Abstract
The glucocorticoid receptor (GR) is a constitutively expressed transcriptional regulatory factor (TRF) that controls many distinct gene networks, each uniguely determined by particular cellular and physiological contexts. The precision of GR-mediated responses seems to depend on combinatorial, context-specific assembly of GR-nucleated transcription regulatory complexes at genomic response elements. In turn, evidence suggests that context-driven plasticity is conferred by the integration of multiple signals, each serving as an allosteric effector of GR conformation, a key determinant of regulatory complex composition and activity. This structural and mechanistic perspective on GR regulatory specificity is likely to extend to other eukaryotic TRFs.
Control of gene transcription is critical for development, physiology and homeostasis. Thus, aberrant transcription regulation commonly drives disease processes. Transcriptional regulatory factors (TRFs) have a critical role in this process by recognizing specific DNA sequences to activate or repress the expression of specific genes. One of the best-characterized metazoan TRFs is glucocorticoid receptor (GR), the founding member of the nuclear receptor superfamily — proteins that evolved to bind specific small lipophilic signalling molecules1.
Expressed in nearly all vertebrate cells, GR directly up- and downregulates thousands of genes distinct to the cell type, governing various aspects of development, metabolism, stress response, inflammation and other key tissue and organismal processes. GR is encoded by the nuclear receptor subfamily 3 group C member 1 (NR3C1) gene, which is located on chromosome 5 (5q31) and is closely related to its paralogues NR3C2, which encodes mineralocorticoid receptor (MR), NR3C3, which encodes progesterone receptor (PR), and NR3C4, which encodes androgen receptor (AR). These four nuclear receptors share a common domain structure consisting of an amino-terminal domain (NTD), a zinc-finger DNA-binding domain (DBD), a hinge region and a carboxy-terminal ligand-binding domain (LBD). Embedded within these domains are regions that confer regulatory activity; for GR, these are denoted as activation function domain 1 (AF1; also known as fra/isactivating domain 1, taul and τ1), tau2 (also known as transactivating domain 2 and τ2) and AF2 (FIG. 1a). While the DBD and LBD are highly conserved between GR, MR, PR and AR, the N-terminal AF1 domains and the surrounding NTD regions within these four proteins are more divergent in sequence and size. In addition, alternative splicing and translational start sites produce multiple isoforms of the NR3C family members. The predominant and best-studied isoform of GR, GRα, is a 777-amino-acid polypeptide in humans. Other less abundant and less well-characterized, but likely functional, isoforms of GR have been described and are reviewed in REFS 2.3. For example, a GRβ isoform carries LBD alterations that change its ligand binding properties and may compete with GRα in certain experimental settings4.
Figure 1 |. GR signalling and DNA binding.
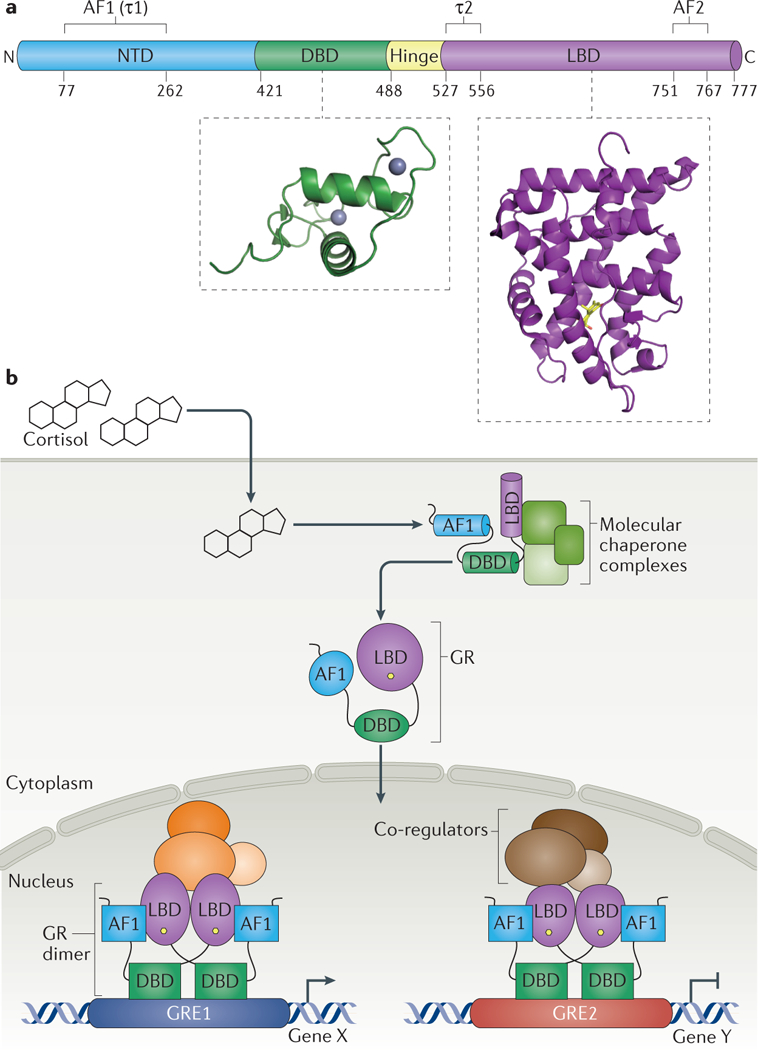
a | Linear domain structure of glucocorticoid receptor (GR). GR comprises: the amino-terminal domain (NTD), DNA-binding domain (DBD), hinge region and ligand-binding domain (LBD). Embedded in these domains are segments that participate, context-specifically, in transcription regulation: activation function domain 1 (AFl,τ1),tau2 (τ2) and AF2. Insets: Crystal structures are shown for a single DBD (green; adapted from RCSB Protein Data Bank identifier (PDB ID): 1R4R17), with coordinated zinc ions shown as grey spheres, and for a LBD (purple) liganded with cortisol (yellow) and complexed with steroid receptor co-regulator 2 (SRC-2) peptide (not shown) (PDB ID: 4P6X61). b | Overview of signalling mediated by the natural GR ligand, cortisol. Activating ligand interacts with monomeric GR associated with molecular chaperone-containing complexes in the cytosol. This induces local and remote allosteric changes that potentiate nuclear transport and other activities. Within the nucleus, GR nucleates multi-component transcription regulatory complexes containing various other transcriptional regulatory factors (TRFs) and transcriptional co-regulators at different glucocorticoid response elements (GREs) to activate or repress transcript ion of particular target genes. GRE1 and GRE2 represent distinct GREs within the genome, Gene X and Gene Y represent the genes under the control of GRE1 and GRE2, respectively.
