Abstract
The present study aimed to identify shared microRNAs (miRNAs) in ovarian cancer (OC) cells and their exosomes using microarray data (accession number GSE103708) available from the Gene Expression Omnibus database, including exosomal samples from 13 OC cell lines and 3 normal ovarian surface epithelial cell lines, and their original cell samples. Differentially expressed miRNAs (DE-miRNAs) were identified using the Linear Models for Microarray data method, and mRNA targets of DE-miRNAs were predicted using the miRWalk2 database. The potential functions of target genes were analyzed using Database for Annotation, Visualization and Integrated Discovery and intersected with known OC-associated pathways downloaded from the Comparative Toxicogenomics Database. The associations between crucial miRNAs and target genes, and their clinical associations, were validated using data from The Cancer Genome Atlas. As a result, 16 upregulated and 6 downregulated DE-miRNAs were shared in OC cell lines and their exosomes compared with normal controls. The target genes of 11 common DE-miRNAs were predicted. Among these DE-miRNAs, a low expression of homosapiens (hsa)-miR-145-5p was significantly correlated with a poor prognosis and higher stages. Although 91 target genes were predicted for hsa-miR-145-5p, only 4 genes [connective tissue growth factor (CTGF), myotubularin-related protein 14, protein phosphatase 3 catalytic subunit alpha and suppressor of cytokine signaling 7] were suggested as risk factors for prognosis. The subsequent Pearson’s correlation analysis validated a significant negative correlation between hsa-miR-145-5p and CTGF (r=−0.1126, P=0.02188). According to the results of the functional analysis, CTGF is involved in the Hippo signaling pathway (hsa04390). In conclusion, decreased expression of hsa-miR-145 in OC and OC-derived exosomes may be a crucial biomarker for the diagnosis and treatment of OC.
Keywords: ovarian cancer, exosomes, microRNA-145-5p, connective tissue growth factor, Hippo signaling pathway
Introduction
Ovarian cancer (OC) is the most common malignant tumor threatening the health of women in China. In 2015, an estimated 52,100 incident cases were diagnosed, with >22,500 mortalities (1). Although patients with OC are generally treated with a series of standard treatments, including surgery, chemotherapy and radiotherapy, the prognosis remains unsatisfactory for the majority of patients, with an overall 5-year survival rate of ~30% (2). Additionally, advanced OC may only be observed at the point of the initial diagnosis, which may be too late for patients to receive optimal treatment. Therefore, a screen of effective diagnostic biomarkers and the development of novel therapeutic strategies are urgently required.
Based on previous evidence, microRNAs (miRNAs/miRs) serve important roles in the development and progression of OC by specifically binding to the 3′-untranslated region (3′-UTR) of target mRNAs to inhibit translation or induce degradation (3). For example, Qin et al (4) observed a significant decrease in the expression of miR-152 in OC specimens and three OC cell lines. The transfection of miR-152 mimic into OC cells inhibited proliferation and migration by targeting the 3′-UTR of the forkhead box protein 1 (FOXP1) and resulted in improved overall survival. Liu et al (5) confirmed the over-expression of miR-26a in human OC specimens and revealed correlations between lymph node metastasis, an advanced International Federation of Gynecology and Obstetrics (FIGO) stage and poor survival. miR-216a promoted the metastatic behaviors and epithelial-mesenchymal transition (EMT) of OC cells by inhibiting its direct downstream target phosphatase and tensin homolog and subsequently regulating the protein kinase B pathway. Therefore, miRNAs may serve as promising biomarkers for the diagnosis of OC and attractive therapeutic targets.
In addition to their intracellular functions, miRNAs are also packaged into membrane-bound vesicles, including exosomes, and released to the extracellular environment to facilitate tumorigenesis and progression (6). For example, in the study by Ying et al (7), OC-derived exosomes released miR-222-3p into macrophages and induced the formation of a tumor-associated macrophage-like phenotype by decreasing the expression of suppressor of cytokine signaling 3 (SOCS3) and activating SOCS3/signal transducer and activator of transcription (STAT)-3 signaling pathway, thereby promoting the growth and metastasis of OC. According to the study by Kanlikilicer et al (8), miR-6126 is released from OC cells via exosomes. miR-6126 significantly reduces tumor growth, invasion and migration in vitro and in vivo by suppressing the expression of integrin-β1. De et al (9) observed the anti-neoplastic effects of an Emblica officinalis extract on OC that were mediated by the upregulation of cellular and exosomal miR-375 expression. Meng et al (10) observed correlations between high levels of exosomal miR-373, miR-200a, miR-200b and miR-200c expression with a higher FIGO stage (III-IV) and a shorter overall survival. Therefore, exosomal miRNAs may also be crucial targets for cancer diagnosis and treatment. Strategies targeting co-expressed miRNAs in cancer cells and their exosomes may represent more effective approaches for inducing cancer remission (11), however, this information has been rarely reported in OC.
The present study aimed to investigate the shared miRNAs in OC cells and their exosomes using microarray data downloaded from Gene Expression Omnibus (GEO) database and validate their clinical significance with The Cancer Genome Atlas (TCGA) datasets. The present study preliminarily identified that downregulated hsa-miR-145 was common in OC and OC-derived exosomes and thus may be a crucial biomarker for the diagnosis and treatment of OC.
Materials and methods
Data collection
The miRNA microarray data (accession number GSE103708) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) on January 23, 2018; the dataset contained exosomal samples isolated from 13 human epithelial OC cell lines (A2780, ES-2, CAOV3, SKOV3, OV-90, OAW42, MCAS, COV362, RMG-1, RMUG-S, KURAMOCHI, NIH-OVCAR3 and A2780cis), 3 normal ovarian surface epithelial cell lines (HOSE1, HOSE2 and HOSE3) and their original cell samples. Each cell type had one biological repeat, resulting in 32 samples.
