Abstract
Gemcitabine and pemetrexed are clinically approved antimetabolites for the therapy of mesothelioma diseases. These drugs are often applied in combination with platinum complexes and other drugs. The activity of antimetabolites depended on the expression levels of certain non-coding RNAs, in particular, of small microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). The development of tumor resistance towards antimetabolites was regulated by non-coding RNAs. An overview of the interplay between gemcitabine/pemetrexed antimetabolites and non-coding RNAs in mesothelioma is provided. Further to this, various non-coding RNA-modulating agents are discussed which displayed positive effects on gemcitabine or pemetrexed treatment of mesothelioma diseases. A detailed knowledge of the connections of non-coding RNAs with antimetabolites will be constructive for the design of improved therapies in future.
Keywords: MicroRNA, Long non-coding RNA, Mesothelioma, Gemcitabine, Pemetrexed, Anticancer drugs
Abbreviations: AKBA, 3-acetyl-11-keto-β-boswellic acid; Bcl-2, B-cell lymphoma 2; DADS, diallyl sulfide; DHA, docosahexaenoic acid; DIM, 3,3‘-diindolylmethane; DMPM, diffuse malignant peritoneal mesothelioma; EGCG, epigallocatechin-3-gallate; EMT, epithelial-mesenchymal transition; HOTAIR, HOX transcript antisense RNA; RA, retinoic acid; I3C, indole-3-carbinol; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MPM, malignant pleural mesothelioma; NaB, sodium butyrate; NSCLC, non-small cell lung cancer; PEG, polyethylene glycole; PEITC, phenethylisothiocyanate; PDCD4, programmed cell death 4; PTEN, phosphatase and tensin homolog; SAHA, suberoylanilide hydroxamic acid; SFN, sulforaphane; TSA, trichostatin A
1. Introduction
Mesothelioma features an aggressive tumor disease with increasing incidence associated with high mortality rates (median survival of ca. 1 year after diagnosis), poor prognosis and ca. 40.000 deaths per year worldwide [1]. Asbestos was identified as the most important carcinogen for mesothelioma development, and in combination with other factors (e.g., SV40 virus infection, genetic disposition such as BAP1 mutation or inactivation) asbestos promoted inflammation processes, enhanced EGFR, VEGFR, Akt, and Notch signaling as well as 5-LOX expression [2,3]. Asbestos-mediated cell necrosis released HMGB1 leading to an inflammation response in macrophages and mesothelial cells and to transformation of mesothelial cells [3]. Altered protein expression is often accompanied by altered levels of certain non-coding RNAs. Mesothelioma cell expression of miRNAs often differed from the expression profiles of non-malignant samples while circulating miRNAs were likewise determined in mesothelioma patients [3,4]. Various miRNAs can serve as prognostic factors, as potential therapeutic targets and as therapeutic agents [4]. Several miRNAs were shown to regulate oncogenes, apoptosis and/or vital signaling pathways [5]. In particular, drug-resistant cancer stem-like cells are strongly regulated by non-coding RNAs [6]. High expression of miR-21-5p, miR-221-3p, and the miR-17-92 cluster (miR-17-5p, miR-20a-5p) associated with drug resistance was observed from short survivors among malignant pleural mesothelioma (MPM, i.e., the most abundant mesothelioma disease) patients when compared with long survivors [7]. MiRNAs have also emerged as anticancer drugs and minicells called TargomiR, which are loaded with miRNA-16 mimic miRNA, displayed an acceptable safety profile and moderate clinical response in a clinical phase 1 trial with MPM patients which can be optimized in combination with other anticancer drugs [8]. Another class of non-coding RNAs dubbed long non-coding RNAs (lncRNAs) is ample in the genome, and lncRNAs have become of distinct importance for the understanding of cancer diseases as well [9]. Many lncRNAs are involved in vital cellular processes such as chromatin remodeling, gene regulation, inhibition of smaller miRNA molecules, and in the regulation of Wnt signaling [[9], [10], [11]].
Indeed, there are only very few treatment options for mesothelioma patients at the moment. The problematic location of the primary tumors near vital organs often render surgery and radiation therapy options while systemic platinum chemotherapy in combination with pemetrexed is given in most cases as first-line therapy of mesothelioma [[12], [13], [14]]. The antimetabolites gemcitabine and pemetrexed were thoroughly investigated in mesothelioma patients and feature valuable tools for MPM chemotherapy [15]. The combination of platinum complexes and pemetrexed with the VEGFR-inhibitor bevacizumab was recently identified as a superior first-line therapy of MPM when compared with the currently mostly applied platinum plus pemetrexed treatment [16,17]. This review provides an overview of the interactions of mesothelioma-relevant non-coding RNAs with the approved antimetabolites gemcitabine and pemetrexed. Various agents with known effects on non-coding RNA activity and expression were discussed which exhibited beneficial effects on gemcitabine or pemetrexed activity against mesothelioma diseases. This work thematically follows a review recently published in this journal about the role of non-coding RNAs concerning the efficacy of approved platinum-based drugs in mesothelioma [18].
2. Antimetabolites and their interactions with non-coding RNAs in mesothelioma
2.1. Gemcitabine and pemetrexed
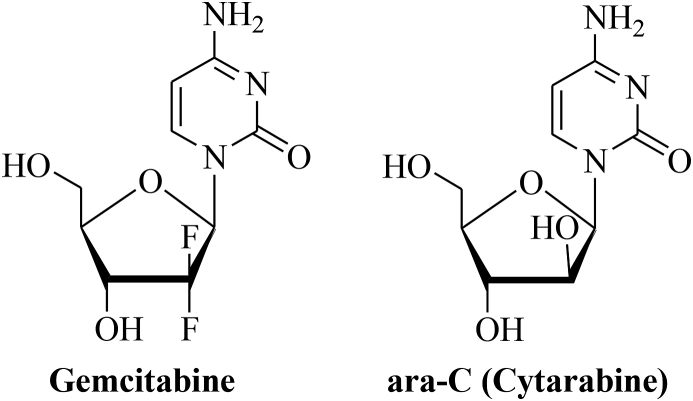
Gemcitabine (Gemzar®) is an antimetabolite applied against a variety of solid tumors including cancers of the lung and the pancreas [19,20]. It is a deoxycytidine analog (2‘-deoxy-2‘,2‘-difluorocytidine-monohydrochloride) similar to the drug ara-C (cytosine arabinoside, cytarabine) (Fig. 1) [21]. But in comparison to ara-C, the optimized drug gemcitabine exhibited increased membrane permeability, extended inhibition of DNA synthesis, and higher affinity to deoxycytidine kinase [21]. Upon activating phosphorylation in the cell, gemcitabine triphosphate featuring a competitive analog of deoxycytidine triphosphate is incorporated in the deoxycytidine sites of the DNA and the addition of another nucleoside to the gemcitabine DNA strand masks the incorporated gemcitabine from DNA repair [20,21]. In this way DNA chain elongation is inhibited and apoptosis is induced. Due to its mild toxicity profile gemcitabine is a suitable candidate for combination therapies and the combination of gemcitabine with DNA-damaging drugs such as alkylating agents, platinum complexes and radiotherapy appeared promising [20,21]. Gemcitabine was approved by the FDA in 1996 for the treatment of advanced pancreatic cancer [22]. Side effects of high-dose gemcitabine (1000 mg/m2) include neutropenia, proteinuria, increased hepatic transaminase levels, nausea, vomiting, and mild skin rash [23]. Resistance factors of gemcitabine treatment include transporter deficiency (reduced hENT1 expression), overexpression of ribonucleotide reductases and of gemcitabine-deactivating cytidine deaminases [22]. In the latter case, 4-(N)-protection of gemcitabine with PEG, squalene or valproic acid as well as with linear carboxylic acids (valeroyl, heptanoyl, lauroyl, stearoyl) were successfully designed in order to overcome deamination [22]. The combination of gemcitabine with cisplatin was widely applied for the treatment of MPM before the combination of platinum complexes with anti-folates emerged as an improved first-line therapy option for MPM patients [24]. Currently, gemcitabine is casually applied as a second-line therapy of MPM [24]. A recent study of gemcitabine plus cisplatin/carboplatin in MPM patients underlined the efficacy and safety of this type of combination therapy for MPM patients [24]. In addition, gemcitabine plus the Vinca alkaloid vinorelbine was tested in a female MPM patient who was treated with pemetrexed before and because this patient showed partial response and tumor regression the gemcitabine plus vinorelbine combination therapy may represent a suitable salvage therapy for patients who are pre-treated with pemetrexed and suffer from a relapse [25].
Fig. 1.
Structures of gemcitabine and ara-C (cytarabine).
Pemetrexed (Alimta®) is a folic acid analog comprising a pyrrolo[2,3-d]pyrimidine scaffold and acts as a folate antimetabolite against cancer (Fig. 2) [26]. It is a potent inhibitor of pyrimidine and purine synthesis and inhibits vital enzymes such as glycinamide ribonucleotide formyltransferase (GARFT), aminoimidazolecarboxamide ribonucleotide formyltransferase (AICARFT) and thymidylate synthase (TS) [[26], [27], [28]]. Accumulation of toxic nucleotides as well as AMPK activation and mTOR inhibition contribute to the cytotoxic activity of pemetrexed [29]. Resistance mechanisms of pemetrexed include overexpression of TS, suppression of the folate transporter SLC19A1 (solute carrier family 19 member 1) and Akt activation [29,30]. Pemetrexed is clinically approved for the therapy of NSCLC and mesothelioma either as single agent or in combination with platinum complexes (cisplatin, carboplatin) [31,32]. It was superior to gemcitabine and showed promising results from various clinical studies with mesothelioma patients [[33], [34], [35]]. The combination of the anti-VEGF antibody bevacizumab with cisplatin plus pemetrexed showed improved results and the bevacizumab-cisplatin-pemetrexed therapy has become the standard therapy for MPM patients in France [36]. As already mentioned above, vinorelbine can be applied for mesothelioma patients pre-treated with pemetrexed [25].
Fig. 2.
Structure of pemetrexed.
2.2. Gemcitabine, non-coding RNAs and mesothelioma
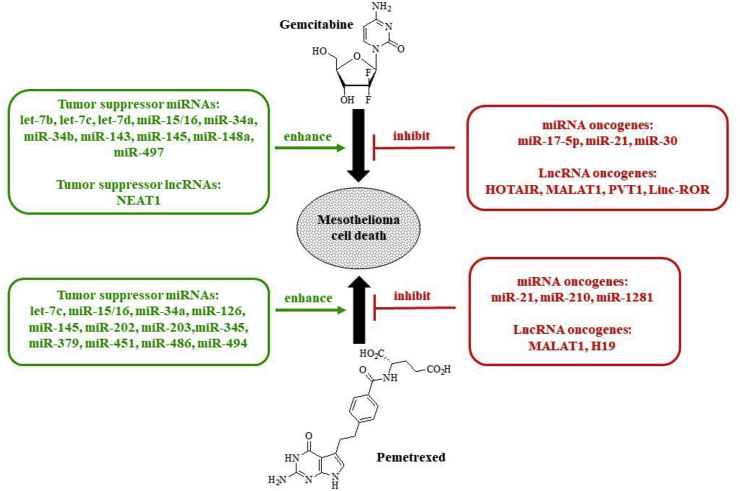
Several miRNAs were identified which regulate cancer cell sensitivity to gemcitabine treatment (Fig. 3). Most of these gemcitabine-related results were obtained from pancreas and lung cancer samples, however, these identified miRNAs also play a crucial role for mesothelioma.
Fig. 3.
Gemcitabine, pemetrexed and non-coding RNAs in mesothelioma.
Let-7 miRNAs feature prominent examples of tumor suppressor microRNAs and let-7b, let-7c, and let-7d, for instance, were recently identified as tumor suppressors in mesothelioma [[37], [38], [39]]. Let-7d is overexpressed in MPM and may play a role concerning enhanced antimetabolite action [39]. Let-7c inhibited migration and invasion through ITGB3 and MAP4K3 targeting in NSCLC [40]. In pancreatic cancer, let-7 (let-7b/c/d) upregulation increased the activity of gemcitabine distinctly and, thus, it is conceivable that let-7 expression has beneficial effects on gemcitabine action in mesothelioma as well [41]. The tumor suppressor miR-16 was downregulated in samples from MPM patients who experienced extrapleural pneumonectomy [42]. Restoration of miR-16 expression by synthetic mimics suppressed anti-apoptotic Bcl-2 and showed in vivo anticancer activity in MPM xenografts [43] CCND1 featured another target of miR-16 and restored miR-16 also sensitized MPM cells to gemcitabine treatment [43].
The downregulation of tumor suppressing miR-34s turned non-malignant mesothelial cells into oncogenic cells [44]. In particular, miR-34a suppression was correlated with bad chemotherapeutic response of diffuse malignant peritoneal mesothelioma (DMPM), and c-MET and AXL were identified as targets of miR-34a in DMPM [45]. It was shown that miR-34a expression sensitized breast cancer cells to gemcitabine in a Linc-ROR-dependent way [46,47]. Tumorigenesis in MPM was correlated with suppression of the tumor suppressor miR-34b/c activity by methylation [48]. Vice versa, enhanced miR-34b/c expression induced antiproliferative effects, G1 phase cell cycle arrest and low motility of MPM cells [48]. In human osteosarcoma cells, increased miR-34b expression caused by treatment with the mTOR-inhibitor sirolimus led to increased gemcitabine activity [49].
The tumor suppressor miR-143 is distinctly suppressed in MPM patients as to analyses of resected and biopsy samples [50]. Recently, miR-143 increased the sensitivity of gemcitabine in bladder cancer cells via suppression of IGF-1R [51]. MiR-145 features another tumor suppressor, which is downregulated in MPM and exerts its activity by suppression of OCT4 and ZEB1 [52]. Indeed, the expression of miR-145 sensitized pancreatic adenocarcinoma cells to gemcitabine [53]. In contrast to that, the tumor suppressor miR-148a is highly expressed in mesothelioma and gemcitabine sensitizing effects of miR-148a were identified in pancreatic cancer models [54,55].
MiR-497 was suppressed in MPM cells and the miR-497 tumor suppressor enhanced gemcitabine activity in pancreatic cancer by downregulation of FGF2 and FGFR1 [56,57].