GR activity is gated by the endogenous steroid hormone cortisol in humans, or by exogenous glucocorticoid drugs, such as dexamethasone. In the absence of ligand, apo-GR is monomeric in the cytoplasm, where it associates with molecular chaperone complexes containing heat shock protein 90 (HSP90) and HSP70 as well as other factors5; this interaction promotes high-affinity hormone binding while inactivating other receptor activities, such as nuclear localization and DNA binding. Glucocorticoid binding provokes conformational changes in GR that activate multiple functional domains, including nuclear-localization sequences within the hinge and LBD regions. After translocation into the nucleus, GR associates with specific genomic glucocorticoid response elements (GREs)6 and nucleates the assembly of transcription regulatory complexes containing GR, other TRFs and co-regulatory factors, which together activate or repress the transcription of glucocorticoid-responsive genes7·8 (FIG. 1b). Although GR forms stable complexes with DNA in vitro, it seems to exchange within seconds in vivo9,10, implying that distinct GR molecules establish initial genomic contact and regulate transcription, and that GR is actively disassembled from chromatin in vivo11,12. The role of these striking in vivo dynamics in the mechanisms of regulation is not known.
GR operates in a context-specific manner — it regulates gene networks that are precisely determined in a given context, yet displays remarkable plasticity as a function of cell type and physiological state (recently reviewed in REF. 13), leading to diverse outcomes. For example, GR-mediated gene expression governs apoptosis in the context of haematopoietic T cells14 but increases adipogenesis, lipolysis and differentiation in adipose cells15. How can both precision and plasticity of GR-regulated transcription be achieved? Addressing this apparent paradox of precision and plasticity is in fact the overarching challenge for all eukaryotic transcription regulation. GR provides a striking framework in which to address this challenge because it is expressed ubiquitously in vertebrate cells and the GR-regulated gene networks are strongly cell type-specific.
Here, we first discuss how GR interacts with the genome in vivo and in vitro, highlighting the context specificity of the in vivo interactions. Second, we document the importance of context in specifying GR activity and discuss how different ‘surfaces’ of this TRF, the accessibility of which is modulated by the context-specific cues, seem to define the ‘regulatory logic’ of GR function. Third, we consider how four classes of signals — DNA binding, ligand binding, post-translational modifications (PTMs) and interaction with other, non-GR TRFs — are integrated to affect GR structure and function. Fourth, we examine multiple classes of co-regulatory factors that associate with GR interaction surfaces to assemble transcription regulatory complexes and impose enzymatic actions that modulate transcription. Finally, we present a model that accounts for the precision and plasticity of eukaryotic transcription regulation. In this model, TRFs act as scaffolds whose conformations are altered through allostery by signalling inputs, and co-regulators serve both as readers that associate with surfaces induced on TRF scaffolds by those allosteric signals and as enzymes that modulate target gene transcription.
GR-genome interactions
TRFs, including GR, obtain a portion of their regulatory specificity by interacting with specific genomic loci. These interactions can be direct GR-DNA contacts, or GR can associate with other transcription regulators that are separately bound to DNA. Specific genomic occupancy by a TRF is typically highly context specific, with patterns of binding differing substantially in distinct cellular and physiological settings. Importantly, locus-specific GR binding is not a sufficient determinant of regulatory activity. In this section, we focus on outlining the direct and indirect interactions that GR establishes with the genome (FIG. 2); see Supplementary information S1 (table) for a summary of experimental techniques referred to in this section.
Figure 2 |. Modes of site-specific GR-genome interactions.
