Abstract
Congenital hypopituitarism (CH) is a relatively rare disease that is characterized by the deficiency of one or more hormones secreted by the pituitary gland, which leads to metabolic disorders, amenorrhea and infertility. However, the underlying molecular mechanisms of CH have not yet been fully elucidated. The present study evaluated the genome-wide methylation level of whole blood DNA in 12 patients with CH and 12 age-matched controls using Illumina Human Methylation 450 array, in order to determine the roles of epigenetic regulation in the pathogenesis of CH. The results demonstrated that the methylation levels of 51 CpG sites were significantly different between the patients with CH and the controls. Functional enrichment analysis identified that the aberrant methylated genes were enriched in gene sets associated with metabolic or cellular process, immune system process and reproduction. In addition, two CpG sites on genes LIM domain kinase 2 (LIMK2) and piwi-like RNA-mediated gene silencing 2 (PIWIL2), which are involved in spermatogenesis and/or testicular development, were identified to be hypermethylated in male patients with CH. The hypermethylation of these sites was further validated in another 40 patients with CH and 40 matched controls with a quantitative bisulfite pyrosequencing method, and the methylation levels of these two loci demonstrated promising diagnostic capacities for CH. The present results suggested that aberrant methylation of genes may be involved in the pathogenesis of CH, and hypermethylation of LIMK2 and PIWIL2 may contribute to the infertility of male patients with CH. Further studies are required to elucidate the underlying mechanisms of the epigenetic regulation of these genes.
Keywords: CH, DNA methylation, epigenetics, spermatogenesis
Introduction
Hypopituitarism has been recognized as a rare disorder with an estimated incidence of 4.2 cases per 100,000 per year and an estimated prevalence of 45.5 cases per 100,000 worldwide (1). It is characterized by the deficiency of one or more of the hormones secreted by the pituitary gland. The disease may result from distinct causes and it may be congenital or acquired. The most common etiology factors for acquired hypopituitarism include pituitary tumors (2), traumatic brain injury (3-5), pituitary irradiation (6), congenital hypopituitarism (CH), perinatal insults (7) and genetic mutations (8). Although the clinical symptoms of this disease are usually unspecific, it may cause life-threatening events and lead to an increased risk of cardiovascular diseases, poor pregnancy potential and infertility for the patients (9,10).
As a heterogeneous entity with various types of etiology factors, mutations in the genes encoding a number of hypothalamo-pituitary transcription factors, including HESX homeobox 1 (HESX1), LIM homeobox 1 (LHX1), PROP paired-like homeobox 1 (PROP1), and POU class 1 homeobox 1 (POU1F1) may lead to CH; however, only 10% of cases are estimated to be caused by genetic factors (11). Prenatal or birth trauma and/or asphyxia and specific types of midline defect syndromes may additionally lead to CH (12); however, their association with CH are usually hard to determine. The underlying mechanisms for the clinical symptoms of patients with CH are due to the deficiency in pituitary hormones, including the growth hormone, adrenocorticotropin (ACTH), prolactin (PRL), thyroid stimulating hormone (TSH), luteinising hormone (LH) and follicle-stimulating hormone (FSH) (13). These deficiencies result in recurrent hypoglycemia and acute adrenal insufficiency with impairments of growth and neurodevelopment (13). Furthermore, due to the lack of gonadotropin hormones, spermatogenesis and/or testicular development are abnormal, resulting in infertility for male patients. In females, successful pregnancy is rare, as hypopituitarism is associated with an increased risk of pregnancy complications, including abortion, anemia, pregnancy-induced hypertension, placental abruption, premature birth and postpartum hemorrhage (14). In the clinical setting, lifelong hormone replacement treatment (HRT) is applied to patients with CH with corticosteroids, levothyroxine, sex hormones and growth hormones; however, the treatments may only partially recover the abnormal development of patients and therefore they remain at high risk of infertility and mortality (15-17). Therefore, the underlying molecular mechanisms for the pathogenesis of CH require further examination.
DNA methylation is a crucial epigenetic modification that influences the structure of the genome and regulates subsequent gene transcription activities (18). Previous studies of targeted gene methylation demonstrated the significant associations between DNA methylation and hormone exposure in early-life, particularly in hormonally-mediated sexual differentiation (19,20). As DNA methylation status may affect gene expression in response to hormones during the lifespan (20), it is possible that DNA methylation may be modified in patients with CH with hormone deficiency, and it may be involved in the development and progression of the disease. Therefore, the epigenome-wide methylation status of the whole blood DNA was detected in patients with CH, in order to identify genes that may be aberrantly methylated in patients with CH compared with control subjects. The present results may provide further insight into the pathogenesis of the disease and may provide novel therapeutic targets for CH treatment and biomarkers for diagnosis.
Materials and methods
Participant recruitment
The discovery set included 12 male patients with CH and 12 male healthy controls matched for age (±3 years) and body-mass index (±1 kg/m2). The participants were recruited at the Ruijin Hospital North (Shanghai, China) between November 2015 and May 2016. Eligible patients were clinically diagnosed with hypopituitarism based on clinical history, symptoms, biochemical and brain magnetic resonance imaging (MRI) tests. Patients with pituitary tumors, traumatic brain injury, pituitary irradiation or a clear diction of dystocia history were excluded. All CH patients had been diagnosed with panhypopituitarism with multiple pituitary hormone deficiency at a young age (~6-12 years old). The MRI scan of all the patients with CH suggested a presence of a transected or interrupted hypothalamic-pituitary stalk or pituitary hypoplasia (21). The patients had received routine cortisone and levothyroxine treatment; however, had not yet received the human chorionic gonadotrophin treatment at the time of recruitment. Controls were selected from local residents receiving annual physical examination, were free of acute or chronic diseases (including diabetes, cancer and cardiovascular diseases), and had no symptom of growth and developmental abnormalities. The validation set included another 40 male patients with CH and 40 age-matched male controls. The recruitment of the validation set followed the same protocols as described for the discovery set, and was conducted between June 2016 and July 2017. Written informed consent was obtained from all the participants, and the study was approved by the Institutional Review Board of Ruijin Hospital North.
Endocrinological hormone tests
The serum hormonal levels were determined to evaluate the hormone secretion activity of the pituitary gland during the drug withdrawal period, using commercial radioimmunoassay kits in the clinic. The blood levels of free thyroxine 3, free thyroxine 4 (FT4), TSH, cortisol, ACTH, LH, FSH, PRL, progesterone (PROG) and testosterone were determined with Beckman DX I800 instruments (Beckman Coulter, Inc., Brea, CA, USA), according to the manufacturer’s protocol in the clinical testing lab.
Genome-wide DNA methylation level analysis
In total, 5 ml blood was provided by each participant during their drug withdrawal period and the genomic DNA was isolated using the QIAamp DNA Blood Mini kit (Qiagen GmbH, Hilden, Germany). Following quality assessment, bisulfite conversion of the genomic DNA was performed using the Zymo EZ DNA Methylation kit (Zymo Research Corp., Irvine, CA, USA). DNA methylation levels of blood were quantified with the Human Methylation450K BeadArray (Illumina, Inc., San Diego, CA, USA) following the manufacturer’s protocol, and the samples of cases and controls were processed simultaneously. The methylation level of a specific CpG site was quantified as a β value ranging from 0 (no methylation) to 1 (full methylation). The raw DNA methylation β values were normalized using the Subset-quantile Within Array Normalization method (22) with the minfi Bioconductor (version 1.26.2) package (www.bioconductor.org). Probes with detection P>0.01 and/or covering potential single nucleotide polymorphism (SNP) sites were excluded from analysis. Student’s t-test was performed to identify the significantly methylated CpG loci between the patients with CH and controls. The top candidate probes were selected based on an adjusted P-value using the Benjamini-Hochberg False Discovery Rate (FDR) procedure with adjusted P<0.05 and β-difference >0.14 (23). The DNA methylation data are available at the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE107737.
Pathway enrichment analysis of the differentially methylated genes was performed with the publicly available tool Protein Analysis Through Evolutionary Relationships (PANTHER; http://www.pantherdb.org/) that provides access to various gene set libraries (24). A pathway was considered as significantly enriched if the FDR P-value was <0.05.