There are also oncogenic miRNAs commonly described as oncomirs that regulate gemcitabine activity aside the tumor suppressor miRNAs mentioned above. The expression of the oncomir miR-17-5p was high in short survivors of MPM [7]. Further to this, the suppression of miR-17-5p restored gemcitabine activity in pancreatic cancer cells by induction of Bim and, thus, miR-17-5p may play a role concerning gemcitabine activity against mesothelioma as well [58]. The oncomir miR-21 represents a well-documented oncogene in various cancers and so it was likewise overexpressed in MPM and suppressed PDCD4 (programmed cell death 4) in MPM [59]. MiR-21 expression led to gemcitabine resistance in breast and pancreatic cancer by upregulation of Akt signaling and suppression of PTEN [60,61]. Interestingly, treatment of pancreatic cancer cells with indole-3-carbinol (I3C) suppressed miR-21 expression via PDCD4 upregulation and overcame gemcitabine resistance in the end [62].
A list of miRNAs involved in gemcitabine resistance and sensitivity with connections to mesothelioma diseases is given in Table 1.
Table 1.
MicroRNA tumor suppressors and oncomirs proven or strongly assumed to be correlated with gemcitabine activity in mesothelioma.
| miRNA | Target(s) | Function | Expressiona |
|---|---|---|---|
| let-7b | Akt, Caspase 3, Twist, β-catenin | tumor suppressor | – |
| let-7c | ITGB3, MAP4K3 | tumor suppressor | lower in short survivors |
| let-7d | – | tumor suppressor | higher |
| miR-15/16 | Bcl-2, CCND1 | tumor suppressor | lower |
| miR-17-5p | Bim | oncomir | higher in short survivors |
| miR-21 | PDCD4, PTEN | oncomir | higher |
| miR-30 | FN1, vimentin, N-cadherin | oncomir | – |
| miR-34a | c-MET/Akt | tumor suppressor | lower |
| miR-34b | Bcl-2 | tumor suppressor | lower |
| miR-143 | IGF-1R | tumor suppressor | lower |
| miR-145 | OCT4 | tumor suppressor | lower |
| miR-148a | – | tumor suppressor | higher |
| miR-497 | FGF2, FGFR1 | tumor suppressor | lower |
Expression in mesothelioma when compared with non-malignant/benign samples or other tumors.
Aside miRNAs, long-non-coding RNAs play a crucial role for the activity of gemcitabine in mesothelioma as well. Patients with high EF177379 (NEAT1) expression had prolonged overall survival times in case they haven't received induction chemotherapy [63]. NEAT1 expression sensitized cholangiocarcinoma to gemcitabine treatment in a BAP1-dependent way [64].
Aside NEAT1, there are further lncRNAs of importance for mesothelioma diseases. HOTAIR (HOX transcript antisense RNA) and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) feature prominent examples [65,66]. Sarcomatoid mesothelioma was accompanied by shorter survival and revealed upregulated HOTAIR and MALAT1 expression [66]. Interestingly, it was shown that gemcitabine treatment induced HOTAIR expression in pancreatic cancer stem cells leading to drug resistance [67]. Similarly, MALAT1 expression led to gemcitabine resistance and enforced the stem cell character of cancer cells by induction of SOX-2 [68]. The suppression of PVT1 increased drug sensitivity in MPM cells [69]. Downregulation of anti-apoptotic BCL2L1, BCL2, ICEBERG (Caspase 1 inhibitor), and BIRC8 (Baculoviral IAP repeat-containing protein 8) as well as induced expression of pro-apoptotic LTB (lymphotoxin beta), BCL2L14 (Bcl-2 like protein 14), FASLG (FS ligand) and TNFRSF1B (tumor necrosis factor receptor superfamily member 1B) were the reasons for the enhanced chemosensitivity [69]. Suppression of EZH2 and PVT1 by curcumin restored gemcitabine activity in pancreatic cancer cells [70]. The role of the lncRNA Linc-ROR has already been mentioned above [46]. A list of long non-coding RNAs that may be involved in cisplatin resistance and sensitivity of mesothelioma is given in Table 2.
Table 2.
Long non-coding RNA (lncRNA) tumor suppressors and oncogenes in mesothelioma proven or strongly assumed to be correlated with gemcitabine activity.
| lncRNA | Target(s) | Function |
|---|---|---|
| NEAT1 | BAP1 | tumor suppressor |
| HOTAIR | – | oncogene |
| MALAT1 | SOX-2 | oncogene |
| PVT1 | LTB, BLC2L14, FASLG, TNFRSF1B, BCL2L1, BCL2, ICEBERG, BIRC8, EZH2 | oncogene |
| Linc-ROR | – | oncogene |
2.3. Pemetrexed, non-coding RNAs and mesothelioma
Pemetrexed owns a salient position in the therapy of mesothelioma and, thus, the identification of the influence of non-coding RNAs on pemetrexed activity is of great importance (Fig. 3). Pemetrexed treatment induced let-7c expression in MPM which is of certain importance since let-7c was identified as a tumor suppressor in MPM [71,72]. In NSCLC, migration and invasion was suppressed by let-7c via regulation of its targets ITGB3 and MAP4K3 [73]. Similar to gemcitabine, restoration of tumor suppressor miR-15/16 and treatment of MPM cells with miR-16 mimic resulted in increased activity of pemetrexed [43]. In comparison with non-neoplastic pleura, the tumor suppressors miR-126 and miR-145 were suppressed in biopsies and resected MPM tumors [71]. Interestingly, MPM samples after pemetrexed plus cisplatin therapy exhibited reduced suppression profiles of miR-126 and miR-145 when compared with the untreated MPM samples which can be a reason for the relatively high efficacy of the cisplatin/pemetrexed combination therapy in MPM patients [71]. Indeed, upregulation of miR-145 enhanced the activity of pemetrexed against MPM cells [52]. Further to this, when compared with chemotherapy-naïve MPM biopsies the expression of the tumor suppressors miR-451 and miR-486-5p was significantly induced by pemetrexed/cisplatin combination therapy [71]. The expression of the tumor suppressor miR-34a was downregulated in diffuse malignant peritoneal mesothelioma (DMPM) [45]. It was shown that miR-34a also regulates a key factor of folate biosynthesis (methylene tetrahydrofolate reductase) and may function as a prognostic marker for the outcome of pemetrexed-based anticancer therapy [74,75]. MiR-203 features another tumor suppressor with relevance concerning MPM and it was downregulated in MPM [76]. In NSCLC samples from pemetrexed/cisplatin-treated patients a high miR-203 expression was correlated with drug sensitivity [77]. MiR-379, a regulator of IL-18, was also suppressed in pemetrexed-resistant MPM cells [78]. In addition, miR-202 was suppressed in asbestos-related lung cancer [79]. Interestingly, pemetrexed treatment of lymphoblastoid cells also downregulated miR-202 expression accompanied by upregulation of the miR-202 target MTHFD2 [80]. It is assumed that the formation of MPM is related to mesothelial hyperplasia. The expression of miR-494 was reduced in mesothelial hyperplasia and linked with FDZ4 expression [81]. In lymphoblastoid cells, induction of SUFU was linked with suppression of miR-494 in pemetrexed-treated cells [81]. In the same cells, pemetrexed also downregulated miR-1281 which is a miRNA upregulated in MPM patients and asbestos-exposed people [80,82].
As already mentioned above, the role of oncomirs such as miR-21 for the establishment of various cancer diseases is well described. MiR-21 was overexpressed in MPM and regulated mesothelin and PDCD4 [59,[83], [84], [85]]. Suppression of miR-21 might sensitize MPM to pemetrexed treatment and combination of pemetrexed with miR-21 antisense oligonucleotides in cationic solid lipid nanoparticles appears promising [86]. The oncomir miR-210 was also downregulated by pemetrexed plus cisplatin therapy when compared with therapy-naïve samples [71].
A list of miRNAs involved in pemetrexed resistance and sensitivity with connections to mesothelioma diseases is provided in Table 3.
Table 3.
MicroRNA tumor suppressors and oncomirs in mesothelioma proven or assumed to be correlated with pemetrexed activity.
| miRNA | Target(s) | Function | Expressiona |
|---|---|---|---|
| let-7c | ITGB3, MAP4K3 | tumor suppressor | lower in short survivors |
| miR-15/16 | Bcl-2, CCND1 | tumor suppressor | lower |
| miR-21 | PDCD4, PTEN, mesothelin | oncomir | higher |
| miR-34a | c-MET/Akt | tumor suppressor | lower |
| miR-126 | – | tumor suppressor | lower |
| miR-145 | OCT4 | tumor suppressor | lower |
| miR-202 | MTHFD2 | tumor suppressor | – |
| miR-203 | DKK1 | tumor suppressor | lower |
| miR-210 | NUPR1, HTRA1, RGS10 | oncomir | higher in short survivors |
| miR-345 | MRP1 | tumor suppressor | higher |
| miR-379 | IL-18 | tumor suppressor | lower |
| miR-451 | PSMB8, MIF, ERCC1 | tumor suppressor | lower |
| miR-486 | ARHGAP5 | tumor suppressor | lower in short survivors |
| miR-494 | FZD4, SUFU | tumor suppressor | lower |
| miR-1281 | – | oncomir | higher |
Expression in mesothelioma when compared with non-malignant/benign samples or other tumors.
Concerning the effects of mesothelioma-correlated long non-coding RNAs on pemetrexed efficacy, a high expression of H19 was accompanied by poor response of NSCLC patients to pemetrexed-cisplatin combination therapy [87]. Similarly, MALAT1 expression was found to be inversely correlated with chemotherapy response including pemetrexed-based therapies [88].
2.4. Suitable non-coding RNA modulating drugs for the combination with gemcitabine or pemetrexed
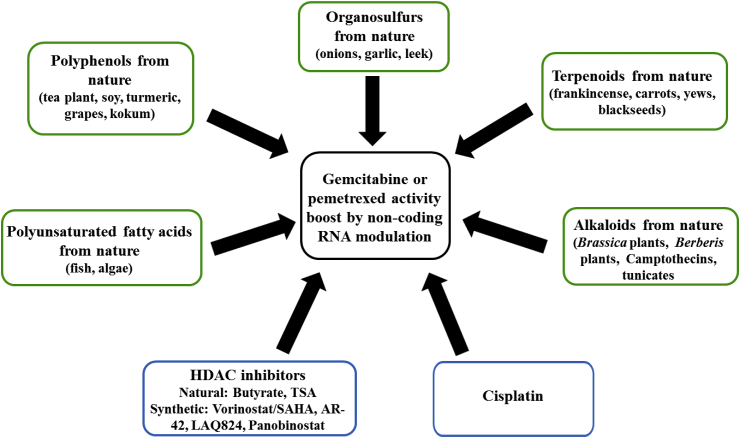
The combination of gemcitabine or pemetrexed with other drugs, which can modify non-coding RNAs strongly associated with gemcitabine or pemetrexed efficacy in mesothelioma diseases, is of particular interest in order to achieve better therapy responses. Suitable combination drugs are either intended to suppress the expression of oncomirs associated with gemcitabine or pemetrexed resistance or to upregulate tumor suppressing non-coding RNAs in order to enhance the activity of gemcitabine and pemetrexed. It became clear that a good deal of possible combination drugs represent natural products or modified natural products. Prominent compound classes are featured by (poly-)phenols, terpenoids, alkaloids, fatty acids and HDAC inhibitors (Fig. 4). Another example features the DNA-interacting drug cisplatin whose manifold non-coding RNA-modulating effects in mesothelioma were presented in a previous review [18]. In short, cisplatin treatment has casually led to the suppression of miR-21 and to the induction of let-7c, miR-34a, miR-145 and miR-451 [18]. Hence, cisplatin can possibly sensitize mesothelioma cells to treatments with gemcitabine or pemetrexed and it is widely applied as a first-line treatment for MPM patients in combination with pemetrexed [18].
Fig. 4.
Non-coding RNA modulating drugs with relevance to gemcitabine or pemetrexed activity.
2.4.1. Phenolic compounds
It has recently been shown that (poly-)phenols can influence the response of various cancers by regulation of non-coding RNA expression. A prominent example is represented by the main catechin constituent of green tea plants (Camellia sinensis), epigallocatechin-3-gallate (EGCG), which is of interest for a wide audience because of the increasing popularity of green tea in the western world as a health-promoting life-style beverage [89]. Adverse effects of EGCG comprise certain liver toxic effects observed from some individuals taking high doses of EGCG or green tea extract as well as inactivating reactions with boronic acid drugs such as bortezomib [90,91].
Concerning a modulation of miRNAs with relevance to gemcitabine and/or pemetrexed activity, EGCG increased the expression of the tumor suppressors let-7b, let-7c, miR-15/16, miR-34a, miR-34b, miR-126 and miR-494 while the oncomirs miR-21, miR-30 and miR-210 were suppressed by EGCG in various cancer cells [[92], [93], [94], [95], [96], [97], [98], [99], [100]]. Concerning lncRNAs, EGCG upregulated the expression of the tumor suppressor NEAT1 by induction of CTR1 in lung cancer cells [101].
Soy (Glycine max) is an integral component of the daily diet of millions of people all over the world and its isoflavone component genistein was identified as a suppressor of the oncomir miR-21. Further flavanoids such as glyceollins, 3,6-dihydroxyflavone (3,6-DHF), and silibinin suppressed miR-21 as well, while genistein, glyceollins, 3,6-DHF, and quercetin were able to induce the expression of the important tumor suppressor miR-34a [[102], [103], [104], [105], [106], [107], [108]]. In addition, miR-16 was upregulated by quercetin and miR-148a by glabridin [[109], [110], [111], [112]]. The well-studied oncogenic lncRNA HOTAIR was distinctly suppressed by genistein [113].
Various bioactive components have been identified from popular Indian spices and one of the most prominent example, curcumin (diferuloylmethane), which is a natural polyphenol from the rhizome of the Indian spice turmeric (Curcuma longa), upregulated miR-15a, miR-16-1, miR-34a, miR-145, and miR-203, and suppressed oncogenic miR-17-5p and miR-21 [[114], [115], [116], [117], [118], [119]]. Curcumin also regulated various lncRNAs. HOTAIR-dependent metastasis formation was blocked and the expression of oncogenic H19 and PVT1 was suppressed by curcumin [70,120,121]. In addition to curcumin, several new (semi-)synthetic curcuminoids such as CDF and EF24 have emerged as more potent anticancer compounds when compared with curcumin, and both CDF and EF24 downregulated miR-21 while CDF additionally suppressed miR-210 and induced miR-143 expression [[122], [123], [124], [125]].
