Abstract
MicroRNAs (miRs) have been reported to serve critical roles in the progression of tumors. However, thus far, the role of miR-34a in breast cancer is largely unknown. Therefore, the present study aimed to investigate the expression and role of miR-34a in breast cancer, and to further explore the underlying molecular mechanism. Reverse transcription-quantitative polymerase chain reaction was performed to detect the level of miR-34a in human breast cancer tissues. In addition, the role of miR-34a in MCF-7 breast cancer cells was investigated by performing miR-34a overexpression or downregulation through transfection with miR-34a mimic or inhibitor, respectively. Next, the viability and invasion of the MCF-7 cells were respectively analyzed by MTT assay and transwell assay, while apoptosis and cell cycle progression were examined by flow cytometry. Furthermore, associated protein levels were measured using western blotting. The results demonstrated that miR-34a was downregulated in human breast cancer tissues in comparison with the adjacent normal tissues. In addition, Notch1 was demonstrated to be a direct target of miR-34a. miR-34a mimic transfection inhibited MCF-7 cell viability, induced cell apoptosis and G1 phase arrest, and prevented cell invasion, while miR-34a inhibitor transfection resulted in the opposite effects. In conclusion, the presented data indicated that miR-34a suppressed breast cancer cell proliferation and invasion, and its effect may partly be exerted by targeting Notch1.
Keywords: breast cancer, microRNA-34a, proliferation, invasion, Notch1
Introduction
Breast cancer is one of the most important malignancies that threaten the life of women worldwide (1,2). In recent years, advances in early diagnosis and systematic treatment of breast cancer have led to a significant reduction in mortality (1,2). However, thus far, breast cancer remains difficult to cure due to the recurrence, metastasis and tolerance to radiotherapy and chemotherapy. According to statistics, breast cancer accounts for ~23% of all cancer cases in women, and mortality due to breast cancer accounts for ~14% of all cancer-associated mortalities in women, while the incidence of breast cancer patients are increasing at an annual rate of >1.3 million (1). Therefore, identifying novel effective diagnostic and treatment methods is currently urgent in the field of breast cancer research.
MicroRNAs (miRNAs), a class of endogenous non-coding single-stranded small RNAs with a length of approximately 18–24 nucleotides, regulate the expression of the endogenous genes at the transcriptional or post-transcriptional levels via specific binding to the 3′-untranslated region (UTR) of target mRNA (3–5). Currently, abnormal expression of miRNAs is observed in almost all the malignant tumors, indicating that miRNAs are closely associated with the occurrence and development of tumors. miR-34a is one of the earliest detectable tumor suppressor miRNAs, located on chromosome 1p36.23 and widely found in all normal tissues, with the exception of the lungs (6). miR-34a has been reported to be significantly decreased in colon cancer, prostate cancer, pancreatic cancer and other tumor tissues (7–9), and to be involved in the regulation of tumor cell proliferation, invasion and apoptosis among other activities (10). As a downstream molecule of p53, miR-34a is considered to be an important tumor suppressor molecule that regulates the expression of multiple target genes and is involved in tumor suppression via forming a network with the relevant regulatory molecules (11).
The Notch pathway is a highly conserved signaling pathway widely expressed in vertebrates and invertebrates, serving an important regulatory role in embryonic development, organ maturation and tumor progression (12). In mammals, the Notch signaling pathway includes four transmembrane receptors (namely Notch1-4) and five ligands (Delta-like 1, 3 and 4; Jagged-1 and −2) (13). Studies have demonstrated that activation of Notch1 or Notch4 in mice induces the formation of spontaneous breast tumors, demonstrating that activation of the Notch pathway is an important cause of breast cancer (14–16).
In the present study, the aim was to investigate the expression and role of miR-34a in breast cancer tissues and cells, as well as to explore the underlying molecular mechanism involved.
