Abstract
Neurodegenerative diseases are among the most common causes of disability worldwide. Although neurodegenerative diseases are heterogeneous in both their clinical features and the underlying physiology, they are all characterised by progressive loss of specific neuronal populations. Recent experimental evidence suggests that long non-coding RNAs (lncRNAs) play important roles in the CNS in health and disease. Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) is an abundant, ubiquitously expressed lncRNA, which forms a scaffold for a specific RNA granule in the nucleus, or nuclear body, the paraspeckle. Paraspeckles act as molecular hubs for cellular processes commonly affected by neurodegeneration. Transcriptomic analyses of the diseased human tissue have revealed altered NEAT1 levels in the CNS in major neurodegenerative disorders as well as in some disease models. Although it is clear that changes in NEAT1 expression (and in some cases, paraspeckle assembly) accompany neuronal damage, our understanding of NEAT1 contribution to the disease pathogenesis is still rudimentary. In this review, we have summarised the available knowledge on NEAT1 involvement in the molecular processes linked to neurodegeneration and on NEAT1 dysregulation in this type of disease, with a special focus on amyotrophic lateral sclerosis. The goal of this review is to attract the attention of researchers in the field of neurodegeneration to NEAT1 and paraspeckles.
Keywords: NEAT1, Paraspeckle, lncRNA, Neurodegeneration, Amyotrophic lateral sclerosis
1. Introduction
Neurodegenerative diseases (NDDs) are one of the most common (and growing) causes of disability worldwide. Today, due to the global extension of lifespan, they represent one of the greatest public health threats. Dementia of various origins (including Alzheimer's disease), Parkinson's, Huntington's and motor neuron diseases all belong to this group, collectively accounting for a significant proportion of age-related morbidity, including cognitive and motor impairment. NDDs are heterogeneous in their clinical signs and in the underlying physiology, however they are all characterised by the progressive loss of specific neuronal populations [1]. No disease-modifying therapy is currently available for any of these conditions, with existing drugs being limited to treating the symptoms rather than the disease process.
Extensive transcriptomic studies fuelled by advanced high-throughput sequencing technologies and comprehensive bioinformatic analysis have dramatically expanded our understanding of the incredibly complicated yet precisely regulated human transcriptome. It is now widely accepted that more than 80% of human genome is dynamically transcribed, in a temporally and spatially regulated manner, and that RNAs not encoding any protein (non-coding RNAs, ncRNAs) play essential roles in presumably almost every aspect of cellular biology [2]. In contrast to the ancient short ncRNAs, long ncRNAs (lncRNAs) appear later in evolution with one third being primate specific; they are believed to be crucial for the evolution of primate brains [3]. It is not surprising therefore that lncRNAs, which generally show unique tissue and cell-type specific expression pattern, are particularly abundant within the central nervous system (CNS), with at least 40% of tissue-specific lncRNAs expressed in the brain [4]. lncRNAs are involved in the regulation of a plethora of neurospecific processes, including neural plasticity, synaptic transmission, neurogenesis, brain development and ageing [5,6]. Nuclear Paraspeckle Assembly Transcript 1 (NEAT1) is a ubiquitous, highly expressed, nuclear-retained regulatory lncRNA with important roles in cellular physiology and pathophysiology. In this review, we will discuss the emerging roles for this lncRNA and the nuclear body it assembles, the paraspeckle, in NDDs, with a special focus on amyotrophic lateral sclerosis.
2. NEAT1: two non-coding transcripts with distinct cellular roles
2.1. Long non-coding RNAs: an introduction
The diverse group of ncRNAs includes ribosomal RNA (rRNA), transfer RNA (tRNA), microRNA (miRNA) and lncRNAs, among others [7]. ncRNAs have an extremely wide range of size distribution, from ∼20 nucleotides to several kilobases, and lncRNAs are defined as those longer than 200 nucleotides. lncRNAs are derived from multiple sources within the genome, including intergenic regions, gene regulatory regions (UTRs, promoters and enhancers) and specific chromosomal regions (telomeres); they can also be derived from the mitochondrial genome [[8], [9], [10]]. These RNAs are subject to post-transcriptional processing and can undergo 5’ capping, polyadenylation, alternative splicing and RNA editing [11,12]. The broad range of functions of lncRNAs includes roles in transcriptional and epigenetic mechanisms via the recruitment of transcription factors and chromatin-modifying complexes to specific nuclear and genomic sites; alternative splicing and other post-transcriptional RNA modifications through the assembly of nuclear domains containing RNA-processing factors; nuclear-cytoplasmic shuttling; and translational control [13,14]. lncRNAs organize nuclear architecture by concentrating in specific domains close to transcription sites and forming lncRNA-protein complexes which scaffold protein components of the transcription machinery and modify chromatin state [15,16]. The abundance of lncRNAs in the nervous system suggested a strong intrinsic association between the CNS function and this class of non-coding transcripts, prompting studies into and rapid discovery of the critical roles lncRNAs play in neurodevelopment, neuronal plasticity as well as diseases of the nervous system [17,18].
2.2. NEAT1: basic facts
Originally named as Nuclear Enriched Abundant Transcript 1, which was subsequently changed to Nuclear Paraspeckle Assembly Transcript 1, NEAT1 is one of the most abundant lncRNAs in the mammalian nucleus [19]. It is transcribed by RNA polymerase II from a genetic locus called familial tumour syndrome multiple endocrine neoplasia (MEN) type I on human chromosome 11 [20], therefore it is also known under the name “MENepsilon/beta”. The locus gives rise to two isoform transcripts, 3.7 kb NEAT1_1 (MENepsilon) and 23 kb NEAT1_2 (MENbeta), which completely overlap in their 5′-end, through distinct RNA processing mechanisms. NEAT1_1 is canonically polyadenylated. In contrast, RNase P recognizes the tRNA-like structure present at the 3′-end of the primary NEAT1_2 transcript and cleaves it to generate a triple helix which stabilises NEAT1_2 [21,22]. Mouse Neat1 isoforms are slightly smaller, 3.1 kb for Neat1_1 and 20 kb for Neat1_2. Unlike other lncRNAs, which commonly lack sequence conservation, NEAT1 is relatively conserved across mammalian species, supporting its important biological function [19]. Indeed, the same structural subdomains in NEAT1 could be identified in 40 out of 64 mammalian species examined, with significant sequence similarity [23]. Moreover, the main structural features of the NEAT1 locus are also conserved in marsupials, despite variability of the primary DNA sequence [24].
Mouse Neat1 transcripts have been shown to be very unstable, with half-lives of ∼30 min and ∼60 min for Neat1_1 and Neat1_2, respectively [25]. Surprisingly, the same study found that human NEAT1 is ∼7 times more stable than mouse Neat1. The relative instability of NEAT1 may be critical for rapid cellular response to and subsequent recovery from stress.
Differential analysis of the two NEAT1 transcripts has proven challenging for three reasons: i) complete 5′-end overlap of the two isoforms; ii) presence of adenine stretches in the non-overlapping region of NEAT1_2, which impedes selective capture of the polyadenylated NEAT1_1 using an oligo-d(T) primer; iii) low NEAT1_2 yield when using conventional RNA extraction methods, due to reduced extractability of this isoform [26].