Various berries and grapes produce the anticancer active stilbene derivative resveratrol which was able to upregulate miR-34a while miR-21 was suppressed by resveratrol [[126], [127], [128], [129], [130]]. In addition, the expression of miR-143 was upregulated by the close resveratrol analog pterostilbene [130]. Concerning lncRNAs, resveratrol upregulated NEAT1 and suppressed MALAT1 [131,132]. Natural bisphenols such as honokiol exhibited distinct anticancer properties and honokiol upregulated miR-34a and miR-143 [[133], [134], [135]]. Emodin features a natural trihydroxy-anthraquinone with anticancer activity and emodin downregulated miR-210 while it upregulated miR-34a, miR-126 and miR-429 expression [[136], [137], [138], [139]]. The polycyclic polyprenylated acylphloroglucinol (PPAP) garcinol features another interesting natural product, which was isolated from plums of the kokum tree (Garcinia indica), and downregulated miR-21 while the expression of tumor suppressing miR-494 was upregulated by garcinol [140,141]. Tumor suppressors were induced by other (poly-)phenols as well. MiR-148a was upregulated by caffeic acid, miR-15b by mangiferin (from mango) and miR-34a by pomegranate extract rich in polyphenolic components [[142], [143], [144]].
Last but not least, the approved phenolic anticancer drug etoposide, a topoisomerase II inhibitor derived from the natural product podophyllotoxin, displayed an induction of the tumor suppressor miR-34a in a p53-dependent way in cancer cells treated with etoposide [[145], [146], [147]]. Thus, a combination of gemcitabine or pemetrexed with etoposide appears promising for the treatment of mesothelioma diseases (see below).
A list of polyphenolic drugs and their effects on non-coding RNAs is given in Table 4.
Table 4.
Polyphenolic drugs with effects on non-coding RNA tumor suppressors (inducing effects) and oncogenes (suppressing effects) including oncomirs and oncogenic lncRNAs in mesothelioma correlated with gemcitabine or pemetrexed activity.
| Drugs | Tumor suppressors | Oncogenes |
|---|---|---|
| EGCG | let-7b, let-7c, miR-15/16, miR-34a, miR-34b, miR-126, miR-494, NEAT1 | miR-21, miR-30, miR-210 |
| Genistein | miR-34a | miR-21, HOTAIR |
| Glyceollins | miR-34a | miR-21 |
| 3,6-DHF | miR-34a | miR-21 |
| Quercetin | miR-16, miR-34a | – |
| Silibinin | – | miR-21 |
| Glabridin | miR-148a | – |
| Curcumin | miR-15a, miR-16-1, miR-34a, miR-145, miR-203 | miR-17-5p, miR-21, HOTAIR, H19, PVT1 |
| CDF | miR-143 | miR-21, miR-210 |
| Resveratrol | miR-34a, NEAT1 | miR-21, MALAT1 |
| Pterostilbene | miR-143 | – |
| Emodin | miR-34a, miR-126, miR-429 | miR-210 |
| Honokiol | miR-34a, miR-143 | – |
| Garcinol | miR-494 | miR-21 |
| Caffeic acid | miR-148a | – |
| Mangiferin | miR-15b | |
| Pomegranate extract | miR-34a | – |
| Etoposide | miR-34a | – |
2.4.2. Terpenoids
Terpenoids represent an important natural product class with distinct potential for the treatment of cancer diseases [148]. Paeoniflorin, a glucosylated monoterpene from Paeonia lactiflora, led to miR-16 upregulation in cancer cells [149]. The PEGylated monoterpene thymoquinone, a natural product isolated from Nigella sativa, induced the expression of the tumor suppressor miR-34a while the sesquiterpene lactone antrocin isolated from fungi upregulated let-7c expression [150,151]. Another sesquiterpene lactone, parthenolide from feverfew (Tanacetum parthenium) induced the expression of miR-15a and miR-16 [152].
The well-investigated diterpene retinoic acid (RA, vitamin A) caused expression upregulation of the tumor suppressors let-7c miR-15/16, and miR-223 [153,154]. In addition, 1,25-dihydroxyvitamin D3 (1,25-D) upregulated miR-15a [155]. The vitamin E analog Δ-tocotrienol induced the expression of miR-34a [156]. Another vitamin E analog called antroquinonol upregulated miR-15/16 [157].
Triterpenes also feature a very interesting natural product class concerning potential anticancer drugs. Several triterpenes such as ursolic acid, cucurbitacin I, and ginsenoside Rh2 downregulated the expression of the oncomir miR-21, and Rh2 upregulated the tumor suppressor miR-148a [[158], [159], [160], [161]]. AKBA (3-acetyl-11-keto-β-boswellic acid) represents another triterpene isolated from Boswellia plants and induced the expression of miR-34a [162,163].
The clinically approved terpenoid anticancer drug paclitaxel (taxol, ex Taxus brevifolia) stabilizes microtubules and blocks mitosis in cancer cells [164]. As a single compound paclitaxel displayed no visible improvement against MPM in a phase II study when compared with standard therapy while curation of a peritoneal mesothelioma patient (a 71-years old woman) was observed in combination with carboplatin [165,166]. A more recent case study with nanoparticle albumin-bound paclitaxel in combination with carboplatin displayed repeated responses in an epitheloid MPM patient (a 76-years old man) [167]. Carboplatin/pemetrexed treatment did not work in this patient and, thus, albumin-bound paclitaxel plus carboplatin would be a suitable alternative for patients with cisplatin/pemetrexed-resistant MPM [167]. Interestingly, miR-34a expression was upregulated by paclitaxel treatment, which may play a role for combinations with other drugs [168]. Further to this, the paclitaxel analog docetaxel increased the expression of miR-34a in epithelial MPM cells [169].
A list of terpenoid drugs and their effects on non-coding RNAs is given in Table 5.
Table 5.
Terpenoid drugs with effects on non-coding RNA tumor suppressors (inducing effects) and oncogenes (suppressing effects) in mesothelioma correlated with gemcitabine or pemetrexed activity.
| Drugs | Tumor suppressors | Oncogenes |
|---|---|---|
| Paeoniflorin | miR-16 | – |
| PEG-Thymoquinone | miR-34a | – |
| Antrocin | let-7c | – |
| Parthenolide | miR-15a, miR-16 | – |
| Retinoic acid | let-7c, miR-15/16, miR-223 | – |
| 1,25-D | miR-15a | – |
| Δ-Tocotrienol | miR-34a | – |
| Antroquinonol | miR-15/16 | – |
| Ursolic acid | – | miR-21 |
| Rh2 | miR-148a | miR-21 |
| AKBA | miR-34a | – |
| Paclitaxel | miR-34a | – |
2.4.3. Alkaloids
There are natural indoles of simple structure featured by 3,3‘-diindolylmethane (DIM) and indole-3-carbinol (I3C, from Brassica vegetables) which can regulate cancer-relevant miRNAs [170]. I3C was able to suppress miR-21 [62,171]. DIM, which is the condensation product of I3C built in the stomach, upregulated the tumor suppressors let-7b, let-7c, let-7d, and miR-34 [[172], [173], [174], [175]].
Camptothecin derivatives (quinoline alkaloids from Camptotheca acuminata) and Vinca alkaloids (indole alkaloids from Catharanthus roseus) represent the most prominent examples of clinically approved anticancer alkaloids and the influence of miRNAs on their anticancer activity has been reviewed [176]. Camptothecin and its water-soluble derivatives irinotecan and topotecan represent quinoline alkaloid topoisomerase I inhibitors and the latter drugs are clinically approved against cancer [177]. Topotecan was able to induce miR-34b expression in cancer cells and might be a promising drug for combinations with gemcitabine [178].
Trabectedin (Ecteinascidin 743, Yondelis®) is a very promising tetrahydroisoquinoline isolated from the Caribbean tunicate Ecteinascidia turbinata showing a unique mode of DNA alkylation and this drug is currently clinically approved for soft tissue sarcoma treatment [179]. Meanwhile, results from clinical trials with epitheloid and sarcomatoid/biphasic MPM patients receiving trabectedin were published suggesting distinct activity of trabectedin against MPM [180,181]. Interestingly, trabectedin downregulated miR-21 expression in cancer cells and an influence by FUS-CHOP was proposed [182]. Hence, the combination of trabectedin with pemetrexed appears promising for the treatment of mesothelioma diseases. In addition, the oncogene miR-21 was suppressed by the isoquinoline alkaloid berberine (ex Berberis aristata) [183]. Another natural isoquinoline called palmatine chloride induced the expression of miR-34a [183,184].
A list of alkaloid drugs and their effects on non-coding RNAs is given in Table 6.
Table 6.
Alkaloid drugs with effects on non-coding RNA tumor suppressors (inducing effects) and oncogenes (suppressing effects) in mesothelioma correlated with gemcitabine or pemetrexed activity.
| Drugs | Tumor suppressors | Oncogenes |
|---|---|---|
| I3C | – | miR-21 |
| DIM | let-7b, let-7c, let-7d, miR-34 | – |
| Topotecan | miR-34b | – |
| Trabectedin | – | miR-21 |
| Berberine | – | miR-21 |
| Palmatine | miR-34a | – |
2.4.4. Miscellaneous natural products
Further natural products that don't belong to the compound classes mentioned above modulated miRNAs in cancers. Vitamin C (ascorbate), for instance, upregulated miR-345 expression [185]. Folic acid suppressed miR-21 expression [186]. Polyunsaturated fatty acids (PUFAs) induced the expression of let-7d and miR-15b [187]. In particular, docosahexaenoic acid (DHA), a vitamin F component (i.e., a polyunsaturated fatty acid/PUFA) from fish oil, was able to downregulate miR-21 while miR-126 expression was induced by DHA [188,189]. Diallyl disulfide (DADS), sulforaphane (SFN) and phenethylisothiocyanate (PEITC) feature natural organosulfur compounds commonly found in onions, garlic and leek. These organosulfurs upregulated various tumor suppressors such as let-7c (by PEITC), miR-34a (by DADS), miR-145 (by SFN), and miR-486 (SFN) [[190], [191], [192], [193], [194], [195]].
A list of miscellaneous natural drugs and their effects on non-coding RNAs is given in Table 7.
Table 7.
Miscellaneous natural drugs with effects on non-coding RNA tumor suppressors (inducing effects) and oncogenes (suppressing effects) in mesothelioma correlated with gemcitabine or pemetrexed activity.
| Drugs | Tumor suppressors | Oncogenes |
|---|---|---|
| Vitamin C | miR-345 | – |
| Folic acid | – | miR-21 |
| PUFAs | let-7d, miR-15b, miR-126 | miR-21 |
| DADS | miR-34a | – |
| SFN | miR-145, miR-486 | – |
| PEITC | let-7c | – |
2.4.5. HDAC inhibitors
Chromosomal DNA in eukaryotic nuclei is bound to highly alkaline histone protein octamers in order to form so-called nucleosomes which represent the main components of chromatin and play a crucial role for chromatin regulation [196]. Acetylation of lysine residues of histones reduces the positive charge and, thus, the lysine acetylation state of histone proteins controls gene expression. Histone acetylation as well as acetylation of other proteins (e.g., tubulin, p53) is regulated by histone deacetylases (HDACs) and histone acetyltransferases (HATs) [197]. There is only limited data available for HAT inhibitors concerning the regulation of non-coding RNAs. The natural HAT inhibitor garcinol and its effects on the regulation of non-coding RNAs are presented in Table 4. The inhibition of HDAC enzymes has shown distinct anticancer effects and some HDAC inhibitors were already clinically approved for the treatment of T cell lymphoma and multiple myeloma [198]. In contrast to HAT inhibitors, manifold data concerning the influence of HDAC inhibitors on non-coding RNA expression is already available.
The sodium salt of butyric acid, sodium butyrate (NaB), was identified as an HDAC inhibitor of very simple structure. NaB induced the expression of the tumor suppressors miR-15/16, miR-126, miR-143, miR-145 and miR-202 [[199], [200], [201], [202]].
The NaB derivative sodium phenylbutyrate (NaPBA) upregulated miR-34a and miR-148a [203]. The natural HDAC inhibitor trichostatin A (TSA) induced let-7b, let-7c, miR-15/16, miR-34a, miR-126, miR-202, miR-203, and miR-486 [[204], [205], [206], [207]]. The synthetic HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, vorinostat), which was approved for the treatment of T cell lymphoma, induced the expression of let-7d, miR-15a, miR-16, and miR-34b while the expression of the lncRNA HOTAIR was suppressed by SAHA [[208], [209], [210], [211]]. In addition, SAHA suppressed miR-17-5p [212]. AR-42 upregulated let-7b, let-7d, miR-15b, and miR-34a while miR-17-5p and miR-21 were suppressed [213]. LAQ824 downregulated miR-21 [214]. Panobinostat (LBH-589) upregulated let-7b, miR-15a and miR-16 [208,215].
It has to be mentioned that HDAC inhibitors can casually also exert negative effects such as suppression of tumor suppressor miRNAs or induction of oncomirs. For instance, NaPBA downregulated miR-34b/c [216]. The approved drug SAHA led to suppression of miR-345 [210,217]. Panobinostat induced the expression of oncogenic miR-31 [218]. A detailed list of miRNAs regulated by HDAC inhibitors in various cancer entities was published [219]. Hence, caution is recommended and more research is necessary in order to find out if HDAC inhibitors represent suitable combination partners for gemcitabine or pemetrexed.
A list of HDAC inhibitors and their effects on non-coding RNAs is given in Table 8.
Table 8.
HDAC inhibitors with effects on non-coding RNA tumor suppressors (inducing effects) and oncogenes (suppressing effects) in mesothelioma correlated with gemcitabine or pemetrexed activity.
| Drugs | Tumor suppressors | Oncogenes |
|---|---|---|
| NaB | miR-15/16, miR-126, miR-202, miR-143, miR-145 | – |
| NaPBA | miR-34a, miR-148a | – |
| TSA | let-7b, let-7c, miR-15/16, miR-34a, miR-126, miR-202, miR-203, miR-486 | – |
| SAHA | let-7d, miR-15a, miR-16, miR-34b | HOTAIR |
| AR-42 | let-7b, let-7d, miR-15b, miR-34a | miR-17-5p, miR-21 |
| LAQ824 | – | miR-21 |
| Panobinostat | let-7b, miR-15a, miR-16 | – |
2.4.6. Pemetrexed as sensitizer for other approved anticancer drugs
Pemetrexed is currently applied as the first-line therapy for MPM patients. As mentioned above, the combination with other drugs can improve the activity of pemetrexed in a non-coding RNA mediated way. Vice versa, pemetrexed itself is a regulator of mesothelioma-relevant miRNAs such as the tumor suppressors let-7c, miR-451 and miR-486-5p as well as the oncomir miR-210 [71,72].