Pyrosequencing analysis
The pyrosequencing method was applied in order to determine the methylation level of candidate CpG sites in the validation set. The bisulfite conversion and cleanup of DNA samples was performed with the EpiTect Fast Bisulfite Conversion kit (Qiagen,GmbH). Fragments covering the target sites were amplified from the bisulfite converted genomic DNA with the biotinylated forward and normal reverse primers, which were designed with PyroMark Assay Design Software (version 2.0; Qiagen, GmbH). The biotinylated polymerase chain reaction (PCR) was performed with EpiTap™ HS polymerase (Takara Biotechnology Co., Ltd., Dalian, China) and the thermocycling conditions were as follows: 98°C for 10 sec, 55°C for 30 sec, 72°C for 30 sec with 35 cycles followed by 72°C for 45 sec and 4°C for hold. The amplicon product was purified and made into single-stranded, which was used as the template in the pyrosequencing reaction. The pyrosequencing was conducted using the Pyro Mark Q24 Advanced (Qiagen, GmbH), according to the manufacturer’s protocol, with the specific sequencing primers (cg01606027, forward PCR primer 5′-TTT TGG TTA TTT GGA TAT GTT TTT TGT GAT-3′, reverse PCR primer 5′-TCA ACC AAA CCA ACT AAT CCC TAA TA-3′, and pyrosequencing primer 5′-GTA AGG AAA TGA TAA TTG TGA A-3′; cg03594078, forward PCR primer 5′-TGT GTA AAT ATT TAA TGG TAG TAA GTG TAG -3′, reverse PCR primer 5′-AAT TTA CTA AAT CCT ACC CCC AAA A-3′ and the pyrosequencing primer 5′-AAT TTT TTG TTT TTT TAG GAA TT-3′). The PyroMark CpG Software 1.0 (Qiagen, GmbH) was used to calculate the CpG methylation levels based on the height of the T and C peaks at the methylation site with the formula C/(C + T) ×100.
Reverse transcription-quantitative (RT-q)PCR in whole blood cells
The white blood cells of the participants in the training cohort were purified from whole blood collected in the EDTA sodium tubes, by centrifugation at 2,000 x g for 15 min at room temperature. The white blood cells were lysed with the TRIzol® reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the total RNA was extracted with the Direct-zol RNA kit (Zymo Research, Corp.). A total of 500 ng total RNA were used to synthesize the cDNA using the PrimeScript 1st strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd.) and incubated at 30°C for 10 min, 42°C for 30 min followed by 70°C for 15 min. The qPCR reaction was performed with the SYBR® Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.). LIM domain kinase 2 (LIMK2) and piwi-like RNA-mediated gene silencing 2 (PIWIL2), The primers were: LIM kinase 2 (LIMK2), forward 5′-GGA TTC CCT CAC CAA CTG GTA-3′ and reverse 5′-AGC CAC CAT AAA AGG CCC TG-3′; piwi-like RNA-mediated gene silencing 2 (PIWIL2), forward 5′-AGC AGG TGG TAT CAG CAG AGA-3′ and reverse 5′-TGT ATT TTG ACG AGG TTC AGT CC-3′; and GAPDH, forward 5′-AAG GTG AAG GTC GGA-3′ and reverse 5′-AAT GAA GGG GTC ATT GAT GG-3′. qPCR was performed at 95°C for 30 sec followed by 95°C for 5 sec and 60°C for 30 sec with 40 cycles. Relative expression of the genes was calculated relative to the mean expression level of LIMK2 and PIWIL2 in the control group normalized by GAPDH with the 2-ΔΔCq method (25).
Statistical analysis
Age, weight, height and body mass index (BMI) of the participants are presented as mean ± standard deviation and the differences between the groups were analyzed with the Student’s t-test. The hormone levels of the participants were presented as median level in together with the lowest to highest range of the hormone, and the differences between the groups were evaluated with the Mann-Whitney test. Differences for the methylation levels of the CpG sites in pyrosequencing analysis and the relative expression of the genes between the groups were evaluated with the Student’s t-test. The receiver-operating characteristic (ROC) curve analysis and the corresponding area under the curve (AUC) were calculated to determine the diagnosis values for the methylation levels of the candidate CpG sites. All analyses were performed with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Baseline characteristics of the participants in the discovery set
To compare the genome-wide methylation levels of the whole blood DNA, first, 12 patients with CH and 12 age-matched controls (±3 years) were recruited as the discovery set. The basic clinical characteristics of the participants are presented in Table I. The mean age for the patients with CH was 21.58±2.71 years and 24.00±3.67 years for the controls (P=0.081). All patients with CH had received stan-dardized cortisone and levothyroxine therapy at the time of enrollment with the median treatment time of 6 years (range 2-10 years), and none of them had received the human chorionic gonadotrophin treatment. All patients had a symptom of oligospermia or azoospermia at the time of recruitment with the Tanner stage between G1 to G3 (Table I). No significant difference was identified between the patients with CH and controls for height, weight and body mass index (Table I). Compared with the controls, the patients with CH had significantly lower levels of the hormones secreted by the pituitary gland. In addition, significantly lower FT4, cortisol and ACTH levels were observed in the patients with CH compared with the controls (Table I). Furthermore, the hormones associated with gonadotropic function, LH, FSH, PROG and TESTO, were additionally at lower levels in the patients with CH compared with the controls (Table I).
Table I.
Characteristics of the paired patients congenital with hypopituitarism and controls in the discovery set.
| Characteristics | Congenital hypopituitarism, n=12 | Controls, n=12 | P-value |
|---|---|---|---|
| Basic information | |||
| Age, years | 21.58±2.71 | 24.00±3.67 | 0.081 |
| Height, cm | 169.0±8.72 | 173.0±5.17 | 0.185 |
| Weight, kg | 65.13±16.94 | 67.75±3.93 | 0.606 |
| BMI, kg/m2 | 22.42±3.95 | 22.70±1.92 | 0.827 |
| Tanner stage | G1/G2/G3 (3/6/3) | - | - |
| Thyroid function | |||
| FT3, pmol/l | 3.70 (2.69-6.34) | 4.22 (3.51-5.06) | 0.434 |
| FT4, pmol/l | 9.67 (6.86-17.19) | 13.52 (11.67-17.08) | 0.018 |
| TSH, µIU/l | 0.69 (0.00-5.94) | 1.80 (0.56-2.76) | 0.369 |
| Corticotropic function | |||
| Cortisol, µg/dl | 0.465 (0.01-5.55) | 12.41 (7.10-21.00) | <0.001 |
| ACTH, pg/ml | 9.09 (5.27-10.96) | 42.00 (20.00-55.00) | <0.001 |
| Gonadotropic function | |||
| LH, mIU/l | 0.11 (0.01-1.26) | 4.05 (1.47-5.29) | <0.001 |
| FSH, µIU/ml | 0.18 (0.02-1.70) | 4.97 (1.80-7.98) | <0.001 |
| PRL, ng/ml | 16.81 (2.21-49.13) | 8.58 (4.27-11.62) | 0.065 |
| E2, pg/ml | 20.00 (20.00-28.60) | 26.46 (20.00-35.40) | 0.087 |
| PROG, ng/ml | 0.10 (0.10-0.20) | 0.12 (0.10-0.79) | 0.017 |
| TESTO, ng/ml | 0.10 (0.10-2.00) | 4.75 (2.25-7.01) | <0.001 |
The data for basic information are presented as the mean ± standard deviation. All other data are presented as the median (range). ACTH, adrenocorticotropic hormone; BMI, body mass index; FT3, free thyroxine 3; FT4, free thyroxine 4; TSH, thyroid stimulating hormone; PRL, prolactin; PROG, progesterone; TESTO, testosterone; LH, luteinising hormone; FSH, follicle-stimulating hormone.