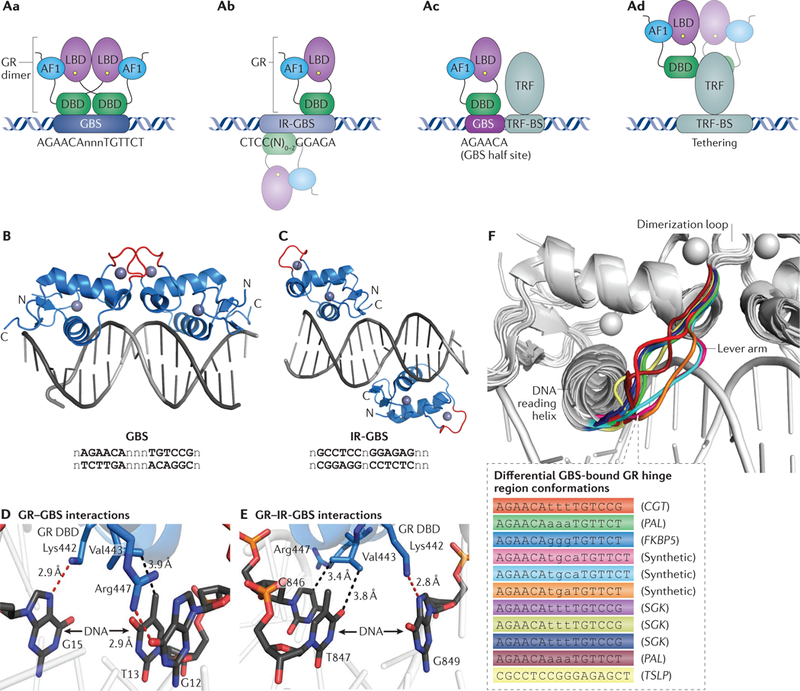
A | Glucocorticoid receptor (GR) associates with specific genomic sites in multiple ways. Aa | Two GR monomers bind a canonical GR-binding seguence (GBS) present in a glucocorticoid response element (GRE) in a head-to-head fashion; dimerization is achieved through interactions in the sister DNA-binding domains (DBDs). Ab | GR bindsto inverted-repeat GBSs (IR-GBSs).The crystal structure of this interaction (see also part Ac) shows two GR monomers bound to opposite sides of the DNA in a head-to-tail fashion; however, negative cooperativity argues that GR may bind as a monomer to IR-GBSs in vivo (thus, the second GR monomer is shown faded). Ac | GR can interact with GBS half sites; these elements typically contain a single hexamer related to the consensus seguence that is palindromic in the full GBS and may operate in conjunction with proximal non-GR transcriptional regulatory factors (TRFs), although there is no evidence for enrichment of particular TRF binding site (TRF-BS) motifs contiguous with the GBSs. Ad | GR can interact at specific genomic sites without directly binding to DNA. Here, GR is tethered to a non-GR TRF through protein-protein interactions. The faded GR monomers depict uncertainty of whether monomeric or dimeric GR binds at tethering elements. B| Overall crystal structure of the DBD of GR with the canonical GBS(RCSB Protein Data Bank identifier (PDB ID: 3FYL18).C| Overall crystal structure of the complex between the DBD of GR and the IR-GBS (PDB ID: 4HN5 (REF. 25)). D | Zoom of structure shown in B.The GR DBD makesthree base-specific contacts within the major groove of the DNA at the GBS.This interaction is mediated by hydrogen bonds (red dashed lines) and van der Waals interactions (black dashed lines). E | Zoom of structure shown in C. The GR DBD makescontactsto an IR-GBSthat are similartothose made in the case of the canonical GBS, with the exception of Arg447, which does not make any contacts with the DNA. F | Superposition of all deposited structures of the interactions between DBD and DNA reveals especially striking conformational differences within the GR lever arm (residues 469–474, a loop connecting the ‘DΝΑ-reading helix’with the dimerization region), demonstrating allosteric modulation of GR by DNA seguence. DNA seguences from each structure are listed and colour coded, the gene the seguence is derived from is listed in parenthesis (PDB IDs listed in the top-down order of the seguences given: 3FYL18, 3G99 (REF. 18), 3G6U18, 1GLU17, 1R4O17, 1R4R17, 3G9P18, 3G9O18, 3G9M18, 3G9J18 and 4HN5 (REF. 25)). CGT, ceramide UDP-galactosyltransferase (also known as UGT8); FKBP5, FK506-binding protein 5 (also known as FKBP1B); LBD, ligand-binding domain; PAL, synthetic palindromic consensus site with AAA spacer; SGK, serum/glucocorticoid-regulated kinase 1; TSLP, thymic stromal lymphopoietin protein receptor (also known as CRLF2).
Direct and indirect binding in vitro
DBDs of GR and other nuclear receptors contain two highly conserved subdomains, each with four Cys residues coordinating a single zinc ion, followed by an amphipathic helix and a peptide loop. The helix of the first subdomain contains the proximal box (P box), bearing three residues that make base-specific contacts in the major groove of the binding sequence. The second subdomain helix makes nonspecific contacts with the DNA helix backbone and minor groove, whereas the peptide loop provides distal box (D box) residues important for GR dimerization16·17.