Doxorubicin (adriamycin) is a well-established anthraquinone topoisomerase II inhibiting anticancer drug [220]. One main resistance factor for doxorubicin treatment is represented by ABC-transporters which are associated with increased drug efflux and detoxification. The expression of the ABC-transporter MDR1 was regulated by miR-451 and re-expression of miR-451 sensitized breast cancer cells to doxorubicin [221]. Liposomised doxorubicin in combination with gemcitabine and carboplatin as third-line therapy exhibited distinct efficacy with a disease control rate of 60%, however, toxicity concerns arose and less toxic treatment options, maybe with pemetrexed instead of gemcitabine, are sought for [222]. Similarly, the water-soluble camptothecin derivative irinotecan, a clinically approved topoisomerase 1 inhibitor, was investigated for any relations with activity promoting miRNAs. Irinotecan in combination with cisplatin may have activity in mesothelioma patients [223]. Indeed, upregulation of miR-451 expression sensitized cancer stem cells to irinotecan treatment by suppression of the ABC-transporter ABCB1 which is responsible for the elimination of irinotecan in resistant cancer cells [224]. Hence, pemetrexed might increase the activity of doxorubicin and irinotecan in resistant cancer forms by induction of miR-451.
The Vinca alkaloid vinorelbine (navelbine) is often applied as a second-line therapy of MPM in case of relapse [225]. Vinorelbine exhibited moderate activity in pemetrexed-pretreated MPM patients [226]. Vinorelbine-resistant cancer cells revealed higher expression levels of miR-210 [227]. Since pemetrexed-based chemotherapy was able to suppress miR-210 expression a combination with vinorelbine appears promising.
Inhibitors of tyrosine kinases such as sunitinib and sorafenib were clinically approved for the treatment of various tumor diseases. In addition, sorafenib displayed considerable activity in a phase 2 trial for pemetrexed/platinum-pretreated malignant mesothelioma [228]. Sorafenib-resistant cancer cells showed low expression levels of the tumor suppressors let-7c and miR-34a [229]. As already mentioned above, pemetrexed-based chemotherapy upregulated let-7c and miR-34a and features a suitable combination option for sorafenib (Fig. 5).
Fig. 5.
Possible activity boost of approved anticancer drugs by pemetrexed-mediated miRNA modulation.
3. Conclusions
Non-coding RNAs can modulate gemcitabine and pemetrexed activity in mesothelioma in various ways that either sensitize or harden cancer cells to antimetabolite treatment. In addition, gemcitabine and pemetrexed actively modified the expression profiles of non-coding RNAs in mesothelioma. It is possible to use the already known facts about the interactions between non-coding RNAs and gemcitabine or pemetrexed in order to identify both problems and chances. Facing these facts, future research efforts should provide more details about the relationship between non-coding RNAs and gemcitabine or pemetrexed treatment in order to improve the therapy of mesothelioma diseases with high mortality rates and to prolong the survival times of mesothelioma patients as well as their quality of life.
References
- 1.Odgerel C.-O., Takahashi K., Sorahan T., Driscoll T., Fitzmaurice C., Yoko-o M., Sawanyawisuth K., Furuya S., Tanaka F., Horie S., van Zandwijk N., Takala J. Estimation of the global burden of mesothelioma deaths from incomplete national mortality data. Occup. Environ. Med. 2017;74:851–858. doi: 10.1136/oemed-2017-104298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romano M., Catalano A., Nutini M., D'Urbano E., Crescenzi C., Claria J., Libner R., Davi G., Procopio A. 5-Lipoxygenase regulates malignant mesothelial cell survival: involvement of vascular endothelial growth factor. FASEB J. 2001;15:2326–2336. doi: 10.1096/fj.01-0150com. [DOI] [PubMed] [Google Scholar]
- 3.Rossini M., Rizzo P., Bononi I., Clementz A., Ferrari R., Martini F., Tognon M.G. New perspectives on diagnosis and therapy of malignant pleural mesothelioma. Front. Oncol. 2018;8:91. doi: 10.3389/fonc.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truini A., Coco S., Genova C., Mora M., Dal Bello M.G., Vanni I., Alama A., Rijavec E., Barletta G., Biello F., Maggioni C., Grossi F. Prognostic and therapeutic implications of microRNA in malignant pleural mesothelioma. MicroRNA. 2016;5:12–18. doi: 10.2174/2211536605666160128151018. [DOI] [PubMed] [Google Scholar]
- 5.Truini A., Coco S., Alama A., Genova C., Sini C., Dal Bello M.G., Barletta G., Rijavec E., Buffarato G., Boccardo F., Grossi F. Role of microRNAs in malignant mesothelioma. Cell. Mol. Life Sci. 2014;71:2865–2878. doi: 10.1007/s00018-014-1584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakamoto N., Honma R., Sekino Y., Goto K., Sentani K., Ishikawa A., Oue N., Yasui W. Non-coding RNAs are promising targets for stem cell-based cancer therapy. Non-coding RNA Res. 2017;2:83–87. doi: 10.1016/j.ncrna.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin R.C.Y., Kirschner M.B., Cheng Y.Y., van Zandwijk N., Reid G. MicroRNA gene expression signatures in long-surviving malignant pleural mesothelioma patients. Genomics Data. 2016;9:44–49. doi: 10.1016/j.gdata.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., Huynh Y., Chrzanowaska A., Fulham M.J., Bailey D.L., Cooper W.A., Kritharides L., Ridley L., Pattison S.T., MacDiarmid J., Brahmbhatt H., Reid G. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1296–1297. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 9.Fang Y., Fullwood M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Dev. Reprod. Biol. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutschner T., Diederichs S., Rna K. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarkou V., Galaras A., Giakountis A., Hatzis P. Crosstalk mechanisms between the WNT signaling pathway and long non-coding RNAs. Non-coding RNA Res. 2018;3:42–53. doi: 10.1016/j.ncrna.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W., Wu X., Wu L., Zhang W., Zhao X. Advances in the diagnosis, treatment and prognosis of malignant pleural mesothelioma. Ann. Transl. Med. 2015;3:182. doi: 10.3978/j.issn.2305-5839.2015.07.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotova S., Wong R.M., Cameron R.B. New and emerging therapeutic options for malignant pleural mesothelioma: review of early clinical trials. Cancer Manag. Res. 2015;7:51–63. doi: 10.2147/CMAR.S72814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C., Niyikiza C., Paoletti P. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 15.Nowak A.K. Chemotherapy for malignant pleural mesothelioma: a review of current management and a look to the future. Ann. Cardiothorac. Surg. 2012;1:508–515. doi: 10.3978/j.issn.2225-319X.2012.10.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zalcman G., Mazieres J., Margery J., Greillier L., Audigier-Valette C., Moro-Sibilot D., Molinier O., Corre R., Monnet I., Gounant V., Rivière F., Janicot H., Gervais R., Locher C., Milleron B., Tran Q., Lebitasy M.-P., Morin F., Creveuil C., Parienti J.-J., Scherpereel A. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomized, controlled, open-label, phase 3 trial. Lancet. 2016;387:1405–1414. doi: 10.1016/S0140-6736(15)01238-6. [DOI] [PubMed] [Google Scholar]
- 17.Ceresoli G.L., Zucali P.A., Mencoboni M., Botta M., Grossi F., Cortinovis D., Zilembo N., Ripa C., Tiseo M., Favaretto A.G., Soto-Parra H., De Vincenzo F., Bruzzone A., Lorenzi E., Gianoncelli L., Ercoli B., Giordano L., Santoro A. Phase II study of pemetrexed and carboplatin plus bevacizumab as first-line therapy in malignant pleural mesothelioma. Br. J. Canc. 2013;109:552–558. doi: 10.1038/bjc.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biersack B. Relations between approved platinum drugs and non-coding RNAs in mesothelioma. Non-coding RNA Res. 2018 doi: 10.1016/j.ncrna.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble S., Goa K.L. Gemcitabine – a review of its pharmacology and clinical potential in non-small cell lung cancer and pancreatic cancer. Drugs. 1997;54:447–472. doi: 10.2165/00003495-199754030-00009. [DOI] [PubMed] [Google Scholar]
- 20.Manegold C. Gemcitabine (Gemzar ®) in non-small cell lung cancer. Expet Rev. Anticancer Ther. 2004;4:345–360. doi: 10.1586/14737140.4.3.345. [DOI] [PubMed] [Google Scholar]
- 21.Huang P., Chubb S., Hertel L.W., Grindey G.B., Plunkett W. Action of 2’,2’-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- 22.Moysan E., Bastiat G., Benoit J.-P. Gemcitabine versus modified gemcitabine: a review of several promising chemical modifications. Mol. Pharm. 2013;10:430–444. doi: 10.1021/mp300370t. [DOI] [PubMed] [Google Scholar]
- 23.Reid J.M., Qu W., Safgren S.L., Ames M.M., Krailo M.D., Seibel N.L., Kuttesch J., Holcenberg J. Phase I trial and pharmacokinetics of gemcitabine in children with advanced solid tumors. J. Clin. Oncol. 2004;22:2445–2451. doi: 10.1200/JCO.2004.10.142. [DOI] [PubMed] [Google Scholar]
- 24.Ak G., Metintas S., Akarsu M., Metintas M. The effectiveness and safety of platinum-based pemetrexed and platinum-based gemcitabine treatment in patients with malignant pleural mesothelioma. BMC Canc. 2015;15:510. doi: 10.1186/s12885-015-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agatsuma T., Koizumi T., Yasuo M., Urushihata K., Tsushima K., Yamamoto H., Hanaoka M., Fukushima M., Honda T., Kubo K. Successful salvage chemotherapy with gemcitabine and vinorelbine in a malignant pleural mesothelioma patient previously treated with pemetrexed. Jpn. J. Clin. Oncol. 2010;40:1180–1183. doi: 10.1093/jjco/hyq101. [DOI] [PubMed] [Google Scholar]
- 26.Shih C., Chen V.J., Gossett L.S., Gates S.B., MacKellar W.C., Habeck L.L., Shackelford K.A., Mendelsohn L.G., Soose D.J., Patel V.F., Andis S.L., Bewley J.R., Rayl E.A., Moroson B.A., Beardsley G.P., Kohler W., Ratnam M., Schultz R.M. LY231514, a pyrrolo[2,3-d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res. 1997;57:1116–1123. [PubMed] [Google Scholar]
- 27.Chattopadhyay S., Moran R.G., Goldman I.D. Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol. Canc. Therapeut. 2007;6:404–417. doi: 10.1158/1535-7163.MCT-06-0343. [DOI] [PubMed] [Google Scholar]
- 28.Racanelli A.C., Rothbart S.B., Heyer C.L., Moran R.G. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation and mammalian target of rapamycin inhibition. Cancer Res. 2009;69:5467–5474. doi: 10.1158/0008-5472.CAN-08-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Righi L., Papotti M.G., Ceppi P., Billé A., Bacillo E., Molinaro L., Ruffini E., Scaglotti G.V., Selvaggi G. Thymidylate synthase but not excision repair cross-complementation group 1 tumor expression predicts outcome in patients with malignant pleural mesothelioma treated with pemetrexed-based chemotherapy. J. Clin. Oncol. 2010;28:1534–1539. doi: 10.1200/JCO.2009.25.9275. [DOI] [PubMed] [Google Scholar]
- 30.Tanino R., Tsubata Y., Harashima N., Harada M., Isobe T. Novel drug-resistance mechanisms of pemetrexed-treated non-small cell lung cancer. Oncotarget. 2018;9:16807–16821. doi: 10.18632/oncotarget.24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson D.M., Keating G.M., Wagstaff A.J. Pemetrexed: a review of its use in malignant pleural mesothelioma and non-small cell lung cancer. Am. J. Cancer. 2004;3:387–399. [Google Scholar]
- 32.Baldwin C.M., Perry C.M. Pemetrexed – a review of its use in the management of advanced non-squameous non-small cell lung cancer. Drugs. 2009;69:2279–2302. doi: 10.2165/11202640-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Goudar R.K. Review of pemetrexed in combination with cisplatin for the treatment of malignant pleural mesothelioma. Therapeut. Clin. Risk Manag. 2008;4:205–211. doi: 10.2147/tcrm.s1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazarika M., White R.M., Jr., Booth B.P., Wang Y.-C., Ham D.Y.L., Liang C.Y., Rahman A., Gobburu J.V.S., Li N., Sridhara R., Morse D.E., Lostritto R., Garvey P., Johnson J.R., Pazdur R. Pemetrexed in malignant pleural mesothelioma. Clin. Canc. Res. 2005;11:982–992. [PubMed] [Google Scholar]
- 35.Reck M., Stahel R.A., von Pawel J., Karthaus M., Korfee S., Serke M., Schuette W.H.-W., Eschbach C., Fink T.H., Leschinger M.I., Mangold C. Pemetrexed in the treatment of malignant mesothelioma: results from an expanded access program in Germany. Respir. Med. 2010;104:142–148. doi: 10.1016/j.rmed.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Brosseau S., Assoun S., Naltet C., Steinmetz C., Gounant V., Zalcman G. A review of bevacizumab in the treatment of malignant pleural mesothelioma. Future Oncol. 2017;13:2537–2546. doi: 10.2217/fon-2017-0307. [DOI] [PubMed] [Google Scholar]
