Abstract
Respiratory disease caused by Mycoplasma ovipneumoniae and Pasteurellaceae poses a formidable challenge for bighorn sheep (Ovis canadensis) conservation. All-age epizootics can cause 10–90% mortality and are typically followed by multiple years of enzootic disease in lambs that hinders post-epizootic recovery of populations. The relative frequencies at which these epizootics are caused by the introduction of novel pathogens or expression of historic pathogens that have become resident in the populations is unknown. Our primary objectives were to determine how commonly the pathogens associated with respiratory disease are hosted by bighorn sheep populations and assess demographic characteristics of populations with respect to the presence of different pathogens. We sampled 22 bighorn sheep populations across Montana and Wyoming, USA for Mycoplasma ovipneumoniae and Pasteurellaceae and used data from management agencies to characterize the disease history and demographics of these populations. We tested for associations between lamb:ewe ratios and the presence of different respiratory pathogen species. All study populations hosted Pasteurellaceae and 17 (77%) hosted Mycoplasma ovipneumoniae. Average lamb:ewe ratios for individual populations where both Mycoplasma ovipneumoniae and Pasteurellaceae were detected ranged from 0.14 to 0.40. However, average lamb:ewe ratios were higher in populations where Mycoplasma ovipneumoniae was not detected (0.37, 95% CI: 0.27–0.51) than in populations where it was detected (0.25, 95% CI: 0.21–0.30). These findings suggest that respiratory pathogens are commonly hosted by bighorn sheep populations and often reduce recruitment rates; however ecological factors may interact with the pathogens to determine population-level effects. Elucidation of such factors could provide insights for management approaches that alleviate the effects of respiratory pathogens in bighorn sheep. Nevertheless, minimizing the introduction of novel pathogens from domestic sheep and goats remains imperative to bighorn sheep conservation.
Introduction
Most large-mammal populations across western North America were decimated following settlement by Europeans, though restoration efforts since the early 20th Century have been particularly successful in reestablishing ungulates in areas where suitable habitat exists [1]. Although management actions have increased bighorn sheep (Ovis canadensis) numbers and distribution, the species’ abundance is still less than 10% of historic levels [2]. Currently, most bighorn sheep populations are small and patchily distributed [3], making them more vulnerable to apparent competition [4], predation [5], inbreeding depression [6], and a variety of other threats that collectively amount to the Allee effect [7]. Moreover, respiratory disease has hindered recovery efforts and dictates most bighorn sheep management policies [8].
Respiratory disease epizootics are typically first expressed as all-age mortality events followed by multiple years of enzootic disease that causes high summer mortality of lambs [9]; though the severity and duration of the disease varies. Outcomes following respiratory disease epizootics in bighorn sheep populations range from full recovery to local extinction [10] and long-term recovery is heavily driven by lamb recruitment [11]. Domestic sheep (Ovis aries) typically host pathogens that cause this disease in bighorn sheep and experiments commingling domestic sheep with bighorn sheep have collectively resulted in a 98% mortality rate for bighorn sheep [12–14]. Evidence also suggests domestic goats (Capra aegagrus hircus) pose a disease risk for bighorn sheep, though the risk appears to be less severe than that posed by domestic sheep [13,15,16]. There is also some evidence that cattle (Bos taurus) pose a disease risk [13,17], though cattle are not a natural host for all pathogens associated with respiratory disease in bighorn sheep [18]. Accordingly, disease management efforts have focused on eliminating contact between bighorn sheep and livestock, particularly domestic sheep and goats.
Respiratory disease in bighorn sheep is polymicrobial, with both Mycoplasma ovipneumoniae (M. ovipneumoniae) and Pasteurellaceae species contributing to disease [18]. The natural host range of M. ovipneumoniae is limited to Caprinae species (including domestic sheep and goats) and this bacterium has been associated with respiratory disease epizootics in bighorn sheep [19,20]. Experimental M. ovipneumoniae inoculation into domestic sheep co-penned with bighorn sheep resulted in epizootic respiratory disease in bighorn sheep, but not domestic sheep [21]. Commingling of bighorn sheep with domestic sheep that had no evidence of exposure to M. ovipneumoniae resulted in lower mortality of bighorn sheep [22]. Pasteurellaceae species including leukotoxigenic Mannheimia haemolytica (M. haemolytica), leukotoxigenic Bibersteinia trehalosi (B. trehalosi), and Pasteurella multocida (P. multocida) are also regularly detected in the lungs of bighorn sheep that have succumbed to respiratory disease [23–27], and direct inoculation of leukotoxigenic M. haemolytica or B. trehalosi produces fatal pneumonia in bighorn sheep, but not domestic sheep [28]. P. multocida has been associated with respiratory disease epizootics [20], but not experimentally shown to cause disease.
Rothman’s sufficient-component cause model of disease [29] suggests there are a minimum set of factors and conditionals that cause disease. The history of respiratory disease in bighorn sheep strongly suggests that pathogens originating from domestic livestock are a necessary component of this disease [30] and experimental evidence suggests that introduction of these pathogens to immunologically naïve bighorn sheep is a very strong risk factor, if not sufficient, for epizootics. Other sufficient causes for respiratory disease might require specific ecological conditions along with resident pathogens already circulating in a population [31]. Conversely, enzootic lamb pneumonia in populations that typically follows epizootics could cease or subside due to extinction of pathogens or removal of other component causes related to interactions between the hosts, pathogens, and environment. The identification of pathogen species linked to respiratory disease in bighorn sheep populations without indication of active disease [18] suggests post-epizootic population recovery may result from removal of non-pathogen component causes of the disease. In such populations, variation in host-pathogen interactions (e.g., increases in pathogen virulence, transmission rates, or the accumulation of susceptible individuals) may periodically lead to conditions sufficient for respiratory disease in adults and/or lambs [32]. It follows that characteristics of a population or its environment can influence susceptibility to disease.
Detection error of diagnostic protocols used to detect pathogens, along with a propensity for research to focus on diseased populations, has affected the ability to understand the etiology of respiratory disease in bighorn sheep [19,25,33]. Only recently have molecular and statistical tools to reduce and measure, respectively, detection error of diagnostic protocols been developed and implemented [34–36]. As such, there has been little ability to accurately monitor resident respiratory pathogen communities in bighorn sheep populations. The relative proportion of epizootics involving the introduction of novel pathogens versus resident pathogens is unknown as are other (non-pathogen) causal components of respiratory disease.
The effects of respiratory disease on demographic rates of bighorn sheep, such as recruitment, adult survival, and population growth have been described [37,9,38,11]. However, the relationship between the presence of the pathogens associated with respiratory disease (Pasteurellaceae or M. ovipneumoniae) and population performance has not been well described. Such an assessment could shed light on the relative strength of pathogen versus ecological components as risk factors for respiratory disease. The presence of respiratory pathogens in bighorn sheep populations with strong demographic performance is consistent with ecological components playing a role in causing this disease. Identification of such components could lead to additional disease management strategies that mitigate the effects of resident or novel respiratory pathogens, thereby increasing resilience of bighorn sheep populations to disease.
We assessed bacterial respiratory pathogen communities in a variety of bighorn sheep populations across Montana and Wyoming and related population characteristics and recruitment rates to the presence of Pasteurellaceae and M. ovipneumoniae. The primary objectives of this study were to: 1) assess how common the respiratory pathogens are among bighorn sheep populations; 2) quantify the probability of presence of undetected pathogens by applying recently developed estimates of detection probability for diagnostic protocols used to detect them [36]; 3) assess whether the presence of any specific pathogen or combination of pathogens was associated with population characteristics or recruitment rates; and 4) assess how frequently populations exhibited characteristics indicative of stability and viability in the presence of a given pathogen. Given the long history of domestic sheep grazing in the two states [30], we predicted the respiratory pathogens would be hosted by the majority of study populations. We suspected the effect of respiratory pathogens on demographic performance depended on ecological factors. This combined with our first prediction led us to predict no association between demographic performance of our study populations and presence of respiratory pathogens.
Methods
Ethics statement
Capture and handling of animals reported herein comply with scientific guidelines and permits acquired from the State of Montana, the State of Wyoming, and the National Park Service. All animal capture and handling protocols were approved by Institutional Animal Care and Use Committees at Montana State University (Permit # 2011–17, 2014–32), Montana Department of Fish, Wildlife, and Parks (Permit # 2016–005), National Park Service (Permit # NPS 2014.A3), or Wyoming Game and Fish Department (Permit # 854).
Study populations
We sampled 22 bighorn sheep populations that occupied varying habitat types across Montana and Wyoming and had varying disease and management histories (Table 1). We defined populations to correspond with the survey and reporting methods of their respective wildlife management agencies. Most study populations were geographically separated from other study populations, but populations in the Absaroka Range of Wyoming (n = 6), the Rocky Mountain Front of Montana (n = 2), and the upper Missouri River Breaks of Montana (n = 2) had less geographic separation from neighboring populations. Topography in the ranges of the study populations varies from rugged badlands, with 300 meters of vertical relief to large mountain ranges with over 2,000 meters of vertical relief. Annual precipitation in the ranges varies from 30 cm to over 100 cm. Inhabited ecological regions included northern Rocky Mountains, northern Rocky Mountain Foothills, northern rolling plains, and central Rocky Mountains [39]. Study populations primarily occupied public lands but many also used private lands to varying degrees.