Data preprocessing and identification of differentially expressed miRNAs (DE-miRNAs)
The GSE Series Matrix and annotations files were retrieved from the Agilent-046064 Unrestricted_Human_miRNA_V19.0_Microarray Array platform GPL18402. The probe names were transferred to gene symbols based on the platform annotation information. When multiple probes were mapped to a given gene, the mean expression value was used. Data were then log2-transformed and quantile normalized using the Robust Multiarray Average function available in the Bioconductor R package (v3.32.5; http://www.biocon-ductor.org/packages/release/bioc/html/limma.html). The DE-miRNAs between OC and normal ovarian surface epithelial cells and exosomes and original cells were identified using the Linear Models for Microarray data method in the Bioconductor R package (v3.32.5; http://www.bioconductor.org/packages/release/bioc/html/limma.html) (12). The significance of differences of DE-miRNAs were assessed using the empirical Bayes moderated t-test. Subsequently, the P-value was corrected for multiple comparisons using the Benjamini-Hochberg (BH) procedure (13). miRNAs with a false discovery rate <0.05 and |logFC(fold change)|>1 were considered differentially expressed. The ability of DE-miRNAs to differentiate the samples was tested using the Euclidean distance-based bidirectional hierarchical clustering analysis in the Pheatmap package (v1.0.8; http://cran.r-project.org/web/packages/pheatmap/index.html), following which a heat map was produced. A Venn diagram (http://bioinfor-matics.psb.ugent.be/webtools/Venn/) was used to visualize the shared DE-miRNAs between different exosomes and original cells.
Target gene prediction
The mRNA targets of DE-miRNAs were predicted using the miRWalk database (v2.0; http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) (14). Then, the miRNA-target gene interaction network was constructed and visualized using Cytoscape software (v3.4; www.cytoscape.org/) (15).
Screening prognosis-associated miRNAs and their target genes
The miRNA and mRNA Seq data from OC samples (Level 3) were obtained from the TCGA database (https://tcga-data.nci.nih.gov/). The expression levels of targeted miRNAs and mRNAs in OC were analyzed using the fragment per kilobase per million mapped reads value from the TCGA data.
A univariate Cox regression analysis was performed to screen prognosis-associated miRNAs/mRNAs using the survival package in R (v2.4; https://cran.r-project.org/web/packages/survival/index.html) (16), with the log-rank P<0.05 set as the threshold. Univariate and multivariate Cox regression analyses were used to screen prognosis-associated clinical characteristics, including age, radiation therapy, stage (17), histological grade (18), neoplasm subdivision, lymphatic invasion, recurrence and survival, and then the potential associations between crucial miRNAs and clinical characteristics were also calculated using the survival package. The Kaplan-Meier curve with the log-rank test was plotted using GraphPad Prism software (v5; GraphPad Software, Inc., La Jolla, CA, USA) to determine significant associations between the expression of miRNAs/mRNAs and patient survival outcomes. Pearson’s correlation coefficients were calculated to assess the correlations between miRNAs and mRNAs. P<0.05 was considered to indicate a statistically significant difference.
Functional enrichment analysis
The enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) (19) pathways of target genes were predicted by searching the Database for Annotation, Visualization and Integrated Discovery online tool (v6.8; http://david.abcc.ncifcrf.gov) (20). P<0.05 was set as the cut-off criterion. In addition, all known OC-associated pathways were also downloaded from the Comparative Toxicogenomics Database (CTD; http://ctd.mdibl.org/) (21). The pathways obtained from these two sources were overlapped to obtain crucial miRNAs involved in OC-associated pathways.
Results
Identification of DE-miRNAs between cancer and normal cells
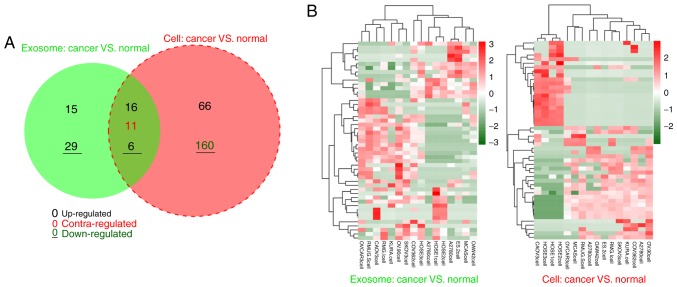
In the exosomal samples, 77 miRNAs were differentially expressed between 13 OC cancer and 3 normal cells, including 37 upregulated and 40 downregulated miRNAs. For the original cell samples, 259 DE-miRNAs were identified between 13 OC cancer and 3 normal cells, including 87 upregulated and 172 downregulated miRNAs. According to the Venn diagram, 22 DE-miRNAs with similar expression trends (16 upregulated and 6 downregulated) were shared between exosomal and original cell samples (Fig. 1; Table I), suggesting that these 22 DE-miRNAs may be particularly crucial for OC development in an exosomal or non-exosomal manner. Therefore, these DE-miRNAs were examined in subsequent analyses.
Figure 1.
Shared differentially expressed miRNAs in 13 ovarian cancer cells and their exosomes compared with normal cells. (A) Venn diagram. (B) Heat map. Contra-regulated in the Venn diagram indicates there was a different expression trend in ovarian cancer cells and their exosomes. Red in the heat map indicates the high expression of miRNAs; the green in the heat map indicates the lower expression of miRNAs. miRNA, microRNA.
Table I.
Shared differentially expressed miRNAs in exosomes and original cells.