Genome-wide methylation analysis of whole blood DNA in CH patients
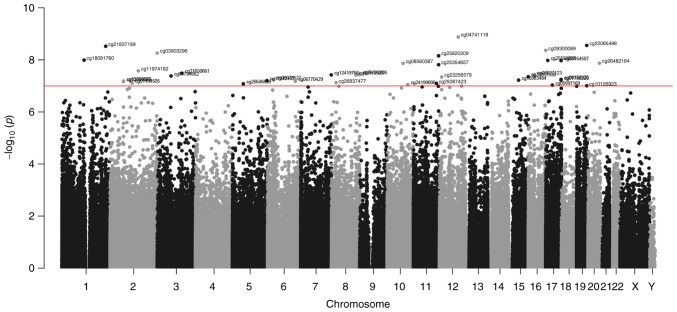
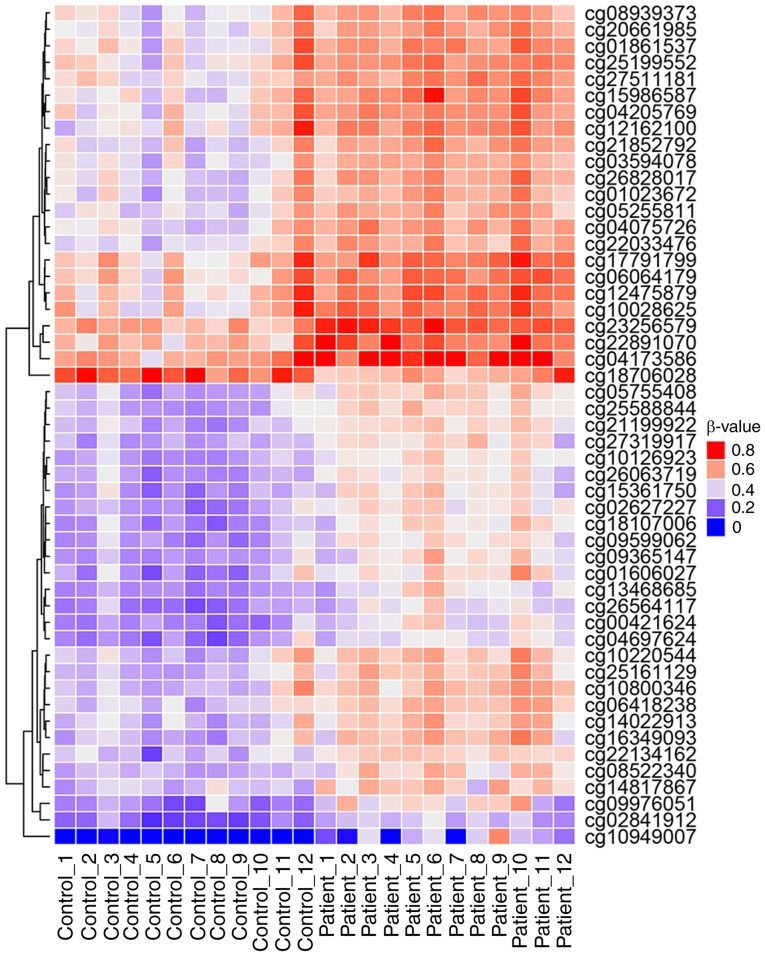
Using the Human Methylation450K BeadArray, the CpG methylation levels of the whole blood DNA in patients with CH and controls were determined. The Manhattan plot demonstrated the CpG sites on each chromosome and their corresponding unadjusted P-values for the differences of meth-ylation levels between patients with CH and the controls (Fig. 1). The selection criteria for the candidate CpG sites are presented in Fig. 2. Following the epigenome-wide methylation analysis of the discovery set, 58,128 differentially methylated probes (DMPs) were identified between the patients with CH and the controls (unadjusted P-value <0.05). Of these, 3,225 DMPs had an FDR adjusted P<0.05 and 64 probes had a β-difference >0.14. Subsequent to excluding probes covering potential SNP sites, 51 significantly aberrant methylated CpG sites were selected for further analysis (Fig. 3). Of these, 50 sites were hypermethylated, whereas, only one CpG site, cg18706028, was hypomethylated in patients with CH (Fig. 3). These 51 DMPs were clustered into five regions according to gene annotation, including 10 DMPs located the in TSS1500Ind, one DMP in TSS200Ind, 27 DMPs located on gene body, three in the 5′-untranslated region (UTR), one in the 3′-UTR region of the gene, and the other 10 DMPs located in the intergenic region (Table II).
Figure 1.
Manhattan plot demonstrating the −log10 P-values and their genomic locations for all CpG probes (total 414,733) included in the methylation analysis. The P-value threshold of 1×10−7 is represented by a horizontal red line and probes over the threshold were labeled.
Figure 2.
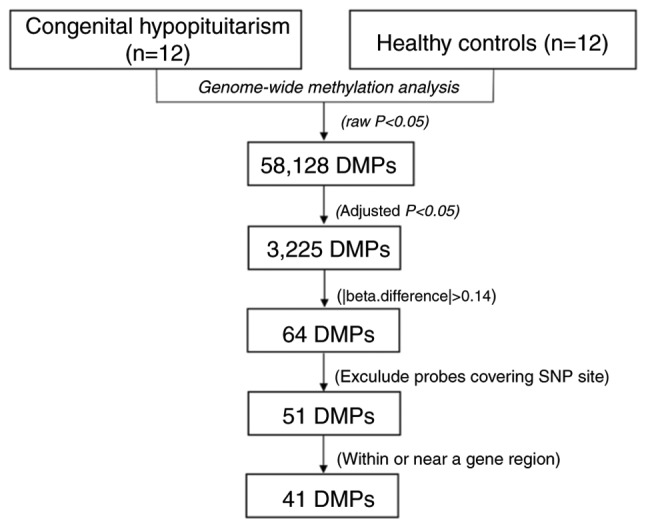
Schematic diagram of the selection procedure for the significantly methylated loci in the discovering set. DMP, differentially methylated probe; SNP, single nucleotide polymorphism.
Figure 3.
Hierarchical clustering analysis for the 51 significantly differentially methylated CpG sites between the patients with congenital hypopituitarism and the controls. Red denotes hypermethylated levels and blue color hypomethylated levels.
Table II.
Differentially methylated positions in the peripheral blood DNA of 12 paired patients with congenital hypopituitarism and controls in the discovery set.