- 37.Truini A., Coco S., Nadal E., Genova C., Mora M., Dal Bello M.G., Vanni I., Alama A., Rijavec E., Biello F., Barletta G., Merlo D.F., Valentino A., Ferro P., Ravetti G.L., Stigliani S., Vigani A., Fedeli F., Beer D.G., Roncella S., Grossi F. Downregulation of miR-99a/let-7c/miR-125b miRNA cluster predicts clinical outcome in patients with unresected malignant pleural mesothelioma. Oncotarget. 2017;8:68627–68640. doi: 10.18632/oncotarget.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohn E.J., Won G., Lee J., Yoon S.W., Lee I., Kim H.J., Kim S.-H. Blockage of epithelial to mesenchymal transition and upregulation of let7b are critically involved in ursolic acid induced apoptosis in malignant mesothelioma cell. Int. J. Biol. Sci. 2016;12:1279–1288. doi: 10.7150/ijbs.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ak G., Tomaszek S.C., Kosari F., Metintas M., Jett J.R., Metintas S., Yildirim H., Dundar E., Dong J., Aubry M.C., Wigle D.A., Thomas C.F., Jr. MicroRNA and mRNA features of malignant pleural mesothelioma and benign asbestos-related pleural effusion. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/635748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., Zheng Q., Ma Y., Zhang J., Wu N., Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., VandenBoom T.G., II, Kong D., Wang Z., Ali S., Philip P.A., Sarkar F.H. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirschner M.B., Cheng Y.Y., Badrian B., Kao S.C., Creaney J., Edelman J.J., Armstrong N.J., Vallely M.P., Musk A.W., Robinson B.W., McCaughan B.C., Klebe S., Mutsaers S.E., van Zandwijk N., Reid G. Increased circulating miR-625-3p: a potential biomarker for patients with malignant pleural mesothelioma. J. Thorac. Oncol. 2012;7:1184–1191. doi: 10.1097/JTO.0b013e3182572e83. [DOI] [PubMed] [Google Scholar]
- 43.Reid G., Pel M.E., Kirschner M.B., Cheng Y.Y., Mugridge N., Weiss J., Williams M., Wright C., Edelman J.J.B., Vallely M.P., McCaughan B.C., Klebe S., Brahmbhatt H., MacDiarmid J.A., van Zandwijk N. Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. 2013;24:3128–3135. doi: 10.1093/annonc/mdt412. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka N., Toyooka S., Soh J., Tsukuda K., Shien K., Furukawa M., Muraoka T., Maki Y., Ueno T., Yamamoto H., Asano H., Otsuki T., Miyoshi S. Downregulation of microRNA-34 induces cell proliferation and invasion of human mesothelial cells. Oncol. Rep. 2013;29:2169–2174. doi: 10.3892/or.2013.2351. [DOI] [PubMed] [Google Scholar]
- 45.El Bizawy R., De Cesare M., Pennati M., Deraco M., Gandellini P., Zuco V., Zaffaroni N. Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-MET an AXL inhibition: persistent activation of ERK and AKT signaling as a possible cytoprotective mechanism. J. Hematol. Oncol. 2017;10:19. doi: 10.1186/s13045-016-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y.M., Liu Y., Wei H.Y., Lv K.Z., Fu P.F. Large intergenic non-coding rna-ror reverses gemcitabine-induced autophagy and apoptosis in breast cancer cells. Oncotarget. 2016;7:59604–59617. doi: 10.18632/oncotarget.10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farooqi A.A., Tabassum S., Ahmad A. MicroRNA-34a: a versatile regulator of myriads of targts in different cancers. Int. J. Mol. Sci. 2017;18:2089. doi: 10.3390/ijms18102089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubo T., Toyooka S., Tsukudam K., Sakaguchi M., Fukazawa T., Soh J., Asano H., Ueno T., Muraoka T., Yamamoto H., Nasu Y., Kishimoto T., Pass H.I., Matsui H., Huh N., Miyoshi S. Epigenetic silencing of microRNA-34b/c plays an important role in the pathogenesis of malignant pleural mesothelioma. Clin. Canc. Res. 2011;17:4965–4974. doi: 10.1158/1078-0432.CCR-10-3040. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y., Zhao R., Tseng K.-F., Li K., Lu Z., Liu Y., Han K., Gan Z., Lin S., Hu H., Min D. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharm. Sin. 2016;37:519–529. doi: 10.1038/aps.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen M., Grauslund M., Ravn J., Sorensen J.B., Andersen C.B., Santoni-Rugiu E. Diagnostic potential of miR-126, miR-143, miR-145, and miR-652 in malignant pleural mesothelioma. J. Mol. Diagn. 2014;16:418–430. doi: 10.1016/j.jmoldx.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Wang H., Li Q., Niu X., Wang G., Zheng S., Fu G., Wang Z. MiR-143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF-1R signaling. Oncol. Lett. 2017;13:435–440. doi: 10.3892/ol.2016.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cioce M., Ganci F., Canu V., Sacconi A., Mori F., Canino C., Korita E., Casini B., Alessandrini G., Cambria A., Carosi M.A., Blandino R., Panebianco V., Facciolo F., Visca P., Volinia S., Muti P., Strano S., Croce C.M., Pass H.I., Blandino G. Protumorigenic effects of miR-145 loss in malignant pleural mesothelioma. Oncogene. 2014;33:5319–5331. doi: 10.1038/onc.2013.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Y., Ge X., Wen Y., Shi Z.-M., Chen Q.-D., Wang M., Liu L.-Z., Jiang B.-H., Lu Y. MiRNA-145 increases therapeutic sensibility to gemcitabine treatment of pancreatic adenocarcinoma cells. Oncotarget. 2016;7:70857–70868. doi: 10.18632/oncotarget.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esebua M., Ding Q., Wang M., Layfield L. MiRNA 148a is highly expressed in malignant mesothelioma. Am. J. Clin. Pathol. 2014;142:A222. [Google Scholar]
- 55.Delpu Y., Lulka H., Sicard F., Saint-Laurent N., Lopez F., Hanoun N., Buscail L., Cordelier P., Torrisani J. The rescue of miR-148a expression in pancreatic cancer: an appropriate therapeutic tool. PloS One. 2013;8 doi: 10.1371/journal.pone.0055513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balatti V., Maniero S., Ferracin M., Veronese A., Negrini M., Ferocci G., Martini F., Tognon M.G. MicroRNAs dysregulated in human malignant pleural mesothelioma. J. Thorac. Oncol. 2011;6:844–851. doi: 10.1097/JTO.0b013e31820db125. [DOI] [PubMed] [Google Scholar]
- 57.Xu J., Wang T., Cao Z., Huang H., Li J., Liu W., Liu S., You L., Zhou L., Zhang T., Zhao Y. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. Oncotarget. 2014;5:6983–6993. doi: 10.18632/oncotarget.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan H., Liu W., Sun W., Wu J., Ji M., Wang Q., Zheng X., Jiang J., Wu C. MiR-17-5p inhibitor enhances chemosensitivity to gemcitabine via upregulating Bim expression in pancreatic cancer cells. Dig. Dis. Sci. 2012;57:3160–3167. doi: 10.1007/s10620-012-2400-4. [DOI] [PubMed] [Google Scholar]
- 59.Nicolè L., Cappellesso R., Sanavia T., Guzzardo V., Fassina A. MiR-21 overexpression and programmed cell death 4 down-regulation features malignant pleural mesothelioma. Oncotarget. 2018;9:17300–17308. doi: 10.18632/oncotarget.24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Z.-H., Tao Z.-H., Zhang J., Li T., Ni C., Xie J., Zhang J.-F., Hu X.-C. MiRNA-21 induces epithelial to mesenchymal transition and gemcitabine resistance via the PTEN/AKT pathway in breast cancer. Tumor Biol. 2016;37:7245–7254. doi: 10.1007/s13277-015-4604-7. [DOI] [PubMed] [Google Scholar]
- 61.Giovannetti E., Funel N., Peters G.J., Del Chiaro M., Erozenci L.A., Vasile E., Leon L.G., Pollina L.E., Groen A., Falcone A., Danesi R., Campani D., Verheul H.M., Boggi U. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 62.Paik W.H., Kim H.R., Park J.K., Song B.J., Lee S.H., Hwang J.-H. Chemosensitivity induced by down-regulation of microRNA-21 in gemcitabine-resistant pancreatic cancer cells by indole-3-carbinol. Anticancer Res. 2013;33:1473–1482. [PubMed] [Google Scholar]
- 63.Wright C.M., Kirschner M.B., Cheng Y.Y., O'Byrne K.J., Gray S.G., Schelch K., Hoda M.A., Klebe S., McCaughan B., van Zandwijk N., Reid G. Long non coding RNAs (lncRNAs) are dysregulated in malignant pleural mesothelioma (MPM) PloS One. 2013;8 doi: 10.1371/journal.pone.0070940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parasramka M., Yan I.K., Wang X., Nguyen P., Matsuda A., Maji S., Foye C., Asmann Y., Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol. Canc. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D'Angelo E., Agostini M. Long non-coding RNA and extracellular matrix: the hidden players in cancer-stroma cross-talk. Non-coding RNA Res. 2018 doi: 10.1016/j.ncrna.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh A.S., Heery R., Gray S.G. In silico and in vitro analyses of lncRNAs as potential regulators in the transition from the epitheloid to sarcomatoid histotype of malignant pleural mesothelioma (MPM) Int. J. Mol. Sci. 2018;19:1297. doi: 10.3390/ijms19051297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L., Dong P., Wang W., Huang M., Tian B. Gemcitabine treatment causes resistance and malignancy of pancreatic cancer stem-like cells via induction of lncRNA HOTAIR. Exp. Ther. Med. 2017;14:4773–4780. doi: 10.3892/etm.2017.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiao F., Hu H., Han T., Yuan C., Wang L., Jin Z., Guo Z., Wang L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015;16:6677–6693. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riquelme E., Suraokar M.B., Rodriguez J., Mino B., Lin H.Y., Rice D.C., Tsao A., Wistuba I.I. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J. Thorac. Oncol. 2014;9:998–1007. doi: 10.1097/JTO.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshida K., Toden S., Ravindranathan P., Han H., Goel A. Curcumin sensitizes pancreatic cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. 2017;38:1036–1046. doi: 10.1093/carcin/bgx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersen M., Grauslund M., Ravn J., Sorensen J.B., Andersen C.B., Santoni-Rugiu E. Diagnostic potential of miR-126, miR-143, miR-145, and miR-652 in malignant pleural mesothelioma. J. Mol. Diagn. 2014;16:418–430. doi: 10.1016/j.jmoldx.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Truini A., Coco S., Nadal E., Genova C., Mora M., Dal Bello M.G., Vanni I., Alama A., Rijavec E., Biello F., Barletta G., Merlo D.F., Valentino A., Ferro P., Ravetti G.L., Stigliani S., Vigani A., Fedeli F., Beer D.G., Roncella S., Grossi F. Downregulation of miR-99a/let-7c/miR-125b miRNA cluster predicts clinical outcome in patients with unresected malignant pleural mesothelioma. Oncotarget. 2017;8:68627–68640. doi: 10.18632/oncotarget.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., Zeng Q., Ma Y., Zhang J., Wu N., Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 74.Shookhoff J.M., Gallicano G.I. A new perspective on neural tube defects: folic acid and microRNA misexpression. Genesis. 2010;48:282–294. doi: 10.1002/dvg.20623. [DOI] [PubMed] [Google Scholar]
- 75.Li X., Wei S., Chen J. Critical appraisal of pemetrexed in the treatment of NSCLC and metastatic pulmonary nodules. OncoTargets Ther. 2014;7:937–945. doi: 10.2147/OTT.S45148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suraokar M., Coombies K., Tsao A., Wistuba I., Zhang Y., Chow C., Kim D., Diao L., Fujimoto J., Mehran R., Wang J., Behrens C. Abstract B37: investigating the potential of miR-203 as a therapeutic candidate and its role in the pathobiology of malignant pleural mesothelioma (MPM) Clin. Canc. Res. 2012;18(3 Suppl) Abstract nr B37. [Google Scholar]
- 77.Cheng R., Lu C., Zhang G., Zhang G., Zhao G. Overexpression of miR-203 increases the sensitivity of NSCLC A549/H460 cell lines to cisplatin by targeting Dickkopf-1. Oncol. Rep. 2017;37:2129–2136. doi: 10.3892/or.2017.5505. [DOI] [PubMed] [Google Scholar]
- 78.Yamamoto K., Seike M., Takeuchi S., Soeno C., Miyanaga A., Noro R., Minegishi Y., Kubota K., Gemma A. MiR-379/411 cluster regulates IL-18 and contributes to drug resistance in malignant pleural mesothelioma. Oncol. Rep. 2014;32:2365–2372. doi: 10.3892/or.2014.3481. [DOI] [PubMed] [Google Scholar]
- 79.Nymark P., Guled M., Borze I., Faisal A., Lahti L., Salmenkivi K., Kettunen E., Anttila S., Knuutila S. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Genes Chromosomes Canc. 2011;50:585–597. doi: 10.1002/gcc.20880. [DOI] [PubMed] [Google Scholar]
- 80.Gamazon E.R., Trendowski M.R., Wen Y., Wing C., Delaney S.M., Huh W., Wong S., Cox N.J., Dolan E. Gene and microRNA perturbations of cellular response to pemetrexed implicate biological networks and enable imputation of response in lung adenocarcinoma. Sci. Rep. 2018;8:733. doi: 10.1038/s41598-017-19004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramírez-Salazar E.G., Salinas-Silva L.C., Vázquez-Manríquez M.E., Gayoss-Gómez L.V., Negrete-Garcia M.C., Ramírez-Rodriguez S.L., Chávez R., Zenteno E., Santillán P., Kelly-García J., Ortiz-Quintero B. Analysis of microRNA expression signatures in malignant pleural mesothelioma, pleural inflammation, and atypical mesothelial hyperplasia reveals common predictive tumorigenesis-related targets. Exp. Mol. Pathol. 2014;97:375–385. doi: 10.1016/j.yexmp.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 82.Birnie K.A., Prêle C.M., Thompson P.J., Badrian B., Mutsaers S.E. Targeting microRNA to improve diagnostic and therapeutic approaches for malignant mesothelioma. Oncotarget. 2017;8:78193–78207. doi: 10.18632/oncotarget.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Santi C., Vencken S., Blake J., Haase B., Benes V., Gemignani F., Landi S., Greene C.M. Identification of miR-21-5p as a functional regulator of mesothelin expression using microRNA capture affinity coupled with next generation sequencing. PloS One. 2017;12 doi: 10.1371/journal.pone.0170999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kirschner M.B., Cheng Y.Y., Armstrong N.J., Lin R.C., Kao S.C., Linton A., Klebe S., McCaughan B.C., van Zandwijk N., Reid G. MiR-score: a novel 6-microRNA signature that predicts survival outcomes in patients with malignant pleural mesothelioma. Mol. Oncol. 2015;9:715–726. doi: 10.1016/j.molonc.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Busacca S., Germano S., De Cecco L., Rinaldi M., Comoglio F., Favero F., Murer B., Mutti L., Pierotta M., Gaudino G. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am. J. Respir. Cell Mol. Biol. 2010;42:312–319. doi: 10.1165/rcmb.2009-0060OC. [DOI] [PubMed] [Google Scholar]