| miRNA | Exosome
|
Original cells
|
||||
|---|---|---|---|---|---|---|
| logFC | P-value | FDR | logFC | P-value | FDR | |
| hsa-miR-202-3p - | 1.14 | 7.29×10−5 | 1.48×10−2 | −2.14 | 1.13×10−2 | 3.92×10−2 |
| hsa-miR-5684 | −1.73 | 5.76×10−5 | 1.17×10−2 | −1.58 | 1.94×10−3 | 8.36×10−3 |
| hsa-miR-376a-3p - | 1.74 | 9.67×10−6 | 1.97×10−3 | −1.80 | 3.36×10−3 | 1.38×10−2 |
| hsa-miR-141-3p | 2.13 | 8.71×10−6 | 1.77×10−3 | 2.01 | 6.05×10−3 | 2.29×10−2 |
| hsa-miR-376c-3p - | 2.07 | 4.41×10−6 | 8.95×10−4 | −2.08 | 6.82×10−4 | 3.32×10−3 |
| hsa-miR-381-3p - | 2.11 | 6.51×10−6 | 1.33×10−3 | −2.35 | 4.88×10−4 | 2.47×10−3 |
| hsa-miR-145-5p - | 2.67 | 1.15×10−8 | 2.35×10−6 | −2.16 | 1.84×10−4 | 1.03×10−3 |
| hsa-miR-378i | 2.79 | 2.54×10−6 | 5.15×10−4 | 2.13 | 2.05×10−3 | 8.70×10−3 |
| hsa-miR-98-5p | 1.286 | 2.42×10−4 | 4.91×10−2 | 5.30 | 4.07×10−6 | 4.21×10−5 |
| hsa-miR-7-5p | 1.48 | 9.31×10−5 | 1.90×10−2 | 4.92 | 6.08×10−5 | 3.89×10−4 |
| hsa-miR-374b-5p | 1.34 | 1.57×10−4 | 3.19×10−2 | 5.59 | 5.44×10−7 | 1.45×10−5 |
| hsa-miR-374a-5p | 1.42 | 1.08×10−4 | 2.20×10−2 | 5.74 | 1.54×10−7 | 8.57×10−6 |
| hsa-miR-301a-3p | 1.56 | 8.62×10−5 | 1.76×10−2 | 5.60 | 5.44×10−7 | 1.45×10−5 |
| hsa-miR-17-3p | 2.06 | 1.84×10−4 | 3.74×10−2 | 5.42 | 1.64×10−6 | 2.15×10−5 |
| hsa-miR-335-5p | 3.09 | 2.31×10−4 | 4.70×10−2 | 4.22 | 1.65×10−3 | 7.19×10−3 |
| hsa-miR-186-5p | 3.34 | 7.16×10−6 | 1.46×10−3 | 3.93 | 6.75×10−3 | 2.53×10−2 |
| hsa-miR-148a-3p | 2.50 | 1.50×10−5 | 3.05×10−3 | 5.27 | 5.01×10−6 | 4.90×10−5 |
| hsa-miR-532-5p | 3.35 | 1.78×10−4 | 3.63×10−2 | 4.30 | 1.21×10−3 | 5.51×10−3 |
| hsa-miR-660-5p | 3.45 | 1.29×10−4 | 2.62×10−2 | 4.43 | 7.61×10−4 | 3.64×10−3 |
| hsa-miR-205-5p | 4.31 | 6.74×10−6 | 1.37×10−4 | 4.48 | 7.61×10−4 | 3.64×10−3 |
| hsa-miR-126-3p | 4.15 | 1.17×10−5 | 2.37×10−3 | 4.85 | 6.98×10−5 | 4.44×10−4 |
| hsa-miR-200c-3p | 4.00 | 7.13×10−7 | 1.45×10−4 | 5.05 | 2.99×10−5 | 2.06×10−4 |
FC, fold change; FDR, false discovery rate; hsa, homo sapiens; miRNA, microRNA.
Comparison of target genes of DE-miRNAs between OC cells and normal controls
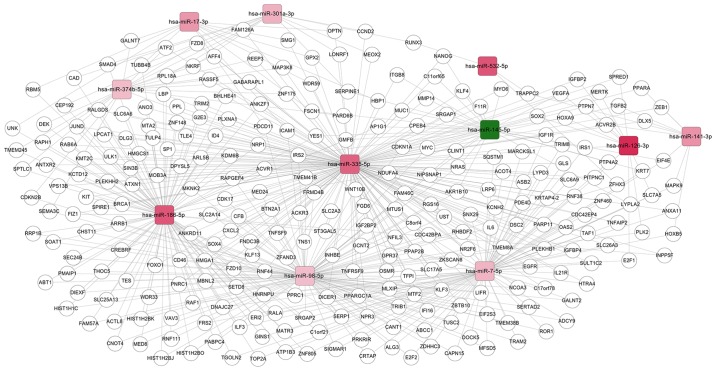
Subsequent to searching the miRWalk 2.0 database, only the target genes of 11 shared DE-miRNAs [upregulated: homo sapiens (has)-miR-126-3p, hsa-miR-141-3p, hsa-miR-17-3p, hsa-miR-186-5p, hsa-miR-301a-3p, hsa-miR-335-5p, hsa-miR-532-5p, hsa-miR-7-5p, hsa-miR-374b-5p, and hsa-miR-98-5p; down-regulated: hsa-miR-145-5p] were obtained. These common miRNAs and their target genes were then used to construct a miRNA-mRNA regulatory network, as demonstrated in Fig. 2. This network included 4,198 nodes (11 miRNAs and 4187 target genes) and 462 edges (interaction associations).
Figure 2.
Regulatory network comprising 11 common differentially expressed miRNAs in ovarian cancer cells and their exosomes, and their target genes. Red squares represent upregulated miRNAs, and green squares represent downregulated miRNAs. The changes in red shades indicated different numbers of target genes (darker shades = higher numbers). hsa, homo sapiens; miRNA, microRNA.