Both NEAT1 isoforms are expressed in the majority of cultured cells, with the exception of embryonic stem cells, which usually acquire paraspeckles after 4–5 days of differentiation [27]. In vivo, Neat1_1 is widely expressed, being present at high levels in most organs and tissues of the adult mouse. In contrast, Neat1_2 is only detected in some cell types in the digestive tract [28]. Both transcripts are also hardly expressed in the mouse embryo, suggesting a limited role for Neat1 in mammalian development [28]. Neat1_2 is undetectable in the adult nervous system [28,29], and human and murine postmitotic neurons cultured in vitro also have low levels of this isoform [30]. Data on NEAT1 expression in other cells in the CNS is scarce. Neat1 levels are low in cultured neuronal/oligodendrocyte progenitors while it becomes upregulated during differentiation [31].
2.3. The paraspeckle
The most well established function of NEAT1_2 is the assembly of the nuclear bodies paraspeckles. Multiple NEAT1_2 molecules line up to construct and maintain the spherical paraspeckle structure [21,32]. Paraspeckles are located in the interchromatin space of mammalian cell nuclei, usually on the border of splicing speckles [33,34]. NEAT1_2 but not NEAT1_1 is an essential component of paraspeckles, therefore the presence of NEAT1_2 is a reliable marker for paraspeckle appearance. Paraspeckle formation is absolutely dependent on RNA polymerase II transcription of NEAT1_2 and on the binding of paraspeckle proteins to this isoform [35]. NEAT1_2 provides a scaffold for >60 protein components and likely multiple RNA components including NEAT1_1 [[36], [37], [38]]. Paraspeckles are currently defined as foci containing both NEAT1_2 and an essential paraspeckle protein (e.g. SFPQ, NONO) [39]. Electron and super-resolution microscopic analyses revealed the orderly spatial arrangement of NEAT1_2 and multiple proteins within the paraspeckle, with a characteristic ‘core-shell’ structure [[39], [40], [41]]. A more recent study has mapped the minimal paraspeckle-forming region to the repeat-rich middle part of NEAT1_2 [42]. Computational analysis suggests that the architectural function of NEAT1_2 within paraspeckles is realised via long-range interactions and requires little structure conservation [23].
The majority of protein components of paraspeckles are RNA-binding proteins [37]. Some of them (SFPQ, NONO, RBM14, hnRNPK) are known to stabilise NEAT1_2, whereas others regulate the ratio of the two isoforms (CPSF6, NUDT21) or maintain secondary paraspeckle structure (FUS, DAZAP1). Another common structural feature of paraspeckle proteins is the presence of low complexity, prion-like domains enabling phase separation/liquid demixing [42,43], which is required for the formation of all types of RNA granules [44]. Removal or disruption of these prion-like domains in core paraspeckle proteins such as FUS and RBM14 is sufficient to dissipate paraspeckles [43,45].
Although paraspeckle proteome is relatively well-established, less information is available on the repertoire of RNA components of paraspeckles. NEAT1_1 is probably the second most abundant RNA within paraspeckles, after NEAT1_2. It has been estimated that each paraspeckle contains 6.5 NEAT1_1 and 53 NEAT1_2 molecules [26], and in stable cell lines, which almost all possess paraspeckles, NEAT1_1 is hardly detected outside these structures. Two structural classes of RNA species have been reported to be enriched in paraspeckles, namely, A-to-I hyperedited transcripts including those with inverted Alu repeats [46]; and miscellaneous of AG-rich RNAs [40]. Most recently, it has been shown that a distinct functional class of transcripts - mRNAs coding for mitochondrial proteins - are recruited to and retained within paraspeckles [47]. In addition, pri-mRNAs being processed by the Microprocessor may constitute a significant proportion of paraspeckle-anchored RNA [48].
It is believed that NEAT1_2/paraspeckles fulfil a number of functions independent of the presence of NEAT1_1. Among them: i) regulation of translation via nuclear retention of A-to-I hyperedited RNAs [27,46,49]; ii) regulation of transcription via sequestration of transcription factors, such as SFPQ [50,51]; iii) modulation of pri-miRNA processing [48]. Since NEAT1_1 is expressed in multiple cell types devoid of paraspeckles in vivo, including neurons [28], it is clear that it also has a variety of NEAT1_2/paraspeckle-independent functions. Recent advances in gene editing, which allow generation of cell lines with isoform-specific knockout [52], should drive forward our understanding of the functional significance of NEAT1_1.
Paraspeckles are stress-responsive nuclear bodies, in that NEAT1_2 upregulation and the increase in the size/number of paraspeckles accompany a number of physiological and pathological stressful conditions, such as differentiation [21] and inhibited proteasome function [50]. However, we are yet to establish the unifying feature(s) of stresses triggering the paraspeckle response.
3. NEAT1/paraspeckles regulate cellular pathways commonly affected in neurodegenerative diseases
Neurological disorders are defined as diseases of the central and peripheral nervous systems, and included within this category are age-related NDDs [53]. NDDs represent a large group of devastating, relentlessly progressive conditions with diverse clinical and pathological expressions, which are typically characterised by preferential loss of certain subsets of neurons in specific anatomical regions [54,55]. Among the hundreds of different NDDs, there are several common ones, including Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD) and Huntington's disease (HD), which collectively affect an estimated 50 million people globally. There is often little family history, and even among cases with familial aggregation, their genetic landscape can be highly heterogeneous. Polygenic/multifactorial nature of these conditions presents an obstacle to selection of specific molecular pathways for developing targeted therapeutics applicable across different disease subtypes.
Experimental evidence accumulated so far suggests that NEAT1 fine-tunes the function of multiple neurodegeneration-associated pathways, including critical ones, such as inflammation and neuronal apoptosis. Moreover, some of these pathways, e.g. mitochondrial signaling and miRNA biogenesis, are specifically regulated by NEAT1_2/paraspeckles. Below, we have summarised current knowledge of the role of NEAT1/paraspeckles in each of these pathways.
Apoptosis and the p53 pathway. Programmed cell death is the primary cause of neuron loss in NDDs, and both apoptosis and necroptosis contribute to this pathological process [56,57]. p53 is an established mediator of apoptosis in response to major insults in neurons [58]. Levels of p53 are generally increased in the affected CNS regions in NDDs, both in patients and in respective in vivo disease models [59]. In vivo studies confirmed that p53 acts as a positive regulator of disease pathogenesis in mouse models of NDDs [60,61].
Experimental data support the anti-apoptotic properties of NEAT1 and/or paraspeckles. Neat1 ablation renders mouse fibroblasts sensitive to cell death induced by proteasome inhibition [50]. Similarly, acute NEAT1 downregulation in human cells potentiates dsRNA cytotoxicity [30]. Furthermore, NEAT1 inhibits apoptosis under conditions of low oxygen and/or glucose [62,63]. Human and mouse NEAT1 are positively regulated by p53, and a p53 binding site on Neat1 promotor containing a consensus p53-binding element has been identified [64,65]. So far, studies of the interplay between NEAT1 and p53 have been carried out mainly in the context of cancer. Precise molecular mechanisms, including neurospecific ones, which underlie the co-operation of NEAT1 with the p53 pathway and other apoptotic factors/pathways still remain to be elucidated.
