Supplemental Digital Content is available in the text
Keywords: autophagy, chloroquine, hydroxychloroquine, meta-analysis
Abstract
Background:
Autophagy is a mechanism which relies on lysosomes for clearance and recycling of abnormal proteins or organelles. Many studies have demonstrated that the deregulation of autophagy is associated with the development of various diseases including cancer. The use of autophagy inhibitors is an emerging trend in cancer treatment. However, the value of autophagy inhibitors remains under debate. Thus, a meta-analysis was performed, aiming to evaluate the clinical value of autophagy-inhibitor-based therapy.
Methods:
We searched for clinical studies that evaluated autophagy-inhibitor-based therapy in cancer. We extracted data from these studies to evaluate the relative risk (RR) of overall response rate (ORR), 6-month progression-free survival (PFS) rate, and 1-year overall survival (OS) rate.
Results:
Seven clinical trials were identified (n = 293). Treatments included 2 combinations of hydroxychloroquine and gemcitabine, 1 combination of hydroxychloroquine and doxorubicin, 1 combination of chloroquine and radiation, 2 combinations of chloroquine, temozolomide, and radiation, and 1 hydroxychloroquine monotherapy. Autophagy-inhibitor-based therapy showed higher ORR (RR: 1.33, 95% confidence interval [CI]: 0.95–1.86, P = .009), PFS (RR: 1.72, 95% CI: 1.05–2.82, P = .000), OS (RR: 1.39, 95% CI: 1.11–1.75, P = .000) values than the therapy without inhibiting autophagy.
Conclusion:
This meta-analysis showed that autophagy-inhibitor-based therapy has better treatment response compared to chemotherapy or radiation therapy without inhibiting autophagy, which may provide a new strategy for the treatment of cancers.
1. Introduction
Since the induction of the term by Christian de Duve in 1963, advances in the understanding of autophagy have come a long way.[1] When Yoshinori Ohsumi was awarded the Nobel Prize for Physiology or Medicine for his work on elucidating the mechanisms of autophagy in 2016, the importance of autophagy in health and disease especially in cancer was recently highlighted. It is thought that autophagy can prevent development of cancer under normal circumstances. However, once cancer is established, the process of autophagy often promotes tumor cell survival and growth.[2,3] The association with autophagy and cancer is complex, thus targeting autophagy in the treatment of cancer is still controversial.[4,5]
Cancer has constituted enormous burden to human.[6] Although chemotherapy, radiotherapy, and surgery have been standard treatments for patients with cancers, outcomes of these treatments including the overall survival (OS) rates of patients are still far from ideal.[7]
It is thought that autophagy is a survival mechanism conserved from yeast to mammals.[8] Autophagy is also a known survival mechanism across several tumor types.[9–11] Many studies have proved that combining various types of anticancer drugs with either pharmacologic or genetic autophagy inhibition can improve antitumor effects.[2,4,12] Fitzwalter and Thorburn found that the process of autophagy can protect cells from undergoing programmed cell death, which may partially explain the association between autophagy and cancer development.[13] However, this protective ability is not always the same, for example, autophagy can either inhibit or promote apoptosis under different cellular contexts in response to similar death stimuli, such as death receptor agonists including CD95 ligand (FASL) and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL; also known as TNFSF10).[14] These opposing effects can be explained by the degradation of different pro-apoptotic or anti-apoptotic regulators by autophagy.[15,16]
At present, chloroquine (CQ) and hydroxychloroquine (HCQ) are the only available autophagy inhibitors in clinical. CQ and HCQ can inhibit autophagy by blocking the fusion of autophagosomes with lysosomes.[17] In addition, CQ also has some autophagy-independent anticancer effects, including sensitizing cancer cells to chemotherapy.[18–20] Several related clinical trials have been conducted to evaluate the safety and value of it.
To date, no meta-analysis has looked at autophagy inhibition therapy. This meta-analysis focused on the efficacy of autophagy inhibitors (CQ and HCQ) in the treatment of patients with cancer, aiming to evaluate the clinical value of autophagy-inhibitor-based therapy in different types of cancer.