Screening prognosis-associated miRNAs and their target genes
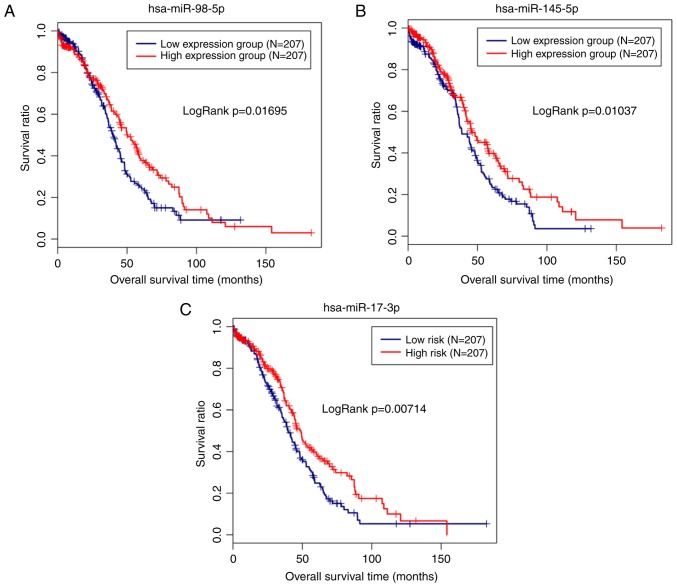
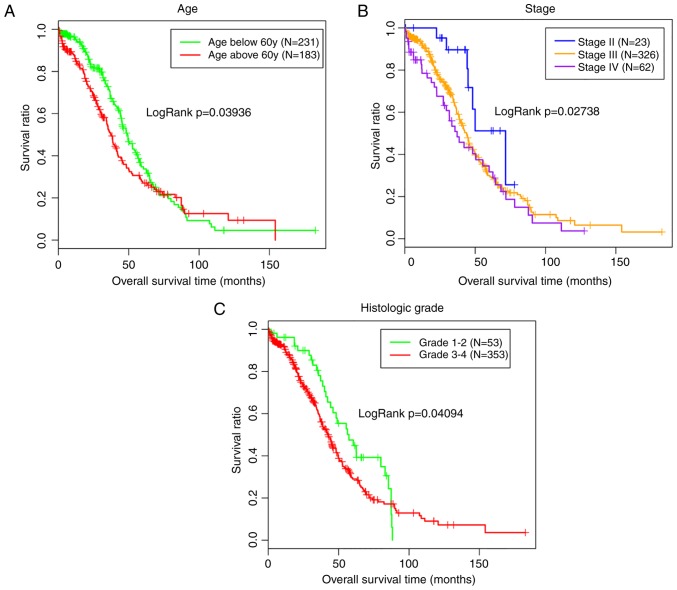
The miRNA/mRNA expression and clinical data associated with OC were also extracted from the TCGA database to validate the clinical importance of the miRNAs identified in the present study. Subsequently, the miRNA and mRNA expression data were matched with the clinical data for 414 patients. The expression of 11 shared DE-miRNAs was extracted from the TCGA data and then combined with the clinical data to screen the prognosis-associated miRNAs. Notably, hsa-miR-98-5p (upregulated; P=0.01046), hsa-miR-145-5p (downregulated; P=0.0178) and hsa-miR-17-3p (upregulated; P=0.0294) were significantly correlated with the survival outcomes of patients with OC. The Kaplan-Meier analysis additionally revealed a poor prognosis for patients with low expression levels of hsa-miR-145-5p, as expected (Fig. 3). Similarly, the subsequent univariate analyses revealed an association between hsa-miR-145-5p expression and the tumor stage (Table II), which was also an independent prognosis factor with survival (Table III; Fig. 4).
Figure 3.
Kaplan-Meier analysis of the associations between miRNAs and overall survival of patients with ovarian cancer. (A) miR-98-5p. (B) miR-145-5p. (C) miR-17-3p. hsa, homo sapiens; miRNA, microRNA.
Table II.
Associations between miRNAs and clinical characteristics using data from The Cancer Genome Atlas data.
| hsa-miR-17-3p
|
hsa-miR-145-5p
|
hsa-miR-98-5p
|
|
|---|---|---|---|
| Clinical characteristics | P-value | P-value | P-value |
| Age (59.44±11.42) | 0.10 | 0.30 | 0.26 |
| Radiation therapy (yes/no) | 0.90 | 0.61 | 6.62×10−3 |
| Neoplasm subdivision (bilateral/left/right) | 0.95 | 0.09 | 0.30 |
| Stage (II/III/IV) | 0.01 | 0.01 | 0.39 |
| Lymphatic invasion (yes/no) | 0.53 | 0.65 | 0.12 |
| Histologic grade (G1-G2/G3-G4) | 0.26 | 0.85 | 0.88 |
| Recurrence (yes/no) | 0.14 | 0.74 | 0.09 |
hsa, homo sapiens; miRNA, microRNA. Data are presented as the mean ± standard deviation. The survival package was used for the statistical analysis.
Table III.
Prognosis-associated clinical characteristics using data from The Cancer Genome Atlas data.
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Radiation therapy (yes/no) | 0.84 | 0.53-1.34 | 0.47 | - | - | - |
| Neoplasm subdivision (bilateral/left/right) | 0.99 | 0.80-1.21 | 0.89 | - | - | - |
| Lymphatic invasion (yes/no) | 1.14 | 0.70-1.86 | 0.59 | - | - | - |
| Recurrence (yes/no) | 1.73 | 1.45-2.20 | 0.21 | - | - | - |
| Age (59.44±11.42) | 1.02 | 1.01-1.03 | 3.78×10−3 | 1.02 | 1.01-1.03 | 2.49×10−3 |
| Stage (II/III/IV) | 1.38 | 1.04-1.82 | 0.03 | 1.36 | 1.02-1.83 | 0.04 |
| Histologic grade (G1-G2/G3-G4) | 1.39 | 0.96-2.00 | 0.04 | 1.35 | 0.93-1.95 | 0.04 |
HR, hazard ratio; CI, confidence interval. Data are presented as the mean ± standard deviation. The survival package was used for the statistical analysis.
Figure 4.
Kaplan-Meier analysis of the associations between the clinical characteristics and survival outcomes of patients with ovarian cancer. (A) Age. (B) Stage. (C) Histological grade.