Table 1. General attributes of the 22 bighorn sheep populations investigated in this study.
| Population | Hunt-Area | Estimated pop. size | All-age epizootics | Origin | Sub-pops1 | Connectivity level | Migratory | Ecological region2 |
|---|---|---|---|---|---|---|---|---|
| Galton | MT-102 | 70 | None | Native | 2 | Limited | Yes | N. Rocky Mtn. |
| Perma-Paradise | MT-124 | 325 | None | Restored | 2 | Isolated | No | N. Rocky Mtn. |
| Petty Creek | MT-203 | 160 | None | Restored | 2 | Isolated | No | N. Rocky Mtn. |
| Lost Creek | MT-213 | 100 | 1991,2010 | Restored | 2 | Limited | Yes | Central Rocky Mtn. |
| Highlands3 | MT-340 | 75 | 1995 | Restored | 3 | Isolated | No | Central Rocky Mtn. |
| Sun Canyon4 | MT-499 | 150 | 1925, 1932, 1984, 2010 | Native | 3 | Meta Population | Yes | Central Rocky Mtn./ N. Rocky Mtn. Fthls |
| Gibson Lake North | MT-423 | 100 | 1925, 1932, 1984, 2010 | Native | 5 | Meta Population | Yes | Central Rocky Mtn./ N. Rocky Mtn. Fthls |
| Fergus | MT-482 | 545 | None | Restored | 4 | Meta Population | No | N. Rolling Plains |
| Choteau-Blaine | MT-680 | 770 | None | Restored | 10 | Meta Population | No | N. Rolling Plains |
| Middle Missouri Breaks | MT-622 | 400 | None | Restored | 3 | Isolated | No | N. Rolling Plains |
| Hilgard5 | MT-302 | 280 | 1997 | Augmented-Native | 3 | Isolated | Yes | Central Rocky Mtn. |
| Upper Yellowstone | MT-3995 | 320 | 2012, 2014 | Native | 8 | Meta Population | Yes | Central Rocky Mtn. |
| Stillwater | MT-500a | 75 | Pre-1920 | Native | 2 | Limited | Yes | Central Rocky Mtn. |
| Clark’s Fork | WY-1 | 600 | None | Native | >10 | Meta Population | Yes | Central Rocky Mtn. |
| Trout Peak | WY-2 | 700 | None | Native | >10 | Meta Population | Yes | Central Rocky Mtn. |
| Wapiti Ridge | WY-3 | 850 | None | Native | >10 | Meta Population | Yes | Central Rocky Mtn. |
| Yount’s Peak | WY-4 | 875 | 2011–2013 | Native | >10 | Meta Population | Yes | Central Rocky Mtn. |
| Franc’s Peak | WY-5 | 710 | 2011–2013 | Native | >10 | Meta Population | Yes | Central Rocky Mtn. |
| Dubois Badlands | WY-22 | None | Native | 2 | Meta Population | Yes | Central Rocky Mtn. | |
| Targhee | WY-6 | 80 | None | Native | 2 | Isolated | No | Central Rocky Mtn |
| Whiskey Mtn. | WY-10 | 850 | 1991 | Native | >5 | Meta Population | Yes | Central Rocky Mtn. |
| Jackson | WY-7 | 425 | 2002, 2012 | Native | 6 | Meta Population | Yes | Central Rocky Mtn. |
1 Subpopulation defined as a group of animals within a population that shares a distinct winter range that may be composed of multiple social groups.
2 Ecological region defined according to USDA Handbook 296 [39]
3. Recent demographic data were not available for the Highlands populations
4. Population was monitored as a single population but inhabits multiple hunting districts.
5. One subpopulation was established from another subpopulation in 2015
Study populations included native populations (n = 14), restored populations (n = 7), and augmented populations (n = 1). Some were part of metapopulations (n = 13), while others had limited connectivity (n = 3) or were mostly isolated (n = 6). Population structure ranged from one subpopulation to over 10 subpopulations. Subpopulations were defined as groups of animals sharing distinct winter ranges that may be composed of multiple social groups. Respiratory disease histories also varied among populations: 11 had no history of confirmed respiratory disease, six had a single all-age epizootic, and five others had multiple all-age epizootics. Estimated abundance in individual populations ranged from 70 to 875 animals (Table 1). Rams were harvested from all study populations with average annual ram harvest ranging from 0.4 to 55. Ewes were harvested from seven study populations (Table A in S1 Appendix) with mean annual ewe harvest from these populations ranging from <1 to 24. Density-reduction translocations had occurred in two study populations and augmentation translocations occurred in two study populations since 2011 (Table A in S1 Appendix).
Demographic and population characteristic data collection
Routine monitoring data from wildlife management agencies provided the most rigorous and consistently collected data available for most populations. Demographic data used in these analyses were primarily collected by Montana Department of Fish, Wildlife & Parks (FWP) and Wyoming Game & Fish Department (WGFD) personnel as part of annual trend counts and age/sex classifications from 2006 to 2017, typically during January-February or March-April. For one study population (Fergus), we used trend counts conducted during the summer (July-August) and classification counts that were collected during the spring (April-May). Classification and trend counts were conducted exclusively during the summer for one population (Choteau-Blaine), which was excluded from the recruitment analysis. Winter classification counts, but not trend counts, were conducted on a near-annual basis for five study populations (Clark’s Fork, Trout Peak, Wapiti Ridge, Yount’s Peak, Franc’s Peak). The majority of population surveys were conducted from an aerial platform, though surveys for several populations with easily accessible winter ranges were conducted from the ground (Sun Canyon, Gibson Lake North, Hilgard, Lost Creek, and Stillwater). Recruitment data were obtained from classification counts of individuals within a consistent area of core winter range across years. If populations were surveyed multiple times within the same season, the maximum number of animals within each age and sex class observed across surveys was recorded as the counts for that year. Biologists overseeing the study populations were queried to censor poor surveys and obtain information regarding population traits including abundance objectives, 10-year trends, number of subpopulations, connectivity level, as well as history of respiratory disease.
Indices of demographic performance
We characterized demographic performance of study populations by mean recruitment rates, whether they were at or above abundance objectives, and whether they had a growing or stable population trend. We indexed recruitment rates as the ratio of lambs to ewes (lamb:ewe ratio) counted during winter or spring classification surveys. If mean lamb:ewe ratios of a population were ≥ 0.2, we considered average recruitment of the population satisfactory [40]; population size was satisfactory if it was at or above the management objective; and population trend was satisfactory if growth was stable or increasing. We split data from populations that experienced all-age epizootics within the time-frame recruitment data were collected into pre-epizootic and post-epizootic categories for analysis if pathogen data were collected both before and after epizootics. If pathogen data were not collected before epizootics, we excluded demographic data preceding the die-off from analysis. We omitted the Highlands population from the demographic analyses because recent demographic data were not available.
Animal capture and sampling
We captured bighorn sheep using chemical immobilization, baited drop nets, or helicopter net-gunning and sampled for presence of M. ovipneumoniae, leukotoxigenic M. haemolytica or Mannheimia glucosida (combined as M. haemolytica as these two species are not reliably differentiated by available diagnostic tests [41]), leukotoxigenic Mannheimia ruminalis or Mannheimia spp. (combined as Mannheimia spp. because the ability to identify Mannheimia ruminalis from other species was not available during initial years of data collection), leukotoxigenic B. trehalosi, lktA (a necessary gene for production of leukotoxin A by Pasteurellaceae), and P. multocida. We assessed presence of Pasteurellaceae by using a sterile polyester-tipped applicator (Puritan #25–806 1PD, Guilford, Maine, USA) to swab the tonsillar crypts (with the aid of a lighted swab extender and tongue depressor). We also assessed the presence of Pasteurellaceae by using a sterile polyester-tipped applicator to swab the nasal cavity in a subset of animals. We assessed presence of M. ovipneumoniae by using polyester-tipped applicators to swab the nasal cavity. We used ix diagnostic protocols were used to detect Pasteurellaceae and we used three to detect M. ovipneumoniae. We opportunistically incorporated diagnostic testing data for Pasteurellaceae and M. ovipneumoniae from lung samples collected from 15 bighorn sheep that died in an all-age respiratory disease epizootic in the Upper Yellowstone Complex during winter 2014/2015.
Diagnostic protocols
We used six different protocols to detect the four Pasteurellaceae species of interest, five of which were previously described and evaluated for sensitivity [36]. These five protocols all entailed collecting tonsil swabs but varied by the number of swabs collected, the transport media and corresponding environmental conditions, the diagnostic test (PCR and/or culture), and/or the diagnostic laboratory that was used. The protocol not previously described entailed collecting nasal swabs rather than tonsil swabs, but was otherwise identical to one of the previously described protocols. Several diagnostic protocols that we used to sample 12 animals in December 2017 deviated slightly from described protocols. Complete descriptions of all Pasteurellaceae protocols, including those used in December 2017, are found in S2 Appendix.
Additionally, we conducted a confirmatory Pasteurellaceae lktA PCR test on a subset of 255 swab samples (from 245 individual animals) submitted to Washington Animal Disease Diagnostic Laboratory (WADDL) for Pasteurellaceae culture. The primary streak zone of culture plates containing beta-hemolytic Pasteurellaceae isolates were preferentially selected for lktA PCR, however random plates were selected for lktA PCR if beta-hemolytic isolates were not detected in a sufficient number of samples. The primers and protocol for this PCR were described by Walsh et al. 2016 [42]. Our sampling approach did not allow evaluation of detection probability for this test.
We used three previously described and evaluated diagnostic protocols to detect M. ovipneumoniae [36] from nasal swabs. Complete descriptions of the M. ovipneumoniae protocols are also found in S2 Appendix.
Lungs collected from pneumonia-killed bighorn sheep in the Upper Yellowstone population were grossly inspected by trained personnel who collected tissue samples and submitted them on dry ice to WADDL for Pasteurellaceae culture, lktA PCR, and M. ovipneumoniae PCR. We did not evaluate detection probability for any of the pathogen species in lung tissue samples.
Updating pasteurellaceae detection probabilities
We updated published estimates of Pasteurellaceae detection probability (i.e., sensitivity) [36] by appending testing data from 248 bighorn sheep sampled during November 2016 to March 2017 to the dataset used in the publication. We sampled these supplemental animals for Pasteurellaceae using the TSB protocol (samples frozen in tryptic soy broth with glycerol) and the TSB-Nasal protocol (see S2 Appendix for complete description of protocols). Detection probability of the TSB-Nasal protocol had not been previously estimated. We could not update detection probabilities for beta-hemolytic B. trehalosi because we did not detect this pathogen in the supplemental animals. We estimated detection probability for all protocols using a single-species, single-season occupancy model [43] that allowed detection probability for each pathogen to vary by protocol and prevalence to vary by population and year. To minimize the ad-hoc procedures required to address boundary issues when pathogen prevalence was estimated to be 0 or 1, we used the RMark package [44] in Program R [45] to access Program Mark’s [46] ability to use the sine link to model pathogen prevalence.