Materials and methods
Human tissue specimens
The study was approved by the Human Ethics Committee Review Board of Wuxi Taihu Hospital (Wuxi, China), and informed consent was obtained from each patient. In total, primary breast cancer tissues and paired adjacent normal tissues were obtained from 30 females aged between 35 to 65 years (mean age, 50 years) who underwent resection surgery of breast tumors in the Wuxi Taihu Hospital between September 2015 and June 2016. None of the patients had received treatment prior to surgery. All patients recruited for the current study were diagnosed with invasive ductal carcinoma according to the morphological criteria detailed in the World Health Organization histologic classification of breast tumors (17). Of the 30 patients, 3, 7 and 20 patients were diagnosed with stage III, II and I invasive ductal carcinoma, respectively. All tissue samples were assessed and diagnosis was confirmed by pathologists. Tumor and adjacent normal tissues were snap-frozen in liquid nitrogen and then stored at −80°C until further use.
Cell culture
The MCF-7 cell line was obtained from American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS). Cells were incubated at 37°C in a humidified atmosphere of 5% CO2, and passaged every 2–3 days.
Cell transfection
At 24 h before cell transfection, MCF-7 cells (5×104 cells per well) were plated into 6-well plates and incubated at 37°C with 5% CO2. Next, miR-34a mimic (item no. P4185-250UCI), mimic control (cat. no. M-03-S), miR-34a inhibitor (item no. P4185-250UCI) or inhibitor control (cat. no. M-04; GenePharma Co., Ltd., Shanghai, China) were transfected into the MCF-7 cells using 30 µl Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as per the manufacturer's protocol. Cells without any treatment were considered as the negative control (NC) group. After incubation for 48 h, these cells were subjected to subsequent experiments. The transfection efficiency was assessed by detecting miR-34a expression using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
MTT assay
At 48 h after transfection, cell viability was detected using an MTT assay. Briefly, MCF-7 cells were harvested, reseeded into a 96-well culture plate (5,000 cells per well) and cultured at 37°C for 24, 48 or 72 h. Subsequently, MTT solution (Amresco, LLC, Solon, OH, USA) was added into each culture well and incubated for another 4 h. Finally, the optical density value was measured at 570 nm by using a SynergyTM 2 Multi-function Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA). Each experiment was repeated at least three times.
Cell cycle distribution assay
At 48 h after transfection, flow cytometry was applied to analyze the MCF-7 cell cycle distribution. Briefly, the transfected MCF-7 cells were collected by trypsinization and then fixed with ethanol overnight at 4°C. Subsequently, the MCF-7 cells were washed by PBS and stained with ribonuclease A (100 µg/ml) and propidium iodide (PI; 50 µg/ml; both Beyotime Institute of Biotechnology, Shanghai, China) in the dark at 4°C for 30 min. Finally, a flow cytometer (FACSCanto II; BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze the cell cycle distribution. Tests were repeated at least three times.
Apoptosis analysis assay
MCF-7 cells were transfected with miR-34a mimic, mimic control, miR-34a inhibitor or inhibitor control, and at 48 h after the transfection, cells were labeled with Annexin V-FITC and PI (Cell Signaling Technology, Inc., Danvers, MA, USA) as per the manufacturer's protocol. Flow cytometry (BD Biosciences) was then applied to analyze cell apoptosis. Tests were repeated three times.
Cell invasion assay
Transwell assay was used to detect the cell invasion ability. Briefly, MCF-7 cells were transfected with miR-34a mimic, mimic control, miR-34a inhibitor or inhibitor control. After 48 h, the cells were harvested, re-suspended in serum-free medium, and then seeded into the upper chamber with a Matrigel-coated membrane matrix. Cell culture medium containing 20% FBS was added to the lower chamber as a chemoattractant, and the cells were incubated for 48 h at 37°C with 5% CO2. Finally, the non-invading cells on the upper surface were removed, while the invasive cells on the bottom surface of the membrane were fixed with methanol for 30 min at room temperature and then stained with hematoxylin for 5 min at room temperature. A microscope was then used to observe the stained cells. Tests were repeated three times.
