Abstract
Acute-on-chronic liver failure (ACLF) is a newly-defined serious syndrome with major features of acute decompensation (AD) of hepatic cirrhosis, liver failure and failure of multiple other organs. To date, the mechanism underlying the development and progression of ACLF remains to be fully elucidated. It has been noted that ACLF is associated with immune dysregulation. However, studies have mainly focused on T-cell responses. The present study aimed to determine the composition and alterations of B-cell receptor (BCR) heavy chain repertoires associated with ACLF using next generation sequencing (NGS). A total of six patients with hepatitis B virus (HBV)-related ACLF and six healthy control subjects were prospectively enrolled in the present study. The B-cell immunoglobulin heavy chain (IGH) repertoires in peripheral blood mononuclear cells (PBMCs) obtained from the patients with HBV-related ACLF and the control subjects were analyzed using NGS, coupled with multiplex polymerase chain reaction, were Illumina sequenced, and were further characterized using the international ImMunoGeneTics database. The distribution of the BCR complementarity-determining region 3 (CDR3) variable (V), diversity (D) and joining (J) and V-J gene segments were found to be comparable between the ACLF and control groups. Of note, the degree of clonal expansion in the ACLF group was significantly higher than that in the control group (P<0.05). Furthermore, a t-test of the distribution ratio of the V, D, J and V-J combinations in patients with ACLF and control subjects revealed differentially expressed genes. In total, six genes were upregulated and 19 genes were downregulated in response to ACLF. The difference between these two groups was statistically significant (P<0.05). The approach used in the present study was feasible and effective for analyzing peripheral B-cell repertoires in HBV-related ACLF. These results provide direct evidence that the BCR repertoire is important in immune responses, autoimmunity and alloreactivity, and that there is a link between the BCR repertoire and HBV-ACLF. Therefore, ACLF-specific BCR CDR3 sequences hold promise for therapeutic benefit to HBV-ACLF in the future.
Keywords: acute-on-chronic liver failure, immune repertoire, next generation sequencing, B cell receptor
Introduction
Acute-on-chronic liver failure (ACLF) has been increasingly recognized as a distinct disease entity. ACLF has been newly-defined as a serious syndrome characterized by the acute decompensation (AD) of hepatic cirrhosis, which encompasses the acute deterioration of hepatic function and involves the dysfunction of multiple other organs in patients with chronic liver disease (1). Based on an analytic study of 1,343 patients, the European Association for the Study of the Liver and Chronic Liver Failure (EASL-CLIF) consortium suggested the following diagnostic criteria for ACLF: i) Liver cirrhosis and AD as defined by the development of ascites, encephalopathy, gastrointestinal hemorrhage and bacterial infection; ii) organ failure assessed according to the Chronic Liver Failure-Sequential Organ Failure Assessment (CLIF-SOFA) score; iii) a 28-day mortality rate >15%. In this CANONIC study, the prevalence of ACLF reached as high as 30.9%, and this considerably relevant syndrome appeared to be different from AD, which was not only based on liver failure, multiple organ failure and high mortality rates, but also on age, precipitating events and systematic inflammation (2). It has been reported that alcoholic liver cirrhosis represents 50-70% of the underlying liver diseases of ACLF in Western countries, whereas hepatitis-associated liver cirrhosis constitutes 10-30% of all ACLF cases. In the majority of Asian countries with high a prevalence of hepatitis B virus (HBV) infection, chronic hepatitis B (CHB) is considered to be the major causative factor for ACLF, accounting for ~70% of ACLF cases, and accounts for as little as 15% of alcohol-related ACLF (2-4).
To date, the mechanism underlying the development and progression of ACLF remain to be fully elucidated. It has been noted that that ACLF is correlated to immune dysregulation. However, studies have mainly focused on T-cell response, despite the fact that human lymphocytes are well known to comprise T- and B-cells, which account for almost 20% of peripheral lymphocytes. These fulfil an important role in the immune response. An effective B-cell immune response is largely dependent on B-cells with diverse B-cell receptors (BCRs), which are capable of specifically recognizing and binding antigens. Each BCR is structurally connected by two heavy chains with variable (V), constant (C), transmembrane and cytoplasmic regions, and two light (L) chains which have V and C regions.
It has been documented that the V region comprises two domains, namely, VH and VL, and possesses three complementarity-determining regions(CDRs): CDR1, CDR2 and CDR3. CDRs are variable in amino acid composition and sequence, and the range can reach as high as 69-612 in the library, allowing the identification of a variety of antigens and the production of specific antibodies. These three CDRs act to identify and jointly determine BCR antigen specificity (5). Similar to the T-cell receptor repertoire, the diversity of the BCR repertoire is possibly formed through the recombination of multiple sets of similar genes to generate functional genes. Immunoglobulin heavy (IGH) chains are encoded by recombination of variable (V), diversity (D) and joining (J) genes, which are formed from sets of V, D and J genes (IGHV and IGHJ), whereas VJ rearrangements of the κ and λ chain V genes (IGKV, IGLV) and J genes (IGKJ, IGLJ) encode the immunoglobulin light chains (6,7). It has long been observed that there is a broad deficit of B-cells in patients who suffer from acute liver diseases, and that there is an association between exposure to circulating antigens and the abnormality of shunting or Kupffer cells, which is presumably the consequence (8).
High-throughput sequencing (HTS) is an advanced genetic tool that enables the investigation of more complex sequencing targets. In the present study, HTS was coupled with multiplex polymerase chain reaction (PCR), Illumina sequencing and the international ImMunoGeneTics database to assess the composition and alteration of BCR heavy chain repertoires associated with ACLF. It is anticipated that the results of the present study may provide direct evidence of the role of BCR heavy chain repertoires in the development and progression of ACLF.