The DBD positions GR at specific genomic sites by at least three classes of GR-DNA interactions and at least one class of GR-protein interaction (FIG. 2A). The best-characterized GR-DNA interaction is through the canonical GR-binding sequence (GBS) (FIG. 2Aa,B,D,F), which is composed of two pseudo-palindromic hexameric AGAACA repeats, separated by a three-base- pair spacer17. The ‘DNA-reading helix’ utilizes the side chains of Arg447, Lys442 and Val443 to make three base-specific contacts within the major groove of each GBS half site (FIG. 2B.D). GR binding in this head-to-head fashion stabilizes interactions between two sister GR DBDs, which promotes GR-GR and GR-DNA interactions, creating positive cooperativity18·19. Five amino acids within the D box provide critical protein-protein contacts, stabilizing the GR DBD dimer on DNA. Mutational disruption of the dimerization loop (D-loop) conformation can affect GR’s regulatory activity20. For example, Ala458 makes a hydrogen bond with Ile483 on the dimer partner, and mutation of the Ala to Thr has been shown to alter GR activity in a gene-specific manner20–22. Although this mutation was initially thought to be devoid of dimerization potential, it does still forms dimers on DNA but with diminished cooperativity19. In addition, a relatively weak LBD-LBD interaction (1.5 μΜ using GR LBD fragments in the presence of ligand and co-regulator peptide)23 may contribute to GR dimerization, but the biological significance of this contact has not been established.
The second class of GR-DNA interaction is a recently discovered inverted-repeat GBS (IR-GBS) (FIG. 2Ab,C,E), which is characterized as an alternative GR-binding motif containing CTCC(N)0–2GGAGA24. Structural studies of the GR DBD-IR-GBS revealed that GR molecules bind these non-identical sites on opposite sides of DNA, separated by a one-base-pair spacer in a head-to-tail fashion25 (FIG. 2C). One monomer makes three contacts within a high-affinity binding site mediated by Lys442 and Val443, which are the same side chains that participate in recognizing the DNA in the GR DBD-GBS structure. Arg447 establishes hydrogen bonds with a guanine in canonical GBS structures, but in the IR-GBS structure Arg447 is prevented from making these base-specific interactions owing to steric clashes with a thymine25. The other monomer uses only Arg447 to make one base-specific contact to a guanine. As the two GR monomers bind to opposite sides of the DNA at IR-GBSs, they are not in direct contact through the DBD dimerization interface.
Biochemical analyses reveal that GR binds IR-GBS elements with negative cooperativity, whereby binding of one monomer dramatically reduces the propensity of a second monomer to bind. This contrasts strongly with the cooperative binding seen at canonical GBSs25. Furthermore, nuclear magnetic resonance (NMR) studies indicate that the D-loop residues display significant changes in chemical environment consistent with dimerization when bound to a canonical GBS, whereas they are unaffected on binding to an IR-GBS26. As with canonical GBSs, it has not been examined whether LBD-LBD interactions participate in GR binding to IR-GBSs. Taken together, we infer from these in vitro studies that monomeric GR most likely binds these elements in vivo. Other 3-keto steroid receptors, MR, PR and AR, can all bind to a canonical GBS, but only GR is able to bind an IR-GBS26. Even the MR DBD, which shares 90% sequence identity with the GR DBD, cannot bind or repress transcription from an IR-GBS, owing to epistatic mutations that occurred during the evolution of the steroid receptor family that limit this function in all steroid receptors except GR26.
In a third class of GR-DNA interactions (FIG. 2Ac), not yet characterized structurally, selective binding of GR to canonical half site DNA sequences (consensus AGAACA) has been reported21,27. This binding may be facilitated by secondary interactions between GR and other non-GR TRFs bound to DNA proximal to the GBS, although there is no evidence for enrichment of particular TRF motifs contiguous with these half site GBSs and the understanding of which TRFs are involved in the regulation of gene expression at such composite GREs is still limited. Finally, GR can occupy specific genomic regions without directly binding DNA (FIG. 2Ad). In this mechanism, known as tethering, the DBD makes protein-protein contacts with other TRFs, such as activator protein 1 (AP-1) or nuclear factor-κΒ (NF-κΒ), specifically bound at their cognate sequence motifs28–31 (FIG. 2Ad).
Contextual genomic occupancy in vivo
The four classes of sequence-specific GR-DNA interactions characterized in vitro (FIG. 2A) occur at high frequency in mammalian genomes. Chromatin immuno- precipitation followed by sequencing (ChIP-Seq)32 is a robust, sensitive and fairly accurate (albeit not to single-nucleotide resolution) method to identify genomic segments occupied by GR in vivo, here termed GR-occupied regions (GORs) (for details of the technique see Supplementary information S1 (table)). It is clear from these approaches that GORs, although numerous, overlie only a small fraction of the potential sites predicted from in vitro studies, and that they are highly context specific. For example, only 0.5% of 11,666 GORs identified in mouse liver cells were found in common in four other mouse cell types examined33·34, and 83% of them were unique to liver35. Although differences in methodological and statistical approaches may complicate meta-analyses, dramatically different GORs among different cell types have been commonly noted13.