2. Materials and methods
2.1. Study design and search strategy
All relevant information of this meta-analysis was identified from published autophagy trials in cancer. We searched for the trials based on the following computerized bibliographic databases: PubMed/Medline, Embase, the Cochrane Central Register of Controlled Trials (CENTRAL), and Google Scholar without any language restrictions. July 2018 was the cut-off date. The following keywords were included: combination therapy, autophagy, inhibitor, hydroxychloroquine, chloroquine. The search algorithm for pubmed was generated as follows: (((((((((((((((Chlorochin) OR Chingamin) OR Khingamin) OR Nivaquine) OR Chloroquine Sulfate) OR Sulfate, Chloroquine) OR Chloroquine Sulphate) OR Sulphate, Chloroquine) OR Aralen) OR Arequin) OR Arechine)) OR “Chloroquine”[Mesh])) AND ((((((((Oxychlorochin) OR Oxychloroquine) OR Hydroxychlorochin) OR Plaquenil) OR Hydroxychloroquine Sulfate) OR Hydroxychloroquine Sulfate (1:1) Salt)) OR “Hydroxychloroquine”[Mesh])) AND ((randomized controlled trial[Publication Type] OR randomized[Title/Abstract] OR placebo[Title/Abstract])), and the search algorithm for Embase was generated as follows: ((“chloroquine”/exp OR “chloroquine” OR “chloroquine”:ab,ti OR “chingamin”:ab,ti OR “khingamin”:ab,ti OR “nivaquine”/exp OR “nivaquine” OR “chloroquine sulfate”:ab,ti OR “sulfate, chloroquine”:ab,ti OR “sulphate, chloroquine”:ab,ti OR “aralen”:ab,ti OR “arequin”:ab,ti OR “arechine”:ab,ti OR “chlorochin”:ab,ti) AND (“hydroxychloroquine”/exp OR “hydroxychloroquine”) OR “oxychlorochin”:ab,ti OR “oxychloroquine”:ab,ti OR “hydroxychlorochin”:ab,ti OR “plaquenil”:ab,ti OR “hydroxychloroquine sulfate”:ab,ti OR “hydroxychloroquine sulfate (1:1) salt”:ab,ti) AND (“randomized controlled trial”/exp OR “randomized controlled trial”). To find potential publications, we reviewed a reference list of related articles for further analysis.
2.2. Inclusion and exclusion criteria
Retrieved articles had to satisfy the following criteria: the study must be a clinical study concerning the efficacy of chloroquine or hydroxychloroquine in the treatment of tumors; the study must report any of the following information: overall response rate (ORR), 1-year OS rate, and 6-month progression-free survival (PFS) rate.
The exclusion criteria were: studies were not related to our research topics or not clinical trials; retrospective studies, letters, editorials, expert opinions; studies lacked necessary data.
2.3. Data extraction
All reviewers independently extracted data with a piloted extraction form, and checked all data carefully. This meta-analysis extracted data including 1st author, published year, number of patients, study design, intervention methods, cancer types, autophagy inhibitor, clinical trial phase, additional treatment, and clinical response. ORR was collected directly or calculated according to CRR and PRR. Data extraction was performed independently by 2 reviewers and differences were resolved by a 3rd reviewer.
2.4. Assessment of the study quality and risk of bias
Cochrane Collaboration's tool was used to evaluate the risk of bias, any controversies were resolved by mutual discussion. Assessment of the study quality was based on the latest 2009 version of the initial Stroke Therapy Academic Industry Roundtable (STAIR) standard. It includes sample-size calculation, inclusion and exclusion criteria, allocation concealment, blinded assessment of outcome, reporting of patients excluded from analysis and reporting potential conflicts of interest and study funding. All reviewers assessed the qualities in all included studies. The “unclear” means the quality was not clear. Details on the risk of bias in fourteen studies are illustrated in Supporting Information Figure S4.
2.5. Statistical analysis
Statistical analysis, forest plots, and detection of publication bias were calculated by Stata/SE 12.0 (Stata Corp, College Station, TX), and we used Review Manager (RevMan5.3; The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark) to assess the risk of bias. Hazard ratios (HRs) were used for evaluation. Publication bias was assessed by Egger test and Begg test. All analyses (OS, ORR, PFS) were performed using a random-effects model (M–H heterogeneity). In addition, we calculated 95% confidence intervals (CIs) for each estimate.