Proteotoxicity. Uncontrollable protein aggregation and inclusion formation is undoubtedly a hallmark of all NDDs [66]. Disease-associated proteins typically become misfolded and form amyloid or non-amyloid deposits in the affected CNS regions. Failure of the three main protein quality control systems, namely the molecular chaperone network, the ubiquitin-proteasome system and the autophagosome-lysosome pathway, will lead to proteostasis collapse in neurons and their rapid demise [[67], [68], [69], [70]].
Heat shock response is one of the main pro-survival pathways acting to maintain cellular protein homeostasis. Heat-shock proteins (HSPs) controlled by heat shock transcription factor 1 (HSF1) are molecular chaperones exerting anti-aggregational activity. Attenuated function of HSPs may be contributory in various NDDs [71], and HSF1 is proposed as a therapeutic target in these conditions [72]. Recently, a heat shock element (HSE) has been identified within NEAT1 promoter region, and HSF1-inducing stimuli have been shown to upregulate NEAT1 and enhance paraspeckle formation [73].
Proteasome inhibition is a well-established trigger of NEAT1 upregulation and paraspeckle hyper-assembly. Both NEAT1 isoforms are accumulated in cells treated with commonly used proteasome inhibitors bortezomib or MG132 [50]. Cells with enlarged paraspeckles may present with up to 50% nucleoplasmic depletion of paraspeckle proteins, many of which are transcription factors. Thus paraspeckles are likely to heavily contribute to the regulation of gene expression under these conditions. However, how exactly disrupted proteasome degradation signals to NEAT1 and paraspeckles is yet to be determined. One possible mechanism is that supressed proteasomal degradation of HSF1 drives the build-up of activated HSF1, which in turn augments NEAT1 synthesis [74].
Inflammatory response. Neuroinflammation, defined as activation and/or proliferation of resident immune cells in the CNS, is one of the central disease mechanisms in NDDs, as reviewed extensively elsewhere [75]. Genetic evidence also supports the major contribution of inflammatory pathways in the disease process. For example, genome-wide association studies (GWAS) found strong links between the inflammatory pathway genes and the risk of AD. As of today, over 20 genes bearing risk alleles have been identified in this pathway, and many of them are microglia-specific [76]. Genetic variants in TREM2, a gene which in the CNS is expressed only by microglia, have been associated with AD, FTD and PD [77].
NEAT1 involvement in the cellular response to pathogens is well established. In fact, Neat1_1 was first discovered as a virus-inducible non-coding RNA (VINC): a transcript undetectable in uninfected mouse brains was produced in response to Japanese encephalitis virus or Rabies virus infection [78]. NEAT1 roles in antiviral response, including via paraspeckles, have been confirmed in subsequent studies [51,79]. In virus-infected or dsRNA-stimulated HeLa cells, upregulated NEAT1 and enlarged paraspeckles act to sequester SFPQ away from promoters of cytokine and chemokine genes, such as IL8 or CCL5, which are repressed by SFPQ in the basal state. Importantly, the function of paraspeckles in this process was addressed using NEAT1_2 overexpression, confirming that the majority of the observed effects are mediated by paraspeckles. Subsequently, we showed that accumulation of endogenous dsRNA also promotes NEAT1 synthesis and paraspeckle assembly in human stable cell lines [30]. Another facet of NEAT1 function in the innate immunity is the regulation of cellular response to abnormal DNA species. Upon dsDNA exposure, NEAT1 binds to HEXIM1 protein leading to remodelling of a specific nuclear complex and activation of the cGAS-STING-IRF3 pathway and interferon signalling [80]. In other studies, NEAT1 was also found to positively regulate the expression of a group of cytokines and chemokines, including IL6 and CXCL10, in human monocytic cells in response to bacterial stimulation [81,82]. However, data from another study indicate that NEAT1 represses IL-1β and TNF-α production in the immortalized mouse neuronal line HT22 [63], pointing to possible cell type and/or species specificity of NEAT1 effects. While it is clear that NEAT1 upregulation is generally associated with positive regulation of immune signalling, currently we lack critical knowledge of the differential NEAT1 expression in the immune cells within the CNS. This information is required to instruct further studies of NEAT1 contribution to neuroinflammatory responses in vitro and in vivo.
Mitochondrial signalling. Defects in mitochondrial function accompany almost all NDDs [83]. Because of high energy demand, neurons are exceptionally sensitive to mitochondrial dysfunction. PD-causative mutations have been identified in the genes encoding proteins involved in mitochondrial quality control, parkin and PINK1 [84].
Recently, an unexpectedly strong mitochondria-paraspeckle crosstalk has been reported [47]. A large group of nuclear encoded genes regulating mitochondrial functions have been found to control NEAT1 levels and paraspeckle abundance via the transcription factor ATF2. Moreover, this effect is reciprocal, because elimination of paraspeckles is sufficient to cause mitochondrial abnormalities, including reduced respiration and ATP production as well as defective mitochondrial fission [47]. One can speculate that the observed effects of mitochondrial stress on paraspeckles could be at least in part mediated by the induction of immune response by damaged mitochondria [85]. Indeed, mitochondrial defects may lead to mtDNA instability, its escape to the cytosol and activation of cGAS-STING-IRF3 signalling responsive to foreign DNA, as described above.
Neurospecific pathways. NEAT1 function has been linked to neuronal excitability and axonal growth/maintenance. Abnormal excitability of the affected neuronal populations has been reported in ALS, AD and PD and likely represents an important disease mechanism common for many NDDs [86]. Hyperexcitability of upper and lower motor neurons has long been considered as one of the early disease signs in ALS [87,88].
NEAT1 is an activity-dependent transcript, as can be inferred from its higher expression in the “active”, high-spiking regions of the human brain and its responsiveness to depolarisation [89,90]. Moreover, since knockdown of NEAT1 in cultured human neurons leads to a significant increase in the expression of ion channel components, NEAT1 may control the transcription of genes encoding this class of proteins and act as a negative regulator of neuronal excitability [89]. The same study also reports that NEAT1 can directly bind to different potassium channel-interacting proteins, for example KCNAB2, important in reducing the excitability of neurons. NEAT1 is acutely downregulated in response to depolarisation, which is proposed to aid the translocation of KCNAB2 from nucleus to cytoplasm. In the cytoplasm, KCNAB2 would modulate the excitatory response through interacting with the membrane channels [89]. Notably, these effects are attributable to NEAT1_1 function, because this isoform is almost solely expressed in cultured postmitotic neurons used in the above experiments.
Unexpectedly, transcriptomic analysis of non-neuronal cells lacking Neat1 expression, Neat1 knockout fibroblasts, revealed that the genes most downregulated in these cells are involved in the nervous system development and function as well as axon guidance [64]. Indeed, studies in primary neuronal cultures showed that Neat1 knockdown decreases axonal growth, whereas Neat1 overexpression has the opposite effect [63].
miRNA biogenesis, chromatin remodelling and circadian rhythms. A number of other cellular pathways, whose dysfunction is less specific yet also relevant to NDDs, are modulated by NEAT1/paraspeckles.