Western blot analysis
At 48 h after transfection, cells were harvested by cell scrapers and washed with PBS. The total cell proteins were extracted using a lysis buffer (Cell Signaling Technology, Inc.) according to the manufacturer's protocol. The concentration of protein samples was determined with a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amount of the protein samples (30 µg/lane) were resolved by 12% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes. Subsequent to blocking with 5% skim milk at room temperature for 2 h, the membranes were blotted overnight at 4°C with primary antibody against Notch1 (cat no. 3608; Cell Signaling Technology, Inc.; dilution, 1:1,000), and then incubated with anti-rabbit horseradish peroxidase-linked IgG secondary antibody at room temperature for 2–3 h (cat no. 7074; Cell Signaling Technology, Inc.; dilution, 1:2,000). GAPDH antibody (cat no. 5174; Cell Signaling Technology, Inc.; dilution, 1:5,000) was also used as the internal control and incubated at room temperature for 2–3 h. Finally, the protein bands were observed using an enhanced chemiluminescence detection system (Super Signal West Dura Extended Duration Substrate; Thermo Fisher Scientific, Inc.).
RT-qPCR analysis
The breast tissues (cancer and adjacent normal), MCF-10A cells (Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China) as normal breast cells and MCF-7 cells were analyzed by PCR to examine miRNA expression. Breast tissues (50–100 mg) cut into pieces were homogenized prior to total RNA extraction. Total RNA from breast tissue homogenates, and MCF-10A and MCF-7 cells was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in line with the manufacturer's protocol. RT was then performed to generate cDNA using a PrimeScript Reverse Transcription Reagent kit (Takara Biotechnology Co., Ltd., Beijing, China) according to the manufacturer's protocol. Subsequently, qPCR was conducted to analyze the cDNA using a TaqMan Universal PCR Master Mix kit (Thermo Fisher Scientific, Inc.). The amplification conditions were as follows: 95°C for 10 min, followed by 38 cycles of 95°C for 10 sec and 58°C for 60 sec. miR-34a expression was normalized to U6 and Notch1 mRNA expression was normalized to GAPDH. The primer sequences for qPCR were as follows: U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′; miR-34a forward, 5′-CGTCACCTCTTAGGCTTGGA-3′ and reverse, 5′-CATTGGTGTCGTTGTGCTCT-3; and Notch1 forward, 5′-GAGGCGTGGCAGACTATGC-3′ and reverse, 5′-CTTGTACTCCGTCAGCGTGA-3′. The 2−ΔΔCq method was used to quantify the relative gene expression levels (18).
Dual-luciferase reporter assay
Bioinformatics analysis was initially conducted with TargetScan software (http://www.targetscan.org) in order to predict the potential targets of miR-34a, and the findings revealed that Notch1 was a potential target of miR-34a. To confirm this prediction, MCF-7 cells were seeded into each well of a 24-well plate (5×104 cells/well). At 24 h later, the cells were co-transfected with Notch1 3′UTR pmirGLO plasmid (containing mutant Notch1 3′UTR or wild-type Notch1 3′UTR) and a miR-34a mimic or mimic control (NC) vector using Lipofectamine® 2000 reagent according to the manufacturer's protocol. Following incubation for another 48 h, the luciferase activity was assessed using the dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA).
Statistical analysis
Data are presented as the mean ± standard deviation. SPSS statistical software (version 17.0; SPSS, Inc., Chicago, IL, USA) was used for all statistical analyses. Student's t-test or one-way analysis of variance followed by Tukey's test was used to compare the data between groups. P<0.05 was considered to denote a statistically significant difference.