Materials and methods
Patients
In the present prospective study, six patients with HBV-related ACLF and six healthy controls (control subjects) were recruited between January and September 2013 at the No. 181 Hospital of Guilin (Guilin, China). The diagnosis of ACLF was pathologically and clinically confirmed on the basis of the diagnostic criteria developed by the Asia-Pacific Association for the Study of the Liver (3,9), which has been accepted for defining this novel clinical entity as follows: ‘An acute hepatic insult manifesting as jaundice and coagulopathy, which is complicated within four weeks by ascites and/or encephalopathy in a patient with previously diagnosed or undiagnosed chronic liver disease’. The patients with HBV-related ACLF were confirmed to have CHB, without any evidence of sepsis/bacterial infection, combined hepatitis C virus infection, or active alcoholism. Therefore, the patients with ACLF in the present study shared a similar etiology, which was HBV infection. However, there were no differences among BCRs associated with the etiologies of ACLF. It is noteworthy that jaundice with serum bilirubin ≥5 mg/dl or 85 µmol/l, and coagulopathy with INR >1.5 or prothrombin activity <40% are required for the final diagnosis of ACLF (3). The patients with HBV-related ACLF had a mean age of 34.12 years old, with an age range of 20-54 years old. The six healthy controls (control subjects) were matched for age, gender and ethnicity, and had no clinical signs of liver disease.
Prior to the present study, written informed consent was obtained from each participant or from their guardians. The present study complied with the Declaration of Helsinki for investigations involving humans, and the study protocol was carefully reviewed and approved by the Ethics Committee of the No. 181 Hospital of Guilin.
Multiplex PCR
Amplification of the BCR CDR3 region by multiplex-PCR. In order to prepare the template library for subsequent analysis using a Genome Analyzer, multiplex-PCR was performed. DNA was extracted from PBMCs using GenFIND DNA (Agencourt; Beckman Coulter, Inc., Brea, CA, USA) extraction kits according to the manufacturer’s protocol. The following were added to a 200-µl PCR tube: 500 ng DNA, Mul-PCR Primer (V primer and J primer) using a QIAGEN multiplex PCR kit. The tubes were placed into a thermal cycler and the PCR cycling program was run with 30 cycles. Agarose gel electrophoresis: Using 2% agarose gel, electrophoresis conditions: 100 V for 2 h. Size selection: 100-190 bp. The amplicons were purified using the QIAquick Gel Extraction kit (Qiagen GmbH, Hilden, Germany). Samples were quantified using Quant-iT PicoGreen (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The rearranged BCR CDR3 regions were amplified using genomic DNA as the template, with 12 forward primers, each specific to the functional BCR-V segment, and four reverse primers, each unique to the BCR-J segment. All the forward and reverse primers harbored the universal forward and reverse primer sequences (forward, 5′-AAT GAT ACG GCG ACC ACC GA-3′ reverse, 5′-CAA GCA GAA GAC GGC ATA CGA-3′) at their 5′ ends, respectively, which are compatible with the GA2 cluster station solid-phase PCR. Following the multiplex-PCR amplification, the targeted PCR products were purified using agarose gel electrophoresis and a QIA quick PCR purification kit, according to the manufacturer’s protocol. The generated library was then quantified using the following two approaches: Determination of the average length on the Agilent 260 bioanalyzer instrument with Agilent DNA 600 reagents, and reverse transcription-quantitative PCR analysis using the TaqMan probe. For multiplex PCR amplification of DNA, 30 ng DNA was mixed with the JH reverse primer and the FR1 forward primer set (0.25 µM each), using 0.5 µl Phusion® High-Fidelity DNA Polymerase (Finnzymes; Thermo Fisher Scientific, Inc.), 1 µl dNTPs (0.25 µM) and 1 µl DTT (0.25 µM), per 50 µl reaction. The following PCR program was used: 3 min at 94°C, 35 cycles of 30 sec at 94°C, 30 sec at 60°C and 1 min at 72°C, with a final extension cycle of 7 min at 72°C on an MJ Thermocycler (10).
The libraries were subsequently amplified with the c-Bot system to create a cluster on the flow cell, followed by paired-end sequencing of the amplified flow cell using an Illumina MiSeq instrument, in which the generally accepted read length of 60 bp was used. In order to ensure analysis quality, any raw reads that contained the low quality sequence and adaptor sequence were filtered, following which clean reads suitable for the subsequent data analysis were obtained. The data processing procedures were summarized as follows: i) Removal of reads with the adapter; ii) elimination of N reads, as N is unable to determine the information for a base with a proportion >6%; iii) removal of reads of relatively low quality (sequence quality value ≤5 bases accounting for >50% of the entire read).
Data analysis
During the development of B-lymphocytes, the V, D and J gene rearrangement of BCR, the V-J fragments, or that between the V-D-J fragments may be randomly inserted into various numbers of nucleotides. By contrast, different numbers of nucleotides may be missed in the V-J fragments or those between the V-D-J fragments. Therefore, a highly diverse CDR3 variable area, which has marked differences in length and amino acid sequence, was formed. This allowed the diversity of B-lymphocytes to be assessed through the length of CDR3. The length of CDR3 in normal individuals is similarly normally distributed. In the present study, systematic analysis of the frequent use of V, D J and V-J fragments was performed. The upregulated and downregulated genes were identified using Student’s t-test to compare the BCR heavy chain V, D, J and V-J fragments. Furthermore, the diversity of the BCR repertoire was assessed using the Simpson index of diversity (Ds) (10) and Shannon-Wiener index (H′) (11) in the present study. The statistical analyses were conducted using GraphPad Prism software (version 4.03; GraphPad Software, San Diego, CA, USA). The Mann-Whitney test was used to analyze the differences between the ACLF and control groups, given the relatively small sample size in the present study. P<0.05 was considered to indicate a statistically significant difference.