| Gene/CpG region | Gene symbol | CpG site | P-value | FDR P-value | β-differencea | Chr | Region |
|---|---|---|---|---|---|---|---|
| TSS1500Ind | C11orf41 | cg08939373 | 0.0002 | 0.0391 | −0.1453 | 11 | - |
| GPR77 | cg15361750 | <0.0001 | 0.0170 | −0.1517 | 19 | - | |
| LIMK2 | cg01606027 | 0.0001 | 0.0277 | −0.1715 | 22 | N_Shore | |
| NEIL3 | cg14022913 | 0.0001 | 0.0255 | −0.1429 | 4 | N_Shore | |
| SLC27A3 | cg00421624 | <0.0001 | 0.0052 | −0.1515 | 1 | N_Shore | |
| PRR4 | cg23256579 | <0.0001 | 0.0011 | −0.1650 | 12 | - | |
| GLRX | cg10949007 | 0.0001 | 0.0281 | −0.2245 | 5 | - | |
| HNRPLL | cg18107006 | <0.0001 | 0.0043 | −0.1682 | 2 | S_Shore | |
| PIWIL2 | cg03594078 | <0.0001 | 0.0132 | −0.1414 | 8 | N_Shore | |
| TSS200Ind | HIF3A | cg22891070 | 0.0001 | 0.0227 | −0.1683 | 19 | S_Shore |
| GENEBODY | AGA | cg09976051 | 0.0001 | 0.0264 | −0.1792 | 4 | N_Shore |
| C20orf3 | cg20661985 | 0.0001 | 0.0267 | −0.1460 | 20 | N_Shelf | |
| CCKBR | cg18706028 | 0.0001 | 0.0305 | 0.1487 | 11 | Island | |
| DOT1L | cg04173586 | 0.0001 | 0.0212 | −0.1632 | 19 | S_Shore | |
| ECT2 | cg10028625 | 0.0003 | 0.0455 | −0.1628 | 3 | - | |
| FLJ13197 | cg21199922 | <0.0001 | 0.0039 | −0.1433 | 4 | - | |
| KCNK5 | cg05255811 | <0.0001 | 0.0113 | −0.1495 | 6 | - | |
| N4BP1 | cg27319917 | <0.0001 | 0.0041 | −0.1410 | 16 | - | |
| NKG7 | cg10126923 | <0.0001 | 0.0013 | −0.1487 | 19 | S_Shelf | |
| PLEK | cg13468685 | <0.0001 | 0.0045 | −0.1444 | 2 | - | |
| PLEKHM1P | cg06064179 | 0.0001 | 0.0335 | −0.1433 | 17 | N_Shelf | |
| PRPSAP2 | cg14817867 | <0.0001 | 0.0096 | −0.1473 | 17 | - | |
| RASA3 | cg10800346 | 0.0001 | 0.0221 | −0.1600 | 13 | - | |
| REV3L | cg09365147 | <0.0001 | 0.0156 | −0.1423 | 6 | - | |
| TAF1B | cg25588844 | <0.0001 | 0.0021 | −0.1572 | 2 | - | |
| THOC7 | cg22134162 | <0.0001 | 0.0067 | −0.1404 | 3 | - | |
| TRIM35 | cg05755408 | <0.0001 | 0.0045 | −0.1580 | 8 | - | |
| VIM | cg26063719 | <0.0001 | 0.0033 | −0.1440 | 10 | S_Shore | |
| WIPI1 | cg25161129 | <0.0001 | 0.0123 | −0.1550 | 17 | N_Shore | |
| FGGY | cg10220544 | 0.0003 | 0.0482 | −0.1429 | 1 | - | |
| KDM1A | cg25199552 | 0.0001 | 0.0333 | −0.1476 | 1 | S_Shelf | |
| RPTOR | cg27511181 | <0.0001 | 0.0035 | −0.1568 | 17 | - | |
| RPTOR | cg06418238 | <0.0001 | 0.0050 | −0.1449 | 17 | - | |
| THADA | cg22033476 | <0.0001 | 0.0198 | −0.1500 | 2 | - | |
| CTSC | cg08522340 | <0.0001 | 0.0038 | −0.1503 | 11 | N_Shelf | |
| DPH5 | cg04205769 | 0.0001 | 0.0333 | −0.1481 | 1 | N_Shelf | |
| PROM1 | cg17791799 | 0.0003 | 0.0456 | −0.1434 | 4 | - | |
| UTR5Ind | AGPAT4 | cg02627227 | <0.0001 | 0.0033 | −0.1600 | 6 | - |
| CPNE3 | cg12162100 | 0.0003 | 0.0493 | −0.1583 | 8 | S_Shelf | |
| SYNE1 | cg02841912 | <0.0001 | 0.0051 | −0.1408 | 6 | N_Shore | |
| UTR3Ind | PITPNB | cg26564117 | <0.0001 | 0.0117 | −0.1436 | 22 | - |
| Intergenic | - | cg15986587 | <0.0001 | 0.0159 | −0.1789 | 19 | N_Shelf |
| - | cg16349093 | <0.0001 | 0.0158 | −0.1776 | 5 | - | |
| - | cg21852792 | <0.0001 | 0.0171 | −0.1654 | 2 | N_Shelf | |
| - | cg04075726 | <0.0001 | 0.0079 | −0.1627 | 2 | - | |
| - | cg26828017 | <0.0001 | 0.0192 | −0.1584 | 16 | - | |
| - | cg01861537 | 0.0001 | 0.0342 | −0.1544 | 11 | - | |
| - | cg09599062 | <0.0001 | 0.0050 | −0.1521 | 3 | - | |
| - | cg12475879 | 0.0002 | 0.0394 | −0.1518 | 17 | S_Shelf | |
| - | cg01023672 | 0.0001 | 0.0383 | −0.1461 | 12 | S_Shelf | |
| - | cg04697624 | <0.0001 | 0.0189 | −0.1409 | 12 | N_Shelf |
β-difference for the congenital hypopituitarism patients minus control. FDR, false discovery rate; Chr, chromosome.
Gene set enrichment analysis of the aberrant methylated genes
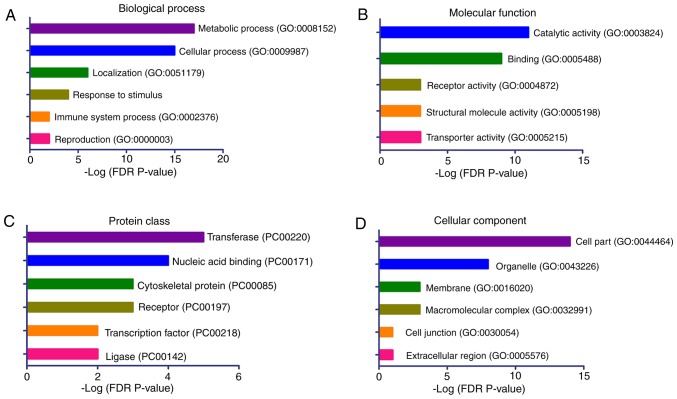
To provide further insights into the biological processes associated with the aberrant methylated genes in the patients with CH, a gene set enrichment analysis was conducted for these genes using the PANTHER classification system. The 41 candidate genes with significant DMPs were included in the analyses (Table II). The results demonstrated that the genes were significantly enriched in the biological processes of ‘metabolic process’, ‘cellular process’, ‘localization’, ‘response to stimulus’, ‘immune system process’ and ‘reproduction’ (Fig. 4A). The significantly enriched molecular functions of these genes were ‘catalytic activity’, ‘binding’, ‘receptor’ and ‘transporter activity’ (Fig. 4B). The top enriched protein classes were ‘transferase’, ‘nucleic acid binding’, ‘cytoskeletal protein’ and ‘receptor’ (Fig. 4C), while the top ranked cellular components were ‘cell part’ and ‘organelle’ (Fig. 4D).
Figure 4.
Gene set enrichment analyses for the aberrantly methylated genes between the patients with congenital hypopituitarism and the controls. (A) Biological process. (B) Molecular function. (C) Protein class. (D) Cellular component. The terms were sorted according to the -log (FDR P-value). FDR, false discovery rate; GO, gene ontology.
