immunopathology of CSU is based on eosinophils and basophil-mediated hypersensitivity reactions characterized by secretion of Th0 cytokines (41). Therefore, CFL1 is abnormally expressed in CSU patients with longer periods of wheals and is probably due to eosinophil changes of function, which needs to be further confirmed.
Tropomyosins are coiled-coil dimmers, one of the major allergenic proteins of crustacean aquatic animals, that is located from head-to-tail polymers along actin filaments and regulate interactions of other proteins, including actin depolymerizing factor (ADF)/cofilins and myosins, with actin (42–45).
There are at least 40 tropomyosin subtypes in eukaryotes, and not all tropomyosins can compete with cofilin for binding to F-actin. The results of this experiment showed that TPM4 expression was consistent with that of CFL1 in the sera of CSU patients with longer duration of wheals, which seemed to be not competitive with F-actin binding. The role of TPM4 and CFL1 in the pathogenesis of CSU and the relationship between them need also to be further studied.
The pathogenesis of CSU remains unclear. As one of the major allergens of crustacean aquatic animals, TPM4 is abnormally expressed in the sera of CSU patients with longer period of wheals. The possible explanation is that the crustacean aquatic animal is one of the main causes in CSU patients with longer duration of the wheals and this result would be further observed in combination with clinical cases.
CD14, one of glycosylphosphatidylinositol-anchored proteins, plays multiple roles in microbial recognition and signaling. It assists to recognize the ligands of TLR1, 2, 3, 4, 6, 7, and 9, and it contributes in many ways to the trigger of the signaling pathways activated in response to LPS (46). Although CD14 protein was expressed in all serum of CSU patients, the expression of CD14 in serum of patients with wheal duration of <2 h was lower than that in healthy controls (ratio of A/D was ~0.54), and higher in sera of patients with long duration of wheals (ratio of B/A was ~1.66). However, the results of further validation did not confirm statistically significant differences between groups of patients with different duration of the wheals and with healthy individuals. CSU is a chronic inflammatory disease caused by the interaction of immune, genetic, environmental and infection factors. The results of ELISA validated that CD14 has no practical significance in the pathogenesis of CSU.
In conclusion, the results of serum identification of CSU patients with different durations of wheals using iTRAQ technique showed that there were different proteins in the serum of CSU patients with different duration. The level of SAA was positively associated with the duration of the wheals in sera of CSU patients, as shown by ELISA. The expression levels of CFL1 and TPM4 were different in sera of CSU patients with different duration of wheals; the expression levels were decreased in the serum of CSU patients with wheal duration of 12–24 h. The results suggest that there is an association between SAA, CFL1 and TPM4 and the duration of wheals in CSU patient, and therefore may be considered as new potential inflammatory biomarkers associated with CSU.
Acknowledgements
We would like to thank Mr. Lei Zhang, who works at IBS Core Facility Platform in Fudan University (Shanghai, China), for his assistance with the iTRAQ labeling.
Funding