Results
BCR heavychainCDR3 sequences from patients with HBV-related ACLF and control subjects
The HTS technique was used to capture the BCR heavychainCDR3 sequences of B-cells prepared from peripheral blood obtained from the recruited patients with ACLF and control subjects. As a result, an average number of 12,243,860.30 in the control group and an average number of 12,299,65.30 in the ACLF group were initially obtained. As the Raw Reads or Sequenced Reads contain low-quality sequences and adaptors sequences, these Raw Reads were filtered to obtain high-quality clean reads with an average number of 6,674,277.80 in the control group and an average number of 6,114,722.70 in the ACLF group, ensuring the quality of the information analysis. These were used for the subsequent data analysis. The total reads/sequences, BCR sequences, in-frame sequences, total IGH CDR3 sequences, unique CDR3 nt sequences, unique CDR3 aa sequences, highly expanded clone (HEC) numbers, and HEC ratios are shown in Table I.
Table I.
Comparison of B-cell receptor statistical data.
| Individual | Total reads | In-frame sequences | Total CDR3 sequences | Unique cdr3 nt sequences | Unique cdr3 aa sequences | HEC number, all |
|---|---|---|---|---|---|---|
| NC-1 | 15,410,305 | 13,317,137 | 12,584,061 | 414,914 | 292,378 | 0 |
| NC-2 | 15,517,826 | 13,016,806 | 12,249,709 | 391,751 | 276,673 | 1 |
| NC-3 | 14,022,700 | 11,415,501 | 6,600,111 | 260,471 | 181,219 | 4 |
| NC-4 | 537,114 | 465,901 | 439,959 | 43,655 | 30,075 | 2 |
| NC-5 | 14,696,122 | 12,483,863 | 11,778,081 | 392,836 | 279,047 | 2 |
| NC-6 | 13,279,095 | 11,231,688 | 6,575,248 | 432,519 | 316,507 | 0 |
| ACLF-1 | 13,494,348 | 11,594,496 | 6,925,522 | 388,512 | 281,302 | 2 |
| ACLF-2 | 11,264,437 | 9,942,837 | 9,479,612 | 452,729 | 329,633 | 0 |
| ACLF-3 | 9,958,559 | 8,099,585 | 7,733,592 | 190,996 | 140,723 | 201 |
| ACLF-4 | 12,978,762 | 9,887,906 | 8,771,599 | 168,652 | 118,150 | 446 |
| ACLF-5 | 12,819,431 | 6,862,593 | 6,206,099 | 316,046 | 228,715 | 6 |
| ACLF-6 | 12,539,789 | 6,300,919 | 9,651,145 | 446,872 | 326,426 | 0 |
CDR3, complementarity-determining region 3; HEC, highly expanded clone; NC, normal control; ACLF, acute-on-chronic liver failure.
Comparison of immune diversity in patients with ACLF and control subjects
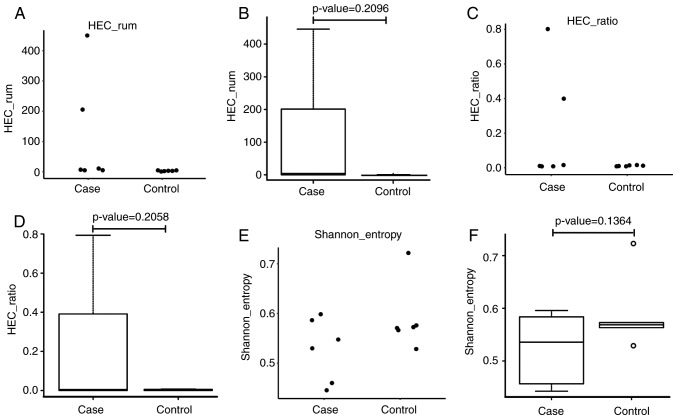
In order to compare the immune diversity between the ACLF and control groups, the normalized Shannon entropy index was applied, which has been well-accepted for quantitatively measuring diversity. In the analysis of immune diversity in these two groups, the index was calculated and rated between 0 and 1, with 1 as the highest diversity and 0 as an indication for no immune diversity. As shown in Fig. 1A-F, immune diversity was marginally higher in the ACLF group, when compared with that in the control subjects, although this difference was not statistically significant (P=0.1364). Subsequently, the HEC in these two groups were examined, and it was found that the HEC was higher in the ACLF group than in the control group, indicating the amplification of abnormal CDR3 sequences. Although differences between these two groups were observed, there was no statistical significance, which was possibly due to the relatively small sample size used in the present study. In comparing the Shannon entropy of the ACLF group with the control group, it was found that the Shannon entropy value distribution of the patients with ACLF was scattered, whereas the value distribution of control subjects was relatively concentrated. As shown in Fig. 1A and B, the Shannon entropy distribution of patients with ACLF was substantially skewed, whereas the Shannon entropy of the control subjects presented with a normal distribution. Although different Shannon entropy distributions were identified in these two groups, the difference was not statistically significant (P=0.2096). The CDR3 HEC was higher in the ACLF patient group, compared with that in the control subjects, however, the difference between these two groups was not statistically significant.
Figure 1.
Comparative analysis of B-cell diversity between the ACLF and control groups. (A) Values in the control group were concentrated, whereas values in the ACLF group were relatively dispersed. (B) A significantly skewed distribution was identified in the ACLF patient group, whereas a substantially normal distribution was identified in the control group (P=0.2096). (C) Scatterplot and (D) box plot of the HEC ratio show that the values in the control group were concentrated and the values in the ACLF group were relatively dispersed (P=0.2058). (E) Scatterplot and (F) box plot of Shannon entropy distribution for patients with ACLF and control subjects, show that the values for the control group were concentrated, whereas the values for the ACLF group were relatively dispersed. HEC, highly expanded clone; ACLF, acute-on-chronic liver failure.