Evaluating freedom from infection
Uncertainty in diagnostic test results is critical information when interpreting results of any pathogen surveillance project. We used a Bayesian hierarchical approach that incorporated uncertainty in detection probability at the sample-level to model presence/absence of undetected pathogens at the population-level (i.e., evaluate freedom from infection [47,48]).We assumed in our modeling approach infection status for each pathogen in the study populations was static throughout the period they were sampled for pathogens unless an all-age epizootic occurred. We justified this assumption based on the relatively short duration of the sampling periods (five years) and the regular persistence of enzootic disease in lambs for multiple years following epizootics [3,11]. We assumed that pathogens were not introduced to populations that did not experience epizootics based on evidence that novel pathogen introduction causes all-age epizootics [21,49] or a sharp decline in recruitment rates [11]. For populations where a specific pathogen was never detected, we estimated the probability the pathogen was truly absent as a function of the pathogen- and protocol-specific detection probabilities (Table C in S2 Appendix), the number of times each protocol was conducted per animal (Tables A and B in S2 Appendix), and the number of animals tested using each protocol (Tables A and B in S2 Appendix). Where multiple protocols were used to detect pathogens in a population, we estimated the overall probability of pathogen absence for that population by grouping sampled animals into cohorts with identical combinations of diagnostic protocols. We modeled probability of pathogen absence assuming no prior knowledge of prevalence or probability of presence (i.e., a uniform prior distribution was used for prevalence and the prior probability of pathogen presence was 0.50). We calculated the probability of pathogen presence as the complement to probability of absence. A complete description of this modeling approach can be found in S2 Appendix. We could not estimate detection probability for several pathogen-protocol combinations, presenting a challenge for estimating probability of pathogen presence when these protocols were employed. We addressed the issue separately depending on the reason the parameter was inestimable (see S2 Appendix for detailed description). We determined that pathogen absence was not reliably assessed in a population when probability of pathogen presence was estimated to be ≥0.10.
Recruitment and pathogen detection analysis
We included the most recent five years of recruitment data for each study population in the recruitment analysis, with the exception of populations where all-age respiratory epizootics have been documented since 2012. In these cases, we treated the population as independent pre- and post-epizootic populations and only included either “population” in the analysis if we assessed the pathogen community before or after the epizootic, respectively. To minimize the extrapolation of recent pathogen data to historic recruitment rates, we restricted the recruitment dataset to the most recent five years for each population. To explore the possibility that novel influential pathogens were introduced to study populations during the time series, we conducted a piecewise regression analysis [50] for each study population using recruitment data dating back to 2006. We used AICc model ranking to assess if and when recruitment rates fundamentally changed, indicating a potential change in the pathogen community (S1 Appendix).
We assessed differences in mean recruitment rates with respect to the detection or non-detection of different respiratory pathogens in the study populations using a Poisson rate model with a random-intercept for population in the “lme4” package [51] in Program R. We used the following model for each respiratory pathogen, using a random intercept for study populations and a binary fixed-effect variable indicating whether the pathogen was detected in each population:
where:
lambsij is lamb count for population i in year j;
ewesij is the ewe count in population i in year j (the offset term);
β0 is the expected lamb count in populations where the pathogen was not detected;
Detectedj is the variable indicating whether the pathogen was detected in population j with a coefficient of β1;
bj is the random intercept of population j;
and εij is the residual error term for the expected number of lambs in population i in year j;
We censored populations from a pathogen’s analysis when that pathogen was not detected and the probability of presence was ≥0.10.
Limited geographic separation of some study populations suggested potential that they could be part of a single population rather than separate populations. To assess consequences of this potential, we conducted a secondary analysis using the same methods described above, but where we treated certain populations in close geographic proximity to each other as a single population. We achieved this by aggregating the adjacent populations’ pathogen sampling and annual demographic data. Populations whose data we aggregated for this secondary analysis included those in the Absaroka Range of Wyoming (WY-1, WY-2, WY-3, WY-4, WY-5, and WY-22) and the Rocky Mountain Front of Montana (MT-499 and MT-423).
Results
Capture and sampling effort
We captured and live-sampled a total of 821 individual bighorn sheep (Female: 724, Male: 93 Unknown: 14) from 22 populations in Montana and Wyoming between November and March of each year from 2012–2017. We captured 744 animals once, 60 animals twice, 15 animals three times, and two animals four times. Fifty-seven (57) animals were lambs at time of capture, 72 animals were yearlings, 773 were two years or older and age was not recorded for 15 animals. The number of animals sampled from a single population ranged from six (Yount’s Peak) to 145 (Hilgard).
Updated detection probability estimates
For two of the three pathogens where detection probability could be updated from previous estimates [36], the estimates for previously reported protocols changed little (Fig A in S2 Appendix); however for P. multocida, the estimated detection probability for the tonsil swab Port-A-Cul protocol decreased from 0.44 (95% CI: 0.06–0.91) to 0.21 (95% CI: 0.01–0.86), and the estimated detection probability for the tonsil swab TSB protocol decreased from 0.13 (95% CI: 0.08–0.20) to 0.02 (95% CI: 0.01–0.05). The new protocol (TSB-Nasal) provided low detection probability for M. haemolytica (0.10, 95%CI: 0.03–0.30) and Mannheimia species (0, 95%CI: inestimable), but provided higher detection probability for P. multocida compared to other culture-based diagnostic protocols (0.31, 95% CI: 0.20–0.45).
Pathogen detection
The median number of pathogen species(excluding lktA) detected per study population was 3.5, and at least one pathogenic agent (including lktA) of interest was detected in each study population;, one species was detected in two study populations, two species were detected in two study populations, three species were detected in five study populations, five species were detected in three study populations, and all five species were detected in eight study populations. In two populations, lktA was detected without detection of any specific leukotoxigenic Pasteurellaceae (Fig 1).
Fig 1. Map of bighorn sheep study populations and detected respiratory pathogen communities.
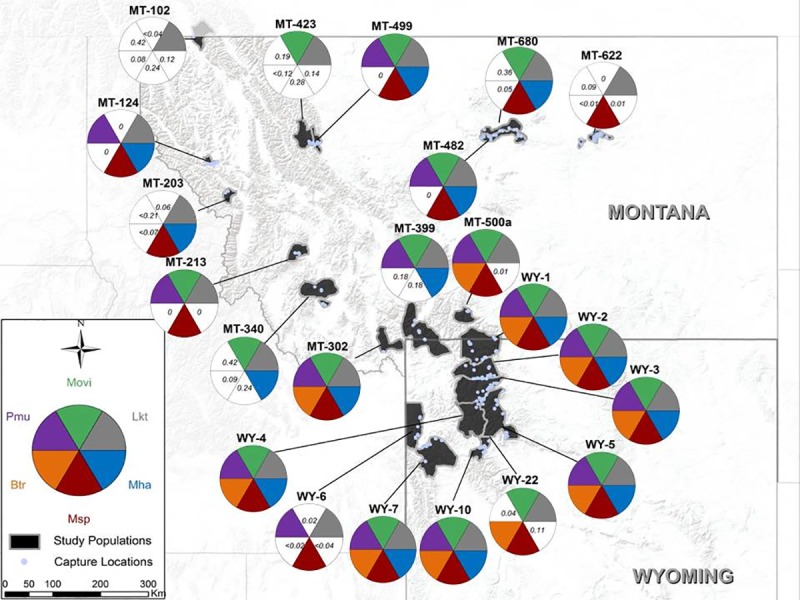
All sections of the pie-charts are fixed to equal size and represent whether the respective pathogens were detected in the study population. The key for pathogen abbreviations are as follows: Movi = Mycoplasma ovipneumoniae, Mha = leukotoxigenic Mannheimia haemolytica/glucosida, Msp = leukotoxigenic Mannheimia spp., Btr = leukotoxigenic Bibersteinia trehalosi, Pmu = Pasteurella multocida, Lkt = Leukotoxin. Where pathogens were not detected, the numbers in the unfilled section indicate the probability the pathogens were present.
Individually, four of the five pathogenic agents were detected in >65% of the study populations. M. ovipneumoniae was detected in 17 of 22 (77%) study populations (Fig 1). Leukotoxigenic M. haemolytica was detected in 15 of 22 (68%) study populations and leukotoxigenic Mannheimia spp. was detected in 18 of 22 (82%) study populations. P. multocida was detected in 15 of 22 (68%) study populations, and leukotoxigenic B. trehalosi was detected in 10 of 22 (45%) study populations including all but one Wyoming study population and two Montana study populations that are adjacent to Wyoming. LktA was detected in all study populations and, therefore, all populations that hosted M. ovipneumoniae also hosted leukotoxigenic Pasteurellaceae (Fig 1). Eighty-eight percent of the 8,460 individual bighorn sheep estimated to exist in the study populations live in populations known to carry M. ovipneumoniae and leukotoxigenic Pasteurellaceae.
Absence of M. haemolytica was reliably assessed (i.e., estimated probability of presence <0.10) in four of the seven populations (57%) where it was not detected, absence of P. multocida was reliably assessed in two of the seven populations (29%) where it was not detected, and absence of B. trehalosi was reliably assessed in 10 of the 12 populations (83%) where it was not detected. Absence of Mannheimia spp. was not reliably assessed in any of the three populations where it was not detected. In contrast, absence of M. ovipneumoniae was reliably assessed in all five populations where it was not detected (Fig 1).
Population attributes and pathogen detection
Biologists indicated nine of 22 (41%) study populations were below abundance objectives (i.e., estimated population size was less than 90% of objective population size for Montana populations or less than 80% of objective population size for Wyoming populations; Table A in S1 Appendix). Biologists indicated eight of 21 (38%) populations for which adequate demographic data were available had declining 10-year population trends, seven (33%) had stable population trends, and six (29%) had increasing population trends. Of the seven populations thought to have stable trends, five (71%) met herd objectives. Mean five-year lamb:ewe ratios were satisfactory (>0.20) for 16 of 20 (80%) populations where requisite data were available (Table A in S1 Appendix).
Greater than 50% of the study populations had satisfactory recruitment and stable or growing 10-year population trends regardless of whether any specific respiratory pathogen was detected (Table 2). In the five populations where M. ovipneumoniae was not detected, there was no history of respiratory disease, and four (80%) were determined to be stable or growing. In comparison, 11 of the 17 (65%) populations where M. ovipneumoniae was detected had a history of respiratory disease, and nine (53%) were considered stable or growing. The populations with Pasteurellaceae had similar attributes where between 53% and 66% had a history of respiratory disease, and between 60% and 71% were classified as stable or increasing populations. Characteristics of populations where any specific Pasteurellaceae was not detected were similar to populations where the species was detected, one exception being that neither population where P. multocida was not detected has a history of respiratory disease (Table 2).