Gene silencing mediated by miRNAs is the major post-transcriptional gene repression mechanism. Altered miRNA levels have been detected in the CNS in multiple NDDs, including AD, PD, HD, ALS and FTD [18,91]. The core Microprocessor component Drosha has been found to be abnormally aggregated in a subtype of ALS caused by repeat expansions in the C9ORF72 gene [92]. Moreover, genetic disruption of miRNA biogenesis in the mammalian nervous system is sufficient to cause a neurodegenerative phenotype. For example, ablation of the endoribonuclease Dicer, which processes miRNA precursors into mature miRNAs, in the adult mouse brain leads to hyperphosphorylation of tau protein and neuronal loss in the hippocampus, the two characteristic features of AD [93]. Furthermore, motor neuron specific knockout of Dicer results in an ALS-like phenotype in mice [94]. A recent study has revealed that NEAT1, as part of paraspeckles, functions to facilitate pri-miRNA processing [48]. This activity of paraspeckles is realised via bringing together multiple components of the nuclear miRNA machinery and scaffolding the Microprocessor complex. NEAT1_2 contains a pseudo pri-miRNA in its 3’ end, which attracts the Microprocessor and increases its local concentration. The role for NEAT1_1 in this process is less clear, however, it may potentiate NEAT1_2 function. A number of NEAT1-binding proteins, namely FUS, TDP-43, SFPQ and EWS, have been reported to modulate miRNA biogenesis [[95], [96], [97], [98]].
NDDs are generally characterised by a misbalance between the activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs) and resulting histone hypoacetylation and repressed chromatin state [99]. Abnormal signalling of CREB-binding protein (CBP), a HAT co-activator, is typical for AD, HD and ALS [100]. According to CHART-Seq results, NEAT1 binds active chromatin sites, whereas stimulation or inhibition of transcription alters the pattern of NEAT1 binding [101]. Furthermore, chromatin isolation by RNA purification (ChIRP) analysis showed that NEAT1 is recruited to gene promoters thereby supporting active chromatin state; it can do so via direct interaction with histone H3 [102]. NEAT1 might serve to recruit chromatin-modifying machinery and thus favour a chromatin landscape for active transcription. It should be noted however that both of the above studies did not distinguish between the two NEAT1 isoforms.
Disturbances of the physiological circadian rhythms, including sleep/awake cycle, core body temperature fluctuations and hormone release are among the earliest symptoms of NDDs [103]. Intriguingly, NEAT1 displays circadian expression patterns, and NEAT1/paraspeckles post-transcriptionally control the daily fluctuations in cellular protein levels through retaining inverted Alu repeat containing mRNAs in the nucleus [104].
In conclusion, since NEAT1_2 is virtually undetectable in the healthy CNS, currently it is of pivotal importance to determine the differential role of NEAT1_1 in the above pathways.
4. NEAT1/paraspeckles are dysregulated in neurodegenerative diseases: evidence from human tissue and disease models
Studies in human post-mortem tissue and experiments in in vitro and in vivo disease models have demonstrated that a neurodegenerative process is generally accompanied by altered NEAT1 levels. Below, we have summarised published information on NEAT1 expression in the affected parts of the CNS in patients and in rodent models of disease (a brief summary is also given in Table 1). The majority of studies report the expression levels of NEAT1 gene without accounting for the existence of the two isoforms with distinct functions. Thus, hereafter, we will refer to NEAT1 gene products as ‘NEAT1‘, clearly stating if the effects are isoform-specific, where and when such data are available.
Table 1.
NEAT1 expression in the CNS of patients and rodent models of neurodegenerative diseases.
| Disease | NEAT levels (and how measured) | Where measured | Proposed role | Proposed mechanism(s) | References |
|---|---|---|---|---|---|
| Amyotrophic lateral sclerosis (ALS) | Up (qRT-PCR, in situ hybridisation). Confirmed NEAT1_2 upregulation. |
Human: spinal motor neurons and glia Rodent: N/A |
Protective |
|
[30,109] |
| Frontotemporal dementia (FTD) | Up (iCLIP, qRT-PCR) |
Human: brain (cortex) Rodent: N/A |
Not known | Since NEAT1 levels were measured indirectly, TDP-43 binding to NEAT1 rather than the actual NEAT1 abundance could increase. | [111,115] |
| Huntington's disease (HD) | Up (microarrays, RNA-Seq, qRT-PCR) |
Human: brain (caudate) Rodent: R6/2 mouse model; brain |
Protective | MeCP2 protein represses NEAT1_2 under normal conditions; in HD, MeCP2 becomes downregulated which leads to NEAT1_2 accumulation. | [[118], [119], [120], [121], [122], [123]] |
| Parkinson's disease (PD) | Up (microarrays) |
Human: brain (substantia nigra) Rodent: MPTP mouse model; midbrain |
Detrimental | NEAT1 stabilises the PD-associated protein PINK1 and promotes PINK1-mediated autophagy. | [127,128] |
| Alzheimer's disease (AD) | Up (microarrays; RNA-Seq, qRT-PCR) |
Human: hippocampus, entorhinal cortex, middle temporal gyrus, posterior cingulate cortex, superior frontal gyrus Rodent: N/A |
Protective | NEAT1 positively regulates miR-15/107 which decreases CDK5R1 expression. | [[133], [134], [135]] |
| Epilepsy | Up (in humans; in rats - acute downregulation). Both isoforms are increased, with different temporal dynamics (qRT-PCR, in situ hybridisation) |
Human: cerebral cortex (neocortex, low and high activity regions) Rodent: rat models of pilocarpine or kainic acid-induced epilepsy (temporal lobe epilepsy) |
Dual |
|
[29,89,90] |
| Traumatic brain injury (TBI) | Up (qRT-PCR) |
Human: cerebral cortex Rodent: N/A |
Protective | Regulation of the inflammatory response following injury. NEAT1 overexpression reduces the production of IL-1β, TNF-α and nitric oxide and protects cells via regulation of apoptosis. | [63,138] |
4.1. Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTD)
ALS is a fatal adult-onset progressive neuromuscular disease affecting motor neurons in the spinal cord and motor cortex. Up to 90% of ALS cases are sporadic (sALS), and the remaining 10% have a familial history (familial ALS, fALS); so far, mutations in more >25 genes have been found to cause fALS and sALS [105,106]. FTD is characterised by frontal lobe pathology and hence decline of cognitive function. There is a significant genetic overlap between ALS and FTD; in addition, the two conditions can co-exist in the same individual, which has led to them being combined into one disease spectrum [107]. Significant proportion of ALS and FTD cases are caused by mutations in genes encoding RNA-binding proteins (RBPs) and/or involve functional deregulation of RBPs, which points to abnormal RNA metabolism as a common pathogenetic mechanism [108].
The first study to implicate NEAT1 in ALS pathogenesis reported enhanced paraspeckle formation in spinal motor neurons in a small cohort of sALS patients [109]. In view of the fact that NEAT1_2 expression is low in the adult nervous system, detection of paraspeckles in motor neurons was somewhat unexpected. Recently, we have confirmed that paraspeckle hyper-assembly is typical for motor neurons in ALS in a different cohort of sALS cases [30]. Furthermore, we also showed that this phenomenon is not restricted to sALS but is also observed in fALS cases caused by the C9ORF72 and TARDBP gene mutations. Mechanistically, augmented paraspeckle formation in ALS can be caused, at least in part, by nuclear depletion of TDP-43, the protein most commonly dysregulated in ALS (aggregated and lost from the nucleus in 95% of sALS cases) [110]. Indeed, TDP-43 binds NEAT1 [111,112], and its downregulation stimulates NEAT1_2 accumulation and paraspeckle assembly in cultured cells [30].