- 86.Kücüktürkmen B., Bozkir A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and miR-21 antisense oligonucleotide to glioblastoma cells. Drug Dev. Ind. Pharm. 2018;44:306–315. doi: 10.1080/03639045.2017.1391835. [DOI] [PubMed] [Google Scholar]
- 87.Wang Q., Cheng N., Li X., Pan H., Li C., Ren S., Su C., Cai W., Zhao C., Zhang L., Zhou C. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558–2567. doi: 10.18632/oncotarget.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei W.-W., Zhou X.-L., Song Y.-J., Yu C.-H., Zhu W.-G., Tong Y.-S. Combination of long noncoding RNA MALAT I and carcinoembryonic antigen for the diagnosis of malignant pleural effusion caused by lung cancer. OncoTargets Ther. 2018;11:2333–2344. doi: 10.2147/OTT.S157551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu F., Wei F., Wang Y., Wu B., Fang Y., Xiong B. EGCG synergizes the therapeutic effect of cisplatin and oxaliplatin through autophagic pathway in human colorectal cancer cells. J. Pharmacol. Sci. 2015;128:27–34. doi: 10.1016/j.jphs.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 90.Younes M., Aggett P., Aguilar F., Crebelli R., Dusemund B., Filipic M., Frutos M.J., Galtier P., Gott D., Gundert-Remy U., Lambré C., Leblanc J.-C., Lillegard I.T., Moldeus P., Mortensen A., Oskarsson A., Stankovic I., Waalkens-Berendsen I., Woutersen R.A., Andrade R.J., Fortes C., Mosesso P., Restani P., Arcella D., Pizzo F., Smeraldi C., Wright M. Scientific opinion on the safety of green tea catechins. EFSA J. 2018;16:5239. doi: 10.2903/j.efsa.2018.5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shah J.J., Kuhn D.J., Orlowski R.Z. Bortezomib and EGCG: no green tea for you? Blood. 2009;113:5695–5696. doi: 10.1182/blood-2009-03-204776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsang J.S., Ebert M.S., van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using genes et signatures. Mol. Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou H., Chen J.X., Yang C.S., Yang M.Q., Deng Y., Wang H. Gene regulation mediated by microRNAs in response to green tea polyphenol EGCG in mouse lung cancer. BMC Genomics. 2014;15(Suppl. 11):S3. doi: 10.1186/1471-2164-15-S11-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Siddiqi I.A., Asim M., Hafeez B.B., Adhami V.M., Tarapore R.S., Mukhtar H. Green tea polyphenol EGCG blunts androgen receptor function in prostate cancer. FASEB J. 2011;25:1198–1207. doi: 10.1096/fj.10-167924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H., Bian S., Yang C.S. Green tea polyphenol EGCG suppresses lung cancer cell growth through upregulating miR-210 expression caused by stabilizing HIF-1α. Carcinogenesis. 2011;32:1881–1889. doi: 10.1093/carcin/bgr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamada S., Tsukamoto S., Huang Y., Makio A., Kumazoe M., Yamashita S., Tachibana H. Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b expression by activating 67-kDa laminin receptor signaling in melanoma cells. Sci. Rep. 2016;6 doi: 10.1038/srep19225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jang J.Y., Lee J.K., Jeon Y.K., Kim C.W. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Canc. 2013;13:421. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang L., Tao C., He A., He X. Overexpression of miR-126 sensitizes osteosarcoma cells to apoptosis induced by epigallocatechin-3-gallate. World J. Surg. Oncol. 2014;12:383. doi: 10.1186/1477-7819-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.An I.-S., An S., Park S., Lee S.N., Bae S. Involvement of microRNAs in epigallocatechin gallate-mediated UVB protection in human dermal fibroblasts. Oncol. Rep. 2013;29:253–259. doi: 10.3892/or.2012.2083. [DOI] [PubMed] [Google Scholar]
- 100.Arola-Arnal A., Bladé C. Proanthocyanidins modulate microRNA expression in human HepG2 cells. PloS One. 2011;6 doi: 10.1371/journal.pone.0025982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang P., Wu X., Wang X., Huang W., Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016;7:43337–43351. doi: 10.18632/oncotarget.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zaman M.S., Sharyari V., Deng G., Thamminana S., Saini S., Majid S., Chang I., Hirata H., Ueno K., Yamamura S., Singh K., Tanaka Y., Tabatabai Z.L., Dahiya R. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PloS One. 2012;7 doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rhodes L.V., Tilghman S.L., Boué S.M., Wang S., Khalili H., Muir S.E., Bratton M.R., Zhang Q., Wang G., Burrow M.E., Collins-Burrow B.M. Glyceollins as novel targeted therapeutic for the treatment of triple-negative breast cancer. Oncol. Lett. 2012;3:163–171. doi: 10.3892/ol.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang H., Hu Y., Yuan L., Xu H., Zhou Y., Zhu J., Zhang Q., Mi M. MicroRNA-34a and microRNA-21 play roles in the chemopreventive effects of 3,6-dihydroxyflavone on 1-methyl-1-nitrosourea-induced breast carcinogenesis. Breast Cancer Res. 2012;14:R80. doi: 10.1186/bcr3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang X. Peng H., Gu Y., Chen J., Yi L., Xie Q., Zhu J., Zhang Q., Mi M. 3,6-Dihydroxyflavone suppresses breast carcinogenesis by epigenetically regulating miR-34a and miR-21. Cancer Prev. Res. 2015;8:509–517. doi: 10.1158/1940-6207.CAPR-14-0357. [DOI] [PubMed] [Google Scholar]
- 106.Cufi S., Bonavia R., Vasquez-Martin A., Oliveras-Ferraros C., Corominas-Faja B., Cuyás E., Martin-Castillo B., Barrajón-Catalán E., Sequra-Carretero A., Joven J., Bosch-Barrera J., Micol V., Menendez J.A. Silibinin suppresses EMT-driven erlotinib resistance by reversing the high miR-21/low miR-200c signature in vivo. Sci. Rep. 2013;3:2459. doi: 10.1038/srep02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang F., Li Y., Mu J., Hu C., Zhou M., Wang X., Si L., Ning S., Li Z. Glabridin inhibits cancer stem cell-like properties of human breast cancer cells: an epigenetic regulation of miR-148a/SMAd2 signaling. Mol. Carcinog. 2016;55:929–940. doi: 10.1002/mc.22333. [DOI] [PubMed] [Google Scholar]
- 108.Lou G., Liu Y., Wu S., Xue J., Yang F., Fu H., Zheng M., Chen Z. The p53/miR-34a/SIRT1 positive feedback loop in quercetin-induced apoptosis. Cell. Physiol. Biochem. 2015;35:2192–2202. doi: 10.1159/000374024. [DOI] [PubMed] [Google Scholar]
- 109.Sonoki H., Sato T., Endo S., Matsunaga T., Yamaguchi M., Yamazaki Y., Sugatani J., Ikari A. Quercetin decreases claudin-2 expression mediated by upregulation of microRNA miR-16 in lung adenocarcinoma. Nutrients. 2015;7:4578–4592. doi: 10.3390/nu7064578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang F., Mu J., Wang X., Ye X., Su L., Ning S., Li Z. The repressive effect of miR-148a on TGF beta-SMADs signal pathway is involved in the glabridin-induced inhibition of the cancer stem cells-like properties in hepatocellular carcinoma cells. PloS One. 2014;9 doi: 10.1371/journal.pone.0096698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiyomaru T., Yamamura S., Fukuhara S., Yoshino H., Kinoshita T., Majid S., Saini S., Chang I., Tanaka Y., Enokida H., Seki N., Nakagawa M., Dahiya R. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PloS One. 2013;8 doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen J., Lin C., Yong W., Ye Y., Huang Z. Calycosin and genistein induce apoptosis by inactivation of HOTAIR/p-Akt signaling pathway in human breast cancer MCF-7 cells, Cell. Physiol. Biochem. 2015;35:722–728. doi: 10.1159/000369732. [DOI] [PubMed] [Google Scholar]
- 113.Gao S.M., Yang J.J., Chen C.Q., Chen J.J., Wang L.Y., Wu J.B., Xing C.Y., Yu K. Pure curcumin decreases the expression of WT1 by upregulation of miR-15a and miR-16-1 in leukemic cells. J. Exp. Clin. Canc. Res. 2012;31:27. doi: 10.1186/1756-9966-31-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Subramaniam D., Ponnurangam S., Ramamoorthy P., Standing D., Battafarano R.J., Anant S., Sharma P. Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PloS One. 2012;7 doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mirgani M.T., Isacchi B., Sadeghizadeh M., Marra F., Bilia A.R., Mowla S.J., Najafi F., Babaei E. Dendrosomal curcumin nanoformulation downregulates pluripotency genes via miR-145 activation in U87MG glioblastoma cells. Int. J. Nanomed. 2014;9:403–417. doi: 10.2147/IJN.S48136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saini S., Aroroa S., Majid S., Shahryari V., Chen Y., Deng G., Yamamura S., Ueno K., Dahiya R. Curcumin modulates microRNA-203-mediated regulation of the Src-Akt axis in bladder cancer. Cancer Prev. Res. 2011;4:1698–1709. doi: 10.1158/1940-6207.CAPR-11-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gandhy S.U., Kim K., Larsen L., Rosengren R.J., Safe S. Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (sp) transcription factors by targeting microRNAs. BMC Canc. 2012;12:564. doi: 10.1186/1471-2407-12-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mudduluru G., George-William J.N., Muppala S., Asangani I.A., Kumarswamy R., Nelson L.D., Allgayer H. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cells. Biosci. Rep. 2011;31:185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- 119.Zhang W., Mai B. MiR-21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non-small cell lung cancer A549 cells. Clin. Transl. Oncol. 2014;16:708–713. doi: 10.1007/s12094-013-1135-9. [DOI] [PubMed] [Google Scholar]
- 120.Pei C.-S., Wu H.-Y., Fan F.-T., Wu Y., Shen C.-S., Pan L.-Q. Influence of curcumin on HOTAIR-mediated migration of human renal cell carcinoma cells. Asian Pac. J. Cancer Prev. APJCP. 2014;15:4239–4243. doi: 10.7314/apjcp.2014.15.10.4239. [DOI] [PubMed] [Google Scholar]
- 121.Kujundzic R.N., Grbesa I., Ivkic M., Katdare M., Gall-Troselj K. Curcumin downregulates H19 gene transcription in tumor cells. J. Cell. Biochem. 2008;104:1781–1792. doi: 10.1002/jcb.21742. [DOI] [PubMed] [Google Scholar]
- 122.Bao B., Ahmad A., Kong D., Ali S., Azmi A.S., Li Y., Banerjee S., Padhye S., Sarkar F.H. Hypoxia induced aggressiveness of prostate cancer cells is linked with deregulated expression of VEGF, IL-6 and miRNAs that are attenuated by CDF. PloS One. 2012;7 doi: 10.1371/journal.pone.0043726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bao B., Ali S., Ahmad A., Azmi A.S., Li Y., Banerjee S., Kong D., Sethi S., Aboukameel A., Padhye S.B., Sarkar F.H., Majumdar A.P.N. Hypoxia-induced aggressiveness of pancreatic cancer cells is due to increased expression of VEGF, IL-6 and miR-21, which can be attenuated by CDF treatment. PloS One. 2012;7 doi: 10.1371/journal.pone.0050165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang C.H., Yue J., Sims M., Pfeffer L.M. The curcumin analog EF24 targets NF-κB and miRNA-21, and has potent anticancer activity in vitro and in vivo. PloS One. 2013;8 doi: 10.1371/journal.pone.0071130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ali S., Ahmad A., Aboukameel A., Ahmed A., Bao B., Banerjee S., Philip P.A., Sarkar F.H. Deregulation of miR-146a expression in a mouse model of pancreatic cancer affecting EGFR signaling. Cancer Lett. 2014;351:134–142. doi: 10.1016/j.canlet.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tili E., Michaille J.J., Alder H., Volinia S., Delmas D., Latruffe N., Croce C.M. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGF-beta signaling pathway in SW480 cells. Biochem. Pharmacol. 2010;80:2057–2065. doi: 10.1016/j.bcp.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu P., Liang H., Xia Q., Li P., Kong H., Lei P., Wang S., Tu Z. Resveratrol induces apoptosis of pancreatic cancer cells by inhibiting miR-21 regulation of Bcl-2 expression. Clin. Transl. Oncol. 2013;15:741–746. doi: 10.1007/s12094-012-0999-4. [DOI] [PubMed] [Google Scholar]
- 128.Kumazaki M., Noguchi S., Yasui Y., Iwasaki J., Shinohara H., Yamada N., Akao Y. Anti-cancer effects of naturally occurring compounds through modulation of signal transduction and miRNA expression in human colon cancer cells. J. Nutr. Biochem. 2013;24:1849–1858. doi: 10.1016/j.jnutbio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 129.Yang S., Li W., Sun H., Wu B., Ji F., Sun T., Chang H., Shen P., Wang Y., Zhou D. Resveratrol elicits anti-colorectal cancer effect by activating miR-34c-KITLG in vitro and in vivo. BMC Canc. 2015;15:969. doi: 10.1186/s12885-015-1958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hagiwara K., Kosaka N., Yoshioka Y., Takahashi R.U., Takeshita F., Ochiya T. Stilbene derivatives promote AGO2-dependent tumour-suppressive micro-RNA activity. Sci. Rep. 2012;2:314. doi: 10.1038/srep00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Q., Sun S., Yu W., Jiang J., Zhuo F., Qiu G., Xu S., Xiang X. Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J. Neuro Oncol. 2015;122:283–292. doi: 10.1007/s11060-015-1718-0. [DOI] [PubMed] [Google Scholar]