LIMK2 and PIWIL2 are hypermethylated in patients with CH
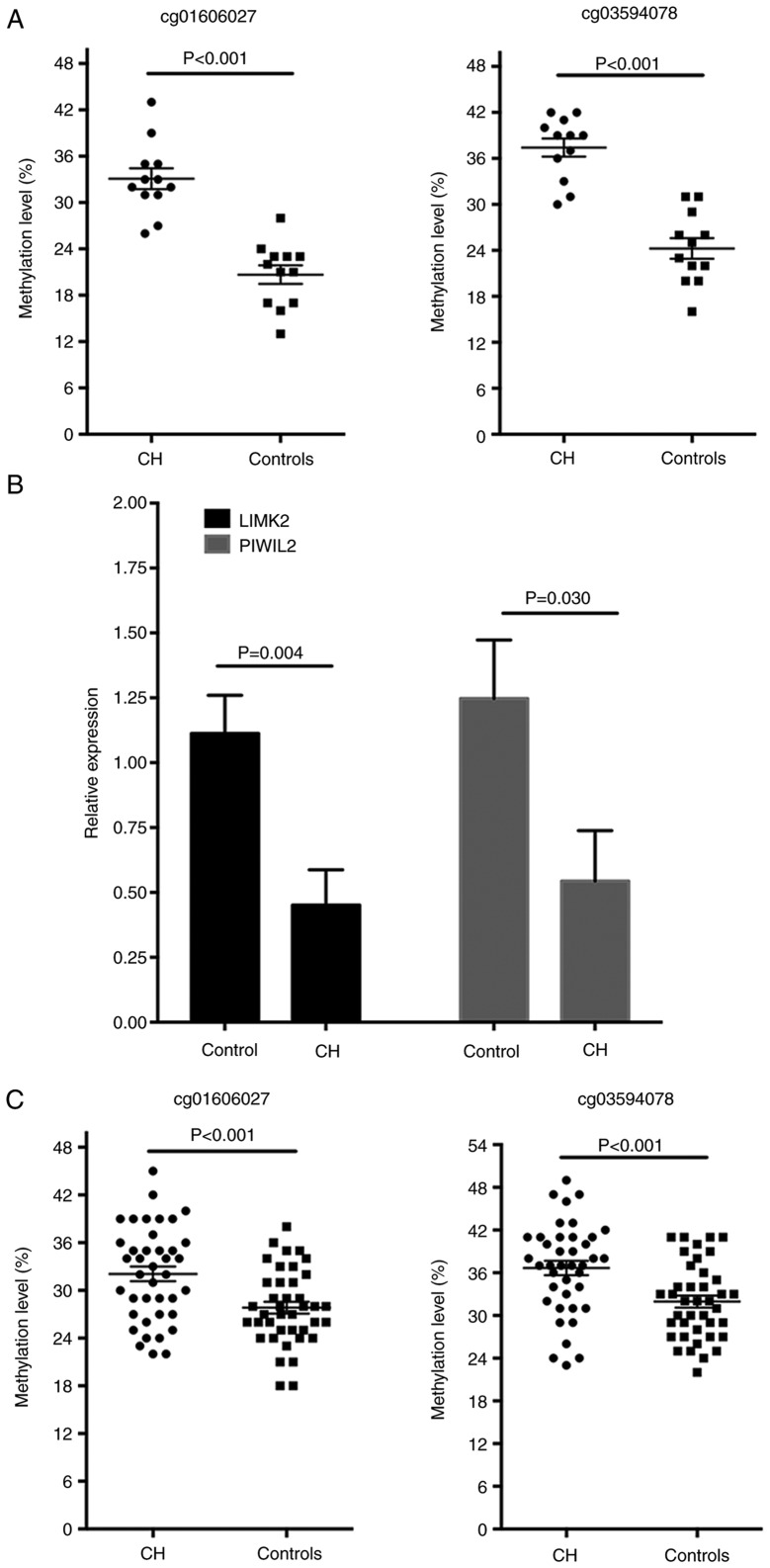
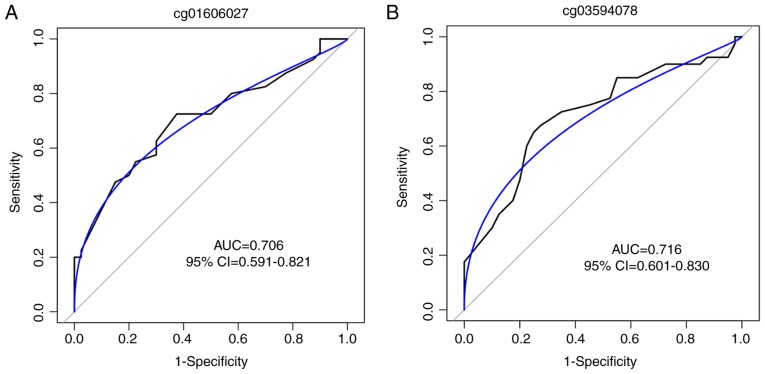
CH in male patients is typically associated with decreased spermatogenesis and testicle development latency (26). The methylation analysis demonstrated that the genes LIMK2 and PIWIL2, which have vital roles in the reproductive system development, were hypermethylated in the TSS1500Ind region (Table II). As the hypermethylation of CpG sites in the promoter may reduce the transcription activities of the target genes, it was hypothesized that the aberrant methylation of these two genes may be involved in the infertility of the male patients with CH. With the pyrosequencing methods, the hypermethylated levels of cg01606027 on LIMK2 and cg03594078 on PIWIL2 of the CH patients in the discovery cohort were validated and it was identified that the methylation level of the two CpG sites was significantly increased in patients with CH (P<0.001 for the two sites; Fig. 5A). In addition, the mRNA expression levels of LIMK2 and PIWIL2 in white blood cells were significantly decreased in the patients with CH compared with the normal controls, as detected by RT-qPCR analysis (Fig. 5B). The methylation levels of these two loci were further determined in another validation set with 40 pairs of patients with CH and controls. The baseline characteristics of the validation set are presented in Table III. No significant difference was observed for the basic characteristics in patients with CH and controls, whereas, the blood levels of TSH, cortisol, ACTH, LH, FSH, PRL and TESTO were significantly lower in the patients with CH compared with controls (Table III). Compared with the controls, the methylation of the cg01606027 on LIMK2 and cg03594078 on PIWIL2 was increased in the patients with CH (P<0.001 for the two sites; Fig. 5C). To estimate the potential diagnostic power of these two methylated CpG sites with regards to differentiating CH samples from controls, the predictive performance of the methylation levels of the two CpG sites for CH disease was examined with the ROC test in the validation cohort. The AUC was 0.706 [95% confidence interval (CI), 0.591-0.821; Fig. 6A] for cg01606027 and 0.716 (95% CI, 0.601-0.830; Fig. 6B) for cg03594078, suggesting that the two loci may hold promise in CH disease diagnosis.
Figure 5.
CpG sites on LIMK2 (cg01606027) and PIWIL2 (cg03594078) are hypermethylated and the expression levels of LIMK2 and PIWIL2 are reduced in patients with CH. (A) Pyrosequencing of CpG sites cg01606027 on LIMK2 and cg03594078 on PIWIL2 suggested that the two sites were hypermethylated in patients with CH compared with controls in the discovery set (n=12). (B) Reverse transcription-quantitative polymerase chain reaction results suggested that the mRNA expression levels of LIMK2 and PIWIL2 were significantly reduced in patients with CH compared with normal controls in the discovery set (n=12). Data are presented as mean ± standard error of the mean. (C) Pyrosequencing suggested that the methylation levels of CpG sites cg01606027 on LIMK2 and cg03594078 on PIWIL2 were increased in patients with CH compared with controls in the validation set (n=40). LIMK2, LIM domain kinase 2; PIWIL2, piwi-like RNA-mediated gene silencing 2; CH, congenital hypopituitarism.
Table III.
Characteristics of the patients with congenital hypopituitarism and controls in the validation set.
| Characteristics | Congenital hypopituitarism, n=40 | Controls, n=40 | P-value |
|---|---|---|---|
| Basic information | |||
| Age, years | 26.49±3.42 | 25.08±3.56 | 0.143 |
| Height, cm | 166.7±8.71 | 171.5±6.34 | 0.093 |
| Weight, kg | 63.28±14.92 | 65.31±7.06 | 0.068 |
| BMI, kg/m2 | 23.10±4.19 | 22.23±2.31 | 0.768 |
| Thyroid function | |||
| FT3, pmol/l | 3.77 (2.57-5.07) | 4.19 (2.32-5.43) | 0.196 |
| FT4, pmol/l | 11.28 (6.96-16.92) | 12.45 (7.34-17.08) | 0.090 |
| TSH, µIU/l | 0.15 (0.00-4.42) | 1.53 (0.56-3.25) | <0.001 |
| Corticotropic function | |||
| Cortisol, µg/dl | 3.01 (0.13-19.13) | 13.69 (7.10-21.21) | <0.001 |
| ACTH, pg/ml | 20.54 (4.65-60.15) | 43.00 (20.00-55.00) | <0.001 |
| Gonadotropic function | |||
| LH, mIU/l | 0.18 (0.01-3.98) | 3.82 (1.27-8.26) | <0.0001 |
| FSH, µIU/ml | 0.72 (0.05-6.16) | 5.04 (1.45-10.36) | <0.0001 |
| PRL, ng/ml | 15.79 (1.63-54.57) | 8.32 (3.56-11.67) | <0.0001 |
| E2, pg/ml | 24.30 (20.00-68.90) | 23.00 (20.00-45.40) | 0.8898 |
| PROG, ng/ml | 0.10 (0.10-0.49) | 0.10 (0.10-0.89) | 0.1731 |
| TESTO, ng/ml | 2.59 (0.10-10.67) | 4.31 (2.25-7.02) | 0.0429 |
The data for basic information are presented as the mean ± standard deviation. All other data are presented as the median (range). ACTH, adrenocorticotropic hormone; BMI, body mass index; FT3, free thyroxine 3; FT4, free thyroxine 4; TSH, thyroid stimulating hormone; PRL, prolactin; PROG, progesterone; TESTO, testosterone; LH, luteinising hormone; FSH, follicle-stimulating hormone.