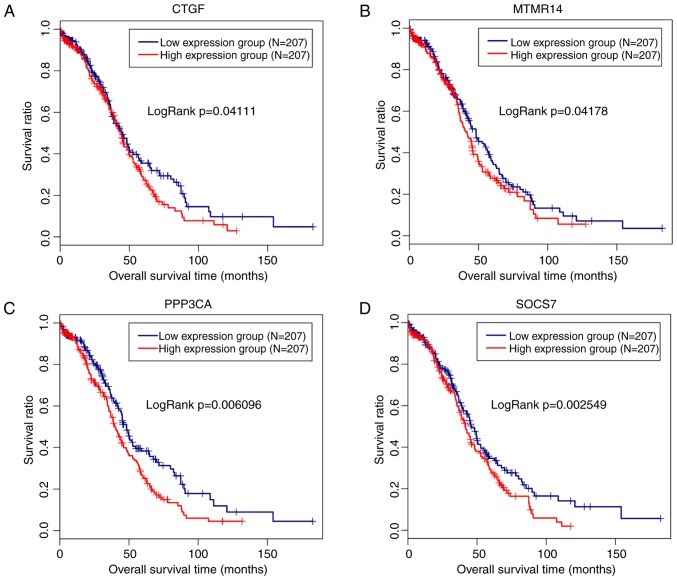
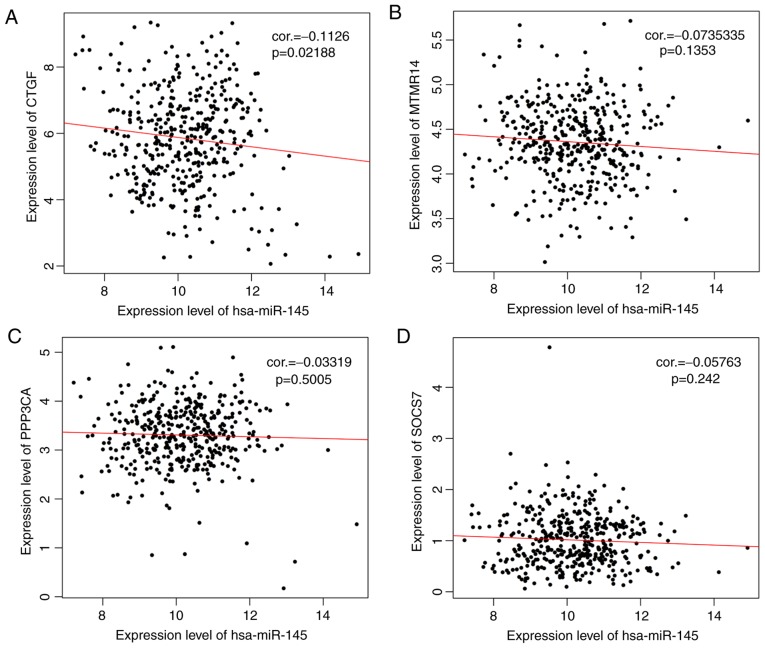
The 91 target genes of hsa-miR-145-5p were included in the screen of prognosis-associated genes. As a result, 11 genes, including ADP ribosylation factor-like GTPase 6 interacting protein 5 (P=0.017), connective tissue growth factor (CTGF; P=0.047), matrix metallopeptidase 12 (P=0.0068), myotubularin-related protein 14 (MTMR14; P=0.03), p21 (RAC1)-activated kinase 4 (P=0.026), protein phosphatase 3 catalytic subunit alpha (PPP3CA; P=0.018), suppressor of cytokine signaling 7 (SOCS7; P=0.0084), STAT1 (P=0.015), tropomyosin 3 (P=0.0015), tetraspanin 6 (P=0.036), vascular endothelial growth factor A (P=0.0049) and AT-rich interaction domain 4B (P=0.015), were obtained, among which 4 genes (CTGF, exp(coef)=1.1; MTMR14, exp(coef)=1.42; PPP3CA, exp(coef)=1.31; SOCS7, exp(coef)=1.43) were suggested to be risk factors for prognosis. Therefore, a Kaplan-Meier survival curve was drawn for these 4 genes to reveal their associations with survival. As anticipated, a poor prognosis was observed for patients expressing high levels of these 4 genes, and the difference was significant (Fig. 5). However, only a significant negative correlation was observed between hsa-miR-145-5p and CTGF (r=−0.1126, P=0.02188) using Pearson’s correlation analysis (Fig. 6).
Figure 5.
Kaplan-Meier analysis of the associations between the target genes of microR-145-5p and survival outcomes of patients with ovarian cancer. (A) CTGF. (B) MTMR14. (C) PPP3CA. (D) SOCS7. CTGF, connective tissue growth factor; MTMR14, myotubularin-related protein 14; PPP3CA, protein phosphatase 3 catalytic subunit alpha; SOCS7, suppressor of cytokine signaling 7.
Figure 6.
Pearson’s correlation analysis of the negative correlations between the target genes and miR-145-5p in patients with ovarian cancer. (A) CTGF. (B) MTMR14. (C) PPP3CA. (D) SOCS7. CTGF, connective tissue growth factor; MTMR14, myotubularin-related protein 14; PPP3CA, protein phosphatase 3 catalytic subunit alpha; SOCS7, suppressor of cytokine signaling 7; hsa, homo sapiens; miRNA, microRNA; cor, correlation coefficient.
According to the functional analysis, the target genes of hsa-miR-145-5p participated in OC development by affecting 26 KEGG pathways, 20 of which overlapped with 56 known OC-associated pathways downloaded from the CTD (Table IV). CTGF was identified to be involved in the Hippo signaling pathway (hsa04390).
Table IV.
Functional enrichment for the target genes of microR-145.