GORs typically reside proximal both to GR-regulated genes and to genes not regulated by GR21,36, so mere proximity is uninformative either about regulatory function or about which genes are regulated by which functional GORs. What is clear is that GORs do not necessarily regulate the gene closest in linear proximity within the genome, and that non-GR-regulated genes commonly reside between a GOR and the nearest GR-regulated gene. Provisional evidence suggests that most GORs reside >10 kb from a glucocorticoid-responsive gene, and that many GORs are 10–100 kb or more upstream or downstream relative to the transcription start sites of genes they are thought to regulate (FIG. 3a), and can reside within introns37. Such action at a distance is well documented, heavily studied and essentially not understood for metazoan transcription regulators.
Figure 3 |. Context-specific GR occupancy and gene regulation.
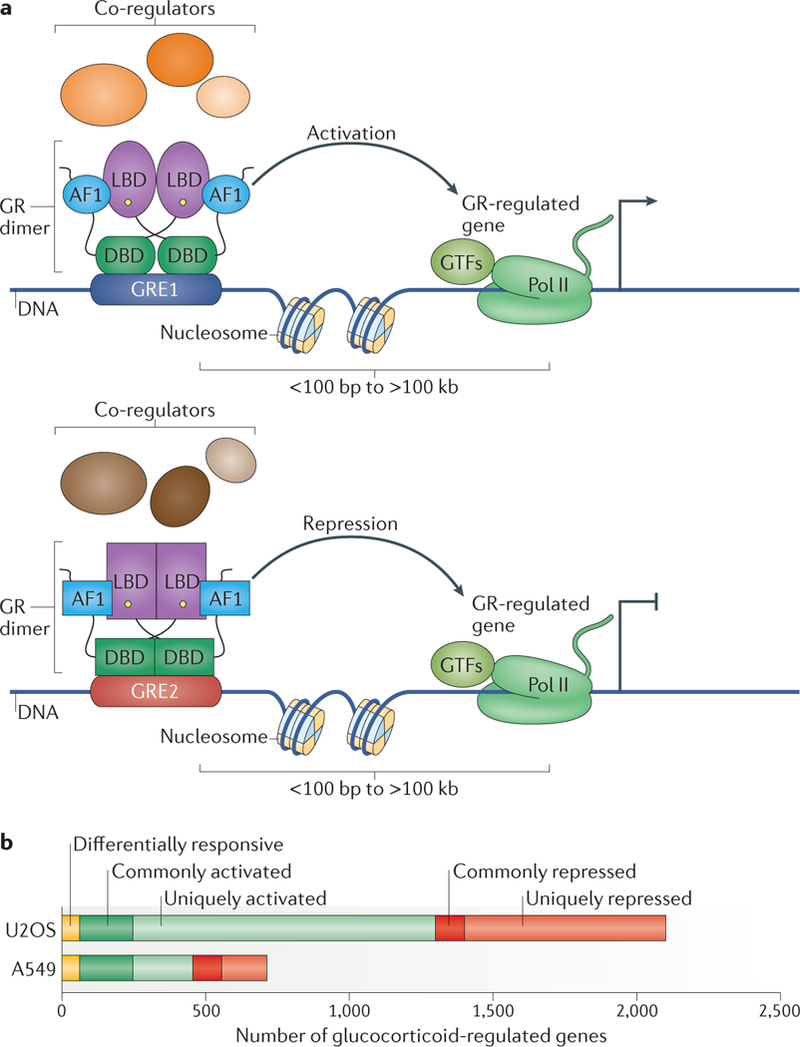
a | Glucocorticoid receptor (GR)-regulated genes are commonly linked to multiple GR-occupied regions (GORs), usually >10 kb from the transcription start site of the regulated gene, one or more of which may be a functional glucocorticoid response element (GRE) for that gene. As shown here, GREs may be near (<100 bp) to or far (>100 kb) from their target genes. b | Illumina Human Ref8 beadchip analysis of glucocorticoid-regulated genes. Genes regulated by 4-hour, 100-nM dexamethasone treatment in lung carcinoma (A549) (K.R.Y., S. Cooper, S.-H. Chen and B. Schiller, unpublished observations) and osteosarcoma (U20S)21 cells. Differentially responsive genes are upregulated in one cell line and downregulated in the other. Common genes are similarly regulated in both cell lines. Unigue genes are regulated in one cell line and not regulated in the other. The abundance of genes within the unigue and differentially regulated classes demonstrates cell-context-specific regulation by GR. Differentially responsive genes demonstrate distinct mechanisms of regulation of target genes in the two cell types. AF1, activation function domain 1; DBD, DNA-binding domain; GTFs, general transcription factors; LBD, ligand-binding domain; Pol II, RNA polymerase II.
How particular GREs and promoters interact, and what drives the specificity of one GRE-promoter interaction over others, is a focus of current debate, but one or more of several proposed gene-looping mechanisms (reviewed in REF. 38) are likely involved.