Figure 6.
ROC curves for the diagnosis of patients with congenital hypopituitarism. ROC curves according to the methylation levels of (A) cg01606027 on LIM domain kinase 2 and (B) cg03594078 on piwi-like RNA-mediated gene silencing 2, in the validation set. The blue line represents the binormal smoothed ROC curve for each CpG site. ROC, receiver-operating characteristic; AUC, area under the curve; CI, confidence interval.
Discussion
In the present study, the genome-wide DNA methylation levels in the peripheral blood of male patients with CH were determined and compared with age-matched controls. A total of 51 DMPs, that may influence gene expression levels involved in CH development and progression, were identified. Pathway enrichment analysis of the hypermethylated and hypomethylated DMP-associated genes demonstrated that these were involved in ‘metabolic process’, ‘immune system process’ and ‘reproduction’. In addition, two hypermethylated CpG loci located on the promoter regions of LIMK2 and PIWIL2, which are closely associated with testiculal development and spermatogenesis, were observed in the patients with CH. These results suggested that epigenetic alterations may contribute to the pathogenesis of CH, and may provide novel therapeutic targets in the future. Furthermore, the methylation levels of the two loci had moderate predictive abilities in patients with CH, suggesting that these may serve as potential diagnostic or therapeutic markers for CH in the future.
CH is a heterogeneous disease with complex causes, which may be acquired or are idiopathic (27,28). Previous studies observed that the most common mutated genes in patients with CH are PROP1, POU1F1, OTX2, HESX1, LHX3, GLI2, CDON, TGIF, SHH and LHX412; however, the prevalence of mutations on these genes was low (29,30). Prenatal and birth trauma or asphyxia may additionally lead to CH; however, their associations with CH are not yet well established. Therefore, the exact pathogenesis of CH remains unclear.
DNA methylation, which is the addition of a methyl group to the cytosine in a CpG dinucleotide, has vital roles in development and cellular differentiation. Aberrant methylation of genes has been demonstrated to contribute to the development and progression of various diseases. Due to the severe deficiency of the hormones secreted by the pituitary gland, HRT is an effective treatment method for patients with CH; however, they still present with increased risk for hypoglycemia, cardiovascular diseases, microphallus, infertility and hepatitis (12). Previous studies demonstrated that the hormone deficiency is significantly associated with DNA methylation levels (31,32). Gonadotropin deficiency results in lower levels of testosterone in male patients with CH (33). With a mouse embryonic neural stem cell (eNSC) model, Bramble et al (34) demonstrated that transcripts in response to testosterone-propionate (TP) treatment are enriched in genes that affect histone modification and DNA methylation. The TP treatment results in a global decrease in 5-methylcytosine abundance in the genomic DNA, and the DNA hypomethylation is maintained in the daughter cell linages of eNSCs post-exposure (34). These results suggested that testosterone deficiency in early life may lead to the aberrant hypermethylation of genes, which contributes to the pathogenesis of neural diseases (34). In addition, DNA methylation levels additionally influence the response to hormone treatment in the clinic. Ouni et al (35) demonstrated that the P2 promoter of the IGF1 gene mediated the responsiveness to growth hormone in short children. Therefore, aberrant epigenetic gene modulation may contribute to the pathogenesis of CH and additionally influence the responsiveness of patients to hormone treatment.
LIMK2 belongs to the serine LIM kinase family (including LIMK1 and LIMK2) and it serves as a regulator of actin dynamics (36). Previous studies demonstrated that LIMK2 has important roles in cell movement, division and structure formation (37,38). A previous study observed that LIMK2 stimulates pathologic cancer cell division by regulating actin filaments (39). In addition, LIMK2 is not only involved in mitosis; however, additionally meiosis, and LIMK2 activity is essential for microtubule center organization and distribution in mouse oocyte meiosis (40,41). Using a mouse knockout model, Takahashi et al (42) identified that LIMK2−/− mice did not demonstrated embryonic lethality or any phenotypic abnormalities in the postnatal growth and development, except for the lack of spermatogenesis in the testis. Compared with the wide-type mice, LIMK2−/− mice were smaller in size and had partial degeneration of spermatogenic cells in the seminiferous tubules (42). The viability of the LIMK2−/− spermatogenic cells was significantly diminished under stress conditions and the potential for germ cells to differentiate in a regenerative state was additionally impaired in the LIMK2−/− testis (42). These results suggested that LIMK2 is critical for the proper progression of spermatogenesis. The present study demonstrated that LIMK2 was hypermethylated and downregulated in the white blood cells of patients with CH compared with the controls, which may partially explain the impaired spermatogenic ability of the patients. LIMK2 may serve as a potential therapeutic target for CH and treatment with testosterone may decrease the methylation of LIMK2 in testicular germ cells. In addition, the methylation levels of LIMK2 may influence the outcome of HRT in the clinic; however, these considerations require further examination in future studies.
PIWIL2, additionally termed MILI, belongs to the PIWI family, a group of argonaute proteins that interact with a class of small piwi-interacting (pi)RNAs, specifically expressed in the testes during spermatogenesis (43). In the mammalian testes, PIWIL2 is involved in the early phases of spermatogenesis, maintaining genome stability by regulating the retrotranspo-sons through DNA methylation silencing in germline cells (44). PIWIL2 is required for germline stem cell self-renewal (45), and PIWIL2 deficiency leads to increased transposon expression, and defects in meiosis and germ cell survival (46). Heyn et al (47) observed that PIWIL2 was hypermethylated in the semen DNA of infertile males, which resulted in the defective production of piRNAs and the hypomethylation of the LINW-1 repetitive sequence. Friemel et al (48) identified that the PIWIL2 was hypermethylated in in the peripheral blood DNA of patients with idiopathic male infertility compared with control males, which is consistent with the results of the present study. These results suggested that DNA methylation of PIWIL2 may regulate gene expression in spermatogenesis and result in abnormal sperm cell development, which may partially contribute to the infertility disorders of male patients with CH in the clinic.
Besides these two loci, CpG sites located in other genes, including hypoxia-inducible factor (HIF)-3α and regulatory-associated protein of mammalian target of rapamycin (mTOR) complex 1 (RPTOR), were additionally noted in the present study; however, their roles in CH development and progression remain unknown. Hypermethylation of HIF-3α has been observed in children with obesity, and a positive association between BMI and DNA methylation level of HIF-3α was identified in a previous study (49). HIF-3α is highly expressed in adipocytes and has vital roles in adipogenesis (50). Pfeiffer et al (51) demonstrated that the methylation levels of CpG site cg22891070, which was hypermethylated in patients with CH, was significantly higher in visceral adipose tissue (VAT) compared with subcutaneous adipose tissue (SAT), and cg22891070 methylation levels were significantly correlated with BMI, and abdominal SAT and VAT area. Therefore, aberrant HIF-3α methylation may contribute to adipose tissue dysfunction, which may lead to the increased risk of obesity-associated diseases in patients with CH. Two CpG sites (cg27511181 and cg06418238) were identified to be hypermethylated in the gene body of RPTOR, which is an important scaffolding protein that recruits the mTOR substrates to rapamycin-senstitive mTOR complex 1 (mTORC1) (52). RPTOR is required for the suppressive function of regulatory T cells (53). Tang et al (54) demonstrated that the CpG site cg06418238 was hypomethylated in the peripheral blood DNA of patients with breast cancer compared with controls. However, the role of RPTOR and its hypermethylation in the development and progression of CH remain unclear. In the future, novel identified epigenetically regulated genes and their roles in CH development and progression require further investigation, which may provide further insight for the mechanisms for the pathogenesis of CH.