2.6. Data availability
All data generated during and/or analyzed in this study are included in this published article (and its supplementary information files).
2.7. Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
2.8. Informed consent
For this type of study, formal consent is not required.
3. Results
3.1. Search results and study characteristics
The procedures and results of study selection are illustrated in “PRISMA Flow Diagram.” Seven studies were included in our meta-analysis after removing retrospective articles and those lacking necessary data.[21–27] These studies were selected to determine the efficacy and difference of two kinds of autophagy inhibitors (CQ and HCQ) in treating cancers. We found multiple types of autophagy inhibition treatment trials, trials included 2 combinations of hydroxychloroquine and gemcitabine, 1 combination of hydroxychloroquine and doxorubicin, 1 combination of chloroquine and radiation, 2 combinations of chloroquine, temozolomide, and radiation, and 1 hydroxychloroquine monotherapy.
3.2. Study characteristics
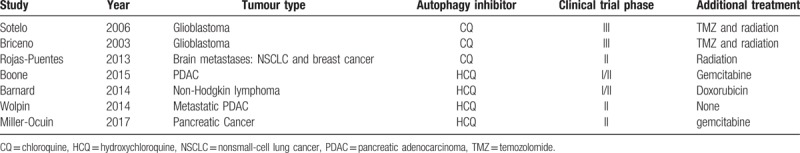
The included studies were published from 2003 to 2017. The detailed characteristics of included studies are shown in Table 1. A total of 293 patients were assessed in these studies, of which 149 patients received CQ or HCQ as autophagy inhibitors, 144 patients received chemotherapy or radiation therapy without autophagy inhibition. A total of 102 patients received combination of autophagy inhibitors and chemotherapeutics like gemcitabine and doxorubicin, a total of 15 patients received combination of chloroquine, temozolomide and radiation, and a total of 10 patients received hydroxychloroquine monotherapy. Patients who did not receive autophagy inhibitors served as the controls. Among these studies, there were 2 phase I/II articles, 3 phase II articles, and 2 phase III articles.
Table 1.
Characteristics of trials included in the meta-analysis.
3.3. Efficacy analysis of autophagy inhibitors in cancer treatment
The ORR, 1-year OS rate, and 6-month PFS rate were used to measure the efficacy of autophagy inhibitors in treating cancers. All seven studies were included in the ORR analysis.[21–27] Six articles were incorporated in the 6-month PFS and 1-year OS rate analysis.[21–26] The 1-year OS rate and 6-month PFS rate of autophagy inhibition therapy were significantly higher than those of therapy without inhibiting autophagy (relative risk [RR]: 1.39, 95% CI: 1.11–1.75, P = .000) and (RR: 1.72, 95% CI: 1.05–2.82, P = .000), respectively. Overall, therapy with autophagy inhibition resulted in a significantly higher ORR compared with therapy without inhibiting autophagy (RR: 1.33, 95% CI: 0.95–1.86, P = .009) (Fig. 1). There was high heterogeneity in the ORR (I2 = 54.2%) and 6-month PFS (I2 = 72.3%) analyses, but heterogeneity in 1-year OS rate was low (I2 = 0.0%). Detailed heterogeneity analyses are shown in Supporting Information Figure S1.
Figure 1.