- 132.Li Q., Liu X., Fu X., Zhang L., Sui H., Zhou L., Sun J., Cai J., Qin J., Ren J., Li Q. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PloS One. 2013;11 doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 133.Avtanski D.B., Nagalingam A., Kuppusamy P., Bonner M.Y., Arbiser J.L., Saxena N.K., Sharma D. Honokiol abrogates leptin-induced tumor progression by inhibiting Wnt1-MTA1-β-catenin signaling axis in a microRNA-34a dependent manner. Oncotarget. 2015;6:16396–16410. doi: 10.18632/oncotarget.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Q., Zhao W., Ye C., Zhuang J., Chang C., Li Y., Huang X., Shen L., Li Y., Cui Y., Song J., Shen B., Eliaz I., Huang R., Ying H., Guo H., Yan J. Honokiol inhibits bladder tumor growth by suppressing EZH2/miR-143 axis. Oncotarget. 2015;6:37335–37348. doi: 10.18632/oncotarget.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hagiwara K., Gailhouste L., Yasukawa K., Kosaka N., Ochiya T. A robust screening method for dietary agents that activate tumour-suppressor microRNAs. Sci. Rep. 2015;5 doi: 10.1038/srep14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma Y.-N., Chen M.-T., Wu Z.-K., Zhao H.-L., Yu H.-C., Yu J., Zhang J.-W. Emodin can induce K562 cells to erythroid differentiation and improve expression of globin genes. Mol. Cell. Biochem. 2013;382:127–136. doi: 10.1007/s11010-013-1726-3. [DOI] [PubMed] [Google Scholar]
- 137.Ren Z., Tong H., Chen L., Yao Y., Huang S., Zhu F., Liu W. miR-211 and miR-429 are involved in emodin's anti-proliferative effects on lung cancer. Int. J. Clin. Med. 2016;9:2085–2093. [Google Scholar]
- 138.Lin S.-Z., Xu J.-B., Ji X., Chen H., Xu H.-T., Hu P., Chen L., Guo J.-Q., Chen M.-Y., Lu D., Wang Z.-H., Tong H.-F. Emodin inhibits angiogenesis in pancreatic cancer by regulating the transforming growth factor-β/drosophila mothers against decapentaplegic pathway and angiogenesis-associated micro-RNAs. Mol. Med. Rep. 2015;12:5865–5871. doi: 10.3892/mmr.2015.4158. [DOI] [PubMed] [Google Scholar]
- 139.Hua J., He Y., Xu Y., Jiang X., Ye W., Pan Z. Emodin prevents intima sickness via Wnt4/Dvl-1/β-catenin signaling pathway mediated by miR-126 in balloon-injured carotid artery rats. Exp. Mol. Med. 2015;47:e170. doi: 10.1038/emm.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Parasramka M.A., Ali S., Banerjee S., Deryavoush T., Sarkar F.H., Gupta S.V. Garcinol sensitizes human pancreatic adenocarcinoma cells to gemcitabine in association with microRNA signatures. Mol. Nutr. Food Res. 2013;57:235–248. doi: 10.1002/mnfr.201200297. [DOI] [PubMed] [Google Scholar]
- 141.Ahmad A., Sarkar S.H., Bitar B., Ali S., Aboukameel A., Sethi S., Li Y., Bao B., Kong D., Banerjee S., Padhye S.B., Sarkar F.H. Garcinol regulates EMT and Wnt signaling pathways in vitro and in vivo, leading to anticancer activity against breast cancer. Mol. Canc. Therapeut. 2012;11:2193–2201. doi: 10.1158/1535-7163.MCT-12-0232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li Y., Jiang F., Chen L., Yang Y., Cau S., Ye Y., Wang X., Mu J., Li Z., Li L. Blockade of TGFβ-SMAD2 by demethylation-activated miR-148a is involved in caffeic acid-induced inhibition of cancer stem cell-like properties in vitro and in vivo. FEBS Open Bio. 2015;5:466–475. doi: 10.1016/j.fob.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiao J., Liu L., Zhong Z., Xiao C., Zhang J. Mangiferin regulates proliferation and apoptosis in glioma cells by induction of microRNA-15b and inhibition of MMP-9 expression. Oncol. Rep. 2015;33:2815–2820. doi: 10.3892/or.2015.3919. [DOI] [PubMed] [Google Scholar]
- 144.Zhou B., Yi H., Tan J., Wue Y., Liu G. Antiproliferative effects of polyphenols from pomegranate rind (Punica granatum L.) on EJ bladder cancer cells via regulation of p53/miR-34a axis. Phytother Res. 2015;29:415–422. doi: 10.1002/ptr.5267. [DOI] [PubMed] [Google Scholar]
- 145.Henwood J.M., Brogden R.N. Etoposide. A review of its pharmacodynamics and pharmacokinetic properties, and therapeutic potential in combination chemotherapy of cancer. Drugs. 1990;39:438–490. doi: 10.2165/00003495-199039030-00008. [DOI] [PubMed] [Google Scholar]
- 146.Hande K.R. Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Canc. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 147.Novello C., Pazzaglia I., Conti A., Quattrini I., Pollino S., Perrego P., Picci P., Benassi M.S. p53-dependent activation of microRNA-34a in response to etoposide-induced DNA damage in osteosarcoma cell lines not impaired by dominant negative p53 expression. PloS One. 2014;9 doi: 10.1371/journal.pone.0114757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang M., Lu J.J., Huang M.Q., Bao J.L., Chen X.P., Wang Y.T. Terpenoids: natural products for cancer therapy. Expet Opin. Invest. Drugs. 2012;21:1801–1818. doi: 10.1517/13543784.2012.727395. [DOI] [PubMed] [Google Scholar]
- 149.Li W., Qi Z., Wei Z., Liu S., Wang P., Chen Y., Zhao Y. Paeoniflorin inhibits proliferation and induces apoptosis of human glioma cells via microRNA-16 upregulation and matrix metalloproteinaase-9 downregulation. Mol. Med. Rep. 2015;12:2735–2740. doi: 10.3892/mmr.2015.3718. [DOI] [PubMed] [Google Scholar]
- 150.Bhattacharya S., Ahir M., Patra P., Mukherjee S., Ghosh S., Mazumdar M., Chattopadhyay S., Das T., Chattopadhyay D., Adhikary A. PEGylated-thymoquinone-nanoparticle mediated retardation of breast cancer cell migration by deregulation of cytoskeletal actin polymerization through miR-34a. Biomaterials. 2015;51:91–107. doi: 10.1016/j.biomaterials.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 151.Yeh C.-T., Huang W.-C., Rao Y.K., Ye M., Lee W.H., Wang L.-S., Tzeng D.T.W., Wu C.-H., Shieh Y.-S., Huang C.-Y.F., Chen Y.-J., Hsiao M., Wu A.T.H., Yang Z., Tzeng Y.-M. A sesquiterpene lactone antrocin from Antrodia camphorata negatively modulates JAK2/STAT3 signaling via microRNA let-7c and induces apoptosis in lung cancer cells. Carcinogenesis. 2013;34:2918–2928. doi: 10.1093/carcin/bgt255. [DOI] [PubMed] [Google Scholar]
- 152.Pickering B.F., Yu D., Van Dyke M.W. Nucleolin protein interacts with microprocessor complex to affect biogenesis of microRNAs 15a and 16. J. Biol. Chem. 2011;286:44095–44103. doi: 10.1074/jbc.M111.265439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rossi A., D’urso O.F., Gatto G., Poltronieri P., Ferracin M., Remondelli P., Negrini M., Caporaso M.G., Bonatti S., Mallardo M. Non-coding RNAs change their expression profile after retinoid induced differentiation of the pro-myelocytic cell line NB4. BMC Res. 2010;3:24. doi: 10.1186/1756-0500-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garzon R., Pichiorri F., Palumbo T., Visentini M., Aqeilan R., Cimmino A., Wang H., Sun H., Volinia S., Alder H., Calin G.A., Liu C.G., Andreeff M., Croce C.M. MicroRNA gene expression during retinoid acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 155.Zitman-Gal T., Green J., Pasmanik-Chor M., Golan E., Bernheim J., Benchetrit S. Vitamin D manipulates miR-181c, miR-20b and miR-15a in human umbilical vein endothelial cells exposed to diabetic-like environment. Cardiovasc. Diabetol. 2014;13:8. doi: 10.1186/1475-2840-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ji X., Wang Z., Geamanu A., Goja A., Sarkar F.H., Gupta S.V. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in non-small cell lung cancer cells. Int. J. Canc. 2012;131:2668–2677. doi: 10.1002/ijc.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kumar V.B., Yuan T.-C., Liou J.-W., Yang C.-J., Sung P.-J., Wenig C.-F. Antroquinonol inhibits NSCLC proliferation by altering PI3K/mTOR proteins and miRNA expression profiles. Mutat. Res. 2011;707:42–52. doi: 10.1016/j.mrfmmm.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 158.Wang J., Li Y., Wang X., Jiang C. Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-β1/miR-21/PDCD4 pathway. Basic Clin. Pharmacol. Toxicol. 2012;111:106–112. doi: 10.1111/j.1742-7843.2012.00870.x. [DOI] [PubMed] [Google Scholar]
- 159.van der Fits L., van Kester M.S., Qin Y., Out-Luiting J.J., Smit F., Zoutman W.H., Willemze R., Tensen C.P., Vermeer M.H. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sézary syndrome. J. Invest. Dermatol. 2011;131:762–768. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 160.Wu N., Wu G.C., Hu R., Li M., Feng H. Ginsenoside Rh2 inhibits glioma cell proliferation by targeting microRNA-128. Acta Pharmacol. Sin. 2011;32:345–353. doi: 10.1038/aps.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.An I.-S., An S., Kwon K.J., Kim Y.J., Bae S. Ginsenoside Rh2 mediates changes in the microRNA expression profile of human non-small cell lung cancer A549 cells. Oncol. Rep. 2013;29:523–528. doi: 10.3892/or.2012.2136. [DOI] [PubMed] [Google Scholar]
- 162.Toden S., Okugawa Y., Buhrmann C., Nattamai D., Anguiano E., Baldwin N., Shakibaei M., Boland C.R., Goel A. Novel evidence for curcumin and boswellic acid-induced chemoprevention through regulation of miR-34a and miR-27a in colorectal cancer. Cancer Prev. Res. 2015;8:431–443. doi: 10.1158/1940-6207.CAPR-14-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Takahashi M., Sung B., Shen Y., Hur K., Link A., Boland C.R., Aggarwal B.B., Goel A. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis. 2012;33:2441–2449. doi: 10.1093/carcin/bgs286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Spencer C.M., Faulds D. Paclitaxel. A review of its pharmacodynamics and pharmacokinetic properties and therapeutic potential in the treatment of cancer. Drugs. 1994;48:794–847. doi: 10.2165/00003495-199448050-00009. [DOI] [PubMed] [Google Scholar]
- 165.van Meerbeeck J., Debruyne C., van Zandwijk N., Postumus P.E., Pennucci M.C., van Breukelen F., Galdermans D., Groen H., Pinson P., van Glabbeke M., van Marck E., Giaccone G. Paclitaxel for malignant pleural mesothelioma: a phase II study of the EORTC lung cancer cooperative group. Br. J. Canc. 1996;74:961–963. doi: 10.1038/bjc.1996.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ogura O., Noguchi T., Nagata K., Noma H., Maemura M., Higashimoto M., Takebayashi Y., Maeda S. A case of malignant peritoneal mesothelioma successfully treated with carboplatin and paclitaxel. Gan To Kagaku Ryoho. 2007;33:1001–1004. [PubMed] [Google Scholar]
- 167.Kanai O., Nakatani K. Fujitam K., Mio T. Repetitive responses to nanoparticle albumin-bound paclitaxel and carboplatin in malignant pleural mesothelioma. Respirol. Case Rep. 2016;4:28–31. doi: 10.1002/rcr2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Frères P., Josse C., Bovy N., Boukerroucha M., Struman I., Bours V., Jerusalem G. Neoadjuvant chemotherapy in breast cancer patients induces miR-34a and miR-122 expression. J. Cell. Physiol. 2015;230:473–481. doi: 10.1002/jcp.24730. [DOI] [PubMed] [Google Scholar]
- 169.Ghawanmeh T., Thunberg U., Castro J., Murray F., Laytragoon-Lewin N. MiR-34a expression, cell cycle arrest and cell death of malignant mesothelioma cells upon treatment with radiation, docetaxel or combination treatment. Oncology. 2011;81:330–335. doi: 10.1159/000334237. [DOI] [PubMed] [Google Scholar]
- 170.Anderton M.J., Manson M.M., Verschoyle R.D., Gescher A., Lamb J.H., Farmer P.B., Steward W.P., Williams M.L. Pharmacokinetics and tissue disposition of indole-3-carbinol and its acid condensation products after oral administration to mice. Clin. Canc. Res. 2004;10:5233–5241. doi: 10.1158/1078-0432.CCR-04-0163. [DOI] [PubMed] [Google Scholar]
- 171.Melkamu T., Zhang X., Tan J., Zeng Y., Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31:252–258. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 172.Kong D., Heath E., Chen W., Cher M.L., Powell I., Heilbrun L., Li Y., Ali S., Sarkar F.H. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PloS One. 2012;7 doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kashat M., Azzouz L., Sarkar S.H., Kong D., Li Y., Sarkar F.H. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- 174.Ahmad A., Ali S., Ahmed A., Ali A.S., Raz A., Sakr W.A., Rahman K.M. 3,3’-Diindolylmethane enhances the effectiveness of herceptin against HER-2/neu-expressing breast cancer cells. PloS One. 2013;8 doi: 10.1371/journal.pone.0054657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li Y., Vandenbloom T.G., II, Kong D., Wang Z., Ali S., Philip P.A., Sarkar F.H. Upregulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Biersack B. Non-coding RNA/microRNA-modulatory dietary factors and natural products for improved cancer therapy and prevention: alkaloids, organosulfur compounds, aliphatic carboxylic acids and water-soluble vitamins. Non-coding RNA Res. 2016;1:51–63. doi: 10.1016/j.ncrna.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zunino F., Paresi G. Camptothecins in clinical development. Expet Opin. Invest. Drugs. 2004;13:269–284. doi: 10.1517/13543784.13.3.269. [DOI] [PubMed] [Google Scholar]
- 178.Boren T., Xiong Y., Hakam A., Wenham R., Apte S., Chan G., Kamath S.G., Chen D.-T., Dressman H., Lancaster J.M. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 2009;113:249–255. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 179.Le V.H., Inai M., Williams R.M., Kan T. Ecteinascidins. A review of the chemistry, biology and clinical utility of potent tetrahydroisoquinoline antitumor antibiotics. Nat. Prod. Rep. 2015;32:328–347. doi: 10.1039/c4np00051j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cortinovis D.L., Hollander L.H., Carlucci L., Grosso F., Ceresoli G.L., Pacchetti I., Zucali P.A., D'Incalci M., Tiseo M., Legramandi L., Canova S., Ugo F., Abbate M.I., Taverna G., Cogliati V., Bidoli P. Trabectedin (T) as a second line treatment option for patients with epithelioid malignant pleural mesothelioma (MPM) in progression following pemetrexed/platin-derivates chemotherapy: ATREUS trial. J. Clin. Oncol. 2017;35:8513. [Google Scholar]