Results
miR-34a is downregulated in breast cancer
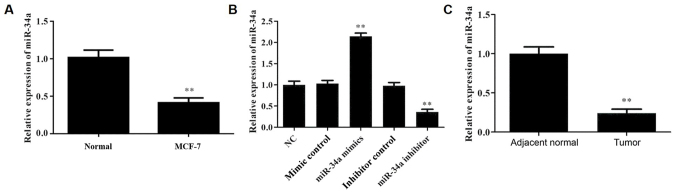
To determine the expression levels of miR-34a in MCF-10A cells and MCF-7 cells, RT-qPCR was performed. The results demonstrated that miR-34a expression was significantly lower in MCF-7 cells compared with normal cells (Fig. 1A). Next, in order to investigate the role of miR-34a in breast cancer, the MCF-7 cell line was used. miR-34a was overexpressed or downregulated in MCF-7 cells by transfections with miR-34a mimic or miR-34a inhibitor, respectively, and the transfection efficiency was detected by RT-qPCR (Fig. 1B). Subsequently, the expression of miR-34a was analyzed by RT-qPCR in breast cancer and adjacent normal breast tissues. As shown in Fig. 1C, compared with the adjacent normal tissues, the level of miRNA-34a in breast cancer tissues was significantly reduced, indicating that miRNA-34a may be involved in breast cancer progression.
Figure 1.
miR-34a expression level in different groups, as detected by reverse transcription-quantitative polymerase chain reaction. Relative expression level of miR-34a in the (A) MCF-10A and MCF-7 cells, (B) MCF-7 cells transfected with miR-34a mimic, mimic control, miR-34a inhibitor or inhibitor control and (C) breast cancer and adjacent normal tissues. **P<0.01 vs. NC group. miR, microRNA; NC, negative control.
miR-34a directly targets Notch1
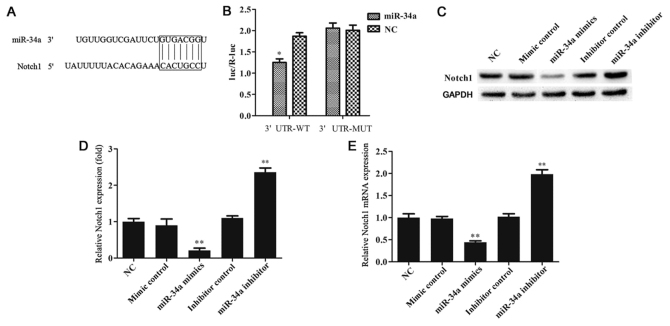
Bioinformatics analysis was conducted with the TargetScan tool to predict the potential targets of miR-34a, and the findings revealed that Notch1 is one of the target genes of miR-34a (Fig. 2A). Previous studies have demonstrated that activation of Notch1 or Notch4 in mice induced the formation of spontaneous breast tumors, demonstrating that activation of the Notch signaling pathway is an important cause of breast cancer (14–16); thus, Notch1 was selected for further investigation in the present study. To confirm this prediction, a dual-luciferase reporter assay was used, and the findings indicated that miR-34a directly targets Notch1 (Fig. 2B). To further reveal whether miR-34a was able to regulate Notch1 expression in MCF-7 cells, the protein and mRNA levels of Notch1 were detected by western blotting and RT-qPCR, respectively. The results indicated that miR-34a negatively regulated Notch1 expression in MCF-7 cells (Fig. 2C-E).
Figure 2.
miR-34a directly targets Notch1. (A) Interaction between miR-34a and 3′UTR of Notch1 was predicted by TargetScan. (B) Luciferase activity of a reporter containing a Notch1 3′UTR-WT or 3′UTR-MUT (with a mutation in the miR-34a binding site). (C) Western blots and (D) quantified expression of Notch1 protein in MCF-7 cells in different groups, expressed as the fold of the NC group. (E) Relative mRNA expression level of Notch1 in MCF-7 cells in different groups. *P<0.05 and **P<0.01 vs. NC group. miR, microRNA; NC, negative control; UTR, untranslated region; WT, wild-type; MUT, mutated.