Table 2. Herd attributes and pathogen detection status for 22 bighorn sheep populations.
| Mycoplasma ovipneumoniae | Mannheimia haemolytica | Mannheimia species | Bibersteinia trehalosi | Pasteurella multocida | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | -4 | + | -4 | + | -4 | + | -4 | + | -4 | |
| # Populations | 17 | 5 | 15 | 4 | 18 | — | 10 | 10 | 15 | 2 |
| Previous all-age respiratory disease epizootics | 65% (11/17) | 0% (0/5) | 53% (8/15) | 50% (2/4) | 44% (8/18) | — | 60% (6/10) | 30% (3/10) | 66% (9/15) | 0% (0/2) |
| Meets abundance objective1 | 53% (9/17) | 60% (3/5) | 67% (10/15) | 50% (2/4) | 61% (11/18) | — | 60% (6/10) | 50% (5/10) | 66% (9/15) | 50% (1/2) |
| Adequate recruitment2 | 66% (10/15) | 100% (4/4) | 93% (13/14) | 75% (3/4) | 82% (14/17) | — | 90% (9/10) | 75% (6/8) | 87% (13/15) | 50% (1/2) |
| Trend stable or growing3 | 56% (9/16) | 75% (3/4) | 71% (10/14) | 50% (2/4) | 59% (10/17) | — | 60% (6/10) | 66% (6/9) | 60% (9/15) | 50% (1/2) |
1. Meeting abundance objective defined as ≥ 80% of listed population objective for Wyoming populations and ≥ 90% of listed population objective for Montana populations pursuant to state and population-specific management goals. Targhee population objectives are no longer defined according to population size or counts.
2. Adequate recruitment defined as average winter lamb:ewe ratios > 0.20 which has been established as a criteria for healthy populations [40].
3. Based on managing biologists assessment of ten-year population trend.
4 Only populations where estimated probability of presence for undetected pathogens was <10% are shown
Recruitment rates and pathogen presence
The average five-year mean lamb:ewe ratio across the study populations was 0.29, and five-year mean lamb:ewe ratios of individual populations ranged from 0.10 to 0.48. Mean five-year lamb:ewe ratios were between 0.20 and 0.30 for six populations, 0.30 and 0.40 for six populations, and 0.40 and 0.50 for four populations (Table A in S1 Appendix). Mean lamb:ewe ratios were less than 0.20 for four populations. Piecewise regression analysis of 2006–2017 classification data identified break-years for 15 of the 20 (75%) study populations with adequate recruitment data. Six of these 15 (40%) populations experienced an increase in lamb:ewe ratios after the break-year and nine (60%) experienced a decrease. Break-years occurred within the recruitment analysis time-series (after 2012) for three populations (Clark’s Fork, Yount’s Peak, and Whiskey Mountain; Table B in S1 Appendix). M. ovipneumoniae and leukotoxigenic Pasteurellaceae were detected prior to the break-year in two of these populations (Clark’s Fork and Whiskey Mountain) and the other population (Yount’s Peak) was not sampled for pathogens prior to the break-year.
Data from the Upper Yellowstone population were split into pre- (<2013) and a post- epizootic (>2014) categories. Ultimately, 90 population-years of recruitment data (lamb:ewe ratios) from 21 populations were available to assess differences in mean lamb:ewe ratios with respect to the detection of each pathogen species. Censoring data from populations where pathogen absence was not reliably assessed reduced the number of records available for analysis to 73 for M. haemolytica, 77 for B. trehalosi, and 77 for P. multocida. Regression analysis was not conducted for Mannheimia spp. because its absence was never reliably assessed; lamb:ewe ratios were summarized for the populations where this pathogen was detected. The frequent co-occurrence of most pathogen species prevented investigation of additive or interactive statistical effects of multiple pathogen species.
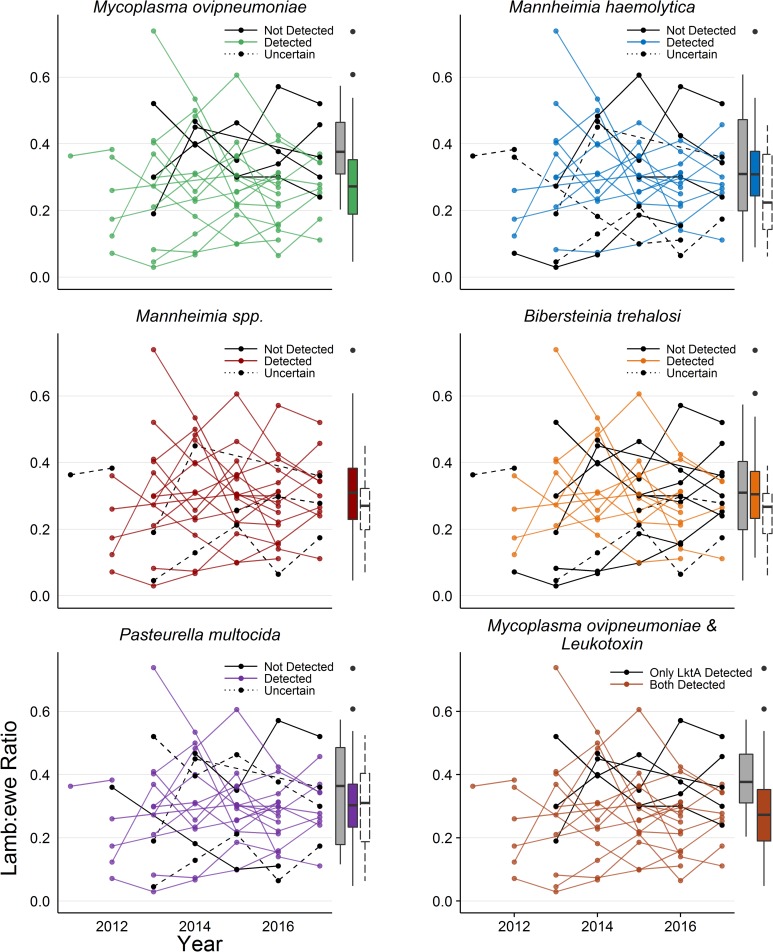
Mean lamb:ewe ratios of individual populations where any specific pathogen was detected ranged from <0.20 to >0.40 (Table 3, Fig 2). For each pathogen species, there were at least four populations that hosted it and had mean lamb:ewe ratios >0.30. There was evidence for an association between detection of M. ovipneumoniae and lamb:ewe ratios (χ21 = 4.49, p-value = 0.03). The estimated mean lamb:ewe ratio was 31% lower (Δ = 0.12) in populations where M. ovipneumoniae was detected than where it was not detected (Table 3, Fig 2). There was no evidence for an association between detection of any Pasteurellaceae species and lamb:ewe ratios (M. haemolytica: χ21 < .01, p-value > 0.99; B. trehalosi: χ21 = 0.83, p-value = 0.77; P. multocida: χ21 = 0.18, p-value = 0.67; Fig 2). Associations between presence of lktA and lamb:ewe ratios could not be explored because lktA was detected in all study populations. Results from the secondary analysis where neighboring populations were treated as a single populations were consistent with the results of the primary analysis, although evidence for a negative association between lamb:ewe ratios and the presence of M. ovipneumoniae was weakened and no longer significant at a 0.05 alpha level (χ21 = 2.67, p-value = 0.10; Table C in S1 Appendix).
Table 3. Model-estimated lamb:ewe ratios and pathogen detection status in 21 bighorn sheep populations.
| Mycoplasma ovipneumoniae | Mannheimia haemolytica | Mannheimia species1 | Bibersteinia trehalosi | Pasteurella multocida1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | -2 | + | -2 | + | -2 | + | -2 | + | -2 | |
| # Populations3 | 16 | 5 | 15 | 4 | 18 | — | 10 | 8 | 15 | 2 |
| Mean lamb:ewe ratio (95% CI) | 0.25 (0.21-0.30) | 0.37 (0.27-0.51) | 0.29 (0.24-0.35) | 0.29 (0.19-0.42) | 0.27 (0.22-0.33) | — | 0.28 (0.23-0.37) | 0.29 (0.21-0.36) | 0.27 (0.23-0.33) | 0.31 (0.18-0.52) |
| Minimum mean population-specific lamb:ewe ratio | 0.13 (MT-213) | 0.31 (WY-6) | 0.15 (MT-499) | 0.13 (MT-213) | 0.12 (MT-213) | — | 0.20 (WY-22) | 0.13 (MT-213) | 0.13 (MT-213) | 0.20 (WY-22) |
| Maximum mean population-specific lamb:ewe ratio | 0.41 (MT-302) | 0.48 (MT-622) | 0.41 (MT-302) | 0.48 (MT-622) | 0.48 (MT-622) | — | 0.41 (MT-302) | 0.48 (MT-622) | 0.41 (MT-302) | 0.48 (MT-622) |
1. Absence of Mannheimia species was never reliably assessed in populations where they were not detected. Estimates obtained from intercept only model.
2. Data from populations where the probability of presence for a particular pathogen was ≥ 0.10 were not used in the regression analysis
3.One population (MT-340) was excluded due to insufficient demographic data
Fig 2. Lamb:ewe ratios and pathogen presence in 21 bighorn sheep populations in Montana and Wyoming.
The y-axes show observed lamb:ewe ratios (lambs:100 ewes) and the x-axes show years the data were collected. Grey boxplots show lamb:ewe ratios of populations where the respective pathogen was not detected but probability of pathogen presence was <10%, colored boxplots show where the respective pathogen was detected and unfilled boxplots show where the respective pathogen was not detected and probability of pathogen presence was ≥10%. The boxplot boxes represent the interquartile range (IQR), whiskers represent range of values in the outer quartiles that are within 1.5 times the IQR from the inter quartiles, and points represent values that are not within 1.5 times the IQR from the inter quartiles.
Discussion
For decades most bighorn sheep populations have been small, isolated, and, consequently, imperiled by a myriad of threats [52,53,3]. Respiratory disease contributes to this predicament by suppressing population growth, range expansion and connectivity among populations. Despite the importance of respiratory disease to the conservation and management of bighorn sheep, its etiology remains unclear due in part to the polymicrobial nature and limited baseline data for pathogen communities hosted by both symptomatic and asymptomatic populations. Integrating what is understood about this disease into management strategies that lead to robust bighorn sheep populations has been challenging. Our findings shed light on approaches that could improve etiological understanding of respiratory disease and develop additional management strategies for bighorn sheep. The negative association between the presence of M. ovipneumoniae and lamb:ewe ratios along with a lack of disease history in populations where this pathogen was not detected provides additional evidence of its importance and confirm it is an important, perhaps necessary, component of epizootic respiratory disease. There was high variability in the demographic traits of populations hosting both M. ovipneumoniae and Pasteurellaceae; the smallest and largest populations in the study were reservoirs for both pathogen groups and a number of populations hosting the pathogens had average lamb:ewe ratios greater than 0.30. These results suggest that the respiratory pathogens necessary for disease are well established in bighorn sheep populations and the effects of these pathogens might be mediated by ecological factors. Though these findings do not confirm ecological factors influence disease expression, such factors could be significant and may be an important consideration in the management of this species given the challenge of maintaining populations free of respiratory pathogens.