Genetic evidence also supports an important role for NEAT1/paraspeckles in ALS pathogenesis. Indeed, out of ∼25 proteins genetically associated with ALS/FTD, eight proteins are paraspeckle components, namely FUS, TDP-43, EWS, TAF15, SFPQ, MATR3, CREST and hnRNP A1, and some of them regulate NEAT1 levels and paraspeckle assembly (Table 2). In addition to the direct negative effect of a mutant protein on NEAT1/paraspeckles, its pathological aggregates can sequester other paraspeckle components. For example, inclusions of FUS protein in ALS caused by FUS gene mutations capture and retain proteins which regulate NEAT1 levels, such as NONO [45], whereas nuclear aggregates formed by mutant CREST can sequester FUS [113]. Furthermore, abnormal nuclear RNA foci formed by the ALS-causative expanded C9ORF72 repeats also sequester paraspeckle proteins hnRNPK, TDP-43 and EWS [114]. Therefore, abnormal levels, distribution, stability and interactions typical for mutant versions of these ALS-linked proteins may disrupt protective NEAT1/paraspeckle mediated signalling in ALS (Fig. 1).
Table 2.
Protein components of paraspeckles (NEAT1 interactors) genetically linked to ALS/FTD.
| Protein | Importance for paraspeckle assembly | Regulation of NEAT1_2 levels | Role in ALS | Role in other neurodegenerative diseases |
|---|---|---|---|---|
| FUS | Essential, >75% loss upon knockdown [37,45] | No or minimal | >50 mutations in fALS and sALS; FUS proteinopathy in these cases [143,144] | FTD (FTLD-FUS) [145] |
| TDP-43 | Depletion enhances paraspeckle assembly [30] | Yes (more NEAT1_2 upon TDP-43 depletion) | >60 mutations in fALS and sALS; TDP-43 proteinopathy in cases with TARDBP and C9ORF72 mutations and in 95% of all sALS cases [110,144,146] | FTD (FTD-TDP) [146]; AD [147] |
| TAF15 | Important, 30–75% loss upon knockdown [37] | No | 6 mutations in 6 unrelated sALS cases and 2 mutations – in 2 fALS cases [148,149] | FTD (FTD-FUS) [150] |
| EWS | Important, 30–75% loss upon knockdown [37] | Yes | 2 mutations in 2 unrelated sALS cases [151] | FTD (FTD-FUS) [150] |
| hnRNPA1 | Important, 30–75% loss upon knockdown [37] | No | 2 mutations in fALS cases; 2 rare variants [152,153] | Multisystem proteinopathy (MSP) [152] |
| CRESTa | ND | ND | 4 mutations in 4 unrelated sALS cases [154,155] | N/A |
| MATR3 | Depletion enhances paraspeckle assembly [156] | Yes (more NEAT1_2 upon MATR3 depletion) | ∼10 mutations in fALS and sALS cases [157,158] | Initially diagnosed myopathy with vocal cord paralysis, diagnosis changed to ‘ALS’ [159] |
| SFPQ | Essential, >75% loss upon knockdown [37] | Yes | 2 mutations in 2 sALS cases [160] | N/A |
neurospecific, effect on paraspeckles in stable cell lines could not be tested.
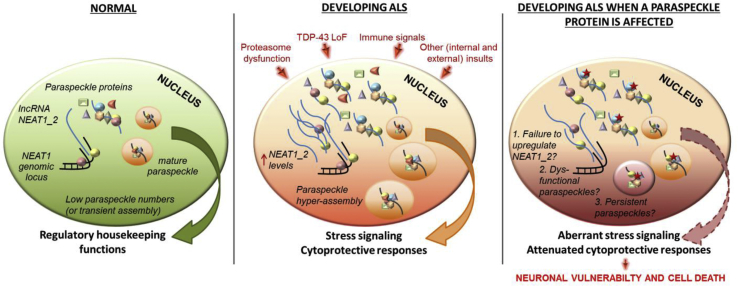
Fig. 1.
NEAT1/paraspeckles in ALS: a working model.
Left panel. Under basal conditions, levels of NEAT1_2 in motor neurons are low and so are the paraspeckle numbers. Paraspeckle assembly might also be transient (“on demand”).
Middle panel. During development of pathological changes typical for ALS, paraspeckle hyper-assembly is triggered by internal and external insults, such as TDP-43 loss of function (LoF), proteostasis collapse and immune response. Subsequent signalling events would enable protective neuronal response to stress and delay neuronal degeneration.
Right panel. In ALS cases with an essential/important paraspeckle protein affected by a mutation (see Table 2), its mutant isoform might negatively impact on protective paraspeckle hyper-assembly. This can be realised through: i) failure to upregulate NEAT1_2 (e.g. if proteins regulating NEAT1_2 levels, such as SFPQ and hnRNP K, are mutated or sequestered into abnormal inclusions/RNA foci); ii) attenuated assembly of paraspeckles or assembly of dysfunctional paraspeckles (e.g. if a structural paraspeckle protein, such as FUS, is mutated); iii) persistence of paraspeckles (e.g. if a mutation confers abnormal stability). Defective paraspeckle response may expedite the development of molecular pathology and accelerate disease onset and progression. A mutant protein is marked by a red star.
NEAT1 is one of the most upregulated transcripts in the brains of patients affected by FTD [111]. It should be noted however that this study measured TDP-43 binding to NEAT1 rather than absolute NEAT1 levels. Another study detected NEAT1 upregulation in the brain in a small cohort of FTD patients using qRT-PCR [115]. Upregulation of NEAT1_2 and paraspeckle formation is not typical for frontal cortex neurons in FTD (our unpublished observations). Studies into differences in NEAT1_2 regulation in motor and cortical neurons may shed light on the reasons behind selective vulnerability of neuronal populations in ALS and FTD.
4.2. Huntington's disease (HD)
HD is a hereditary NDD, of which 90% are familial cases caused by CAG trinucleotide repeat expansion in the HTT gene. HTT encodes a ubiquitously expressed protein with largely unknown functions. HD selectively affects GABAergic medium spiny neurons in the caudate nucleus and putamen, causing a series of symptoms including chorea, cognitive dysfunction and psychiatric disturbances. Healthy individuals have less than 36 CAG-repeats in HTT alleles, while affected individuals possess longer CAG repeats, with the length positively correlated with the severity of the phenotype [116]. Mutant huntingtin may cause neurodegeneration via a combination of transcriptional dysregulation, impaired clearance and toxicity of misfolded proteins, mitochondrial dysfunction, and oxidative stress [117].
Re-analysis of microarray data for the caudate from 44 HD cases and 36 controls [118] showed significant NEAT1 upregulation in the patient cohort [119]. Microarray analysis of another cohort of HD patients further confirmed elevated NEAT1 levels in the caudate of affected individuals [120]. RNA-Seq analysis of prefrontal cortex from 20 HD and 49 control individuals also found moderate, ∼20% upregulation of NEAT1 in HD [121]. A recent study, specifically focused on NEAT1_2, detected 3-fold increase of this isoform in the brain of HD patients by qRT-PCR [122]. Consistently, NEAT1 is overexpressed in the brains of R6/2 transgenic mice, a well-established HD model [120,123]. In cell models of HD, overexpressing mutant huntingtin, exogenous NEAT1_1 protected cells against H2O2-induced toxicity [120], whereas NEAT1_2 knockdown decreased cell survival [122].