The determinants of GOR context specificity are not fully understood, but some positive and negative correlates have been uncovered. For example, DNase I-hypersensitive sites (DHSs) (see Supplementary information S1 (table)), which are thought to identify ‘open chromatin’ regions depleted of nucleosomes but occupied by TRFs and other non-nucleosomal proteins, were found in whole-genome analyses in multiple cell types to pre-exist at up to 95% of GORs formed on hormone treatment34. DHSs vastly out-number GORs (DHSs comprise ~2.5% of the genome, whereas GORs comprise 0.02–0.05% of the genome in the cell types tested)34, and it is interesting to speculate that DHSs at eventual GORs reflect non-GR TRFs pre-bound at eventual composite GREs. Similar results were obtained using formaldehyde-assisted isolation of regulatory elements and deep sequencing (FAIRE seq; see Supplementary information S1 (table)). In this case, pre-existing FAIRE seq signal indicative of open chromatin increased on glucocorticoid stimulation, suggesting that GR interacts with pre-existing open chromatin and then further alters the chromatin environment39. By contrast, negative regulatory DNA sequence (NRS) motifs were identified proximal to a subset of GBSs as anti-correlates to GR genomic occupancy. Here, GBSs that were not occupied by GR via ChIP-Seq experiments were found to have an over-representation of proximal NRS motifs40. Interestingly, NRSs did not affect DNase I sensitivity. Rather, NRSs seemed to be enriched in para-speckle proteins, which were speculated to repress transcription by blocking GR from binding to its canonical site.
GRE context and regulatory logic
The context specificity of GORs demonstrates that DHSs, recognition sequences (GBSs) identified in vitro and non-GR TRFs with which GR interacts are insufficient to localize or predict the localization of GR in vivo. A dramatic example is the IR-GBS-containing GORs, which are widespread and highly occupied in mouse fibroblasts24 but are detected only sparingly in certain other cell types41–43. The relationship of a GOR to a functional GRE is similarly complicated by context. Transient reporter assays, in which a plasmid bearing a genomic fragment underlying a GOR is inserted adjacent to a minimal promoter and reporter (typically luciferase) gene and is transfected into a GR-expressing cell line (see Supplementary information S1 (table)), have been widely used to assess GOR regulatory capacity, and therefore to infer GRE activity. However, the transient conditions for a newly introduced plasmid are surely different from the chromosomal and cellular settings of the endogenous GOR, and the strong context specificity of GR-mediated gene regulation raises concerns about the validity of GRE activity inferred using that approach.
Genome editing using zinc-finger nucleases or transcription activator-like effector nucleases (TALENs) provides, in principle, routes for assaying GRE activity in situ. Unfortunately, the technical complexity of these methods, together with incomplete appreciation of the overriding importance of context, has left these approaches largely unused. As a result, only one GRE has been validated at its endogenous locus in vivo. In that case, a short deletion introduced into the first intron of the mouse circadian clock gene period circadian homo- logue 2 (Per2) fortuitously covered a GOR ~25kb down-stream from the transcription start site and produced allele-specific loss of glucocorticoid-mediated induction of Per2 expression in mesenchymal stem cells44. With the emergence of clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein (Cas) technologies45, precise genome editing will rapidly become the ‘gold standard’ for validation of GRE activity in any chosen cell type or physiological context. Interestingly, underscoring the importance of context, the Per2 GRE identified in mouse mesenchymal stem cells seems not to be functional in human lung carcinoma cells, as assessed by CRISPR-driven deletion (K. Ehmsen, personal communication).
As mentioned above, the genes and gene networks regulated by GR within different cell types are distinct. For example, FIG. 3b shows the vastly different sets of target genes regulated by the synthetic glucocorticoid dexamethasone in two different cancer cell lines. Most GR target genes are unique to one cell line, demonstrating striking cell-context specificity. Indeed, 62 genes regulated in both cells lines are differentially responsive (activated in one cell line, repressed in the other), showing that even genes regulated by GR in both cell lines can be controlled by distinct mechanisms.
Genetic strategies have also been used to identify GR surfaces that are engaged in the regulation of gene expression in different contexts. For instance, a triple point mutant, denoted 30iiB (in which Glul98 is changed to Lys, Phel99 to Leu and Trp213 to Arg), abrogated activation by the AF1 domain, and single point mutants within the DBD dimerization domain (Ala458 to Thr) and AF2 (Glu755 to Arg) knocked out activation by those domains individually for genes tested in U2OS cells. These three surfaces, together with a naturally occurring splice variant, GRγ, which inserts a single amino acid (Arg452) into the lever arm region of the DBD, were found in U2OS cells to function in gene-specific patterns. For example, GR-mediated activation of one set of genes depended solely on AF2, whereas another set used a combination of AFl, DBD and AF2 (REFS 21,46,47). As each altered GR region contains protein interaction domains, it is reasonable to assume that the different patterns of domain use reflect assembly of distinct regulatory complexes at the GREs controlling the different sets of genes.
Beginning with context determinants defined in the genome and in GR, what can be inferred about the regulatory logic that confers such specificity yet permits facile plasticity when conditions are altered? By manipulating ligand dose, duration of treatment and other parameters that affect relative levels of GR activity, one study uncovered ‘toggle switch’-like behaviour in U2OS cell GREs in which a set of genes was activated at low levels of GR activity and underwent a dramatic shift to repression at a transition point as GR activity increased48. This stereotyped regulatory mode has been called incoherent type 1 feed-forward loop (I1-FFL) logic49 and suggests that GREs that regulate these differentially responsive genes initiate two arms of a regulatory circuit, yielding net activation or repression as the level of GR activity is altered by various context determinants. Different FFL network motifs initiated by GR have been described in macrophages50. These findings imply that some, perhaps many, GREs may contain molecular switches that readily confer plasticity in a highly context-sensitive manner. Thus, the GRE seems to emerge as the regulatory logic module, driven by multiple signals and conferring specificity while enabling plasticity. Below, we suggest a conceptual and mechanistic basis for such a regulatory logic for GR, and likely for other eukaryotic TRFs.