We detected M. ovipneumoniae, leukotoxigenic Pasteurellaceae, and Pasteurella multocida in most (13 of 16) populations where the presence of each was reliably assessed (Fig 1). Study populations in the Absaroka Mountains of Wyoming (WY-1, WY-2, WY-3, WY-4, WY-5, and WY-22), the Rocky Mountain Front of Montana (MT-499, MT-423) and the upper Missouri Breaks in Montana (MT-482, MT-680) exist in close proximity to one another and may not have independent pathogen communities due to connectivity. If these populations were aggregated into one population for each region, the number of populations where pathogen presence was reliably assessed would decrease to 12, with the abovementioned pathogens detected in nine of these populations. This proportion is similar to the proportion of bighorn sheep populations across six other U.S. states that were reported to be exposed to M. ovipneumoniae [14]. Assessment of pathogen communities in additional populations in other regions is needed to confirm whether the common presence of respiratory pathogens in our study populations is representative of most bighorn sheep populations.
The high proportion of study populations already exposed to these pathogen species does not indicate that future pathogen introduction is not a concern. Recent evidence suggests that acquired immunity of bighorn sheep to M. ovipneumoniae is strain-specific [49]. Exposure to new respiratory pathogen strains introduced from domestic livestock, neighboring bighorn sheep populations, translocated bighorn sheep, and even sympatric mountain goats (Oreamnos americanus) [54] could negatively affect any bighorn sheep population. Management agencies face the challenge of simultaneously minimizing the introduction of novel pathogens to bighorn sheep populations while also maintaining population connectivity and viable abundances.
Until recently, respiratory pathogen surveillance efforts had little ability to detect the fastidious pathogens associated with respiratory disease in bighorn sheep [19,25] and current protocols are still imperfect [35,36]. Our findings suggest a number of growing or robust populations that have been used as source populations for translocations may have harbored respiratory pathogens that were subsequently introduced to recipient populations or geographic regions, unbeknownst to wildlife managers. Recent work also suggests that recovery of pneumonic populations is unlikely to be enhanced by introduction of animals naïve to the pathogen strains that those populations carry [49,55]. Thus, a body of evidence suggests that augmenting struggling bighorn sheep populations provides little conservation benefit. The consequence of mixing resident pathogen communities should factor into future decisions to translocate bighorn sheep.
The common detection of M. ovipneumoniae and Pasteurellaceae indicates that resident pathogens are a plausible explanation for some proportion of respiratory disease epizootics. Spontaneous respiratory disease epizootics have been previously reported in captive bighorn sheep [32] and numerous epizootics in free-ranging bighorn sheep have been attributed to a shift to unfavorable ecological conditions that triggered increased virulence or transmission of resident pathogens [56,57]. Epizootics in populations already hosting Pasteurellaceae and M. ovipneumoniae might be caused by introduction of novel pathogen strains or changes in the host, pathogens, or environment that lead to increased virulence or transmission of resident pathogens. Identifying pathogens to the strain level will be necessary to determine which mechanism causes epizootics in such populations. If epizootics are frequently caused by resident pathogens, a broader set of preventative measures are needed to reduce the occurrence of respiratory disease. Our approach to evaluate freedom from infection can be used to establish a framework that quantifies the relative role novel and resident pathogens play in causing epizootics.
Despite the negative association between lamb:ewe ratios and presence of M. ovipneumoniae, approximately half of the study populations with average lamb:ewe ratios greater than 0.30 hosted M. ovipneumoniae. This suggests that M. ovipneumoniae may be necessary for population-limiting respiratory disease in bighorn sheep, but it may not be sufficient. We found no correlation between presence of any Pasteurellaceae species we isolated and several indices of population health (i.e., disease history, population trend, achievement of population objective, and recruitment rates). Previous research has also noted little association of Pasteurellaceae species with respiratory disease in bighorn sheep populations [18]. However, leukotoxigenic Pasteurellaceae have been experimentally shown to cause pneumonia in bighorn sheep but not domestic sheep [12,28,58,59], Pasteurellaceae are commonly hosted by healthy domestic sheep [60], bighorn sheep typically develop pneumonia when co-penned with healthy domestic sheep [13], and Pasteurellaceae are commonly involved in the pathology of pneumonia in bighorn sheep [17,26,27]. These lines of evidence suggest that Pasteurellaceae pose a disease risk to bighorn sheep, particularly in populations where M. ovipneumoniae is present. Given poor resolution [41] and detection probability [35,36] of diagnostic protocols used to detect Pasteurellaceae along with the potential for ecological factors to influence disease expression, these pathogens should not be dismissed as a component cause of respiratory disease based on a lack of correlative evidence.
The demographic variability we observed in populations hosting Pasteurellaceae and M. ovipneumoniae could be explained by undetected differences in pathogen communities. Variation in virulence among pathogen strains that we were unable to distinguish could explain the strong demographic rates of some populations. Differences in virulence could be inherent in the strains or due to attenuation after years of persistence in bighorn sheep populations. Similarly, it is also possible that bacteria were misidentified by the diagnostic tests we used, resulting in false positive errors at the population level. This is most likely to have occurred with M. haemolytica, which tests often fail to distinguish from Mannheimia glucosida [41,42], but false positives could have occurred for other Pasteurellaceae or M. ovipneumoniae. However, high specificity has been reported for the diagnostic tests we used to identify M. ovipneumoniae and most Pasteurellaceae agents [42], suggesting that false positive errors were likely uncommon at the population-level for most pathogens except M. haemolytica. Although not examined in this study, transmissible sinus tumors have also recently been discovered in bighorn sheep and are associated with increased shedding of Pasteurellaceae and M. ovipneumoniae [61,62]. It is hypothesized that sinus tumors exacerbate upper respiratory tract infections and increase pathogen transmission. Sinus tumors may play an under-recognized role in bighorn sheep respiratory disease and explain some of the variability in demographic rates that we observed among our study populations. Pathogen prevalence in adult females may also explain some variability in recruitment rates but was not assessed in this investigation because it is estimated with poor precision [36]. Lastly, unidentified pathogens could also contribute to disease severity.
If variation in demographic rates of populations hosting both Pasteurellaceae and M. ovipneumoniae is not explained by unobserved differences in pathogen communities or population structure, then the variation of demographic rates, and presumably disease expression, might be mediated by ecological factors. Strong demographic rates of some populations might be temporary states until conditions sufficient for disease expression are met. Certain environments might be less likely to produce conditions sufficient for disease caused by resident pathogens, leading to differential performance among populations with the same respiratory pathogen community and innate immunological potential. Nutrition [63,64], mineral availability [65], stress [56,66,67], migratory behavior [68], and predation [69] have all been shown or hypothesized to influence disease dynamics, sometimes in complex or counterintuitive ways. Given variable population-management histories and over a century of exposure to domestic sheep experienced by many populations, selection may also have increased disease resilience of individuals in certain study populations. Although there are no obvious signs of genetic selection for resistance of bighorn sheep against respiratory disease, it is likely that extensive translocation efforts have interfered with selective processes [1,70]. In populations that have not been augmented it is more likely that selection for disease resistance would have occurred and been maintained.
The scale of our observations could have influenced our results. The larger populations in the study found to host M. ovipneumoniae and Pasteurellaceae are composed of numerous subpopulations which were not all intensively sampled for respiratory pathogens. Our summarization of demographic and pathogen data at the population-level in these cases assumes that each sub-population is exposed to the respiratory pathogens detected across the entire population. Although we cannot validate this assumption, synchronized respiratory disease epizootics across the Sun River metapopulation in Montana (see [71]) suggest it is not unreasonable. Nevertheless, complex population structure of the large study populations could interact with the coarse scale of data collection to mask poor recruitment rates localized in some subpopulations. For example, heterogeneous lamb survival across subpopulations of a bighorn sheep population with enzootic lamb pneumonia has been documented [72]. An emergent property of such asynchrony within populations hosting M. ovipneumoniae and Pasteurellaceae may be the portfolio effect: an ecological phenomenon where asynchronous and volatile components of a system cumulate to a stable system as whole [73]. If this occurs despite the presence of respiratory pathogens in a population, management strategies that increase the structure of populations occupying a particular area could dampen the effects of respiratory disease on bighorn sheep numbers in that area. Such an approach would not diminish the effects of respiratory disease at the subpopulation level, but it may lead to more stability in the number of bighorn sheep occupying a particular area.
There were several limitations of this study not previously discussed. We evaluated freedom from pathogen infection at the population-level assuming there was homogenous mixing of pathogens within populations and that the prior probability of infection was 0.50. These assumptions may not have been met as most populations are comprised of subpopulations and we detected most pathogens in over 50% of the study populations. Violations of these assumptions would have overestimated the probability that undetected pathogens were absent. The estimates of detection probability used in the freedom from infection analysis were calculated assuming perfect specificity; false positive errors could bias these estimates in either direction depending on the underlying cause of the errors. However, most pathogens were detected in most populations and previous studies have determined the diagnostic tests we used have high specificity, indicating that these issues had little potential to affect the overall conclusions of this work. Future applications can build upon our findings to obtain a more informed prior probability of infection. Due to constraints of investigating a large number of populations, our demographic data were limited and we could not collect cause-specific mortality data to assess the proximate drivers of lamb recruitment in the study populations. Survey data collected on a near-annual basis by wildlife management agencies served as the most consistent data source available across the populations. Although longer demographic time-series were available, we limited the data to recent years to maintain the relevance of the demographic data to the pathogen data that we collected. The use of coarse data over a large number of populations limited our ability to investigate the fine-scale processes underlying the patterns we observed. These limitations resulted from a trade-off that allowed for broad inferences at the cost of fine resolution data. Lastly, the non-random selection of study populations limits the extrapolation of these findings, which may not be reflective of other bighorn sheep populations.
Recommendations and conclusions
This study found that bacterial pathogens associated with respiratory disease are common among the bighorn sheep populations that were studied. Presence of M. ovipneumoniae in a population increased risk of poor lamb recruitment, likely explained by enzootic lamb pneumonia. However, populations hosting M. ovipneumoniae and Pasteurellaceae were the largest of those surveyed and were often stable or growing. The common presence of respiratory pathogens and the variability among populations that host them confirms plausibility for related hypotheses that: 1) population or environmental characteristics can provide increased resilience in the face of respiratory disease; and 2) respiratory disease epizootics may be caused by either introduction of novel pathogens or increased virulence or transmission of resident pathogens. Although this study lacks the geographic and temporal scale to test these hypotheses, it provides baseline data and recommendations for future studies to test them. These recommendations include:
Continue to improve characterization of respiratory pathogen communities by working to adopt diagnostic protocols that provide strong ability to detect pathogens in bighorn sheep populations and identify different strains within pathogen species. Statistical tools should be used to quantify uncertainty in negative test results.