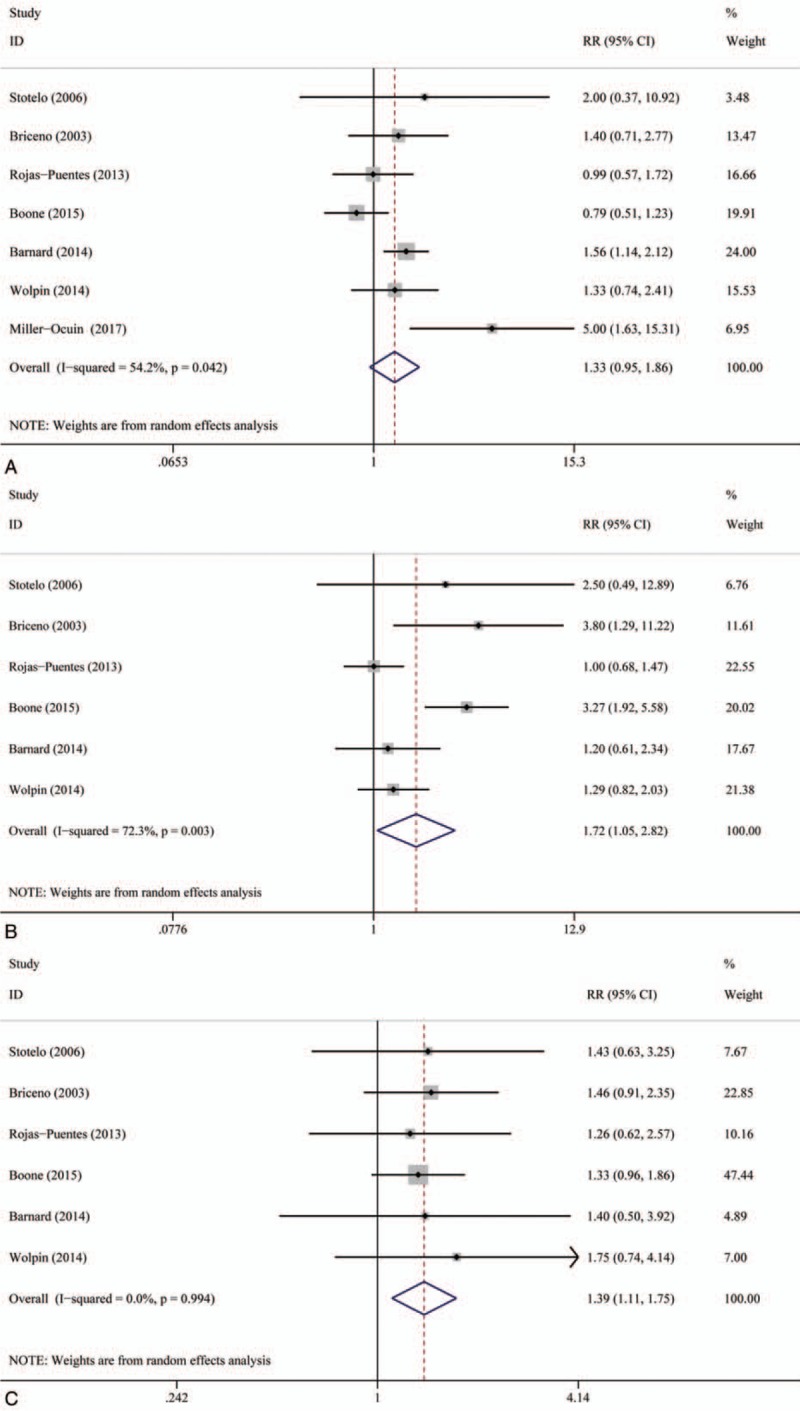
Meta-analysis of overall response rate (ORR), 6-month progression-free survival (PFS), and 1-year overall survival (OS) rate. (A) Overall response rate (ORR); (B) 6-month PFS (c) 1-year OS.
3.4. Subgroup analysis of autophagy inhibitors in cancer treatment
Although CQ and HCQ both can inhibit the process of autophagy, their mechanisms of effecting the survival and development of cancer cells are not the same. Thus, we conducted a subgroup analysis of 2 types of autophagy inhibitors to evaluate whether 2 of them have significant difference in contributing the treatment outcome. We found that CQ and HCQ both can significantly improve ORR, 1-year OS rate, and 6-month PFS rate. HCQ-based therapy can better benefit 1-year OS rate and 6-month PFS rate than CQ-based therapy, and CQ-based therapy can better benefit ORR than HCQ-based therapy (Fig. 2).
Figure 2.