This study was supported by the research project of Shanghai Pudong New Area Science and Technology Bureau (no. PKJ2015-Y28) and the key research program of Shanghai Pudong New Area Health and Family Planning Commission (no. PWZzk2017-28).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YC and SX conceived and designed this study. WK and HC analyzed the data and interpreted the results. YX acquired the data, performed the ELISA assays and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Pudong New Area People's Hospital (Shanghai, China). Signed informed consents were obtained from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zuberbier T, Aberer W, Asero R, Bindslev-Jensen C, Brzoza Z, Canonica GW, Church MK, Ensina LF, Giménez-Arnau A, Godse K, et al. European Academy of Allergy and Clinical Immunology; Global Allergy and Asthma European Network; European Dermatology Forum; World Allergy Organization: The EAACI/GA(2) LEN/EDF/WAO Guideline for the definition, classification, diagnosis, and management of urticaria: The 2013 revision and update. Allergy. 2014;69:868–887. doi: 10.1111/all.12313. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein JA, Lang DM, Khan DA, Craig T, Dreyfus D, Hsieh F, Sheikh J, Weldon D, Zuraw B, Bernstein DI, et al. The diagnosis and management of acute and chronic urticaria: 2014 update. J Allergy Clin Immunol. 2014;133:1270–1277. doi: 10.1016/j.jaci.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 3.Lapi F, Cassano N, Pegoraro V, Cataldo N, Heiman F, Cricelli I, Levi M, Colombo D, Zagni E, Cricelli C, et al. Epidemiology of chronic spontaneous urticaria: Results from a nationwide, population-based study in Italy. Br J Dermatol. 2016;174:996–1004. doi: 10.1111/bjd.14470. [DOI] [PubMed] [Google Scholar]
- 4.Maurer M, Weller K, Bindslev-Jensen C, Giménez-Arnau A, Bousquet PJ, Bousquet J, Canonica GW, Church MK, Godse KV, Grattan CE, et al. Unmet clinical needs in chronic spontaneous urticaria. A GA2LEN task force report. Allergy. 2011;66:317–330. doi: 10.1111/j.1398-9995.2010.02496.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhong H, Song Z, Chen W, Li H, He L, Gao T, Fang H, Guo Z, Xv J, Yu B, et al. Chronic urticaria in Chinese population: A hospital-based multicenter epidemiological study. Allergy. 2014;69:359–364. doi: 10.1111/all.12338. [DOI] [PubMed] [Google Scholar]
- 6.Rabelo-Filardi R, Daltro-Oliveira R, Campos RA. Parameters associated with chronic spontaneous urticaria duration and severity: A systematic review. Int Arch Allergy Immunol. 2013;161:197–204. doi: 10.1159/000346896. [DOI] [PubMed] [Google Scholar]
- 7.Asero R. D-dimer: A biomarker for antihistamine-resistant chronic urticaria. J Allergy Clin Immunol. 2013;132:983–986. doi: 10.1016/j.jaci.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 8.Altrichter S, Boodstein N, Maurer M. Matrix metalloproteinase-9: A novel biomarker for monitoring disease activity in patients with chronic urticaria patients? Allergy. 2009;64:652–656. doi: 10.1111/j.1398-9995.2008.01799.x. [DOI] [PubMed] [Google Scholar]
- 9.Grzanka A, Machura E, Misiolek M, Polaniak R, Kasperski J, Kasperska-Zajac A. Systemic inflammatory response and calcification markers in patients with long lasting moderate-severe chronic spontaneous urticaria. Eur J Dermatol. 2015;25:26–28. doi: 10.1684/ejd.2014.2474. [DOI] [PubMed] [Google Scholar]
- 10.Kolkhir P, André F, Church MK, Maurer M, Metz M. Potential blood biomarkers in chronic spontaneous urticaria. Clin Exp Allergy. 2017;47:19–36. doi: 10.1111/cea.12870. [DOI] [PubMed] [Google Scholar]
- 11.Takahagi S, Mihara S, Iwamoto K, Morioke S, Okabe T, Kameyoshi Y, Hide M. Coagulation/fibrinolysis and inflammation markers are associated with disease activity in patients with chronic urticaria. Allergy. 2010;65:649–656. doi: 10.1111/j.1398-9995.2009.02222.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim JH, Lee HY, Ban GY, Shin YS, Park HS, Ye YM. Serum clusterin as a prognostic marker of chronic spontaneous urticaria. Medicine (Baltimore) 2016;95:e3688. doi: 10.1097/MD.0000000000003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii K, Usuki A, Kan-No Y, Ohgou N. Elevation of circulating thrombin-antithrombin III complex and fibrin degradation products in urticaria: A laboratory finding unrelated to intravascular coagulopathy. J Dermatol. 2008;35:308–310. doi: 10.1111/j.1346-8138.