Develop a monitoring program for a large and diverse set of bighorn sheep populations where a sufficient number of animals (n = 30) [36] can be sampled to assess respiratory pathogen communities. Collect baseline respiratory pathogen data from populations with and without indication of respiratory disease and resample populations following disease epizootics using comparable methods to assess the novel and resident pathogen hypotheses.
Characterize attributes of study populations that could influence expression of respiratory disease at the population-level, such as genetics, habitat, nutrition, physiological status, predation, migratory behavior, or population structure.
Coordinate and standardize these efforts across states and provinces to collect adequate and comparable data from as many contrasting bighorn sheep populations as possible [74].
Continue to maintain and improve upon policies of separation between bighorn sheep and domestic sheep and goats to prevent the transmission of novel pathogens.
We echo recent recommendations for definitions of wildlife health to emphasize resilience and sustainability in the face of numerous anthropogenic and environmental threats rather than the absence of pathogens or disease [75]. Although disease does influence viability, a population that is currently free from disease but is small and isolated with little potential to expand is not necessarily any more viable than one that is currently affected by disease, but has a broad distribution and connectivity with other populations. Our findings suggest that bighorn sheep populations can host a full suite of respiratory pathogens and be healthy under a modern definition of wildlife health. Elucidating the mechanisms driving the patterns we described may aid management of this species.
Supporting information
• Table A in S1 Appendix. Summary of demographic data for 21 bighorn sheep study populations in Montana and Wyoming that were investigated as part of this study.
• Table B in S1 Appendix. Break-years and mean lamb:ewe ratios before and after the break year for each population as estimated by piecewise regression. Lamb:ewe ratios for each population with requisite data were iteratively split into “before” and “after” groups for every year in the dataset and a Poisson regression model to estimate average lamb:ewe ratios for the “before” and “after” groups was run for each year-based grouping. The break-year was chosen as the year-based grouping with the lowest AICC score. Break-years were estimated based on before and after recruitment-rates to detect potential years where respiratory pathogens may have been introduced or gone extinct from study populations. Populations whose recruitment rate declined following a break-year are highlighted in red and populations whose recruitment rate increased following a break-year are highlighted in blue. Years of documented all-age respiratory disease epizootics since 2006 are also shown for reference.
• Table C in S1 Appendix. Comparative results of recruitment and pathogen detection analyses when adjacent study populations were treated as separate populations (primary analysis) vs. aggregated into a single population (secondary analysis).
(DOCX)
• Table A in S2 Appendix. Animals sampled by population, year, and set of diagnostic protocols that were used to detect Pasteurellaceae pathogens. The numbers under diagnostic protocols indicate the number of times the specified protocol was conducted per individual. Different rows within the same population and year represent “cohorts” of animals that were sampled for respiratory pathogens using the same suite of diagnostic protocols.
• Table B in S2 Appendix. Animals sampled by population, year, and set of diagnostic protocols that were used to detect Mycoplasma ovipneumoniae. The numbers under diagnostic protocols indicate the number of times the specified protocol was conducted per individual. Different rows within the same population and year represent “cohorts” of animals that were sampled for respiratory pathogens using the same suite of diagnostic protocols.
• Table C in S2 Appendix. Detection probability parameters for Pasteurellaceae and Mycoplasma ovipneumoniae diagnostic protocols updated and adopted from Butler et al.2017. Bolded beta distributions indicate distributions that were used to model probability of pathogen presence.
• Fig A in S2 Appendix. Estimated detection probabilities and 95% confidence intervals for five respiratory pathogens in bighorn sheep. One set of protocols was used to detect the four Pasteurellaceae organisms (shaded) and a separate set was used to detect Mycoplasma ovipneumoniae (not shaded). Detection probabilities for Mannheimia haemolytica, Mannheimia spp., and Bibersteinia trehalosi are for beta hemolytic or leukotoxigenic strains. Protocols that used fee-for-service diagnostic tests are indicated with an asterisk (*) in the legend and above the upper confidence limit. The protocol not previously evaluated is shown in red. The most up to date detection probability estimates are shown with full opacity and previous estimates are dodged left and partially transparent.
• Fig B in S2 Appendix. Locations of captures and Mycoplasma ovipneumoniae detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig C in S2 Appendix. Locations of captures and leukotoxigenic Bibersteinia trehalosi detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig D in S2 Appendix. Locations of captures and leukotoxigenic Mannheimia haemolytica detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig E in S2 Appendix. Locations of captures and leukotoxigenic Mannheimia species detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig F in S2 Appendix. Locations of captures and Pasteurella multocida detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig G in S2 Appendix. Locations of captures and Leukotoxin A detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
(DOCX)
Acknowledgments
This effort would not be possible without the collective effort of dozens of dedicated wildlife professionals. From Montana Department of Fish, Wildlife and Parks, we would like to thank the following staff who facilitated capturing and sampling bighorn sheep populations: Julie Cunningham, Brent Lonner, Shawn Stewart, Justin Paugh, Sonja Andersen, Bruce Sterling, Ben Jimenez, Liz Bradley, Vanna Boccadori, Ray Vinkey, Scott Thompson, and Tim Thier. We thank Keri Carson and Neil Anderson for provision and organization of sampling supplies as well as skilled sample collection and handling. From Wyoming Game and Fish Department, we thank Doug Brimeyer, Kyle Lash, John Stephens, and Chris Timberlake for facilitating capturing and sampling of bighorn sheep populations. We thank Confederated Salish & Kootenai Tribes for permitting capture of bighorn sheep within their jurisdiction and Stillwater Mining Company for permitting operations on private property. From Montana State University we thank Blake Lowrey and Ethan Lula for help collecting samples, submitting samples, and managing data that were used in this study. From the U.S. Forest Service, we thank Andy Pils for his facilitation of animal captures on the Shoshone National Forest. We appreciate the professional capture services provided by Quicksilver Air Inc., Native Range Capture Services, and Leading Edge Aviation Inc. This manuscript was improved by input from Marco Festa-Bianchet and two anonymous reviewers.
Data Availability
The data for this study are contained within the supporting information files and also at the Figshare database: https://figshare.com/s/a4d4fb9e21fcb3153f37.
Funding Statement
This study was funded by the US Fish and Wildlife Service through the Pittman- Robertson Federal Aid in Wildlife Restoration Act (grant #s W-159-R & W-166-SI; https://wdfw.wa.gov/grants/wildlife_restoration/), the Wyoming Wildlife Foundation (through the Wyoming Governor's Big Game License Coalition; http://wyomingwildlifefoundation.org), Montana Department of Fish Wildlife & Parks (http://fwp.mt.gov), Wyoming Game and Fish Department (https://wgfd.wyo.gov), National Park Service (Yellowstone and Grand Teton National Parks; https://nps.gov), Montana and Wyoming chapters of the Wild Sheep Foundation (https://www.wildsheepfoundation.org), and Canon Inc. USA (through Yellowstone Park Foundation; https://www.yellowstone.org/). Additional funding and scholarships were provided by Montana State University (https://montana.edu) and the Kevin Hurley Wild Sheep Biology Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Picton HD, Lonner TN. Montana’s wildlife legacy: decimation to restoration. Media Works Pub; 2008. [Google Scholar]
- 2.Toweill DE, Geist V. Return of royalty: Wild sheep of North America. Boone and Crockett Club; 1999. [Google Scholar]
- 3.Butler C, Garrott RA, Rotella JJ. Correlates of recruitment in Montana bighorn sheep populations [Internet]. Helena, Montana; 2013 Jul. Available: http://www.wafwa.org
- 4.Johnson HE, Hebblewhite M, Stephenson TR, German DW, Pierce BM, Bleich VC. Evaluating apparent competition in limiting the recovery of an endangered ungulate. Oecologia. 2013;171: 295–307. 10.1007/s00442-012-2397-6 [DOI] [PubMed] [Google Scholar]
- 5.Festa-Bianchet M, Coulson T, Gaillard J- M, Hogg JT, Pelletier F. Stochastic predation events and population persistence in bighorn sheep. Proc R Soc B Biol Sci. 2006;273: 1537–1543. 10.1098/rspb.2006.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogg JT, Forbes SH, Steele BM, Luikart G. Genetic rescue of an insular population of large mammals. Proc R Soc B Biol Sci. 2006;273: 1491–1499. 10.1098/rspb.2006.3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer AM, Dennis B, Liebhold AM, Drake JM. The evidence for Allee effects. Popul Ecol. 2009;51: 341 10.1007/s10144-009-0152-6 [Google Scholar]
- 8.Brewer C, Bleich VC, Foster J, Hosch-Hebdon T, McWhirter D, Rominger E, et al. Bighorn sheep conservation challenges: Management strategies for the 21st Century. Cheyenne, Wyoming, USA: Western Association of Fish and Wildlife Agencies; 2014. July pp. 1–32. [Google Scholar]
- 9.Cassirer EF, Sinclair ARE. Dynamics of pneumonia in a bighorn sheep metapopulation. J Wildl Manag. 2007;71: 1080–1088. 10.2193/2006-002 [Google Scholar]
- 10.Carlsen T, Erickson G. Montana bighorn sheep conservation strategy [Internet]. Helena, Montana; 2010 Jan. Available: http://fwp.mt.gov/fishAndWildlife/management/bighorn/plan.html#plan
- 11.Manlove K, Cassirer EF, Cross PC, Plowright RK, Hudson PJ. Disease introduction is associated with a phase transition in bighorn sheep demographics. Ecology. 2016;97: 2593–2602. 10.1002/ecy.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence PK, Shanthalingam S, Dassanayake RP, Subramaniam R, Herndon CN, Knowles DP, et al. Transmission of Mannheimia haemolytica from domestic sheep (Ovies aries) to bighorn sheep (Ovis canadensis): Unequivocol demonstration with green fluorescent protein-tagged organisms. J Wildl Dis. 2010;46: 706–717. 10.7589/0090-3558-46.3.706 [DOI] [PubMed] [Google Scholar]