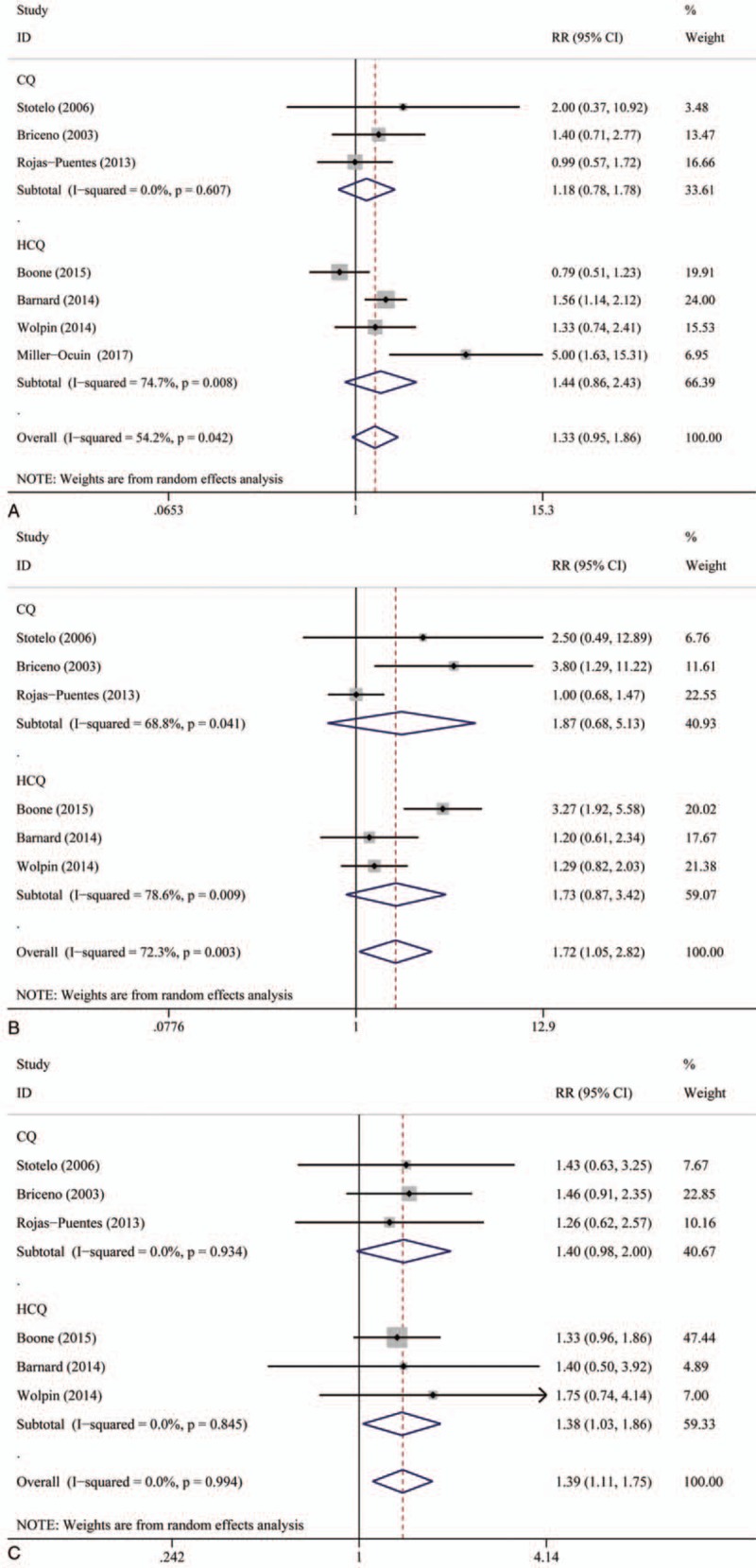
(A) Subgroup analysis of overall response rate (ORR). (B) Subgroup analysis of 6-month progression-free survival (PFS). (C) Subgroup analysis of 1-year overall survival (OS) by type of autophagy inhibitors.
We performed further analyses to evaluate the efficacy of different types of therapy combination. The result of this analysis is shown in Figure 3. We found that adding autophagy inhibitors to radiation did not improve ORR and 6-month PFS rate significantly. Subgroup analysis of autophagy-inhibitor-based combination type and cancer type are shown in Figures 3 and 4, respectively.
Figure 3.

(A) Subgroup analysis of overall response rate (ORR). (B) Subgroup analysis of 6-month progression-free survival (PFS). (C) Subgroup analysis of 1-year overall survival (OS) by type of cancer.