Effect of miR-34a on MCF-7 cell viability
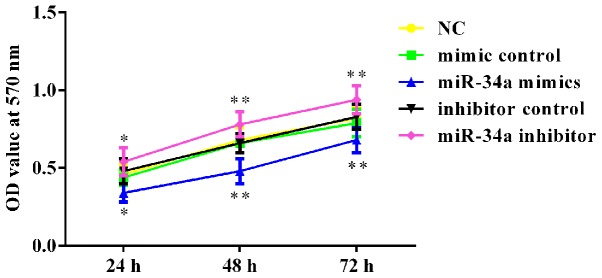
To investigate the effects of miR-34a on breast cancer cell viability, an MTT assay was performed. As shown in Fig. 3, the viability was significantly reduced in miR-34a mimic-transfected cells, while the viability of miR-34a inhibitor-transfected cells was notably increased following transfection at three time points. No statistically significant differences were detected between the NC and the mimic control or inhibitor control groups (Fig. 3).
Figure 3.
miR-34a inhibits MCF-7 cell viability. At 48 h after cell transfection, the viability of MCF-7 cells was analyzed by an MTT assay. *P<0.05 and **P<0.01 vs. NC group. miR, microRNA; NC, negative control; OD, optical density.
Effect of miR-34a on MCF-7 cell cycle distribution and apoptosis
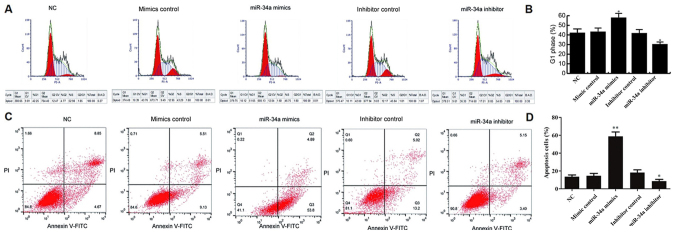
To investigate the effects of miR-34a on breast cancer cell cycle distribution and apoptosis, flow cytometry was applied. The results demonstrated that, at 48 h after transfection with miR-34a mimic, MCF-7 cells were significantly arrested in G1 phase when compared with the NC group. In addition, miR-34a inhibitor markedly declined the G1 phase arrest in MCF-7 cells as compared with that observed in the NC group. No significant difference was identified between the NC and the mimic control or inhibitor control groups (Fig. 4A and B).
Figure 4.
miR-34a induces MCF-7 cell apoptosis and G1 phase arrest. At 48 h after cell transfection, the cell cycle distribution and apoptosis of MCF-7 cells were determined by flow cytometry. (A) Cell cycle distribution and (B) the percentage of cells in G1 phase. (C) Apoptosis of MCF-7 cells in different groups. Q2 and Q3 indicate the early and late apoptosis, respectively. (D) The percentage of apoptotic cells. *P<0.05 and **P<0.01 vs. NC group. miR, microRNA; NC, negative control.
Furthermore, the data revealed that, compared with the NC group, miR-34a mimic significantly induced MCF-7 cell apoptosis (early and late apoptosis). By contrast, cell apoptosis was notably inhibited by miR-34a inhibitor. No significant differences were detected between the NC and the mimic control or inhibitor control groups (Fig. 4C and D).
Effect of miR-34a on MCF-7 cell invasion
At 48 h after cell transfection, the MCF-7 cell invasion ability was determined. The findings suggested that, compared with the NC group, MCF-7 cell invasion ability was significantly inhibited by miR-34a mimic, while miR-34a inhibitor notably promoted the MCF-7 cell invasion ability. However, no significant differences were observed between the NC and the mimic control or inhibitor control groups (Fig. 5).
Figure 5.