| Term | P-value | Genes |
|---|---|---|
| hsa05219:Bladder cancer | 3.91×10−7 | NRAS, CDKN1A, VEGFA, MDM2, CDK4, MYC, MMP1 |
| hsa04550:Signaling pathways regulating | 5.82×10−5 | NRAS, IGF1R, NANOG, POU5F1, SOX2, MYC, FZD7, KLF4 |
| pluripotency of stem cells | ||
| hsa05200:Pathways in cancer | 8.73×10−5 | NRAS, IGF1R, CDKN1A, HDAC2, VEGFA, MDM2, STAT1, |
| CDK4, MYC, FZD7, MMP1, TPM3 | ||
| hsa04151:PI3K-Akt signaling pathway | 1.45×10−4 | NRAS, IGF1R, CDKN1A, EIF4E, ITGB8, IFNB1, VEGFA, |
| MDM2, CDK4, IRS1, MYC | ||
| hsa05220:Chronic myeloid leukemia | 1.57×10−4 | NRAS, CDKN1A, HDAC2, MDM2, CDK4, MYC |
| hsa05205:Proteoglycans in cancer | 5.32×10−4 | NRAS, IGF1R, CDKN1A, NANOG, VEGFA, MDM2, MYC, |
| FZD7 | ||
| hsa05161:Hepatitis B | 5.72×10−4 | NRAS, CDKN1A, IFNB1, TIRAP, STAT1, CDK4, MYC |
| hsa05214:Glioma | 1.19×10−3 | NRAS, IGF1R, CDKN1A, MDM2, CDK4 |
| hsa05202:Transcriptional misregulation in cancer | 1.24×10−3 | IGF1R, ERG, CDKN1A, FLI1, HDAC2, MDM2, MYC |
| hsa04115:p53 signaling pathway | 1.33×10−3 | PPM1D, CDKN1A, SERPINE1, MDM2, CDK4 |
| hsa05218:Melanoma | 1.65×10−3 | NRAS, IGF1R, CDKN1A, MDM2, CDK4 |
| hsa05206:MicroRNAs in cancer | 4.11×10−3 | NRAS, CDKN1A, IRS2, PAK4, VEGFA, MDM2, IRS1, MYC |
| hsa04919:Thyroid hormone signaling pathway | 9.03×10−3 | NRAS, HDAC2, MDM2, STAT1, MYC |
| hsa04110:Cell cycle | 1.20×10−2 | CDKN1A, HDAC2, MDM2, CDK4, MYC |
| hsa04360:Axon guidance | 1.31×10−2 | NRAS, PAK4, ROBO2, PPP3CA, SRGAP1 |
| hsa05216:Thyroid cancer | 1.84×10−2 | NRAS, MYC, TPM3 |
| Shsa04390:Hippo signaling pathway | 2.32×10−2 | CTGF, SOX2, SERPINE1, MYC, FZD7 |
| hsa05215:Prostate cancer | 2.54×10−2 | NRAS, IGF1R, CDKN1A, MDM2 |
| hsa05166:HTLV-I infection | 3.66×10−2 | NRAS, CDKN1A, PPP3CA, CDK4, MYC, FZD7 |
| hsa05169:Epstein-Barr virus infection | 4.78×10−2 | CDKN1A, HDAC2, MDM2, MYC, MAP2K6 |
hsa, homo sapiens; PI3K, phosphoinositide 3-kinase; Akt, protein kinase B; p53, tumor protein 53; HTLV-I, human T-lymphotropic virus type 1.
Discussion
In the present study, the miRNA expression profiles of OC cells and their exosomes compared with normal ovarian surface epithelial cell lines were examined using a microarray. A total of 22 miRNAs were co-expressed in exosomes and the OC cells from which they were derived. Among these miRNAs, the downregulation of hsa-miR-145-5p and its negatively regulated target gene CTGF were additionally demonstrated to be associated with the prognosis of patients with OC by affecting the Hippo signaling pathway, and therefore they may be potentially important diagnostic biomarkers and therapeutic targets for OC.
Based on extensive evidence, miR-145-5p functions as a tumor suppressor gene in various cancers, including OC. For example, Zhang et al (22) observed the downregulation of miR-145-5p in gastric cancer tissues compared with the adjacent normal tissues. Low expression of miR-145-5p was significantly associated with lymph node metastasis, metastasis stage and distant metastasis, ultimately leading to poorer overall survival. In the study by Ozen et al (23), overexpression of miR-145-5p inhibited proliferation and decreased the migration of prostate cancer cells. Similarly, miR-145-5p is also an important target for non-small cell lung cancer (NSCLC) as miR-145-5p overexpression suppressed the EMT in NSCLC cells, which is an important biological process associated with cancer migration and metastasis (24). Upon transfection of miR-145-5p, the angiogenesis ability of SW480 colon carcinoma cells was significantly inhibited (25). Concomitantly, the overexpression of miR-145 significantly suppressed the proliferation, migration and invasion of OC cells and inhibited tumor growth and metastasis in vivo (26,27). Furthermore, tumor protein 53 (TP53) upregulated the expression of miR-145 to enhance its tumor suppressor roles (28-30). However, TP53 mutations occur in ~95% of high-grade serous OC (31). Therefore, miR-145 may be specifically down regulated in advanced OC. Consistent with these studies, miR-145 was expressed at lower levels in all 13 OC cell lines and its downregulation was significantly associated with a shorter survival and higher tumor stage in the present study.
Although miR-145 serves crucial roles in the development of cancer, the majority of studies have focused on the original cells, and the expression of miR-145 in exosomes and its effect on OC remains largely unknown. However, one previous study revealed that the exposure of pancreatic ductal adenocarcinoma to miR-145-5p-enriched tumor-associated stroma exosomes cells decreased cell viability in a dose-dependent manner (32). To the best of our knowledge, the present study is the first to demonstrate the significant downregulation of miR-145-5p expression in OC exosomes, indicating that the enhanced secretion of miR-145 via exosomes may be an underlying approach to treating OC (33,34). In addition, exosomes secreted from primary tumors are transferred and released to the peripheral circulation (35-37). Therefore, the expression of exosomal miRNAs detected in cancer cells is generally similar to their levels in blood. This hypothesis has been verified in the study by Hannafon et al (38), in which miR-1246 and miR-21 were first detected at significantly higher levels in exosomes from breast cancer cells and then validated in patient-derived orthotopic xenograft models, and mouse and human plasma exosomal samples. According to their analysis of the receiver operating characteristic curves, the diagnostic accuracy (area under the curve) of the combination of plasma exosomal miR-1246 and miR-21 levels was 72.66% for breast cancer. Consistent with these data, we hypothesize that the exosomal miR-145-5p identified in OC cells may also be a potential biomarker for the detection of malignant OC.