Analysis of CDR3 length distribution between patients with ACLF and control subjects
It is well-recognized that CDR3 length distributions in normal subjects are approximately normally distributed. In the present study, CDR3 length distributions were analyzed in patients with ACLF (X1A, TX-1A,TY-1A,TA-1A,TB-1A and TC-1A) and control subjects (K1A, K2A, K4A, K6A, K7A and W1A). The results are shown in Fig. 2A and B. The CDR3 normal length distribution of patients with ACLF was compared with that of control subjects. Through Gaussian fitting, the normal CDR3 length distribution of each sample was obtained. Furthermore, the normal distribution between the ACLF and control groups was compared, and it was found that the CDR3 length distribution of control subjects tended to be more normally distributed, when compared with that of patients with ACLF (Fig. 2C and D).
Figure 2.
CDR3 length distribution for patients with ACLF and control subjects. (A) CDR3 length distribution for each of the six control subjects. (B) CDR3 length distribution for each of the six patients with ACLF. (C) Scatterplot analysis of the distribution of CDR3 length in patients with ACLF, vs. control subjects. (D) Boxplot analysis of the CDR3 length distribution in the ACLF group, compared with that in the control group. CDR3, complementarity-determining region 3; ACLF, acute-on-chronic liver failure.
Comparison of usage frequency of the V, D, and J gene segments in the BCR heavy chain
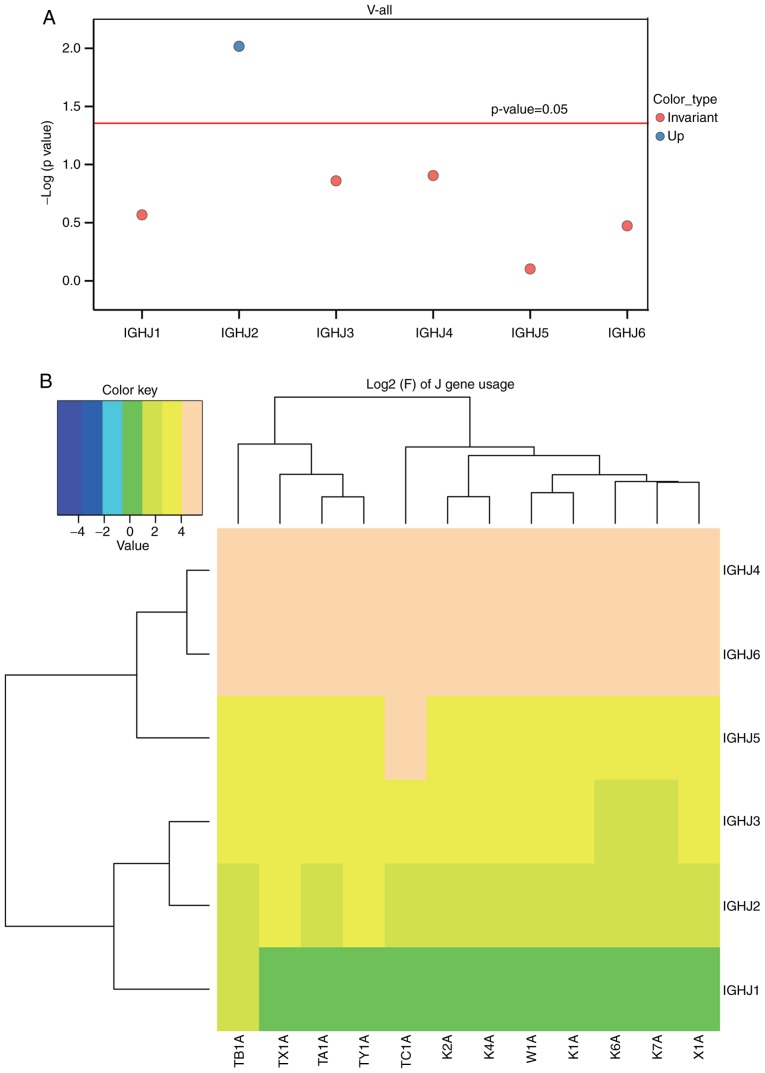
In order to compare the usage frequency of the V, D and J gene segments in the BCR heavy chain, a usage frequency distribution histogram and hierarchical clustering heat map were generated according to each usage frequency in the ACLF and control groups. The differential expression, including the upregulation and downregulation of the individual gene fragments, were then identified in the ACLF group, and compared with the control group. A t-test was used to analyze the usage frequency and distribution ratio of the V, D, J and V-J genes in the six patients with ACLF (X1A, TX-1A, TY-1A, TA-1A, TB-1A and TC-1A) and six control subjects (K1A, K2A, K4A, K6A, K7A and W1A). It was found that the usage of the IGHV gene, which is a well-accepted assessment for BCR repertoire structure, exhibited lower frequencies in the majority or all IGHV genes in the control subjects, when compared with that in the patients with ACLF. Furthermore, these clonal populations had significantly higher frequencies of a single IGHV gene or group of IGHV genes (12). Based on the usage frequency distribution histogram of the V gene of the BCR heavy chain and clustering heat map (Fig. 3A and B), and the results obtained from the t-test for the distribution ratio of the V gene of patients with ACLF and control subjects (Fig. 3; Table II), it was found that the IGHV3-30 gene was significantly upregulated (P<0.05), whereas the IGHV3-74 and IGHV2-70 genes were significantly downregulated (P<0.05). Furthermore, a distribution histogram of the D region usage frequency of the BCR heavy chain and the D sub-genotype of each frequency clustering heat map were generated (Fig. 4A and B). The t-test for the distribution ratio of the D gene in the six patients with ACLF and six control subjects allowed for the identification of the differential gene expression between these two groups (Fig. 4; Table III). As a result, three significantly downregulated genes, IGHD4/OR15-4b, IGHD4/OR15-4a and IGHD2-8, were identified in the ACLF group, compared with the control group (P<0.05). Similarly, a distribution histogram of the J region usage frequency of BCR heavy chain and the J sub-genotype of each frequency clustering heat-map was established (Fig. 5A and B). At-test was then performed for the distribution ratio of the J gene of the six patients with ACLF and six control subjects (Fig. 5, Table IV). The IGHJ2 gene was identified to be significantly upregulated in the ACLF group, when compared with the control group (P<0.05), whereas the IGHJ1, IGHJ5, IGHJ3, IGHJ4 and IGHJ6 genes remained statistically unchanged between these two groups.