Figure 4.
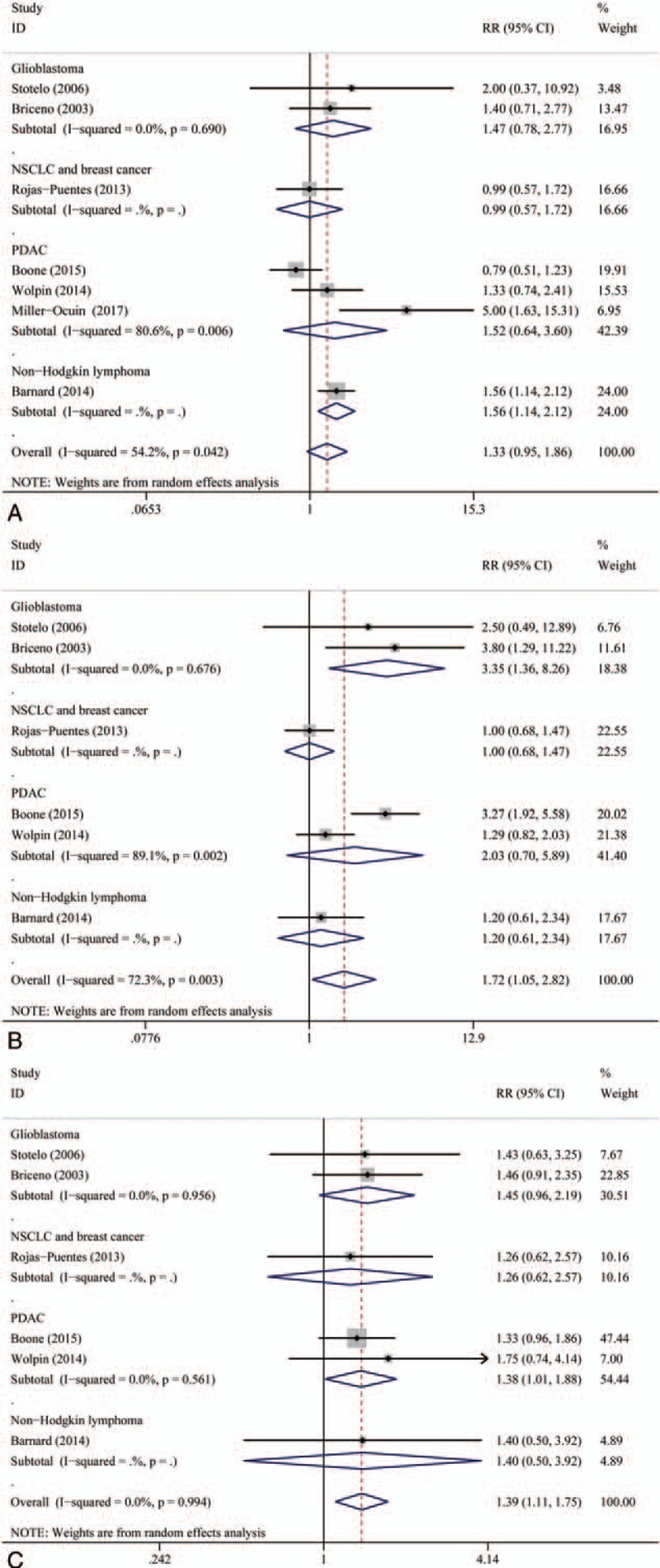
(A) Subgroup analysis of overall response rate (ORR). (B) Subgroup analysis of 6-month progression-free survival (PFS). (C) Subgroup analysis of 1-year overall survival (OS) by type of combination.
3.5. Sensitivity analysis and publication bias
All sensitivity analysis associated with the meta-analyses performed in this study indicated a stable results. No sensitivity analysis shows positive results. Detailed sensitivity analysis is shown in Supporting Information Figure S2.
Our publication bias was based on both Begg test and Egger test. In Begg test, there was no publication bias. The P-values were .230 for ORR, .260 for 6-month PFS, .452 for 1-year OS. In Egger test, the P-values were .515 for ORR, .289 for 6-month PFS, .387 for 1-year OS. The Begg graphs are shown in Figure 5, and the Egger graphs are shown in Supporting Information Figure S3.