miRNAs are considered to participate in disease development by regulating target genes through interactions with complementary sequences in the 3′UTR. Several studies have explored the mechanisms of miR-145-5p in cancer. For example, Matsushita et al (39) predicted putative binding sites for miR-145-5p in 1,735 genes following a search of the microRNA.org database, and additionally confirmed that ubiquitin-like with PHD and ring finger domains 1 may be an indirect target of miR-145-5p to regulate bladder cancer cell aggressiveness in subsequent dual luciferase reporter assays. Wu et al (27) revealed a negative regulatory effect of miR-145 on levels of the ribosomal protein S6 kinase, polypeptide 1 (P70S6K1) and mucin 1, cell surface associated (MUC1) proteins in OC cells. Overexpression of p70S6K1 and MUC1 restored the colony formation and invasion abilities that were inhibited by miR-145. As indicated in the study by Chen et al (40), transfection of an miR-145 agomiR (activator) into OC cells downregulated the expression of its direct target tripartite motif containing 2 and subsequently induced apoptosis. However, the mechanisms of miR-145 in OC are not well understood. In the present study, 91 target genes of miR-145 were screened using the miRwalk2.0 database, and the subsequent clinical correlation analysis using TCGA data suggested that the Hippo signaling pathway-associated gene CTGF may be an important target of miR-145, due to its significant association with a poor prognosis and negative correlation with miR-145 expression. The data from the present study appear to be consistent with previous studies, as high CTGF expression promotes the migration and peritoneal adhesion of OC cells, resulting in a poor prognosis; these changes were abrogated by a human monoclonal antibody against CTGF FG-3019 (41). CTGF is a key downstream intermediate in the Hippo-YAP1/TEAD cascade that controls cell growth and initiates OC formation (42-44). CTGF was indicated to be a direct target of miR-145 using a dual luciferase reporter gene assay, and miR-145 negatively regulated CTGF to affect the proliferation, migration and invasion of esophageal squamous cell carcinoma (45) and malignant glioma cells transfected with miR-145 mimics and CTGF small interfering RNA (46). Nevertheless, to the best of our knowledge, studies of this nature in OC have not been described, requiring additional investigation. Notably, we hypothesized that the upregulation of CTGF in OC cells due to the downregulation of miR-145 may be one potential contributor to the decreased sorting of miR-145 into exosomes and subsequent transfer to acceptor cells, including cancer associated fibroblasts, endothelial cells and tumor-associated macrophages, to inhibit the pro-tumor environment (47). Furthermore, CTGF may also be a crucial target of exosomal miR-145 in the tumor environment. Low expression of miR-145 in exosomes was insufficient to block the transcription of CTGF, leading to its overexpression, acceleration of tumor cell invasion and the carcinogenesis of adjacent normal ovarian epithelial cells (48,49).
The present study had certain limitations: Firstly, 13 OC cell lines were used to screen crucial miRNAs in the present study. Due to the presence of underlying differences in phenotype, the expression of miRNAs varied among these cell lines, which lead to unexpected and paradoxical results for specific miRNAs, including miR-98 and miR-17. According to the prognosis analysis, miR-98 and miR-17 should be expressed at lower levels in OC, but in fact they were upregulated in OC and exosomes. Secondly, the TCGA data did not include a normal control, and potential deviations in the identified correlations between the identified miRNAs and clinical characteristics may have occurred. Thirdly, although the analysis preliminarily revealed a negative correlation between miR-145 and its target gene CTGF in clinical samples, in vitro and in vivo experiments using OC models are required. Fourthly, the loss of exosomal miR-145 as a potentially important factor contributing to OC pathogenesis was identified, but the mechanism underlying its formation and whether modification of exosomes by transfection with miR-145 decrease oncogenesis by affecting the tumor environment remain unknown (33,34,50). Fifthly, although previous studies have suggested that the levels of exosomal miRNAs in cancer cells are consistent with their levels in the peripheral circulation (35-38), the expression of exosomal miR-145 in plasma samples from patients with OC requires additional verification to confirm its underlying value as a biomarker.
According to the data from the present study, the decreased expression of hsa-miR-145 in OC and OC-derived exosomes may be required for the development of OC via the targeted modulation of a downstream intermediate in the Hippo signaling pathway, CTGF. Therefore, upregulation of hsa-miR-145 in exosomes and cells to inhibit CTGF expression may be a potential therapeutic approach for OC.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The microarray data GSE103708 were downloaded from the GEO database in NCBI (http://www.ncbi.nlm.nih.gov/geo/).
Authors’ contributions
WH, QH and XX conceived the design of the original study. WH and YF conducted the statistical analysis and drafted the manuscript. ZS, YY and YZ were involved with the interpretation of the data. QH and XX participated in critical revisions of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Cliby WA, Powell MA, Alhammadi N, Chen L, Miller JP, Roland PY, Mutch DG, Bristow RE. Ovarian cancer in the United States: Contemporary patterns of care associated with improved survival. Gynecol Oncol. 2015;136:11–17. doi: 10.1016/j.ygyno.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 4.Qin W, Xie W, He Q, Sun T, Meng C, Yang K, Luo Y, Yang D. MicroRNA-152 inhibits ovarian cancer cell proliferation and migration and may infer improved outcomes in ovarian cancer through targeting FOXP1. Exp Ther Med. 2018;15:1672–1679. doi: 10.3892/etm.2017.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Pan Y, Han X, Liu J, Li R. MicroRNA-216a promotes the metastasis and epithelial-mesenchymal transition of ovarian cancer by suppressing the PTEN/AKT pathway. Onco Targets Ther. 2017;10:2701–2709. doi: 10.2147/OTT.S114318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neviani P, Fabbri M. Exosomic microRNAs in the tumor microenvironment. Front Med. 2015;2:47. doi: 10.3389/fmed.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, Chen X, Wang X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7:43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanlikilicer P, Rashed MH, Bayraktar R, Mitra R, Ivan C, Aslan B, Zhang X, Filant J, Silva AM, Rodriguez-Aguayo C, et al. Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Res. 2016;76:7194–7207. doi: 10.1158/0008-5472.CAN-16-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De A, Powers B, De A, Zhou J, Sharma S, Van VP, Bansal A, Sharma R, Sharma M. Emblica officinalis extract downregulates pro-angiogenic molecules via upregulation of cellular and exosomal miR-375 in human ovarian cancer cells. Oncotarget. 2016;7:31484–31500. doi: 10.18632/oncotarget.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X, Müller V, Mildelangosch K, Trillsch F, Pantel K, Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7:16923–16935. doi: 10.18632/oncotarget.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin X, Yu S, Xu X, Bo S, Feng J. Comparative analysis of microRNA expression profiles between A549, A549/DDP and their respective exosomes. Oncotarget. 2017;8:42125–42135. doi: 10.18632/oncotarget.15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Du S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [DOI] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 14.Dweep H, Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 15.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 16.Therneau TM. A package for survival analysis in S. R package version 2. 2014;37:7. [Google Scholar]