Figure 3.
Analysis of the usage frequency of V gene segments in the B-cell receptor heavy chain complementarity-determining region 3 region in patients with ACLF and control subjects. (A) Upregulated genes are denoted in blue and downregulated genes are denoted in red. In the t-test, positive values indicate upregulated genes, whereas negative values indicate downregulated genes. (B) Clustering heat map of the V gene sub-types of the six patients with ACLF and six control subjects. ACLF, acute-on-chronic liver failure.
Table II.
Results of the t-test for the distribution ratio of the V gene.
| Type | Control | Case | P-value | T-value |
|---|---|---|---|---|
| IGHV3-74 | 2.191677 | 1.526667 | 0.009973 | −0.26749 |
| IGHV2-70 | 1.486667 | 1.266667 | 0.046805 | −2.26805 |
| IGHV3-30 | 8.946667 | 11.93333 | 0.024998 | 2.645089 |
IGHV, immunoglobulin heavy variable.
Figure 4.
Analysis of the usage frequency of D gene segments in the B-cell receptor heavy chain complementarity-determining region 3 region in patients with ACLF and control subjects. (A) Downregulated genes are shown in red (P<0.05). (B) Clustering heat map of the D gene sub-types of patients with ACLF and control subjects. ACLF, acute-on-chronic liver failure.
Table III.
Results of the t-test for the distribution ratio of the D gene.
| Type | Control | Case | P-value | T-value |
|---|---|---|---|---|
| IGHD4/OR15-4b | 0.35 | 0.158333 | 0.001349 | −5.11743 |
| IGHD4/OR15-4a | 0.293333 | 0.118333 | 0.001483 | −4.87766 |
| IGHD2-8 | 1.393333 | 0.731667 | 0.002883 | −3.929 |
IGHD, immunoglobulin heavy diversity.
Figure 5.
Analysis of the usage frequency of J gene segments in the B-cell receptor heavy chain complementarity-determining region 3 region in patients with ACLF and control subjects. (A) Significantly downregulated genes are denoted in red (P<0.05), and genes with no significant changes are denoted in black. (B) Clustering heat map of the J gene sub-types in patients with ACLF and control subjects. ACLF, acute-on-chronic liver failure; IGHJ, immunoglobulin heavy joining.
Table IV.
Results of the t-test for the distribution ratio of the J gene.
| Type | Control | Case | P-value | T-value |
|---|---|---|---|---|
| IGHJ2 | 2.776667 | 4.56 | 0.009614 | 3.624858 |
| IGHJ4 | 45.35 | 39.975 | 0.124539 | −1.72416 |
| IGHJ3 | 6.173333 | 7.073333 | 0.137946 | 1.661918 |
| IGHJ1 | 1.206667 | 1.495 | 0.270726 | 1.197279 |
| IGHJ6 | 31.35833 | 34.225 | 0.337043 | 1.008973 |
| IGHJ5 | 13.13167 | 12.66667 | 0.79099 | −0.27929 |
IGHJ, immunoglobulin heavy joining.
Subsequently, a bubble chart for the expression of the combinations of each V and J gene segment was established (Fig. 6), and different colors were used for different genes. The differential composition of the V-J combinations in the patients with ACLF (X1A, TX-1A, TY-1A, TA-1A, TB-1A and TC-1A) were then compared with those in the control subjects (K1A, K2A, K4A, K6A, K7A and W1A). Subsequently, a t-test for the distribution ratio of the combinations of the six patients with ACLF and six control subjects was performed, and the negative P-values were plotted (Fig. 6; Table V). As observed in the alterations in gene expression, the upregulated genes are denoted in blue, downregulated genes are denoted in red, and non-significant change combinations are denoted in green (Fig. 6). Furthermore, four significantly upregulated combination genes were found, including (IGHV3-30, IGHJ2),(IGHV3-30, IGHJ3), (IGHV3-30,IGHJ6) and (IGHV4-59,IGHJ2) (P<0.05), whereas 14 combination genes were identified to be significantly downregulated, including (IGHV3-49,IGHJ5), (IGHV3-15,IGHJ5), (IGHV3-11,IGHJ5), (IGHV3-20,IGHJ6), (IGHV3-74,IGHJ5), (IGHV6-1,IGHJ4), (IGHV3-74,IGHJ4), (IGHV2-5,IGHJ5), (IGHV3-72,IGHJ4), (IGHV5-51,IGHJ4), (IGHV3-23,IGHJ4), (IGHV3-20,IGHJ5), (IGHV3-33,IGHJ5) and (IGHV2-70,IGHJ5) (P<0.05).
Figure 6.
Comparison of the combinations of IGHJ and IGHV genes between patients with ACLF and control subjects. Upregulated genes are denoted in blue, downregulated genes are denoted in red. If there was no significant change, green was used. Increased numbers of IGHJ and IGHV combinations were observed in the ACLF group, compared with the control group. ACLF, acute-on-chronic liver failure; IGHJ, immunoglobulin heavy joining; IGHV, immunoglobulin heavy variable.
Table V.