Figure 5.

Publication bias assessed by Begg test. (A) ORR. (B) 6-Month progression-free survival (PFS). (C) 1-Year overall survival (OS).
4. Discussion
To the best of our knowledge, this is the 1st meta-analysis to assess the efficacy of autophagy inhibitors and to compare the difference between 2 types of autophagy inhibitors in the treatment of patients with cancer, focusing on the use of autophagy inhibitors in cancer clinical trials. The current trials have some limitations, but we think outcomes can still provide insights into autophagy inhibition therapy.
Our meta-analysis indicates that adding autophagy inhibitors to the treatment of patients with cancer can contribute higher ORR, 1-year OS rate, and 6-month PFS rate. After all autophagy-inhibitor-based combination therapy evaluated, we found that combination of autophagy inhibitor and gemcitabine yielded the best ORR, and combination of autophagy inhibitor, temozolomide, and radiation yielded the best 6-month PFS rate. While the combination of autophagy inhibitor with radiation did not contribute to significant improvement of ORR and 6-month PFS rate, improvements in 1-year OS rate were documented in all combinations.
Currently, CQ and HCQ are the only available drugs in clinical. Several trials on autophagy inhibition have been conducted and their outcomes have also been published. However, the clinical value of targeting autophagy therapy is still controversial. Hence, we performed a subgroup analysis to evaluate the clinical value of targeting autophagy by CQ and HCQ. This meta-analysis showed that CQ and HCQ both can significantly improve ORR, 1-year OS rate, and 6-month PFS rate. HCQ-based therapy can better benefit 1-year OS rate and 6-month PFS rate than CQ-based therapy, and CQ-based therapy can better benefit ORR than HCQ-based therapy.
In terms of caner type, we found that autophagy inhibitor can lead to the best ORR in patients with non-Hodgkin lymphoma. In patients with glioblastoma, autophagy inhibitor can lead to the best 6-month PFS and 1-year OS. However, autophagy inhibitor did not contribute to significant improvement of ORR and 6-month PFS in patients with nonsmall-cell lung cancer or breast cancer.
It is thought that pharmacologic inhibitors of autophagy can be novel cancer therapeutic agents. Some previous studies proved that autophagy is associated with various physiologic mechanisms including apoptosis, cancer metabolism, and drug resistance.[28–36] Results of this meta-analysis further support previous related studies.
Some certain limitations of the current meta-analysis need to be considered. The studies we included evaluated the treatment agents in various types of cancer with different etiologies and disease course. Different types of cancer may have some effect on the results of this meta-analysis. Two inhibitors (chloroquine or hydroxychloroquine) were evaluated in all cancer types. It may be different in different cancer types. In addition, some studies have reported only short-term follow-up and lack of long-term outcomes. Because of the data limitation, we did not evaluate the safety of autophagy inhibition used in the cancer treatment.
This meta-analysis found that autophagy inhibition by using CQ and HCQ can significantly enhance ORR, 1-year OS rate, and 6-month PFS rate. Combination of autophagy inhibitor and gemcitabine yielded the best ORR, and combination of autophagy inhibitor, temozolomide, and radiation yielded the best 6-month PFS rate. Autophagy inhibitor can lead to the best survival benefit in patients with glioblastoma. The application of autophagy inhibitor in glioblastoma should be paid more attention to, and the efficiency and safety of different autophagy-inhibitor-based combination therapy should be investigated further.
Acknowledgment
The authors thank the data provided by the authors of included trials.
Author contributions
Conceptualization: Ran Xu, Chen Xu, Jing Zhu.
Data curation: Ran Xu, Chen Xu, Jing Zhu.
Formal analysis: Ran Xu, Chen Xu, Jing Zhu.
Funding acquisition: Ran Xu, Chen Xu, Jing Zhu.
Investigation: Ran Xu, Chen Xu, Jing Zhu.
Methodology: Ran Xu, Chen Xu.
Project administration: Ran Xu, Chen Xu, Jing Zhu.
Resources: Ran Xu, Chen Xu.