4.3. Parkinson's disease (PD)
PD is characterised by a distinctive set of movement disorders including bradykinesia, tremor, rigidity and postural instability; psychiatric symptoms are present in a subset of patients. PD primarily affects the nigrostriatal dopaminergic system. The affected neurons develop Lewy bodies – neuronal inclusions composed of α-synuclein; this protein is also mutated in several forms of PD [124]. Other genes associated with PD include PINK1, DJ-1, LRRK2, and Parkin [125]. Similar to HD, mitochondrial damage, oxidative stress, excitotoxicity, protein misfolding and aggregation, and impaired protein clearance have all been implicated in PD [126].
Meta-analysis of a publicly available microarray dataset for substantia nigra (151 PD patients and 130 healthy controls) showed ∼1.5 upregulation of NEAT1 in PD patients [127]. Two in vivo studies report upregulation of NEAT1 in the midbrain of an MPTP mouse model of PD [128,129]. Contrary to the proposed protective effect of NEAT1 in other NDDs, in vitro and in vivo studies suggested that NEAT1 upregulation might play a detrimental role in PD. Neat1 knockdown prior to the MPTP injection in mice significantly increased survival of dopaminergic neurons [128]. NEAT1 knockdown also protected MPP + treated SH-SY5Y neuroblastoma cells from apoptosis [128,129].
4.4. Alzheimer's disease (AD)
AD is the most common NDD presenting with a progressive loss of cortical and hippocampal neurons and irreversible decline of cognitive and behavioural functions; 95% cases are sporadic [130,131]. AD is characterised by two distinctive pathological hallmarks: accumulation of extracellular neuritic plaques composed primarily of Aβ1−42, a cleavage product of the amyloid precursor protein (APP), and intracellular collections of neurofibrillary tangles composed of hyperphosphorylated species of tau protein [55]. The three main causative genes (APP, PSEN1, and PSEN2) and one common genetic risk factor (APOEε4 allele) have been identified. Large-scale GWAS studies found multiple additional risk loci for late-onset AD, with the associated genes mapping to the three principal pathways: cholesterol and lipid metabolism; immune system and inflammatory response; and endosomal vesicle cycling [132].
Microarray analysis of 87 AD and 74 control subjects showed NEAT1 overexpression across five different brain regions of AD patients, namely entorhinal cortex, hippocampus, middle temporal gyrus, posterior cingulate cortex and the superior frontal gyrus [133]. Statistically significant, ∼2.5-fold upregulation of NEAT1 in the hippocampus has been found in a small cohort of late-onset AD patients using RNA-Seq [134]. Another study also reported a 3-fold increase in NEAT1 in the temporal cortex and hippocampus of AD patients as measured by qRT-PCR [135].
4.5. Other diseases characterised by neurodegenerative process
A number of conditions outside the classical age-related NDD group are also characterised by progressive neurodegeneration and associated cognitive and physical impairement. Among them, NEAT1 dysregulation has been detected in epilepsy, traumatic brain injury (TBI) and drug addiction. Chromosomal 11q13 region containing NEAT1 locus was previously implicated in idiopathic seizures [136]. NEAT1 was found to be upregulated in the cerebral cortex in a small cohort of patients with temporal lobe epilepsy, specifically in high activity as compared to low activity regions [89,90]. However, in pilocarpine or kainic acid induced rat epilepsy models, Neat1 was transiently downregulated [89]. Authors propose that chronic upregulation of NEAT1 in neurons and inability to reduce its levels on demand may cause insensitivity to physiological activity rhythms and result in abnormal neuronal excitability. In another study, also in a mouse model of kainic acid induced seizures, the two Neat1 isoforms exhibited different post-seizure dynamics in the hippocampus, with acute Neat1_1 upregulation and delayed but prolonged Neat1_2 upregulation [29]. In this model, Neat1_2 was likely elevated specifically in neurons, whose density is very high in the hippocampus. Since Neat1_2 is undetectable in the brain at the basal state, its induction by activity changes, with a temporal pattern distinct from that of Neat1_1, is very intriguing and warrants further, in-depth studies.
TBI is characterised by neuronal loss and also recognised as a risk factor for NDDs [137]. NEAT1 levels in the cerebral cortex increase significantly following TBI, being nearly 3-fold higher one day after the injury [138]. Interestingly, NEAT1 has also been found upregulated in the brain of heroin users [139].
5. Conclusions and therapeutic implications
The hypothesis that NEAT1 and paraspeckles have important pathogenetic connections to age-related NDDs is gaining more credence. Yet we are just beginning to paint the picture of NEAT1 distribution, regulation and functions in the nervous system and of how they could be affected in NDDs. Carefully planned in vitro and in vivo experiments are still required to unambiguously establish whether, and if so, under which conditions NEAT1 isoforms may be neuroprotective. To begin to appreciate the spectrum of cellular effects of NEAT1 in NDDs, we need to elucidate: 1) cell-specific differences in NEAT1 expression in the CNS and whether this expression pattern is recapitulated in cultured cells; 2) NEAT1 roles in the normal nervous system at molecular, cellular, tissue and behavioural levels; 3) molecular mechanisms and temporal dynamics of NEAT1 regulation in neuronal populations affected in different NDDs; 4) differential roles for NEAT1_1 and NEAT1_2 in the protective neuronal responses. Although no gross neuropathological changes have been reported in the Neat1 knockout mouse model [28], thorough analysis of the CNS function of these mice complemented with studies of isolated primary neurons should provide important clues to the neurospecific-roles of NEAT1. Needless to say, we also need to carry out meta-analyses of available transcriptomic data from patients with different NDDs with a focus on lncRNAs.
Chemical modulation of NEAT1 expression as well as the use of antisense oligonucleotides (ASOs) to switch between the isoforms may become viable therapeutic approaches for this aetiologically diverse group of diseases. ASOs are currently being explored as possible therapeutic options for monogenic NDDs [140]. As a word of caution however, ASOs may affect NEAT1 and paraspeckles in an unconventional way. Indeed, phosphorothioate(PS)-modified ASOs form intranuclear structures which recruit paraspeckle proteins, ultimately leading to rapid NEAT1 decay [141]. Furthermore, the commonly used 2′-Fluoro modification of PS-ASOs have been found to cause degradation of the NEAT1-stabilising proteins NONO and SFPQ and to downregulate NEAT1 [142]. Therefore, further studies are needed to rule out possible impact of ASOs on NEAT1 levels.
Acknowledgements
The work was supported by grants from Medical Research Foundation (fellowship to TAS) and Motor Neurone Disease Association (senior non-clinical fellowship to TAS). HA is a recipient of Cardiff University/China Scholarship Council PhD studentship.