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- 14.Kasperska-Zajac A, Grzanka A, Machura E, Misiolek M, Mazur B, Jochem J. Increased serum complement C3 and C4 concentrations and their relation to severity of chronic spontaneous urticaria and CRP concentration. J Inflamm (Lond) 2013;10:22. doi: 10.1186/1476-9255-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg KC, Fritz Y, Johnston A, Foster AM, Baliwag J, Gudjonsson JE, Schlatzer D, Gokulrangan G, McCormick TS, Chance MR, et al. Proteomics of skin proteins in psoriasis: From discovery and verification in a mouse model to confirmation in humans. Mol Cell Proteomics. 2015;14:109–119. doi: 10.1074/mcp.M114.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Q, Byrum SD, Moreland LE, Mackintosh SG, Kannan A, Lin Z, Morgan M, Stack BC, Jr, Cornelius LA, Tackett AJ, et al. A proteomic study of human merkel cell carcinoma. J Proteomics Bioinform. 2013;6:275–282. doi: 10.4172/jpb.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meral O, Uysal H. Comparative proteomic analysis of fibrosarcoma and skin fibroblast cell lines. Tumour Biol. 2015;36:561–567. doi: 10.1007/s13277-014-2672-8. [DOI] [PubMed] [Google Scholar]
- 18.Erler A, Hawranek T, Krückemeier L, Asam C, Egger M, Ferreira F, Briza P. Proteomic profiling of birch (Betula verrucosa) pollen extracts from different origins. Proteomics. 2011;11:1486–1498. doi: 10.1002/pmic.201000624. [DOI] [PubMed] [Google Scholar]
- 19.Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, Schlievert PM, Reisdorph NA, Leung DY. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol. 2011;127:186–193. doi: 10.1016/j.jaci.2010.10.033. e1-193.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm T, Rutishauser D, Kai-Larsen Y, Lyutvinskiy Y, Stenius F, Zubarev RA, Agerberth B, Alm J, Scheynius A. Protein biomarkers in vernix with potential to predict the development of atopic eczema in early childhood. Allergy. 2014;69:104–112. doi: 10.1111/all.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neil SE, Sitkauskiene B, Babusyte A, Krisiukeniene A, Stravinskaite-Bieksiene K, Sakalauskas R, Sihlbom C, Ekerljung L, Carlsohn E, Lötvall J. Network analysis of quantitative proteomics on asthmatic bronchi: Effects of inhaled glucocorticoid treatment. Respir Res. 2011;12:124. doi: 10.1186/1465-9921-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto T, Miyazaki Y, Shirahama R, Tamaoka M, Inase N. Proteome analysis of bronchoalveolar lavage fluid in chronic hypersensitivity pneumonitis. Allergol Int. 2012;61:83–92. doi: 10.2332/allergolint.11-OA-0315. [DOI] [PubMed] [Google Scholar]
- 23.Kasperska-Zajac A. Acute-phase response in chronic urticaria. J Eur Acad Dermatol Venereol. 2012;26:665–672. doi: 10.1111/j.1468-3083.2011.04366.x. [DOI] [PubMed] [Google Scholar]
- 24.Kasperska-Zając A, Grzanka A, Czecior E, Misiolek M, Rogala B, Machura E. Acute phase inflammatory markers in patients with non-steroidal anti-inflammatory drugs (NSAIDs)-induced acute urticaria/angioedema and after aspirin challenge. J Eur Acad Dermatol Venereol. 2013;27:1048–1052. doi: 10.1111/j.1468-3083.2012.04486.x. [DOI] [PubMed] [Google Scholar]
- 25.Wedi B, Raap U, Wieczorek D, Kapp A. Urticaria and infections. Allergy Asthma Clin Immunol. 2009;5:10. doi: 10.1186/1710-1492-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabroe RA, Grattan CE, Francis DM, Barr RM, Black Kobza A, Greaves MW. The autologous serum skin test: A screening test for autoantibodies in chronic idiopathic urticaria. Br J Dermatol. 1999;140:446–452. doi: 10.1046/j.1365-2133.1999.02707.x. [DOI] [PubMed] [Google Scholar]
- 27.Dreyfus DH. Autoimmune disease: A role for new anti-viral therapies? Autoimmun Rev. 2011;11:88–97. doi: 10.1016/j.autrev.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Kasperska-Zajac A, Brzoza Z, Rogala B. Sex hormones and urticaria. J Dermatol Sci. 2008;52:79–86. doi: 10.1016/j.jdermsci.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Kasperska-Zajac A, Brzoza Z, Rogala B. Serum concentration of dehydroepiandrosterone sulphate in female patients with chronic idiopathic urticaria. J Dermatol Sci. 2006;41:80–81. doi: 10.1016/j.jdermsci.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Takeda T, Sakurai Y, Takahagi S, Kato J, Yoshida K, Yoshioka A, Hide M, Shima M. Increase of coagulation potential in chronic spontaneous urticaria. Allergy. 2011;66:428–433. doi: 10.1111/j.1398-9995.2010.02506.x. [DOI] [PubMed] [Google Scholar]