There were a number of limitations of the present study. The sample size in the discovery set was relatively small. Other CpG sites that may be aberrantly methylated in patients with CH may not have been identified due to the statistical power limitations. The methylation status of the genomic DNA was derived from the whole blood, and its consistence with the DNA methylation levels in the target cells of the pituitary gland or testes are largely unknown. Whether LIMK2 and PIWIL2 were hypermethylated in sperm cells and their roles in the infertility of patients with CH require examination in the future. The mechanisms for the aberrant methylation levels of the target genes remain unknown. Whether the hypermethylation or hypomethylation of the CpG sites was caused by perinatal damage or hormone deficiency, and the molecular mechanisms for the epigenetic regulation of these genes under stress require further investigation in future studies. The effect of HRT in the epigenetic regulation of the aberrantly methylated genes in patients with CH and whether the methylated loci may regulate the outcome of HRT requires further study.
In conclusion, the present study determined the genome-wide DNA methylation levels of the peripheral whole blood DNA in male patients with CH and demonstrated that 51 CpG sites were significantly hypermethylated or hypomethylated in patients with CH compared with normal controls. Two CpG sites cg01606027 and cg03594078 located at the coding region of the genes LIMK2 and PIWIL2, that are essential for spermatogenesis, were hypermethylated, which may contribute to azoospermia or oligozoospermia in male patients with CH. These results suggested that the aberrant epigenetic regulated genes may contribute to the pathogenesis of CH, and that they may serve as novel therapeutic targets or diagnosis biomarkers for CH in the future.
Acknowledgments
Not applicable.
Funding
The present study was financially supported by grants from the National Key R&D Program of China (grant nos. 2017YFC0907001 and 2018ZX10302205), the Science and Technology Commission of Shanghai Municipality (grant no. 16411966800; Shanghai, China), the Shanghai Municipal Commission of Health and Family Planning (grant nos. 20164Y0250 and 201840160; Shanghai, China), the Science and Technology Commission of Jiading District (grant nos. JDKW-2017-W09 and JDKW-2017-W11; Shanghai, China), the interdisciplinary funding of Shanghai Jiao Tong
University (grant no. YG2017QN57; Shanghai, China) and the Ruijin Hospital North for Young Talents (grant nos. 2017RCPY-A01 and 2017RCPY-C01; Shanghai, China).
Availability of data and materials
The Human Methylation 450K DNA methylation data are available at the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE107737.
Authors’ contributions
XF and PC drafted the manuscript and coordinated the study. JC and JL performed the array analysis. XF, CC, EX and PC conducted the validation experiments. JL and PC performed the bioinformatics analysis. XF and PC performed the polymerase chain reaction experiments. XF, CC, EX and JC acquired the samples and information of the participants. XF, JL and PC designed and supervised the study, and revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all the participants, and the study was approved by the Institutional Review Board of Ruijin Hospital North (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- ACTH
adrenocorticotropin
- AUC
area under the curve
- BMI
body mass index
- CI
confidence interval
- CH
congenital hypopituitarism
- DMP
differentially methylated probe
- FDR
false discovery rate
- FSH
follicle-stimulating hormone
- HRT
hormone replacement treatment
- LH
luteinising hormone
- MRI
magnetic resonance imaging
- PRL
prolactin
- PROG
progesterone
- ROC
receiver-operating characteristic
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- SAT
subcutaneous adipose tissue
- TSH
thyroid stimulating hormone
- VAT
visceral adipose tissue
References
- 1.Regal M, Paramo C, Sierra SM, Garcia-Mayor RV. Prevalence and incidence of hypopituitarism in an adult Caucasian population in northwestern Spain. Clin Endocrinol (Oxf) 2001;55:735–740. doi: 10.1046/j.1365-2265.2001.01406.x. [DOI] [PubMed] [Google Scholar]
- 2.Agha A, Sherlock M, Brennan S, O’Connor SA, O’Sullivan E, Rogers B, Faul C, Rawluk D, Tormey W, Thompson CJ. Hypothalamic-pituitary dysfunction after irradiation of nonpituitary brain tumors in adults. J Clin Endocrinol Metab. 2005;90:6355–6360. doi: 10.1210/jc.2005-1525. [DOI] [PubMed] [Google Scholar]
- 3.Bondanelli M, De Marinis L, Ambrosio MR, Monesi M, Valle D, Zatelli MC, Fusco A, Bianchi A, Farneti M, degli Uberti EC. Occurrence of pituitary dysfunction following traumatic brain injury. J Neurotrauma. 2004;21:685–696. doi: 10.1089/0897715041269713. [DOI] [PubMed] [Google Scholar]
- 4.Herrmann BL, Rehder J, Kahlke S, Wiedemayer H, Doerfler A, Ischebeck W, Laumer R, Forsting M, Stolke D, Mann K. Hypopituitarism following severe traumatic brain injury. Exp Clin Endocrinol Diabetes. 2006;114:316–321. doi: 10.1055/s-2006-924254. [DOI] [PubMed] [Google Scholar]
- 5.Cuesta M, Hannon MJ, Crowley RK, Behan LA, Tormey W, Rawluk D, Delargy M, Agha A, Thompson CJ. Symptoms of gonadal dysfunction are more predictive of hypopituitarism than nonspecific symptoms in screening for pituitary dysfunction following moderate or severe traumatic brain injury. Clin Endocrinol. 2016;84:92–98. doi: 10.1111/cen.12874. [DOI] [PubMed] [Google Scholar]
- 6.Darzy KH, Shalet SM. Hypopituitarism after cranial irradiation. J Endocrinol Invest. 2005;28(Suppl 5):S78–S87. [PubMed] [Google Scholar]
- 7.Craft WH, Underwoood LE, Van Wyk JJ. High incidence of perinatal insult in children with idiopathic hypopituitarism. J Pediatr. 1980;96:397–402. doi: 10.1016/S0022-3476(80)80680-9. [DOI] [PubMed] [Google Scholar]
- 8.Nystrom HF, Saveanu A, Barbosa EJ, Barlier A, Enjalbert A, Glad C, Palming J, Johannsson G, Brue T. Detection of genetic hypopituitarism in an adult population of idiopathic pituitary insufficiency patients with growth hormone deficiency. Pituitary. 2011;14:208–216. doi: 10.1007/s11102-010-0278-8. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson JW, Holden N, Hills RK, Wheatley K, Clayton RN, Bates AS, Sheppard MC, Stewart PM. Association between premature mortality and hypopituitarism. West Midlands Prospective Hypopituitary Study Group. Lancet. 2001;357:425–431. doi: 10.1016/S0140-6736(00)04006-X. [DOI] [PubMed] [Google Scholar]
- 10.Kohno H, Kuromaru R, Ueyama N, Miyako K. Premature mortality and hypopituitarism. Lancet. 2001;357:1973–1974. doi: 10.1016/S0140-6736(00)05036-4. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Rodriguez E, Quinteiro C, Barreiro J, Marazuela M, Pereiro I, Peino R, Cabezas-Agricola JM, Dominguez F, Casanueva FF, Bernabeu I. Pituitary stalk dysgenesis-induced hypopituitarism in adult patients: Prevalence, evolution of hormone dysfunction and genetic analysis. Neuroendocrinology. 2011;93:181–188. doi: 10.1159/000324087. [DOI] [PubMed] [Google Scholar]
- 12.Geffner ME. Hypopituitarism in childhood. Cancer Control. 2002;9:212–222. doi: 10.1177/107327480200900304. [DOI] [PubMed] [Google Scholar]
- 13.Braslavsky D, Mendez MV, Prieto L, Keselman A, Enacan R, Gruneiro-Papendieck L, Jullien N, Savenau A, Reynaud R, Brue T, et al. Pilot neonatal screening program for central congenital hypothyroidism: Evidence of significant detection. Horm Res Paediatr. 2017;88:274–280. doi: 10.1159/000480293. [DOI] [PubMed] [Google Scholar]
- 14.Du X, Yuan Q, Yao Y, Li Z, Zhang H. Hypopituitarism and successful pregnancy. Int J Clin Exp Med. 2014;7:4660–4665. [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton RN. Mortality, cardiovascular events and risk factors in hypopituitarism. Growth Horm IGF Res. 1998;8(Suppl A):S69–S76. doi: 10.1016/S1096-6374(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 16.Kung AW, Zhong YY, Lam KS, Wang C. Induction of spermatogenesis with gonadotrophins in Chinese men with hypogonadotrophic hypogonadism. Int J Androl. 1994;17:241–247. doi: 10.1111/j.1365-2605.1994.tb01249.x. [DOI] [PubMed] [Google Scholar]