References
- 1.Dugger B.N., Dickson D.W. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol. 2017;9(7) doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatr. 2014;19(4):410–416. doi: 10.1038/mp.2013.196. [DOI] [PubMed] [Google Scholar]
- 4.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs J.A. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88(5):861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 6.Ng S.Y. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. : TIG (Trends Genet.) 2013;29(8):461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Qureshi I.A., Mehler M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13(8):528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H., Yang L., Chen L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. : TIG (Trends Genet.) 2017;33(8):540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 12.Kapranov P. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 13.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Quinodoz S., Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24(11):651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L.L., Carmichael G.G. Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 2010;22(3):357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riva P., Ratti A., Venturin M. The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 2016;13(11):1219–1231. doi: 10.2174/1567205013666160622112234. [DOI] [PubMed] [Google Scholar]
- 18.Salta E., De Strooper B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 2017;18(10):627–640. doi: 10.1038/nrn.2017.90. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson J.N. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guru S.C. A 2.8-Mb clone contig of the multiple endocrine neoplasia type 1 (MEN1) region at 11q13. Genomics. 1997;42(3):436–445. doi: 10.1006/geno.1997.4783. [DOI] [PubMed] [Google Scholar]
- 21.Sunwoo H. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilusz J.E. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018;46(7):3742–3752. doi: 10.1093/nar/gky046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornelis G. Functional conservation of the lncRNA NEAT1 in the ancestrally diverged marsupial lineage: evidence for NEAT1 expression and associated paraspeckle assembly during late gestation in the opossum Monodelphis domestica. RNA Biol. 2016;13(9):826–836. doi: 10.1080/15476286.2016.1197482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark M.B. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chujo T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017;36(10):1447–1462. doi: 10.15252/embj.201695848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa S. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bluthgen N. Profiling the MAPK/ERK dependent and independent activity regulated transcriptional programs in the murine hippocampus in vivo. Sci. Rep. 2017;7:45101. doi: 10.1038/srep45101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shelkovnikova T.A. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018;13(1):30. doi: 10.1186/s13024-018-0263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer T.R. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki Y.T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U. S. A. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen J.S. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12(1):1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 34.Fox A.H. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12(1):13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 35.Mao Y.S. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011;13(1):95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamazaki T., Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci. (Online) 2015;7:1–41. doi: 10.2741/715. [DOI] [PubMed] [Google Scholar]
- 37.Naganuma T. Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31(20):4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemson C.M. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox A.H. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018;43(2):124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 40.West J.A. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016;214(7):817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Souquere S. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell. 2010;21(22):4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70(6):1038–1053 e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Hennig S. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. JCB (J. Cell Biol.) 2015;210(4):529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeynaems S. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28(6):420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shelkovnikova T.A. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum. Mol. Genet. 2014;23(9):2298–2312. doi: 10.1093/hmg/ddt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L.L., DeCerbo J.N., Carmichael G.G. Alu element-mediated gene silencing. EMBO J. 2008;27(12):1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018;20(10):1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 48.Jiang L. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017;24(10):816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prasanth K.V. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 50.Hirose T. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell. 2014;25(1):169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imamura K. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Li R. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23(6):872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pringsheim T., Fiest K., Jette N. The international incidence and prevalence of neurologic conditions: how common are they? Neurology. 2014;83(18):1661–1664. doi: 10.1212/WNL.0000000000000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Przedborski S., Vila M., Jackson-Lewis V. Neurodegeneration: what is it and where are we? J. Clin. Invest. 2003;111(1):3–10. doi: 10.1172/JCI17522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erkkinen M.G., Kim M.O., Geschwind M.D. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb Perspect Biol. 2018;10(4) doi: 10.1101/cshperspect.a033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venderova K., Park D.S. Programmed cell death in Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2(8) doi: 10.1101/cshperspect.a009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattson M.P. Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 2000;1(2):120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 58.Culmsee C., Mattson M.P. p53 in neuronal apoptosis. Biochem. Biophys. Res. Commun. 2005;331(3):761–777. doi: 10.1016/j.bbrc.2005.03.149. [DOI] [PubMed] [Google Scholar]
- 59.Chang J.R. Role of p53 in neurodegenerative diseases. Neurodegener. Dis. 2012;9(2):68–80. doi: 10.1159/000329999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alves da Costa C. Presenilin-dependent gamma-secretase-mediated control of p53-associated cell death in Alzheimer's disease. J. Neurosci. 2006;26(23):6377–6385. doi: 10.1523/JNEUROSCI.0651-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bae B.I. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47(1):29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Choudhry H. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34(34):4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong J. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav. Immun. 2017;65:183–194. doi: 10.1016/j.bbi.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Mello S.S. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31(11):1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adriaens C. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 66.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 67.Chen B. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3(8):a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ihara Y., Morishima-Kawashima M., Nixon R. The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(8) doi: 10.1101/cshperspect.a006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 71.San Gil R. The heat shock response in neurons and astroglia and its role in neurodegenerative diseases. Mol. Neurodegener. 2017;12(1):65. doi: 10.1186/s13024-017-0208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neef D.W., Jaeger A.M., Thiele D.J. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 2011;10(12):930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lellahi S.M. The long non-coding RNA NEAT1 and nuclear paraspeckles are upregulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 2018 doi: 10.1074/jbc.RA118.004473. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raychaudhuri S. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 2014;156(5):975–985. doi: 10.1016/j.cell.2014.01.055. [DOI] [PubMed] [Google Scholar]
- 75.Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 76.Sims R. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat. Genet. 2017;49(9):1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jay T.R., von Saucken V.E., Landreth G.E. TREM2 in neurodegenerative diseases. Mol. Neurodegener. 2017;12(1):56. doi: 10.1186/s13024-017-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saha S., Murthy S., Rangarajan P.N. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87(Pt 7):1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Q. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4(1):e00596–12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morchikh M. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell. 2017;67(3):387–399 e5. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 81.Zhang F. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 82.Chen D.D. NEAT1 contributes to ox-LDL-induced inflammation and oxidative stress in macrophages through inhibiting miR-128. J. Cell. Biochem. 2018 doi: 10.1002/jcb.27541. [DOI] [PubMed] [Google Scholar]
- 83.Johri A., Beal M.F. Mitochondrial dysfunction in neurodegenerative diseases. J. Pharmacol. Exp. Therapeut. 2012;342(3):619–630. doi: 10.1124/jpet.112.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narendra D.P., Youle R.J. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxidants Redox Signal. 2011;14(10):1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.West A.P. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saxena S., Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71(1):35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 87.Vucic S., Nicholson G.A., Kiernan M.C. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain. 2008;131(Pt 6):1540–1550. doi: 10.1093/brain/awn071. [DOI] [PubMed] [Google Scholar]