Allosteric effectors of GR
If, as outlined above, regulatory specificity is determined by the assembly of distinct regulatory complexes, each attuned to a particular gene, cell and physiological context, what are the molecular determinants of context, and how are their effects communicated to GR? Extending a few examples into a general conclusion, current data suggest that four classes of cellular signals interact with GR, allosterically altering its conformation. The first two classes, the hormonal ligands and covalent PTMs, represent the endpoints of various signal transduction pathways that communicate physiological context to the receptor. A third signal class is the extensive array of DNA sequences bound specifically by GR; these differ at different GREs, and thus these signals impart gene-specific context to the receptor. Finally, the particular cell-specific repertoire of TRFs, including the variability in levels of their expression and activities, are reflections of cell type. Thus, the differences in the expression of TRFs that interact with GR convey cell-specific, allosteric signals. As described above, GR-TRF interactions occur at composite GREs and their special subset, the tethering GREs.
DNA-binding sequences
High-throughput studies have identified thousands of proteins that interact with DNA51, and TRFs are the most common group with sequence-specific DNA-binding activity. Specific DNA sequences not only serve as platforms for binding but also seem to act as direct allosteric effectors of TRFs18,19,52.
Although the oligomeric state of GR (monomeric, dimeric or higher order) at different points in signalling in vivo is a matter of current debate42,43,53,54, it is established that full-length, ligand-bound GR is a monomer in solution in vitro, even at high concentrations55·56. At canonical GBSs, DNA binding increases the local concentration and favourable orientation of protein- protein interactions between two DBDs, resulting in productive dimerization17. Indeed, not only does GR make sequence-specific interactions with each hexameric half site of the DNA but also the three-base-pair spacer between the half sites, at least at certain GBSs, drives cooperative DNA binding and dimerization18. Moreover, allosteric changes provoked by one half site sequence can be transduced intermolecularly across the dimer interface, conformationally altering the dimer partner19. Spacer sequences can alter DNA shape, resulting in conformational changes that originate from the DNA- reading helix, allosterically propagate through the lever arm and alter the conformation of the receptor’s D box, ultimately affecting GR transcriptional activity19. Such conformational distinctions can be seen in comparisons of different GR DBD-GBS structures (FIG. 2F). Finally, recent work has shown that sequences at the +8 and −8 positions flanking the GBS that alter DNA conformation also affect GR DBD structure, as assessed by NMR analysis in vitro and using a zinc-finger nuclease-generated, genomically integrated GBS-reporter system and endogenous GR in vivo 57. Collectively, these studies suggest that DNA sequence-specific conformational states of GR result in the generation or stabilization of distinct patterns of GR surfaces, which serve as interaction platforms, driving alternative transcriptional outcomes.
Additionally, NMR analyses revealed allosteric communications between the dimerized GR monomers at canonical GBSs19, and molecular dynamics simulations (for further details of the method, see Supplementary information S1 (table)) suggested the existence of allosteric regulation between monomers at IR-GBSs (although this might not be relevant in vivo, as IR-GBS elements may be occupied by only monomeric GR in the in vivo scenario26). In both cases, DNA acts as a ligand to impart allosteric changes that could affect affinity for co-regulators and ultimately regulatory out-comes, implying that the allosteric transitions extend into the NTD and LBD. So far, structural studies of GR in the context of allosteric modulation by DNA have been limited to isolated domains, but work with a full-length vitamin D receptor (VDR) and retinoid X receptor (RXR) heterodimer confirms allosteric communication throughout nuclear receptor complexes on DNA binding58. Indeed, hydrogen-deuterium exchange mass spectrometry (HDX-MS) on the full-length, ligand-bound VDR-RXR-DNA complex revealed conformation changes on binding of ligands, DNA and co-regulators. Changes within the VDR DNA-binding sequence (VBS) read through the VDR DBD had far-reaching intramolecular allosteric effects that altered the solvent accessibility of regions within the sister LBD of the complexed RXR molecule58. Future work with full- length GR will be essential to fully describe the allosteric consequences of differential GBS binding.
LBD-binding ligands
The NR3C family members, GR, MR, PR and AR, evolved from a common ancestor and share high sequence similarity within their LBDs; however, key architectural differences of each LBD produce strict ligand selectivity59. Differences in the aromatization of the A ring of the ligand (see FIG. 4a for ligand ring naming and carbon numbering), driven in part by the 3-oxo group interaction with polar residues within NR3 family LBDs distinguish endogenous NR3C family member 3-keto ligands from related NR3A family oestrogen receptor (ER) 3-hydroxy-specific ligand recognition59,60 (FIG. 4a). The first crystal structure of the GR LBD complexed with a ligand (dexarnethasone) displayed an intricate network of polar and nonpolar interactions, which together define ligand selectivity — every polar atom within dexarnethasone directly interacts with the GR LBD23.
Figure 4 |. GR-ligand interactions.