- 13.Wehausen JD, Kelley ST, Ramey RR. Domestic sheep, bighorn sheep, and respiratory disease: a review of the experimental evidence. Calif Fish Game. 2011;97: 7–24. Available: http://www.bio.sdsu.edu/faculty/kelley/47.pdf [Google Scholar]
- 14.Cassirer EF, Manlove KR, Almberg ES, Kamath PL, Cox M, Wolff P, et al. Pneumonia in bighorn sheep: Risk and resilience. J Wildl Manag. 2017; n/a-n/a. 10.1002/jwmg.21309 [Google Scholar]
- 15.Heinse LM, Hardesty LH, Harris RB. Risk of pathogen spillover to bighorn sheep from domestic sheep and goat flocks on private land. Wildl Soc Bull. 2016;40: 625–633. 10.1002/wsb.718 [Google Scholar]
- 16.Besser TE, Cassirer EF, Potter KA, Foreyt WJ. Exposure of bighorn sheep to domestic goats colonized with Mycoplasma ovipneumoniae induces sub-lethal pneumonia. PLoS ONE. 2017;12: e0178707 10.1371/journal.pone.0178707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfe LL, Diamond B, Spraker TR, Sirochman MA, Walsh DP, Machin CM, et al. A bighorn sheep die-off in southern Colorado involving a Pasteurellaceae strain that may have originated from syntopic cattle. J Wildl Dis. 2010;46: 1262–1268. Available: http://www.bioone.org/doi/abs/10.7589/0090-3558-46.4.1262 [DOI] [PubMed] [Google Scholar]
- 18.Besser TE, Frances Cassirer E, Highland MA, Wolff P, Justice-Allen A, Mansfield K, et al. Bighorn sheep pneumonia: Sorting out the cause of a polymicrobial disease. Prev Vet Med. 2013;108: 85–93. 10.1016/j.prevetmed.2012.11.018 [DOI] [PubMed] [Google Scholar]
- 19.Besser TE, Cassirer EF, Potter KA, VanderSchalie J, Fischer A, Knowles DP, et al. Association of Mycoplasma ovipneumoniae infection with population-limiting respiratory disease in free-ranging Rocky Mountain bighorn Sheep (Ovis canadensis canadensis). J Clin Microbiol. 2008;46: 423–430. 10.1128/JCM.01931-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besser TE, Highland M, Baker K, Cassirer EF, Anderson NJ, Ramsey JM, et al. Causes of pneumonia epizootics among bighorn sheep, western United States, 2008–2010. Emerg Infect Dis. 2012;18: 406–414. 10.3201/eid1803.111554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besser TE, Cassirer EF, Potter KA, Lahmers K, Oaks JL, Shanthalingam S, et al. Epizootic pneumonia of bighorn sheep following experimental exposure to Mycoplasma ovipneumoniae. PLoS ONE. 2014;9: e110039 10.1371/journal.pone.0110039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besser TE, Cassirer EF, Yamada C, Potter KA, Herndon C, Foreyt WJ, et al. Survival of bighorn sheep (Ovis canadensis) commingled with domestic sheep (Ovis aries) in the absence of Mycoplasma ovipneumoniae. J Wildl Dis. 2012;48: 168–172. 10.7589/0090-3558-48.1.168 [DOI] [PubMed] [Google Scholar]
- 23.Foreyt WJ. Fatal Pasteurella haemolytica pneumonia in bighorn sheep after direct contact with clinically normal domestic sheep. Am J Vet Res. 1989;50: 341–344. [PubMed] [Google Scholar]
- 24.George JL, Martin DJ, Lukacs PM, Miller MW. Epidemic pasteurellosis in a bighorn sheep population coinciding with the appearance of a domestic sheep. J Wildl Dis. 2008;44: 388–403. 10.7589/0090-3558-44.2.388 [DOI] [PubMed] [Google Scholar]
- 25.Shanthalingam S, Goldy A, Bavananthasivam J, Subramaniam R, Batra SA, Kugadas A, et al. PCR assay detects Mannheimia haemolytica in culture-negative pneumonic lung tissues of bighorn sheep (Ovis canadensis) from outbreaks in the western USA, 2009–2010. J Wildl Dis. 2014;50: 1–10. 10.7589/2012-09-225 [DOI] [PubMed] [Google Scholar]
- 26.Wood ME, Fox KA, Jennings-Gaines J, Killion HJ, Amundson S, Miller MW, et al. How respiratory pathogens contribute to lamb mortality in a poorly performing bighorn sheep (Ovis canadensis) herd. J Wildl Dis. 2016; 10.7589/2016-05-097 [DOI] [PubMed] [Google Scholar]
- 27.Grigg JL, Wolfe LL, Fox KA, Killion HJ, Jennings-Gaines J, Miller MW, et al. Assessing Timing and Causes of Neonatal Lamb Losses in a Bighorn Sheep (Ovis canadensis canadensis) Herd via Use of Vaginal Implant Transmitters. J Wildl Dis. 2017;53: 596–601. 10.7589/2016-10-239 [DOI] [PubMed] [Google Scholar]
- 28.Dassanayake RP, Shanthalingam S, Subramaniam R, Herndon CN, Bavananthasivam J, Haldorson GJ, et al. Role of Bibersteinia trehalosi, respiratory syncytial virus, and parainfluenza-3 virus in bighorn sheep pneumonia. Vet Microbiol. 2013;162: 166–172. 10.1016/j.vetmic.2012.08.029 [DOI] [PubMed] [Google Scholar]
- 29.Rothman KJ. Causes. Am J Epidemiol. 1976;104: 587–592. Available: http://aje.oxfordjournals.org.proxybz.lib.montana.edu/content/104/6/587 [DOI] [PubMed] [Google Scholar]
- 30.Buechner HK. The bighorn sheep in the United States, its past, present, and future. Wildl Monogr. 1960; 3–174. [Google Scholar]
- 31.Rachowicz LJ, Hero J- M, Alford RA, Taylor JW, Morgan JAT, Vredenburg VT, et al. The novel and endemic pathogen hypotheses: Competing explanations for the origin of emerging infectious diseases of wildlife. Conserv Biol. 2005;19: 1441–1448. 10.1111/j.1523-1739.2005.00255.x [Google Scholar]
- 32.Miller M, Hobbs N, Williams E. Spontaneous pasteurellosis in captive Rocky-Mountain bighorn sheep (Ovis-canadensis canadensis)—Clinical, laboratory, and epizootiological observations. J Wildl Dis. 1991;27: 534–542. 10.7589/0090-3558-27.4.534 [DOI] [PubMed] [Google Scholar]
- 33.Miller DS, Hoberg E, Weiser G, Aune K, Atkinson M, Kimberling C. A review of hypothesized determinants associated with bighorn sheep (Ovis canadensis) die-offs. Vet Med Int. 2012;2012: 1–19. 10.1155/2012/796527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safaee S, Weiser GC, Cassirer EF, Ramey RR, Kelley ST. Microbial diversity in bighorn sheep revealed by culture-independent methods. J Wildl Dis. 2006;42: 545–555. 10.7589/0090-3558-42.3.545 [DOI] [PubMed] [Google Scholar]
- 35.Walsh DP, Wolfe LL, Vieira MEP, Miller MW. Detection probability and Pasteurellaceae surveillance in bighorn sheep. J Wildl Dis. 2012;48: 593–602. 10.7589/0090-3558-48.3.593 [DOI] [PubMed] [Google Scholar]
- 36.Butler CJ, Edwards WH, Jennings-Gaines JE, Killion HJ, Wood ME, McWhirter DE, et al. Assessing respiratory pathogen communities in bighorn sheep populations: Sampling realities, challenges, and improvements. PLoS ONE. 2017;12: e0180689 10.1371/journal.pone.0180689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer FJ, Williams E, Miller MW, Zeigenfuss LC. Population growth, fecundity, and survivorship in recovering populations of bighorn sheep. Restor Ecol. 2000;8: 75–84. 10.1046/j.1526-100x.2000.80067.x [Google Scholar]
- 38.Cassirer EF, Plowright RK, Manlove KR, Cross PC, Dobson AP, Potter KA, et al. Spatio-temporal dynamics of pneumonia in bighorn sheep. J Anim Ecol. 2013;82: 518–528. 10.1111/1365-2656.12031 [DOI] [PubMed] [Google Scholar]
- 39.USDA handbook 296: land resource regions and major land resource areas of the United States, the Caribbean, and the Pacific Basin. United States Department of Agriculture, Natural Resource Conservation Service; 2006.
- 40.Drew ML, Edwards WH, Fox KA, Griffin CM, Gonzales BJ, Jennings-Gaines JE, et al. WAFWA wildlife health committee 2014: Bighorn sheep herd health monitoring recommendations. Western Association of Fish and Wildlife Agencies; 2015. [Google Scholar]
- 41.Miller MW, Hause BM, Killion HJ, Fox KA, Edwards WH, Wolfe LL. Phylogenetic and epidemiologic relationships among Pasteurellaceae from Colorado bighorn sheep herds. J Wildl Dis. 2013;49: 653–660. 10.7589/2012-11-274 [DOI] [PubMed] [Google Scholar]
- 42.Walsh DP, Cassirer EF, Bonds MD, Brown DR, Edwards WH, Weiser GC, et al. Concordance in diagnostic testing for respiratory pathogens of bighorn sheep. Wildl Soc Bull. 2016;40: 634–642. 10.1002/wsb.721 [Google Scholar]
- 43.MacKenzie DI, Nichols JD, Lachman GB, Droege S, Andrew Royle J, Langtimm CA. Estimating site occupancy rates when detection probabilities are less than one. Ecology. 2002;83: 2248–2255. Available: http://www.esajournals.org/doi/abs/10.1890/0012-9658(2002)083%5B2248:ESORWD%5D2.0.CO%3B2 [Google Scholar]
- 44.Laake J, Rakhimberdiev BA, Turek D. Package “RMark.” 2015; Available: http://140.247.115.226/yum-rep/CRAN/web/packages/RMark/RMark.pdf
- 45.R Core Team. R: a language and environment for statistical computing. R Foundation for Statisical Development; 2016. [Google Scholar]
- 46.White GC, Burnham KP. Program MARK: survival estimation from populations of marked animals. Bird Study. 1999;46: S120–S139. 10.1080/00063659909477239 [Google Scholar]
- 47.Hanson TE, Johnson WO, Gardner IA, Georgiadis MP. Determining the infection status of a herd. J Agric Biol Environ Stat. 2003;8: 469–485. Available: http://www.jstor.org.proxybz.lib.montana.edu/stable/1400669 [Google Scholar]
- 48.Branscum AJ, Gardner IA, Johnson WO. Bayesian modeling of animal- and herd-level prevalences. Prev Vet Med. 2004;66: 101–112. 10.1016/j.prevetmed.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 49.Cassirer EF, Manlove KR, Plowright RK, Besser TE. Evidence for strain-specific immunity to pneumonia in bighorn sheep. J Wildl Manag. 2016; n/a-n/a. 10.1002/jwmg.21172 [Google Scholar]
- 50.Crawley MJ. Regression. The R Book. John Wiley & Sons, Ltd; 2012. pp. 449–497. Available: http://onlinelibrary.wiley.com/doi/10.1002/9781118448908.ch10/summary [Google Scholar]
- 51.Bates DM. lme4: Mixed-effects modeling with R. URL Httplme4 R-Forge R-Proj Orgbook. 2010; Available: http://lme4.0.r-forge.r-project.org/lMMwR/lrgprt.pdf
- 52.Risenhoover KL, Bailey JA, Wakelyn LA. Assessing the Rocky Mountain bighorn sheep management problem. Wildl Soc Bull 1973–2006. 1988;16: 346–352. Available: http://www.jstor.org/stable/3782116 [Google Scholar]
- 53.Singer FJ, Bleich VC, Gudorf MA. Restoration of bighorn sheep metapopulations in and near western national parks. Restor Ecol. 2000;8: 14–24. 10.1046/j.1526-100x.2000.80062.x [Google Scholar]
- 54.Wolff PL, Besser TE, Nelson D, Ridphat J, McMullen K, Munoz-Gutierrez J, et al. Mountain goats at the livestock-wildlife interface: A susceptible species. Proceedings Bienniel Symposium of the Northern Wild Sheep and Goat Council. Fort Collins, CO, USA: Northern Wild Sheep and Goat Council; 2014. p. 13 Available: http://media.nwsgc.org/proceedings/NWSGC-2014/013_Wolff-et-al_2014.pdf [Google Scholar]
- 55.Plowright RK, Manlove K, Cassirer EF, Cross PC, Besser TE, Hudson PJ. Use of exposure history to identify patterns of immunity to pneumonia in bighorn sheep (Ovis canadensis). Festa-Bianchet M, editor. PLoS ONE. 2013;8: e61919 10.1371/journal.pone.0061919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Festa-Bianchet M. A pneumonia epizootic in bighorn sheep, with comments on preventative management. Proceedings Bienniel Symposium of the Northern Wild Sheep and Goat Council. Northern Wild Sheep and Goat Coucil; 1988. pp. 66–76.