Results of the t-test for the distribution ratio of the V-J gene combinations.
| V-type | J-type | Control | Case | P-value | T-value |
|---|---|---|---|---|---|
| IGHV3-49 | IGHJ5 | 0.026666 | 0.006666 | 0.000815 | −5.070925 |
| IGHV3-15 | IGHJ5 | 0.0716666 | 0.025 | 0.005040 | −3.838859 |
| IGHV3-11 | IGHJ5 | 1.3766666 | 0.995 | 0.00639811 | −3.6823573 |
| IGHV6-1 | IGHJ4 | 0.446666 | 0.278333 | 0.012146 | −3.235252 |
| IGHV3-20 | IGHJ6 | 0.1933333 | 0.63333 | 0.0118824 | −3.0769355 |
| IGHV3-74 | IGHJ5 | 0.3833333 | 0.193333 | 0.0119918 | −3.062569 |
| IGHV3-74 | IGHJ4 | 1.0833333 | 0.6483333 | 0.0158769 | −2.9282990 |
| IGHV3-33 | IGHJ5 | 0.1466666 | 0.045 | 0.035139 | −2.747301 |
| IGHV3-20 | IGHJ5 | 0.0833333 | 0.03 | 0.034568 | −2.651974 |
| IGHV2-5 | IGHJ5 | 0.1183333 | 0.058333 | 0.026997 | −2.617230 |
| IGHV3-72 | IGHJ4 | 0.05 | 0.013333 | 0.028606 | −2.607250 |
| IGHV3-23 | IGHJ4 | 4.5783333 | 3.253333 | 0.033451 | −2.52706 |
| IGHV5-51 | IGHJ4 | 2.0316666 | 1.346666 | 0.032790 | −2.515948 |
| IGHV2-70 | IGHJ5 | 0.3716666 | 0.21 | 0.039841 | −2.362483 |
| IGHV3-30 | IGHJ3 | 0.5366666 | 0.743333 | 0.041738 | 2.340706 |
| IGHV3-30 | IGHJ2 | 0.1733333 | 0.483333 | 0.041722 | 2.637409 |
| IGHV3-30 | IGHJ6 | 2.78 | 4.225 | 0.017769 | 2.879356 |
| IGHV4-59 | IGHJ2 | 0.2283 | 0.52 | 0.014176 | 3.497063 |
IGHJ, immunoglobulin heavy joining; IGHV, immunoglobulin heavy variable.
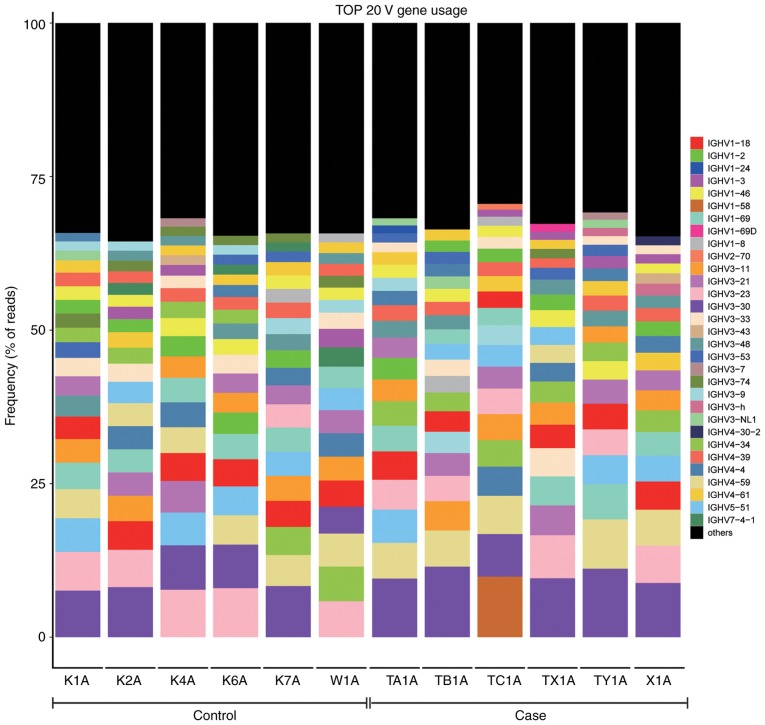
Comparison of the top 20 BCR heavy chain V region genes used between the ACLF and control groups
The top 20 frequently used genes of the BCR heavy chain V subtypes are listed in Fig. 7, in which the percentage of each sample V subtype in the top 20 frequently used genesis shown.
Figure 7.
Identification and characterization of the most frequently used BCR heavy chain V gene segments. The Y axis represents the top 20 most frequently used BCR heavy chain V gene subtypes, and the remaining V sub-types are denoted in black. BCR, B-cell receptor; IGHV, immunoglobulin heavy variable.
CDR3amino acid sequences commonly used in the ACLF and control groups
In the present study, three amino acid sequences commonly used in all patients with ACLF (n=6) and control subjects (n=6) were found, which were AR, FDY and MDV. The nine cloned numbers for patients with ACLF in the CDR3amino acid sequences are listed in Table VI, and the total number of CDR3 clones are shown in the last row. In addition, the three cloned numbers for control subjects in the CDR3amino acid sequences are shown in Table VII, and the total number of all CDR3 clones are shown in the last row.
Table VI.
Number of CDR3 amino acid sequences for the clones of patients with ACLF.
| CDR3 | TA1A | TB1A | TC1A | TX1A | TY1A | X1A | |
|---|---|---|---|---|---|---|---|
| AR | 2.08×10−5 | 6.54×10−6 | 5.17×10−7 | 1.04×10−4 | 9.01×10−6 | 1.02×10−5 | |
| DY | 3.97×10−6 | 6.96×10−6 | 2.72×10−6 | 2.28×10−7 | 4.12×10−6 | 6.84×10−6 | |
| FDY | 3.34×10−6 | 3.69×10−6 | 7.76×10−7 | 1.14×10−7 | 5.88×10−7 | 1.76×10−6 | |
| GMDV | 6.91×10−6 | 3.38×10−6 | 9.05×10−7 | 2.39×10−6 | 4.21×10−6 | 1.09×10−5 | |
| LDV | 7.95×10−8 | 3.16×10−7 | 9.05×10−7 | 1.03×10−6 | 4.90×10−7 | 3.11×10−7 | |
| MDV | 5.48×10−6 | 1.48×10−6 | 3.49×10−6 | 8.89×10−6 | 3.23×10−6 | 6.94×10−6 | |
| YGMDV | 1.11×10−6 | 3.06×10−6 | 1.81×10−6 | 1.25×10−6 | 2.06×10−6 | 2.69×10−6 | |
| YYGMDV | 1.19×10−6 | 7.38×10−7 | 7.76×10−7 | 1.03×10−6 | 1.27×10−6 | 3.11×10−7 | |
| YYYGMDV | 1.59×10−7 | 8.44×10−7 | 1.29×10−7 | 5.70×10−7 | 2.74×10−6 | 2.07×10−7 | |
| Total | 12,584,061 | 9,479,612 | 7,733,592 | 8,771,599 | 10,206,099 | 9,651,145 | |
CDR3, complementarity-determining region 3.