- 17.Bulow B, Hagmar L, Eskilsson J, Erfurth EM. Hypopituitary females have a high incidence of cardiovascular morbidity and an increased prevalence of cardiovascular risk factors. J Clin Endocrinol Metab. 2000;85:574–584. doi: 10.1210/jcem.85.2.6346. [DOI] [PubMed] [Google Scholar]
- 18.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy MM, Nugent BM. Epigenetic contributions to hormonally-mediated sexual differentiation of the brain. J Neuroendocrinol. 2013;25:1133–1140. doi: 10.1111/jne.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nugent BM, Schwarz JM, McCarthy MM. Hormonally mediated epigenetic changes to steroid receptors in the developing brain: Implications for sexual differentiation. Horm Behav. 2011;59:338–344. doi: 10.1016/j.yhbeh.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto G, Netchine I, Sobrier ML, Brunelle F, Souberbielle JC, Brauner R. Pituitary stalk interruption syndrome: A clinical-biological-genetic assessment of its pathogenesis. J Clin Endocrinol Metab. 1997;82:3450–3454. doi: 10.1210/jcem.82.10.4295. [DOI] [PubMed] [Google Scholar]
- 22.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile Within Array Normalization for illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. The adaptive control of the false discovery rate in multiple hypotheses testing. J Behav Educ Statist. 2000;25:60–83. doi: 10.3102/10769986025001060. [DOI] [Google Scholar]
- 24.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41(Database Issue):D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Prabhakar VK, Shalet SM. Aetiology, diagnosis, and management of hypopituitarism in adult life. Postgrad Med J. 2006;82:259–266. doi: 10.1136/pgmj.2005.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffle R, Klammt J. Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract Res Clin Endocrinol Metab. 2011;25:43–60. doi: 10.1016/j.beem.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Kelberman D, Rizzoti K, Lovell-Badge R, Robinson IC, Dattani MT. Genetic regulation of pituitary gland development in human and mouse. Endocr Rev. 2009;30:790–829. doi: 10.1210/er.2009-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi M, Nagasaki K, Fujiwara I, Ishii T, Amano N, Asakura Y, Muroya K, Hasegawa Y, Adachi M, Hasegawa T. Heterozygous defects in PAX6 gene and congenital hypopituitarism. Eur J Endocrinol. 2015;172:37–45. doi: 10.1530/EJE-14-0255. [DOI] [PubMed] [Google Scholar]
- 30.Romero CJ, Nesi-Franca S, Radovick S. The molecular basis of hypopituitarism. Trends Endocrinol Metab. 2009;20:506–516. doi: 10.1016/j.tem.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, Ren M, Li L, Zhu Y, Chu Z, Zhu Z, Ruan Q, Lou W, Zhang H, Han Z, et al. Perinatal testosterone exposure potentiates vascular dysfunction by ERβ suppression in endothelial progenitor cells. PLoS One. 2017;12:e0182945. doi: 10.1371/journal.pone.0182945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huen K, Harley K, Kogut K, Rauch S, Eskenazi B, Holland N. DNA methylation of LINE-1 and Alu repetitive elements in relation to sex hormones and pubertal timing in Mexican-American children. Pediatr Res. 2016;79:855–862. doi: 10.1038/pr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantalone KM, Faiman C. Male hypogonadism: more than just a low testosterone. Cleve Clin J Med. 2012;79:717–725. doi: 10.3949/ccjm.79a.11174. [DOI] [PubMed] [Google Scholar]
- 34.Bramble MS, Roach L, Lipson A, Vashist N, Eskin A, Ngun T, Gosschalk JE, Klein S, Barseghyan H, Arboleda VA, Vilain E. Sex-specific effects of testosterone on the sexually dimorphic transcriptome and epigenome of embryonic neural stem/progenitor cells. Sci Rep. 6(36916):2016. doi: 10.1038/srep36916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouni M, Belot MP, Castell AL, Fradin D, Bougneres P. The P2 promoter of the IGF1 gene is a major epigenetic locus for GH responsiveness. Pharmacogenomics J. 2016;16:102–106. doi: 10.1038/tpj.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Funakoshi H, Nakamura T. LIM-kinase as a regulator of actin dynamics in spermatogenesis. Cytogenet Genome Res. 2003;103:290–298. doi: 10.1159/000076815. [DOI] [PubMed] [Google Scholar]
- 37.Croft DR, Crighton D, Samuel MS, Lourenco FC, Munro J, Wood J, Bensaad K, Vousden KH, Sansom OJ, Ryan KM, et al. p53-mediated transcriptional regulation and activation of the actin cytoskeleton regulatory RhoC to LIMK2 signaling pathway promotes cell survival. Cell Res. 2011;21:666–682. doi: 10.1038/cr.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sumi T, Hashigasako A, Matsumoto K, Nakamura T. Different activity regulation and subcellular localization of LIMK1 and LIMK2 during cell cycle transition. Exp Cell Res. 2006;312:1021–1030. doi: 10.1016/j.yexcr.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 39.Heng YW, Lim HH, Mina T, Utomo P, Zhong S, Lim CT, Koh CG. TPPP acts downstream of RhoA-ROCK-LIMK2 to regulate astral microtubule organization and spindle orientation. J Cell Sci. 2012;125:1579–1590. doi: 10.1242/jcs.096818. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Zhu Y, Cao Y, Wang Q, Du J, Tian J, Liang Y, Ma W. LIM kinase activity is required for microtubule organising centre positioning in mouse oocyte meiosis. Reprod Fertil Dev. 2017;29:791–804. doi: 10.1071/RD15406. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi T, Koshimizu U, Abe H, Obinata T, Nakamura T. Functional involvement of Xenopus LIM kinases in progression of oocyte maturation. Dev Biol. 2001;229:554–567. doi: 10.1006/dbio.2000.9999. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi H, Koshimizu U, Miyazaki J, Nakamura T. Impaired spermatogenic ability of testicular germ cells in mice deficient in the LIM-kinase 2 gene. Dev Biol. 2002;241:259–272. doi: 10.1006/dbio.2001.0512. [DOI] [PubMed] [Google Scholar]
- 43.Yuan ZH, Zhao YM. The regulatory functions of piRNA/PIWI in spermatogenesis. Yi Chuan. 2017;39:683–691. doi: 10.16288/j.yczz.17-245. [DOI] [PubMed] [Google Scholar]
- 44.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J, Esteller M, Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One. 2012;7:e47892. doi: 10.1371/journal.pone.0047892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friemel C, Ammerpohl O, Gutwein J, Schmutzler AG, Caliebe A, Kautza M, von Otte S, Siebert R, Bens S. Array-based DNA methylation profiling in male infertility reveals allele-specific DNA methylation in PIWIL1 and PIWIL2. Fertil Steril. 2014;101:1097–1103.e1. doi: 10.1016/j.fertnstert.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 49.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Hedman AK, Sandling JK, Li LA, Irvin MR, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatanaka M, Shimba S, Sakaue M, Kondo Y, Kagechika H, Kokame K, Miyata T, Hara S. Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biol Pharm Bull. 2009;32:1166–1172. doi: 10.1248/bpb.32.1166. [DOI] [PubMed] [Google Scholar]
- 51.Pfeiffer S, Kruger J, Maierhofer A, Bottcher Y, Kloting N, El Hajj N, Schleinitz D, Schon MR, Dietrich A, Fasshauer M, et al. Hypoxia-inducible factor 3A gene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Sci Rep. 2016;6:27969. doi: 10.1038/srep27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 53.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Q, Holland-Letz T, Slynko A, Cuk K, Marme F, Schott S, Heil J, Qu B, Golatta M, Bewerunge-Hudler M, et al. DNA methylation array analysis identifies breast cancer associated RPTOR, MGRN1 and RAPSN hypomethylation in peripheral blood DNA. Oncotarget. 2016;7:64191–641202. doi: 10.18632/oncotarget.11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Human Methylation 450K DNA methylation data are available at the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE107737.