- 57.Ryder TJ, Williams ES, Mills KW, Bowles KH, Thorne E. Effect of pneumonia on population size and lamb recruitment in Whiskey Mountain bighorn sheep. Proceedings Bienniel Symposium of the Northern Wild Sheep and Goat Council. 1992. pp. 136–146.
- 58.Dassanayake RP, Shanthalingam S, Herndon CN, Subramaniam R, Lawrence PK, Bavananthasivam J, et al. Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Vet Microbiol. 2010;145: 354–359. 10.1016/j.vetmic.2010.04.011 [DOI] [PubMed] [Google Scholar]
- 59.Dassanayake RP, Shanthalingam S, Herndon CN, Lawrence PK, Frances Cassirer E, Potter KA, et al. Mannheimia haemolytica serotype A1 exhibits differential pathogenicity in two related species, Ovis canadensis and Ovis aries. Vet Microbiol. 2009;133: 366–371. 10.1016/j.vetmic.2008.07.015 [DOI] [PubMed] [Google Scholar]
- 60.Jaworski MD, Hunter DL, Ward ACS. Biovariants of isolates of Pasteurella from domestic and wild ruminants. J Vet Diagn Invest. 1998;10: 49–55. 10.1177/104063879801000109 [DOI] [PubMed] [Google Scholar]
- 61.Fox KA, Rouse NM, Huyvaert KP, Griffin KA, Killion HJ, Jennings-Gaines J, et al. Bighorn sheep (Ovis canadensis) sinus tumors are associated with coinfections by potentially pathogenic bacteria in the upper respiratory tract. J Wildl Dis. 2015;51: 19–27. 10.7589/2014-05-130 [DOI] [PubMed] [Google Scholar]
- 62.Fox KA, Wootton S, Marolf A, Rouse N, LeVan I, Spraker T, et al. Experimental transmission of bighorn sheep sinus tumors to bighorn sheep (Ovis canadensis canadensis) and domestic sheep. Vet Pathol. 2016; 10.1177/0300985816634810 [DOI] [PubMed] [Google Scholar]
- 63.Brunner FS, Schmid-Hempel P, Barribeau SM. Protein-poor diet reduces host-specific immune gene expression in Bombus terrestris. Proc R Soc B. 2014;281: 20140128 10.1098/rspb.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Downs CJ, Stewart KM, Dick BL. Investment in constitutive immune function by North American elk experimentally maintained at two different population densities. PLoS ONE. 2015;10: e0125586 10.1371/journal.pone.0125586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poppenga RH, Ramsey J, Gonzales BJ, Johnson CK. Reference intervals for mineral concentrations in whole blood and serum of bighorn sheep (Ovis canadensis) in California. J Vet Diagn Invest. 2012;24: 531–538. 10.1177/1040638712441936 [DOI] [PubMed] [Google Scholar]
- 66.Kraabel BJ, Miller MW. Effect of simulated stress on susceptibility of bighorn sheep neutrophils to Pasteurella haemolytica leukotoxin. J Wildl Dis. 1997;33: 558–566. 10.7589/0090-3558-33.3.558 [DOI] [PubMed] [Google Scholar]
- 67.Stothart MR, Bobbie CB, Schulte-Hostedde AI, Boonstra R, Palme R, Mykytczuk NCS, et al. Stress and the microbiome: linking glucocorticoids to bacterial community dynamics in wild red squirrels. Biol Lett. 2016;12: 20150875 10.1098/rsbl.2015.0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hall RJ, Altizer S, Bartel RA. Greater migratory propensity in hosts lowers pathogen transmission and impacts. White A, editor. J Anim Ecol. 2014;83: 1068–1077. 10.1111/1365-2656.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krumm CE, Conner MM, Hobbs NT, Hunter DO, Miller MW. Mountain lions prey selectively on prion-infected mule deer. Biol Lett. 2010;6: 209–211. 10.1098/rsbl.2009.0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: Which pattern of gene flow is most common? Evolution. 2014;68: 1–15. 10.1111/evo.12258 [DOI] [PubMed] [Google Scholar]
- 71.Sells SN, Mitchell MS, Nowak JJ, Lukacs PM, Anderson NJ, Ramsey JM, et al. Modeling risk of pneumonia epizootics in bighorn sheep. J Wildl Manag. 2015;79: 195–210. 10.1002/jwmg.824 [Google Scholar]
- 72.Manlove KR, Cassirer EF, Cross PC, Plowright RK, Hudson PJ. Costs and benefits of group living with disease: a case study of pneumonia in bighorn lambs (Ovis canadensis). Proc R Soc B Biol Sci. 2014;281: 20142331–20142331. 10.1098/rspb.2014.2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindler DE, Armstrong JB, Reed TE. The portfolio concept in ecology and evolution. Front Ecol Environ 2015. 13: 257–263. 10.1890/140275 [Google Scholar]
- 74.Wild Sheep Working Group—Disease Management Venture. In: Western Association of Fish and Wildlife Agencies [Internet]. [cited 23 Jul 2018]. Available: https://www.wafwa.org/committees___groups/wild_sheep_working_group/disease_management_venture/?usessl=1
- 75.Stephen C. Toward a modernized definition of wildlife health. J Wildl Dis. 2014;50: 427–430. 10.7589/2013-11-305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
• Table A in S1 Appendix. Summary of demographic data for 21 bighorn sheep study populations in Montana and Wyoming that were investigated as part of this study.
• Table B in S1 Appendix. Break-years and mean lamb:ewe ratios before and after the break year for each population as estimated by piecewise regression. Lamb:ewe ratios for each population with requisite data were iteratively split into “before” and “after” groups for every year in the dataset and a Poisson regression model to estimate average lamb:ewe ratios for the “before” and “after” groups was run for each year-based grouping. The break-year was chosen as the year-based grouping with the lowest AICC score. Break-years were estimated based on before and after recruitment-rates to detect potential years where respiratory pathogens may have been introduced or gone extinct from study populations. Populations whose recruitment rate declined following a break-year are highlighted in red and populations whose recruitment rate increased following a break-year are highlighted in blue. Years of documented all-age respiratory disease epizootics since 2006 are also shown for reference.
• Table C in S1 Appendix. Comparative results of recruitment and pathogen detection analyses when adjacent study populations were treated as separate populations (primary analysis) vs. aggregated into a single population (secondary analysis).
(DOCX)
• Table A in S2 Appendix. Animals sampled by population, year, and set of diagnostic protocols that were used to detect Pasteurellaceae pathogens. The numbers under diagnostic protocols indicate the number of times the specified protocol was conducted per individual. Different rows within the same population and year represent “cohorts” of animals that were sampled for respiratory pathogens using the same suite of diagnostic protocols.
• Table B in S2 Appendix. Animals sampled by population, year, and set of diagnostic protocols that were used to detect Mycoplasma ovipneumoniae. The numbers under diagnostic protocols indicate the number of times the specified protocol was conducted per individual. Different rows within the same population and year represent “cohorts” of animals that were sampled for respiratory pathogens using the same suite of diagnostic protocols.
• Table C in S2 Appendix. Detection probability parameters for Pasteurellaceae and Mycoplasma ovipneumoniae diagnostic protocols updated and adopted from Butler et al.2017. Bolded beta distributions indicate distributions that were used to model probability of pathogen presence.
• Fig A in S2 Appendix. Estimated detection probabilities and 95% confidence intervals for five respiratory pathogens in bighorn sheep. One set of protocols was used to detect the four Pasteurellaceae organisms (shaded) and a separate set was used to detect Mycoplasma ovipneumoniae (not shaded). Detection probabilities for Mannheimia haemolytica, Mannheimia spp., and Bibersteinia trehalosi are for beta hemolytic or leukotoxigenic strains. Protocols that used fee-for-service diagnostic tests are indicated with an asterisk (*) in the legend and above the upper confidence limit. The protocol not previously evaluated is shown in red. The most up to date detection probability estimates are shown with full opacity and previous estimates are dodged left and partially transparent.
• Fig B in S2 Appendix. Locations of captures and Mycoplasma ovipneumoniae detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig C in S2 Appendix. Locations of captures and leukotoxigenic Bibersteinia trehalosi detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig D in S2 Appendix. Locations of captures and leukotoxigenic Mannheimia haemolytica detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig E in S2 Appendix. Locations of captures and leukotoxigenic Mannheimia species detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig F in S2 Appendix. Locations of captures and Pasteurella multocida detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
• Fig G in S2 Appendix. Locations of captures and Leukotoxin A detections. Locations were obtained from coordinates of animal capture locations. Where capture coordinates were not available, coordinates were approximated to match landmarks associated with the capture site.
(DOCX)
Data Availability Statement
The data for this study are contained within the supporting information files and also at the Figshare database: https://figshare.com/s/a4d4fb9e21fcb3153f37.