- 88.Wainger B.J. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7(1):1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barry G. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017;7:40127. doi: 10.1038/srep40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lipovich L. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192(3):1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karnati H.K. miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheim. Dis. : JAD. 2015;48(3):563–580. doi: 10.3233/JAD-150395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Porta S. Drosha inclusions are new components of dipeptide-repeat protein aggregates in FTLD-TDP and ALS C9orf72 expansion cases. J. Neuropathol. Exp. Neurol. 2015;74(4):380–387. doi: 10.1097/NEN.0000000000000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hebert S.S. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 2010;19(20):3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 94.Haramati S. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. U. S. A. 2010;107(29):13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morlando M. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31(24):4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawahara Y., Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109(9):3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ouyang H. The RNA binding protein EWS is broadly involved in the regulation of pri-miRNA processing in mammalian cells. Nucleic Acids Res. 2017;45(21):12481–12495. doi: 10.1093/nar/gkx912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bottini S. Post-transcriptional gene silencing mediated by microRNAs is controlled by nucleoplasmic Sfpq. Nat. Commun. 2017;8(1):1189. doi: 10.1038/s41467-017-01126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saha R.N., Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13(4):539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mirabella A.C., Foster B.M., Bartke T. Chromatin deregulation in disease. Chromosoma. 2016;125(1):75–93. doi: 10.1007/s00412-015-0530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.West J.A. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chakravarty D. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hood S., Amir S. Neurodegeneration and the circadian clock. Front. Aging Neurosci. 2017;9:170. doi: 10.3389/fnagi.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torres M. Circadian RNA expression elicited by 3'-UTR IRAlu-paraspeckle associated elements. Elife. 2016;5 doi: 10.7554/eLife.14837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hardiman O. The changing picture of amyotrophic lateral sclerosis: lessons from European registers. J. Neurol. Neurosurg. Psychiatry. 2017;88(7):557–563. doi: 10.1136/jnnp-2016-314495. [DOI] [PubMed] [Google Scholar]
- 106.Renton A.E., Chio A., Traynor B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taylor J.P., Brown R.H., Jr., Cleveland D.W. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao F.B., Almeida S., Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 2017;36(20):2931–2950. doi: 10.15252/embj.201797568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nishimoto Y. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013;6(1):31. doi: 10.1186/1756-6606-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mackenzie I.R.A., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9(10):995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 111.Tollervey J.R. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14(4):452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Polymenidou M. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14(4):459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kukharsky M.S. Calcium-responsive transactivator (CREST) protein shares a set of structural and functional traits with other proteins associated with amyotrophic lateral sclerosis. Mol. Neurodegener. 2015;10:20. doi: 10.1186/s13024-015-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee Y.B. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5(5):1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsuiji H. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol. Med. 2013;5(2):221–234. doi: 10.1002/emmm.201202303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89(5):910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 117.Labbadia J., Morimoto R.I. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem. Sci. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hodges A. Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 119.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 120.Sunwoo J.S. Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol. Neurobiol. 2017;54(2):1577–1586. doi: 10.1007/s12035-016-9928-9. [DOI] [PubMed] [Google Scholar]
- 121.Labadorf A.T., Myers R.H. Evidence of extensive alternative splicing in post mortem human brain HTT transcription by mRNA sequencing. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141298. e0141298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng C. The long noncoding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease-gene dependent toxicities. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy331. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chanda K. Altered levels of long NcRNAs Meg3 and Neat1 in cell and animal models of Huntington's disease. RNA Biol. 2018:1–16. doi: 10.1080/15476286.2018.1534524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Venda L.L. alpha-Synuclein and dopamine at the crossroads of Parkinson's disease. Trends Neurosci. 2010;33(12):559–568. doi: 10.1016/j.tins.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2(1):a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maiti P., Manna J., Dunbar G.L. Current understanding of the molecular mechanisms in Parkinson's disease: targets for potential treatments. Transl. Neurodegener. 2017;6:28. doi: 10.1186/s40035-017-0099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mariani E. Meta-analysis of Parkinson's disease transcriptome data using TRAM software: whole substantia nigra tissue and single dopamine neuron differential gene expression. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0161567. e0161567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yan W. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018;496(4):1019–1024. doi: 10.1016/j.bbrc.2017.12.149. [DOI] [PubMed] [Google Scholar]
- 129.Liu Y., Lu Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson's disease induced by MPTP/MPP+ via regulation of gene expression. Clin. Exp. Pharmacol. Physiol. 2018;45(8):841–848. doi: 10.1111/1440-1681.12932. [DOI] [PubMed] [Google Scholar]
- 130.Reitz C., Brayne C., Mayeux R. Epidemiology of alzheimer disease. Nat. Rev. Neurol. 2011;7(3):137–152. doi: 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Masters C.L. Alzheimer's disease. Nat Rev Dis Primers. 2015;1:15056. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- 132.Moustafa A.A. Genetic underpinnings in Alzheimer's disease - a review. Rev. Neurosci. 2018;29(1):21–38. doi: 10.1515/revneuro-2017-0036. [DOI] [PubMed] [Google Scholar]
- 133.Puthiyedth N. Identification of differentially expressed genes through integrated study of Alzheimer's disease affected brain regions. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0152342. e0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Annese A. Whole transcriptome profiling of Late-Onset Alzheimer's Disease patients provides insights into the molecular changes involved in the disease. Sci. Rep. 2018;8(1):4282. doi: 10.1038/s41598-018-22701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spreafico M. Multiple layers of CDK5R1 regulation in Alzheimer's disease implicate long non-coding RNAs. Int. J. Mol. Sci. 2018;19(7) doi: 10.3390/ijms19072022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hempelmann A. Exploration of the genetic architecture of idiopathic generalized epilepsies. Epilepsia. 2006;47(10):1682–1690. doi: 10.1111/j.1528-1167.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 137.Gupta R., Sen N. Traumatic brain injury: a risk factor for neurodegenerative diseases. Rev. Neurosci. 2016;27(1):93–100. doi: 10.1515/revneuro-2015-0017. [DOI] [PubMed] [Google Scholar]
- 138.Zhong J. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016;1646:589–600. doi: 10.1016/j.brainres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 139.Michelhaugh S.K. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 2011;116(3):459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schoch K.M., Miller T.M. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94(6):1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shen W., Liang X.H., Crooke S.T. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42(13):8648–8662. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shen W. 2'-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 2015;43(9):4569–4578. doi: 10.1093/nar/gkv298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deng H.X. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann. Neurol. 2010;67(6):739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lattante S., Rouleau G.A., Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum. Mutat. 2013;34(6):812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- 145.Neumann M. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132(Pt 11):2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Neumann M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 147.Wilson A.C. TDP-43 in aging and Alzheimer's disease - a review. Int. J. Clin. Exp. Pathol. 2011;4(2):147–155. [PMC free article] [PubMed] [Google Scholar]
- 148.Couthouis J. A yeast functional screen predicts new candidate ALS disease genes. Proc. Natl. Acad. Sci. U. S. A. 2011;108(52):20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ticozzi N. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(3):285–290. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 150.Neumann M. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain : J. Neurol. 2011;134(Pt 9):2595–2609. doi: 10.1093/brain/awr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Couthouis J. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2012;21(13):2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim H.J. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Q. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology. 2016;87(17):1763–1769. doi: 10.1212/WNL.0000000000003256. [DOI] [PubMed] [Google Scholar]
- 154.Chesi A. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16(7):851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Teyssou E. Genetic analysis of SS18L1 in French amyotrophic lateral sclerosis. Neurobiol. Aging. 2014;35(5):1213e9–1213e12. doi: 10.1016/j.neurobiolaging.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 156.Banerjee A. Nuclear poly(A) binding protein 1 (PABPN1) and Matrin3 interact in muscle cells and regulate RNA processing. Nucleic Acids Res. 2017;45(18):10706–10725. doi: 10.1093/nar/gkx786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnson J.O. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 2014;17(5):664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leblond C.S. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2016;37:209e17–209e21. doi: 10.1016/j.neurobiolaging.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 159.Senderek J. Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am. J. Hum. Genet. 2009;84(4):511–518. doi: 10.1016/j.ajhg.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thomas-Jinu S. Non-nuclear pool of splicing factor SFPQ regulates axonal transcripts required for normal motor development. Neuron. 2017;94(4):931. doi: 10.1016/j.neuron.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]