- 181.Cortinovis D.L., Hollander L.H., Floriani I.C., Grosso F., Marinello A., Ceresoli G.L., Pacchetti I., Zucali P.A., D'Incalci M., Canova S., Abbate M.I., Ugo F., Vukcaj S., Bidoli P. Activity and safety of trabectedin in patients with sarcomatoid/biphasic malignant pleural mesothelioma (MPM) J. Clin. Oncol. 2015;33:7561. [Google Scholar]
- 182.Uboldi S., Calura E., Beltrame L., Nerini I.F., Marchini S., Cavalieri D., Erba E., Chiorino G., Ostano P., D'Angelo D., D'Incalci M., Romualdi C. A systems biology approach to characterize the regulatory networks leading to trabectedin resistance in an in vitro model of myxoid liposarcoma. PloS One. 2012;7 doi: 10.1371/journal.pone.0035423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu S., Fang Y., Shen H., Xu W., Li H. Berberine sensitizes ovarian cancer cells to cisplatin through miR-21/PDCD4 axis. Acta Biochim. Biophys. Sin. 2013;45:756–762. doi: 10.1093/abbs/gmt075. [DOI] [PubMed] [Google Scholar]
- 184.Hagiwara K., Gailhouste L., Yasukawa K., Kosaka N., Ochiya T. A robust screening method for dietary agents that activate tumour-suppressor microRNAs. Sci. Rep. 2015;5 doi: 10.1038/srep14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Venturelli S., Sinnberg T.W., Berger A., Noor S., Levesque M.P., Böcker A., Niessner H., Lauer U.M., Bitzer M., Garbe C., Busch C. Epigenetic impacts of ascorbate on human metastatic melanoma cells. Front. Oncol. 2014;4 doi: 10.3389/fonc.2014.00227. Article 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pogribny I.P., Tryndyak V.P., Ross S.A., Beland F.A. Differential expression of microRNAs during hepatocarcinogenesis induced by methyl deficiency in rats. Nutr. Rev. 2008;66:S33–S35. doi: 10.1111/j.1753-4887.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- 187.Davidson L.A., Wang N., Shah M.S., Lupton J.R., Ivanov I., Chapkin R.S. n-3 Polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis. 2009;30:2077–2084. doi: 10.1093/carcin/bgp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mandal C.C., Gosh-Choudhury T., Dey N., Choudhury G.G., Ghosh-Choudhury N. MiR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897–1908. doi: 10.1093/carcin/bgs198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Faragó N., Fehér L.Z., Kitajka K., Das U.N., Puskás L.G. MicroRNA profile of polyunsaturated fatty acid treated glioma cells reveal apoptosis-specific expression changes. Lipids Health Dis. 2011;10:173. doi: 10.1186/1476-511X-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Izzotti A., Calin G.A., Steele V.E., Cartiglia C., Longobardi M., Croce C.M., de Flora S. Chemoprevention of cigarette smoke-induced alterations of microRNA expression in rat lungs. Cancer Prev. Res. 2010;3:62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xiao X., Chen B., Liu X., Liu P., Zheng G., Tang F. Yem H., Xie X. Diallyl sulfide suppresses SRC/Ras/ERK signaling-mediated proliferation and metastasis in human breast cancer by up-regulating miR-34a. PloS One. 2014;9 doi: 10.1371/journal.pone.0112720. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 192.G. Wang, G. Liu, Y. Ye, Y. Fu, X. Zhang, Upregulation of miR-34a by diallyl disulfide suppresses invasion and induces apoptosis in SCG-7901 cells through inhibition of the PI3K/Akt signaling pathway, Oncol. Lett. 11 (016) 2661-2667. [DOI] [PMC free article] [PubMed] [Retracted]
- 193.Slaby O., Sachlova M., Brezkova V., Hezova R., Kovarikova A., Bischofová S., Sevcikova S., Bienertova-Vasku J., Vasku A., Svoboda M., Vyzula R. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutr. Canc. 2013;65:247–254. doi: 10.1080/01635581.2013.756530. [DOI] [PubMed] [Google Scholar]
- 194.Tang H., Kong Y., Guo J., Tang Y., Xie X., Yang L., Su Q., Xie X. Diallyl disulfide suppresses proliferation and induces apoptosis in human gastric cancer through Wnt-1 signaling pathway by up-regulation of miR-200b and miR-22. Cancer Lett. 2013;340:72–81. doi: 10.1016/j.canlet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 195.Shan Y., Zhang L., Bao Y., Li B., He C., Gao M., Feng X., Xu W., Zhang X., Wang S. Epithelial-mesenchymal transition, a novel target of sulforaphane via COX-2/MMP2/Snail, ZEB1 and miR-200c/ZEB1 pathways in human bladder cancer cells. J. Nutr. Biochem. 2013;24:1062–1069. doi: 10.1016/j.jnutbio.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 196.Marino-Ramírez L., Kann M.G., Shoemaker B.A., Landsman D. Histone structure and nucleosome stability. Expert Rev. Proteomics. 2005;2:719–729. doi: 10.1586/14789450.2.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bolden J.E., Peart M.M.J., Johnstone R.R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 198.Eckschlager T., Plch J., Stiborova M., Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017;18:1414. doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ferreira A.C., Robaina M.C., Rezende L.M., Severino P., Klumb C.E. Histone deacetylase inhibitor prevents cell growth in Burkitt's lymphoma by regulating PI3K/Akt pathways and leads to upregulation of miR-143, miR-145, and miR-101. Ann. Hematol. 2014;93:983–993. doi: 10.1007/s00277-014-2021-4. [DOI] [PubMed] [Google Scholar]
- 200.Hu S., Dong T.S., Dalal S.R., Wu F., Bissonnette M., Kwon J.H., Chang E.B. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PloS One. 2011;6 doi: 10.1371/journal.pone.0016221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chen C.Q., Chen C.S., Chen J.J., Zhou L.P., Xu H.L., Jin W.W., Wu J.B., Gao S.M. Histone deacetylase inhibitor trichostatin A increases the expression of Dleu2/miR-15a/16-1 via HDAC3 in non-small cell lung cancer. Mol. Cell. Biochem. 2013;383:137–148. doi: 10.1007/s11010-013-1762-z. [DOI] [PubMed] [Google Scholar]
- 202.Ferreira A.C., Robaina M.C., Rezende L.M., Severino P., Klumb C.E. Histone deacetylase inhibitor prevents cell growth in Burkitt's lymphoma by regulating PI3K/Akt pathways and leads to upregulation of miR-143, miR-145, and miR-101. Ann. Hematol. 2014;93:983–993. doi: 10.1007/s00277-014-2021-4. [DOI] [PubMed] [Google Scholar]
- 203.Saito Y., Liang G., Egger G., Friedman J.M., Chuang J.C., Coetzee G.A., Jones P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 204.Hsieh T.H., Hsu C.Y., Tsai C.F., Long C.Y., Wu C.H., Wu D.C., Lee J.N., Chang W.C., Tsai E.M. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol. Ther. 2015;23:656–666. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rhodes L.V., Nitschke A.M., Segar H.C., Martin E.C., Driver J.L., Elliott S., Nam S.Y., Li M., Nephew K.P., Burow M.E., Collins-Burow B.M. The histone deacetylase inhibitor trichostatin A alters microRNA expression profiles in apoptosis-resistant breast cancer cells. Oncol. Rep. 2012;27:10–16. doi: 10.3892/or.2011.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rauhala H.E., Jalava S.E., Isotalo J., Bracken H., Lehmusvaara S., Tammela T.L., Oja H., Visakorpi T. MiR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int. J. Canc. 2010;127:1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 207.Seol H.S., Akiyama Y., Shimada S., Lee H.J., Kim T.I., Chun S.M., Singh S.R., Jang S.J. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014;353:232–241. doi: 10.1016/j.canlet.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sampath D., Liu C., Vasan K., Sulda M., Puduvalli V.K., Wierda W.G., Keating M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood. 2012;119:1162–1172. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang X., Chen X., Lin J., Lwin T., Wright G., Moscinski L.C., Dalton W.S., Seto E., Wright K., Sotomayor E., Tao J. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene. 2012;31:3002–3008. doi: 10.1038/onc.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schiffgen M., Schmidt D.H., von Rucker A., Muller S.C., Ellinger J. Epigenetic regulation of microRNA expression in renal cell carcinoma. Biochem. Biophys. Res. Commun. 2013;436:79–84. doi: 10.1016/j.bbrc.2013.05.061. [DOI] [PubMed] [Google Scholar]
- 211.Stickertson J.A.B., Grunnet M., Bardram L., Federspiel B., Friis-Hansen L. MiR-449 and HDAC modulates HOTAIR expression which predicts poor prognosis in resected gastro-esophageal adenocarcinomas and doubles proliferation rate in HFE145 cells. J. Gastric Discord. Ther. 2017;3 [Google Scholar]
- 212.Kretzner L., Scuto A., Dino P.M., Kowolik C.M., Wu J., Ventura P., Jove R., Forman S.J., Yen Y., Kirschbaum M.H. Combining histone deacetylase inhibitor vorinostat with aurora kinase inhibitors enhances lymphoma cell killing with repression of c-Myc, hTERT, and microRNA levels. Cancer Res. 2011;71:3912–3920. doi: 10.1158/0008-5472.CAN-10-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Balch C., Naegeli K., Nam S., Ballard B., Hyslop A., Melki C., Reilly E., Hur M.W., Nephew K.P. A unique histone deacetylase inhibitor alters microRNA expression and signal transduction in chemoresistant ovarian cancer cells. Cancer Biol. Ther. 2012;13:681–693. doi: 10.4161/cbt.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Scott G.K., Mattie M.D., Berger C.E., Benz S.C., Benz C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- 215.Di Fazio P., Montalbano R., Neureiter D., Alinger B., Schmidt A., Merkel A.L., Quint K., Ocker M. Downregulation of HMGA2 by the pan-deacetylase inhibitor panobinostat is dependent on has-let-7b expression in liver cancer cell lines. Exp. Cell Res. 2012;318:1832–1843. doi: 10.1016/j.yexcr.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 216.Mazar J., DeBlasio D., Govindarajan S.S., Zhang S., Perrera R.J. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011;585:2467–2476. doi: 10.1016/j.febslet.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 217.Lee E.M., Shin S., Cha H.J., Yoon Y., Bae S., Jung J.H., Lee S.M., Lee S.J., Park I.C., Jin Y.W., An S. Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression profiles in A549 human non-small cell lung cancer cells. Int. J. Mol. Med. 2009;24:45–50. doi: 10.3892/ijmm_00000204. [DOI] [PubMed] [Google Scholar]
- 218.Cho J.-H., Dimri M., Dimri G.P. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cell senescence. J. Biol. Chem. 2015;290:10555–10567. doi: 10.1074/jbc.M114.624361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ali S.R., Humphreys K.J., McKinnon R.A., Michael M.Z. Impact of histone deacetylase inhibitors on microRNA expression and cancer therapy: a review. Drug Dev. Res. 2015;76:296–317. doi: 10.1002/ddr.21268. [DOI] [PubMed] [Google Scholar]
- 220.Edwardson D., Chewchuk S., Parissenti A.M. Resistance to anthracyclines and taxanes in breast cancer. In: Ahmad A., editor. Breast Cancer Metastasis and Drug Resistance. Springer Science + Business Media; New York: 2013. pp. 227–247. [Google Scholar]
- 221.Kovalchuk O., Filkowski J., Meservy J., Ilnytskyy Y., Tryndyak V.P., Chekhun V.F., Pogribny I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Canc. Therapeut. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 222.de Lima V.A., Sorensen J.B. Third-line chemotherapy with carboplatin, gemcitabine and liposomised doxorubicin for malignant pleural mesothelioma. Med. Oncol. 2015;32:11. doi: 10.1007/s12032-014-0458-x. [DOI] [PubMed] [Google Scholar]
- 223.Fennell D.A., Steele J.P., Shamash J., Evans M.T., Wells P., Sheaff M.T., Rudd R.M., Stebbing J. Efficacy and safety of first- or second-line irinotecan, cisplatin, and mitomycin in mesothelioma. Cancer. 2007;109:93–99. doi: 10.1002/cncr.22366. [DOI] [PubMed] [Google Scholar]
- 224.Bitarte N., Bandres E., Boni V., Zarate R., Rodriguez J., Gonzales-Huarriz M., Lopez I., Sola J.J., Alonso M.M., Fortes P., Garcia-Foncillas J. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cell. 2011;29:1661–1671. doi: 10.1002/stem.741. [DOI] [PubMed] [Google Scholar]
- 225.Stebbing J., Powles T., McPherson K., Shamash J., Well P., Sheaff M.T., Slater S., Rudd R.M., Fennell D., Steele J.P. The efficacy and safety of weekly vinorelbine in relapsed malignant pleural mesothelioma. Lung Canc. 2009;63:94–97. doi: 10.1016/j.lungcan.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 226.Zucali P.A., Perrino M., Lorenzi E., Ceresoli G.L., De Vincenzo F., Simonelli M., Gianoncelli L., De Santis R., Giordano L., Santoro A. Vinorelbine in pemetrexed-pretreated patients with malignant pleural mesothelioma. Lung Canc. 2014;84:265–270. doi: 10.1016/j.lungcan.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 227.Zhong S., Ma T., Zhang X., Lv M., Chen L., Tang J., Zhao J. MicroRNA expression profiling and bioinformatics analysis of dysregulated microRNAs in vinorelbine-resistant breast cancer cells. Gene. 2015;556:113–118. doi: 10.1016/j.gene.2014.11.046. [DOI] [PubMed] [Google Scholar]
- 228.Papa S., Popat S., Shah R., Prevost A.T., Lal R., McLennan B., Cane P., Lang-Lazdunski L., Viney Z., Dunn J.T., Barrington S., Landau D., Spicer J. Phase 2 study of sorafenib in malignant mesothelioma previously treated with platinum-containing chemotherapy. J. Thorac. Oncol. 2013;8:783–787. doi: 10.1097/JTO.0b013e31828c2b26. [DOI] [PubMed] [Google Scholar]
- 229.Tang S., Tan G., Jiang X., Han P., Zhai B., Dong X., Qiao H., Jiang H., Sun X. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget. 2016;7:73257–73269. doi: 10.18632/oncotarget.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]