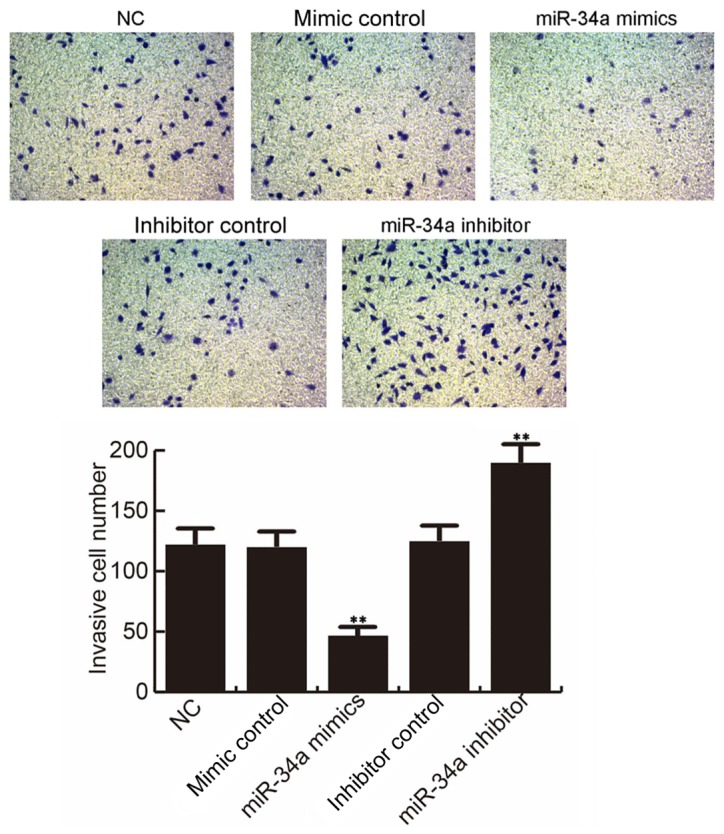
miR-34a prevents MCF-7 cell invasion. At 48 h after cell transfection, the invasion ability of MCF-7 cells was determined by a cell invasion assay, and the data were analyzed. **P<0.01 vs. NC group. miR, microRNA; NC, negative control.
Discussion
In recent years, great progress has been made in the investigation of miR-34a in breast cancer (19–21). Previous studies have demonstrated that multiple miRNAs are abnormally expressed in breast cancer and are closely associated with the development of this tumor (22,23). The study of miRNAs is important for the diagnosis, treatment and prognosis prediction of breast cancer (24). Therefore, in the present study, the expression and role of miR-34a in breast cancer were investigated.
Initially, the level of miR-34a in human breast cancer and adjacent normal tissues was detected in the present study, and the results revealed that miR-34a was significantly decreased in breast cancer tissues. Next, to further investigate the role of miR-34a in the development of breast cancer, the MCF-7 cell line was used to perform in vitro investigation. The bioinformatics prediction tool TargetScan was used to predict the potential targets of miR-34a, and the results indicated that Notch1 is a target of miR-34a. Notch1 pathway is considered to be an important pathway regulating the proliferation of breast cancer cells. Inhibition of Notch1 pathway in breast cancer cells and stem cells has been reported to effectively inhibit cell proliferation and promote apoptosis (25). The underlying mechanism may be associated with the decrease of the expression of nuclear factor-κB, cyclin B1 and B-cell lymphoma-2, which are located downstream of Notch1 (26). It has also been reported that miR-34a is able to inhibit the proliferation of a variety of tumor cells. For instance, Ma et al (27) found that miR-34a inhibited the proliferation and promoted the apoptosis of H1299 cells by targeting transforming growth factor β receptor II. Gougelet et al (28) also observed that miR-34a reduced the proliferation ability of primary cultured hepatocarcinoma cells by inhibiting cyclin D1 and hepatocyte nuclear factor-4α expression levels. The current study demonstrated that miR-34a was able to negatively regulate Notch1 expression in MCF-7 cells, and miR-34a mimic transfection inhibited MCF-7 cell viability, and induced cell apoptosis and G1 phase arrest.