Table VII.
Number of CDR3 amino acid sequences for the clones of control subjects.
| CDR3 | K1A | K2A | K4A | K6A | K7A | W1A |
|---|---|---|---|---|---|---|
| AR | 2.08×10−5 | 1.12×10−5 | 9.15×10−6 | 4.55×10−6 | 6.79×10−6 | 2.84×10−7 |
| FDY | 3.34×10−6 | 3.02×10−6 | 6.51×10−6 | 6.82×10−6 | 1.44×10−6 | 1.13×10−6 |
| MDV | 5.48×10−6 | 4.90×10−6 | 1.23×10−6 | 2.27×10−6 | 1.02×10−6 | 1.38×10−5 |
| Total | 12,584,061 | 12,249,709 | 10,600,111 | 439,959 | 11,778,081 | 10,575,248 |
CDR3, complementarity-determining region 3.
Discussion
ACLF has been recognized as a distinct syndrome from AD, and the development of ACLF has been found to occur under the setting of systematic inflammation (2). However, to date, the mechanism underlying the development and progression of ACLF remains to be fully elucidated (13). In particular, the role of the immune system remains unclear. In the present study, NGS was used to characterize the polymorphisms of the BCR CDR3 gene segments in PBMCs prepared from patients with HBV-ACLF (n=6) and control subjects (n=6). The main novel findings were follows: i) Patients with ACLF exhibited a significantly higher extent of clonal expansion for B-cells, when compared with control subjects; ii) patients with ACLF presented with a skewed usage of BCR CDR3 V, D, J and V-J gene segments, whereas control subjects had a close-to-normal distribution; iii) the BCR heavy chain CDR3 of the V, D, J and V-J combinations of subfamily genes were abnormally expressed in response to ACLF; and iv) there was a significant association between the BCR CDR3 repertoire and ACLF.
It has been reported that a large proportion of B-cell clones in the BCR sequence are at lower frequencies, and these clones indicate its inability for clonal expansion (14). In the present study, certain HECs were identified in each individual BCR heavy chain CDR3, and these clones were able to form in a complex environment with antigen or pathogen stimulation. Furthermore, clonal expansion was observed in patients with ACLF and control subjects, and a higher extent of clonal expansion was present in the ACLF group, when compared with the control group.
Extensive studies have shown that the V, D and J gene segments of BCRs are rearranged in the development of B-lymphocytes, during which different numbers of nucleotides can be randomly inserted into or deleted from the V-J gene segments or V-D-J gene segments. Therefore, a variable CDR3 region is formed with diversity in its length and sequence, in which the CDR3 sequence is able to determine a unique BCR clone type, allowing for the assessment of the clonal diversity of B-lymphocytes through the length of CDR3 (15,16). In the present study, the normal distribution of CDR3 lengths was compared between patients with ACLF and control subjects. The head of the distribution tended to be skewed in patients with ACLF, whereas the distribution was close to normal in control subjects; the difference between these two groups was not statistically significant. In the comparative analysis of Shannon entropy and HEC between patients with ACLF and control subjects, there were differences between these two groups with respect to number and proportion. However, the difference was not statistically significant, which was possibly due to the small sample size. A Student’s t-test was used to analyze the frequency usage of the BCR heavy chain V, D, J and V-J segments, and the following six upregulated genes were identified: IGHV3-30, IGHJ2, (IGHV3-30, IGHJ2), (IGHV3-30, IGHJ3), (IGHV3-30, IGHJ6) and (IGHV4-59, IGHJ2). Furthermore, 19 downregulated genes were identified: IGHV3-74, IGHV2-70, IGHD4/OR15-4b, IGHD4/OR15-4a, IGHD2-8, (IGHV3-49, IGHJ5), (IGHV3-15, IGHJ5), (IGHV3-11, IGHJ5), (IGHV3-20, IGHJ6), (IGHV3-74, IGHJ5), (IGHV6-1, IGHJ4), (IGHV3-74, IGHJ4), (IGHV2-5, IGHJ5), (IGHV3-72, IGHJ4), (IGHV5-51, IGHJ4), (IGHV3-23, IGHJ4), (IGHV3-20, IGHJ5), (IGHV3-33, IGHJ5) and (IGHV2-70, IGHJ5). The identified dysregulation of the BCR heavy chain CDR3 V, D, J and V-J combinations of the sub-family gene may be involved in the development and progression of ACLF. It was hypothesized that the upregulated genes IGHV3-30,IGHJ2, (IGHV3-30, IGHJ2), (IGHV3-30, IGHJ3), (IGHV3-30, IGHJ6) and (IGHV4-59, IGHJ2) are involved in certain BCR-specific clonal proliferations of B-lymphocytes and undermine BCR diversity. This has been shown to be essential in maintaining the normal immune system in control subjects. The more subtypes of immune proteins present, the more effective the immune system against various pathogens. By contrast, the lower the number of subtypes an immune protein has, the more the individual becomes susceptible to diseases. In the present study, the downregulated genes IGHV3-74, IGHV2-70, IGHD4/OR15-4b, IGHD4/OR15-4a, IGHD2-8, (IGHV3-49, IGHJ5), (IGHV3-15, IGHJ5), (IGHV3-11, IGHJ5), (IGHV3-20, IGHJ6), (IGHV3-74, IGHJ5), (IGHV6-1, IGHJ4), (IGHV3-74, IGHJ4), (IGHV2-5, IGHJ5), (IGHV3-72, IGHJ4), (IGHV5-51, IGHJ4), (IGHV3-23, IGHJ4), (IGHV3-20, IGHJ5), (IGHV3-33, IGHJ5) and (IGHV2-70, IGHJ5) may be associated with inhibition of the clonal proliferation of certain specific B lymphocytes of BCRs. It is noteworthy that a number of the above subtypes were significantly reduced in the patients with ACLF, compared with the control subjects, indicating that B-lymphocytes were significantly reduced inpatients with ACLF.