- 17.Shimizu Y, Kamoi S, Amada S, Akiyama F, Silverberg SG. Toward the development of a universal grading system for ovarian epithelial carcinoma: Testing of a proposed system in a series of 461 patients with uniform treatment and follow-up. Cancer. 1998;82:893–901. doi: 10.1002/(SICI)1097-0142(19980301)82:5<893::AID-CNCR14>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Kosary CL. FIGO stage, histology, histologic grade, age and race as prognostic factors in determining survival for cancers of the female gynecological system: An analysis of 1973-87 SEER cases of cancers of the endometrium, cervix, ovary, vulva, and vagina. Semin Surg Oncol. 1994;10:31–46. doi: 10.1002/ssu.2980100107. [DOI] [PubMed] [Google Scholar]
- 19.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The comparative toxicogenomics database: Update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wen X, Hu XL, Cheng LZ, Yu JY, Wei ZB. Downregulation of miR-145-5p correlates with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2016;20:3026–3030. [PubMed] [Google Scholar]
- 23.Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ, Ittmann M. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Invest. 2015;33:251–258. doi: 10.3109/07357907.2015.1025407. [DOI] [PubMed] [Google Scholar]
- 24.Chang Y, Yan W, Sun C, Liu Q, Wang J, Wang M. miR-145-5p inhibits epithelial-mesenchymal transition via the JNK signaling pathway by targeting MAP3K1 in non-small cell lung cancer cells. Oncol Lett. 2017;14:6923–6928. doi: 10.3892/ol.2017.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thuringer D, Jego G, Berthenet K, Hammann A, Solary E, Garrido C. Gap junction-mediated transfer of miR-145-5p from microvascular endothelial cells to colon cancer cells inhibits angiogenesis. Oncotarget. 2016;7:28160–28168. doi: 10.18632/oncotarget.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei Y, Li Y, Yang Q, Liu J, Wei JJ, et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget. 2014;5:10816–10829. doi: 10.18632/oncotarget.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Xiao ZH, Wang K, Liu W, Hao Q. MiR-145 is down-regulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun. 2013;441:693–700. doi: 10.1016/j.bbrc.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 29.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29:613–639. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- 31.Brachova P, Thiel KW, Leslie KK. The consequence of oncomorphic TP53 mutations in ovarian cancer. Int J Mol Sci. 2013;14:19257–19275. doi: 10.3390/ijms140919257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Belsare S, Zhang DY, Beveridge M, Rinaldi C, Trevino JG, Schmittgen TD, Hughes SJ. Stroma-derived extracellular vesicles deliver tumor-suppressive miRNAs to pancreatic cancer cells. Oncotarget. 2017;9:5764–5777. doi: 10.18632/oncotarget.23532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:4711–4716. doi: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Taylor DD, 1, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 36.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;5:383–390. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Ostenfeld MS, Jeppesen D, Morth JP, Khanh HB, Dan T, Borre M, Dyrskjøt L, Ørntoft T. Abstract 3387: Secreted exosomes from cultured bladder cells are enriched for distinct miRNAs detected in circulation of metastatic bladder cancer patients. Cancer Res. 2012;72(Suppl):S3387. doi: 10.1158/1538-7445.AM2012-3387. [DOI] [Google Scholar]
- 38.Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M, Seki N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7:28460–28487. doi: 10.18632/oncotarget.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Dong C, Law PT, Chan MT, Su Z, Wang S, Wu WK, Xu H. MicroRNA-145 targets TRIM2 and exerts tumor-suppressing functions in epithelial ovarian cancer. Gynecol Oncol. 2015;139:513–519. doi: 10.1016/j.ygyno.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Kim MJ, Gloss BS, Rajmohan M, Chang DK, Colvin EK, Jones MD, Samuel Y, Howell VM, Brown LM, Wong CW, et al. Connective tissue growth factor as a novel therapeutic target in high grade serous ovarian cancer. Oncotarget. 2015;6:44551–44562. doi: 10.18632/oncotarget.6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F, et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:n92. doi: 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pobbati AV, Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14:390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan G, Cao X, Dai Q, Zhang B, Huang J, Xiong S, Zhang Y, Chen W, Yang J, Li H. A novel role for microRNA-129-5p in inhibiting ovarian cancer cell proliferation and survival via direct suppression of transcriptional co-activators YAP and TAZ. Oncotarget. 2015;6:8676–8686. doi: 10.18632/oncotarget.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Q, Zhang HY, Zhong BL, Wang XJ, Zhang B, Chen H. MicroRNA-145 inhibits cell migration and invasion and regulates epithelial-mesenchymal transition (EMT) by targeting connective tissue growth factor (CTGF) in esophageal squamous cell carcinoma. Med Sci Monit. 2016;22:3925–3934. doi: 10.12659/MSM.897663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E, et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS One. 2013;8:e54652. doi: 10.1371/journal.pone.0054652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan G, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, Li X, Chen J, Liu K, Li C, Zhu G. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.CAN-15-1625. [DOI] [PubMed] [Google Scholar]
- 49.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trivedi M, Talekar M, Shah P, Ouyang Q, Amiji M. Modification of tumor cell exosome content by transfection with wt-p53 and microRNA-125b expressing plasmid DNA and its effect on macrophage polarization. Oncogenesis. 2016;5:e250. doi: 10.1038/oncsis.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray data GSE103708 were downloaded from the GEO database in NCBI (http://www.ncbi.nlm.nih.gov/geo/).