Notch1 pathway is considered to be involved in the regulation of breast cancer cell invasion and migration. Recently, Zhang et al (29) reported that upregulation of Notch1 in the breast cancer cell line MCF-7 induced epithelial-mesenchymal transition, and promoted cell invasion and migration. In addition, Notch signaling activation promoted the ability of cell migration and growth (30). miR-34a is also an important regulatory factor involved in tumor invasion and metastasis. Lai et al (31) reported that miR-34a was able to inhibit colon cancer cell migration through regulating the sirtuin 1/p53 pathway. Liang et al (32) also indicated that the inhibitory effect of miR-34a on the invasion and migration of prostate cancer cells was associated with the targeted regulation of lymphoid enhancer binding factor 1. Finally, Yu et al (33) demonstrated that miR-34a inhibited the invasion, migration and angiogenesis of bladder cancer by targeting CD44 in vitro and in vivo. In the present study, the results revealed that miR-34a mimic prevented MCF-7 cell invasion, which is consistent with the findings of the aforementioned previous studies.
In conclusion, the data of the current study proved the low expression level of miR-34a in human breast cancer tissues as compared with the adjacent normal tissues. In addition, overexpression of miR-34a was found to inhibit breast cancer cell viability and invasion, as well as induce cell apoptosis and G1 phase arrest. miR-34a may exerts its role, at least partly, by targeting Notch1, and this miRNA may be a promising therapeutic target for the treatment of breast cancer. However, only MCF7 cells were examined in current study; therefore, similar results in other breast cancer cells would be required to establish the role of miR-34a in the suppression of breast cancer cell proliferation and invasion.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XR conceptualized and developed the study design, and performed most of the experiments. LW and LM cultured the cells and performed the cell transfections. XR, HZ and YY participated in western blot analysis. LW, HZ and YY acquired and analyzed the data. XR, XX, LW and LM conducted the statistical analysis, interpreted the data and wrote the manuscript. XR and XX made comments, suggested appropriate modifications and made corrections. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Human Ethics Committee Review Board of Wuxi Taihu Hospital, and informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Pang RT, Leung CO, Lee CL, Lam KK, Ye TM, Chiu PC, Yeung WS. MicroRNA-34a is a tumor suppressor in choriocarcinoma via regulation of Delta-like1. BMC Cancer. 2013;13:25. doi: 10.1186/1471-2407-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang CY, Qiu SL, Sun F, Li W, Wang Z, Yue B, Wu X, Yan D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Corcoran C, Rani S, O'Driscoll L. miR-34a is an Intracellular and exosomal predictive biomarker for response to docetaxelwith clinical relevance to prostate cancer progression. Prostate. 2014;74:1320–1334. doi: 10.1002/pros.22848. [DOI] [PubMed] [Google Scholar]
- 9.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: A new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 12.Yin L, Velazquez OC, Liu ZJ. Notch signaling: Emerging molecular targets for cancer therapy. Biochem Pharmacol. 2010;80:690–701. doi: 10.1016/j.bcp.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 14.Shao S, Zhao X, Zhang X, Luo M, Zuo X, Huang S, Wang Y, Gu S, Zhao X. Notch1 signaling regulates the epithelial inverted question markmesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol Cancer. 2015;14:28. doi: 10.1186/s12943-015-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naik S, MacFarlane M, Sarin A. Notch4 signaling confers susceptibility to TRAIL-induced apoptosis in breast cancer cells. J Cell Biochem. 2015;116:1371–1380. doi: 10.1002/jcb.25094. [DOI] [PubMed] [Google Scholar]