In conclusion, an assay to determine the entire diversity of the B-cell repertoire at a sequence-level resolution in patients with HBV-ACLF was successfully developed. The findings suggested that the development of HBV-ACLF is linked with the BCR heavy chain CDR3 repertoire. Future insightful investigations are required to better understand the role of the BCR repertoire in immune responses, autoimmunity and alloreactivity, and in driving the development and progression of HBV-ACLF.
Acknowledgments
My deepest gratitude goes first and foremost to Professor Weiguo Sui, my supervisor, for his constant encouragement and guidance. He has walked me through all the stages of the writing of this thesis. Without his consistent and illuminating instruction, this thesis could not have reached its present form. Second, I would like to express my heartfelt gratitude to Professor Yong Dai, who instructed and helped me a lot in the past four years. Last my thanks would go to my beloved family for their loving considerations and great confidence in me all through these years. I also owe my sincere gratitude to my friends who gave me their help and time in listening to me and helping me work out my problems during the difficult course of the thesis.
Funding
This study was supported by the Technology Plan of Shenzhen, Guangdong (grant no. JCYJ20160422150329190), the China Postdoctoral Science Foundation (grant no. 2017M610575) and the Guilin Scientific Research and Technology Development Program (grant no. 20170117-1).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
WS and YD conceived and designed the study. QY conducted the experiments. LL, SL and HC participated in the completion of the experiments. QY and LW analyzed the data. LW wrote the manuscript. WS and JZ revised the manuscript. All the authors read and approved the final paper.
Ethics approval and consent to participate
Written informed consent was obtained from each participant or from their guardians. The present study complies with the Declaration of Helsinki for investigations involving humans, and the study protocol was carefully reviewed and approved by the Ethics Committee of the No. 181 Hospital of Guilin (Guilin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jalan R, Gines P, Olson JC, Mookerjee RP, Moreau R, Garcia-Tsao G, Arroyo V, Kamath PS. Acute-on chronic liver failure. J Hepatol. 2012;57:1336–1348. doi: 10.1016/j.jhep.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 2.Moreau R, Jalan R, Gines P, Pavesi M, Angeli P, Cordoba J, Durand F, Gustot T, Saliba F, Domenicali M, et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144:1426–1437. doi: 10.1053/j.gastro.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 3.Sarin SK, Kumar A, Almeida JA, Chawla YK, Fan ST, Garg H, de Silva HJ, Hamid SS, Jalan R, Komolmit P, et al. Acute-on-chronic liver failure: Consensus recommendations of the Asian Pacific Association for the study of the liver (APASL) Hepatol Int. 2009;3:269–282. doi: 10.1007/s12072-008-9106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson JC, Kamath PS. Acute-on-chronic liver failure: Concept, natural history, and prognosis. Curr Opin Crit Care. 2011;17:165–169. doi: 10.1097/MCC.0b013e328344b42d. [DOI] [PubMed] [Google Scholar]
- 5.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder HW, Jr, Cavacini L. Structure and function of immunoglobulins. The J Allergy Clin Immunol. 2010;125:S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 8.Nouri-Aria KT, Alexander GJ, Portmann BC, Hegarty JE, Eddleston AL, Williams R. T and B cell function in alcoholic liver disease. J Hepatol. 1986;2:195–207. doi: 10.1016/S0168-8278(86)80078-2. [DOI] [PubMed] [Google Scholar]
- 9.Sarin SK, Kedarisetty CK, Abbas Z, Amarapurkar D, Bihari C, Chan AC, Chawla YK, Dokmeci AK, Garg H, Ghazinyan H, et al. Acute-on-chronic liver failure: consensus recommendations of the Asian Pacific Association for the Study of the Liver (APASL) 2014. Hepatol Int. 2014;8:453–471. doi: 10.1007/s12072-014-9580-2. [DOI] [PubMed] [Google Scholar]
- 10.Londoño JC, Philipp SE. A reliable method for quantification of splice variants using RT-qPCR. BMC Mol Biol. 2016;17:8. doi: 10.1186/s12867-016-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Keylok CJ. Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos. 2005;109:203–207. doi: 10.1111/j.0030-1299.2005.13735.x. [DOI] [Google Scholar]
- 13.Wang FS, Zhang Z. Host immunity influences disease progression and antiviral efficacy in humans infected with hepatitis B virus. Expert Rev Gastroenterol Hepatol. 2009;3:499–512. doi: 10.1586/egh.09.50. [DOI] [PubMed] [Google Scholar]
- 14.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honjo T. Immunoglobulin Genes. Annu Rev Immunol. 1983;1:499–528. doi: 10.1146/annurev.iy.01.040183.002435. [DOI] [PubMed] [Google Scholar]
- 16.Foreman AL, Van de Water J, Gougeon ML, Gershwin ME. B cells in autoimmune diseases: Insights from analyses of immunoglobulin variable (Ig V) gene usage. Autoimmun Rev. 2007;6:387–401. doi: 10.1016/j.autrev.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.