- 31.Jansen PM, Boermeester MA, Fischer E, de Jong IW, van der Poll T, Moldawer LL, Hack CE, Lowry SF. Contribution of interleukin-1 to activation of coagulation and fibrinolysis, neutrophil degranulation, and the release of secretory-type phospholipase A2 in sepsis: Studies in nonhuman primates after interleukin-1 α administration and during lethal bacteremia. Blood. 1995;86:1027–1034. [PubMed] [Google Scholar]
- 32.Sodin-Šemrl S, Žigon P, Čučnik S, Kveder T, Blinc A, Tomšič M, Rozman B. Serum amyloid A in autoimmune thrombosis. Autoimmun Rev. 2006;6:21–27. doi: 10.1016/j.autrev.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Migita K, Izumi Y, Jiuchi Y, Kozuru H, Kawahara C, Izumi M, Sakai T, Nakamura M, Motokawa S, Nakamura T, et al. Effects of Janus kinase inhibitor tofacitinib on circulating serum amyloid A and interleukin-6 during treatment for rheumatoid arthritis. Clin Exp Immunol. 2014;175:208–214. doi: 10.1111/cei.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogan S, Atakan N. Is serum amyloid A protein a better indicator of inflammation in severe psoriasis? Br J Dermatol. 2010;163:895–896. doi: 10.1111/j.1365-2133.2010.09907.x. [DOI] [PubMed] [Google Scholar]
- 35.Niemi K, Baumann MH, Kovanen PT, Eklund KK. Serum amyloid A (SAA) activates human mast cells which leads into degradation of SAA and generation of an amyloidogenic SAA fragment. Biochim Biophys Acta. 2006;1762:424–430. doi: 10.1016/j.bbadis.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Niemi K, Teirilä L, Lappalainen J, Rajamäki K, Baumann MH, Öörni K, Wolff H, Kovanen PT, Matikainen S, Eklund KK. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol. 2011;186:6119–6128. doi: 10.4049/jimmunol.1002843. [DOI] [PubMed] [Google Scholar]
- 37.Krause K, Metz M, Makris M, Zuberbier T, Maurer M. The role of interleukin-1 in allergy-related disorders. Curr Opin Allergy Clin Immunol. 2012;12:477–484. doi: 10.1097/ACI.0b013e3283574d0c. [DOI] [PubMed] [Google Scholar]
- 38.Büyüköztürk S, Gelincik AA, Genç S, Koçak H, Oneriyidogan Y, Erden S, Dal M, Colakoglu B. Acute phase reactants in allergic airway disease. Tohoku J Exp Med. 2004;204:209–213. doi: 10.1620/tjem.204.209. [DOI] [PubMed] [Google Scholar]
- 39.Lorenzo GD, Mansueto P, Melluso M, Candore G, Cigna D, Pellitteri ME, Salvo AD, Caruso C. Blood eosinophils and serum eosinophil cationic protein in patients with acute and chronic urticaria. Mediators Inflamm. 1996;5:113–115. doi: 10.1155/S0962935196000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pazdrak K, Young TW, Straub C, Stafford S, Kurosky A. Priming of eosinophils by GM-CSF is mediated by protein kinase C betaII-phosphorylated L-plastin. J Immunol. 2011;186:6485–6496. doi: 10.4049/jimmunol.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng W, Wang J, Zhu W, Xu C, He S. Upregulated expression of substance P in basophils of the patients with chronic spontaneous urticaria: Induction of histamine release and basophil accumulation by substance P. Cell Biol Toxicol. 2016;32:217–228. doi: 10.1007/s10565-016-9330-4. [DOI] [PubMed] [Google Scholar]
- 42.Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci. 2015;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- 43.von der Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manstein DJ, Mulvihill DP. Tropomyosin-mediated regulation of cytoplasmic myosins. Traffic. 2016;17:872–877. doi: 10.1111/tra.12399. [DOI] [PubMed] [Google Scholar]
- 45.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanoni I, Granucci F. Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol. 2013;3:32. doi: 10.3389/fcimb.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.