- 16.Bakrania AK, Variya BC, Rathod LV, Patel SS. DEAE-Dextran coated paclitaxel nanoparticles act as multifunctional nano system for intranuclear delivery to triple negative breast cancer through VEGF and NOTCH1 inhibition. Eur J PharmBiopharm. 2018;122:37–38. doi: 10.1016/j.ejpb.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO Classification of Tumours of the Breast. 4th. Lyon: IARC Press; 2012. [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, Li D, Yang C, Ji JS. MicroRNA-34a Inhibition of the TLR signaling pathway via CXCL10 suppresses breast cancer cell invasion and migration. Cell Physiol Biochem. 2018;46:1286–1304. doi: 10.1159/000489111. [DOI] [PubMed] [Google Scholar]
- 20.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Canc Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia Y, Deng XW, Cao MJ, Liu S, Zhang X, Xiao X, Shen S, Hu Q, Sheng W. Nanodiamond-based layer-by-layer nanohybrids mediate targeted delivery of miR-34a for triple negative breast cancer therapy. RSC Adv. 2018;8:13789–13797. doi: 10.1039/C8RA00907D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulationin human breast cancer. Cancer Res. 2015;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA. 2012;109:3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshino I, Matsubara H. MicroRNAs in cancer diagnosis and therapy: From bench to bedside. Surg Today. 2013;43:467–478. doi: 10.1007/s00595-012-0392-5. [DOI] [PubMed] [Google Scholar]
- 25.Suman S, Das TP, Damodaran C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br J Cancer. 2013;109:2587–2596. doi: 10.1038/bjc.2013.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan H, Zhou W, He W, Liu X, Ding Q, Ling L, Zha X, Wang S. Genistein inhibits MDA-MB-231 triple-negative breast cancer cell growth by inhibiting NF-κB activity via the Notch-1 pathway. Int J Mol Med. 2012;30:337–343. doi: 10.3892/ijmm.2012.990. [DOI] [PubMed] [Google Scholar]
- 27.Ma ZL, Hou PP, Li YL, Wang DT, Yuan TW, Wei JL, Zhao BT, Lou JT, Zhao XT, Jin Y, Jin YX. MicroRNA-34a inhibits the proliferation and promotes the apoptosis of non-small cell lung cancer H1299 cell line by targeting TGFβR2. Tumour Biol. 2015;36:2481–2490. doi: 10.1007/s13277-014-2861-5. [DOI] [PubMed] [Google Scholar]
- 28.Gougelet A, Sartor C, Bachelot L, Godard C, Marchiol C, Renault G, Tores F, Nitschke P, Cavard C, Terris B, et al. Antitumour activity of an inhibitor of miR-34a in liver cancer with beta-catenin-mutations. Gut. 2016;65:1024–1034. doi: 10.1136/gutjnl-2014-308969. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Zhao X, Shao S, Zuo X, Ning Q, Luo M, Gu S, Zhao X. Notch1 induces epithelial-mesenchymal transition and the cancer stem cell phenotype in breast cancer cells and STAT3 plays a key role. Int J Oncol. 2015;46:1141–1148. doi: 10.3892/ijo.2014.2809. [DOI] [PubMed] [Google Scholar]
- 30.Bolos V, Mira E, Martinez-Poveda B, Luxán G, Cañamero M, Martínez-A C, Mañes S, de la Pompa JL. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. 2013;15:R54. doi: 10.1186/bcr3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai M, Du G, Shi R, Yao J, Yang G, Wei Y, Zhang D, Xu Z, Zhang R, Li Y, et al. miR-34a inhibits migration and invasion by regulating the SIRT1/p53 pathway in human SW480 cells. Mol Med Rep. 2015;11:3301–3307. doi: 10.3892/mmr.2015.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang J, Li Y, Daniels G, Sfanos K, De Marzo A, Wei J, Li X, Chen W, Wang J, Zhong X, et al. LEF1 targeting EMT in prostate cancer invasion is regulated by miR-34a. Mol Cancer Res. 2015;13:681–688. doi: 10.1158/1541-7786.MCR-14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G, Yao W, Xiao W, Li H, Xu H, Lang B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting CD44. J Exp Clin Cancer Res. 2014;33:779. doi: 10.1186/s13046-014-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.