Software: Ran Xu, Chen Xu, Ziyi Ji, Jing Zhu.
Supervision: Ran Xu, Chen Xu, Jing Zhu.
Validation: Ran Xu, Jing Zhu.
Visualization: Ran Xu, Chen Xu, Jing Zhu.
Writing – original draft: Ran Xu.
Writing – review & editing: Ran Xu, Chen Xu, Ziyi Ji.
Supplementary Material
Footnotes
Abbreviations: CI = confidence interval, CQ = chloroquine, HCQ = hydroxychloroquine, NSCLC = nonsmall-cell lung cancer, ORR = overall response rate, OS = overall survival, PDAC = pancreatic adenocarcinoma, PFS = progression-free survival, RR = relative risk, TMZ = temozolomide.
All authors gave final approval for submission of the manuscript.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931–7. [DOI] [PubMed] [Google Scholar]
- [2].Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012;12:401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther 2011;131:130–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Towers CG, Thorburn A. Therapeutic targeting of autophagy. EBioMedicine 2016;14:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [7].DeVita VT, Jr, Chu E. A history of cancer chemotherapy. Cancer Res 2008;68:8643–53. [DOI] [PubMed] [Google Scholar]
- [8].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res 2014;24:24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Altman JK, Szilard A, Goussetis DJ, et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin Cancer Res 2014;20:2400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Masui A, Hamada M, Kameyama H, et al. Autophagy as a survival mechanism for squamous cell carcinoma cells in endonuclease G-mediated apoptosis. PLoS One 2016;11:e0162786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tan Q, Wang M, Yu M, et al. Role of autophagy as a survival mechanism for hypoxic cells in tumors. Neoplasia 2016;18:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [12].Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Mol Pharmacol 2014;85:830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fitzwalter BE, Thorburn A. Recent insights into cell death and autophagy. FEBS J 2015;282:4279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gump JM, Staskiewicz L, Morgan MJ, et al. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol 2014;16:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thorburn J, Andrysik Z, Staskiewicz L, et al. Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep 2014;7:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Goodall ML, Fitzwalter BE, Zahedi S, et al. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev Cell 2016;37:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang YP, Hu LF, Zheng HF, et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin 2013;34:625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maycotte P, Aryal S, Cummings CT, et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 2012;8:200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eng CH, Wang Z, Tkach D, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci U S A 2016;113:182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Maes H, Kuchnio A, Peric A, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell 2014;26:190–206. [DOI] [PubMed] [Google Scholar]
- [21].Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med 2006;144:337–43. [DOI] [PubMed] [Google Scholar]
- [22].Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg Focus 2003;14:e3. [DOI] [PubMed] [Google Scholar]
- [23].Rojas-Puentes LL, Gonzalez-Pinedo M, Crismatt A, et al. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiat Oncol 2013;8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Boone BA, Bahary N, Zureikat AH, et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann Surg Oncol 2015;22:4402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Barnard RA, Wittenburg LA, Amaravadi RK, et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy 2014;10:1415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wolpin BM, Rubinson DA, Wang X, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist 2014;19:637–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].MillerOcuin JL, Bahary NS, Singhi AD. Inhibition of autophagy improves pathologic and biomarker response to preoperative Gemcitabine/nab-paclitaxel in potentially resectable Pancreatic cancer: a phase II randomized controlled trial Annals of surgical oncology. 2017. [Google Scholar]
- [28].Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 2015;524:361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rabinowitz JD, White E. Autophagy and metabolism. Science 2010;330:1344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rao S, Tortola L, Perlot T, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014;5:3056. [DOI] [PubMed] [Google Scholar]
- [32].Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene 2016;35:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rosenfeldt MT, O’Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013;504:296–300. [DOI] [PubMed] [Google Scholar]
- [34].Ross SJ, Critchlow SE. Emerging approaches to target tumor metabolism. Curr Opin Pharmacol 2014;17:22–9. [DOI] [PubMed] [Google Scholar]
- [35].Valencia T, Kim JY, Abu-Baker S, et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell 2014;26:121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].White E. The role for autophagy in cancer. J Clin Invest 2015;125:42–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during and/or analyzed in this study are included in this published article (and its supplementary information files).