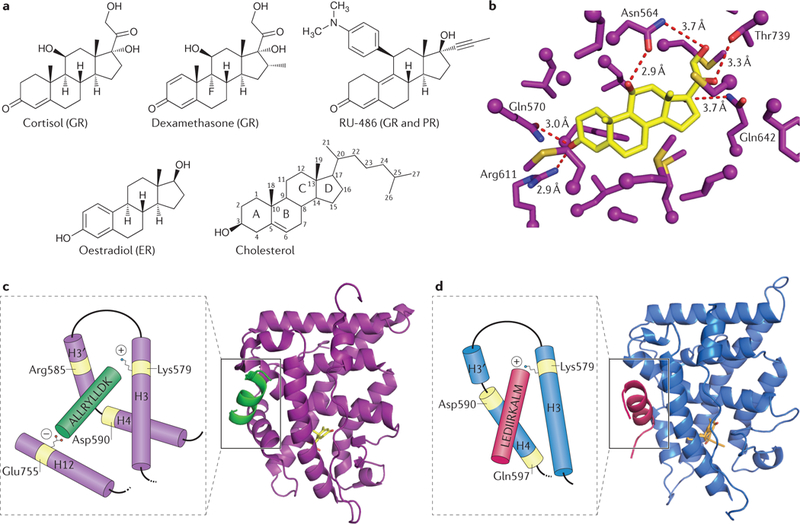
a | Cortisol, dexamethasone and RU-486 are three glucocorticoid receptor (GR) ligands; RU-486 also binds to progesterone receptor (PR); oestradiol, which binds to oestrogen receptor (ER) but not GR, is shown for comparison; cholesterol is shown to provide the sterol carbon numbering convention. b | Ligand-binding pocket (purple) of the ligand-binding domain (LBD) of GR bound to its endogenous ligand, cortisol (yellow). Residues within 4.2 Å are shown, and hydrogen bonds are depicted by dashed red lines. This structure highlights the intricate network of interactions between GR and its ligand, as well as the amount of unoccupied space within the ligand-binding pocket with the ligand bound. This is a zoom of the ligand binding pocket from structure shown in panel c. c | Overall structure of the GR LBD (purple) bound to cortisol (yellow) and the steroid receptor co-regulator 2 (SRC-2) peptide (green) (RCSB Protein Data Bank identifier (PDB ID): 4P6X61).The inset cartoon represents how the LXXLL motif (the so called nuclear receptor box) of the co-regulator peptide interacts with GR. The peptide is held in place by a conserved charge clamp interaction; the positively charged Lys579 residue (+) and negatively charged Glu755 residue (−) on helix 3 (H3) and helix 12 (H12), respectively, mediate this interaction. There is an additional charge clamp that occurs through the residues Arg585 and Asp590. This is referred to as the active conformation, and is associated with specific co-regulator binding. d | Overall structure of the GR LBD bound to RU-486 (yellow) and the nuclear receptor co-repressor (NCoR) co-regulator peptide (pink) (PDB ID: 3H52 (REF. 164)). The inset cartoon represents how the extended co-repressor nuclear receptor (CoRNR) boxes of the co-regulator peptide interact with GR. The extended peptide makes only one of the conserved interactions, through Lys579, and H12 is displaced. This is considered an inactive form of the LBD. Differences in GR conformations presented in parts c and d indicate that different ligands can promote the formation of alternative protein surfaces on GR that in turn differentially affect co-regulator binding.
Despite strong binding specificity, dexarnethasone occupies only ~65% of the GR ligand-binding pocket (leaving >200 Å3 excess volume within the 590-Å3 binding pocket23); similarly, the endogenous ligand cortisol binds specifically but fails to fill the binding pocket61 (FIG. 4b). The additional volume within the binding pocket offers potential space for interaction with alternative modulatory ligands. Moreover, LBD structures bound to cortisol and an alternative GR ligand, RU-486, (FIG. 4c,d) reveal extensive structural malleability within the LBD, which seems to enable interaction with a wide range of potential ligands62 that could confer different allosteric changes, resulting in ligand-specific alterations in regulatory outcomes. The capacities of functional GR ligands to fill only a portion of the binding pocket or to alter the shape of the pocket challenge and, at the same time, liberate the concept and practice of ligand design. Using the dexamethasone-occupied pocket as a guide, arylpyrazole compounds were developed as nonsteroidal GR ligands and shown indeed to differ from dexarnethasone in their phenotypic and molecular actions in several target cell types63. Notably, selective GR modulators (SGRMs) that preserve the anti-inflammatory and immunosuppressive actions of standard glucocorticoids but do not show the adverse effects that accompany chronic glucocorticoid therapeutic regimens have long been sought without success. Hence, discovery of molecules that achieve pre-selected, context-specific regulatory outcomes has proven difficult; we suggest in our concluding remarks a targeted approach.
In summary, GR ligands are critical physiological and pharmacological context inputs, conferring allosteric transitions64 that affect GR-associated molecular chaperone affinities or functions and activate GR nuclear- localization signals and DNA-binding activity. Even subtle modifications of GR ligand chemistry or dose can affect gene-specific GOR formation48·63, or regulatory complex composition and/or function (for example, altered histone acetyltransferase (HAT) activity) without affecting GOR formation itself63.
PTMs
Covalent PTMs of TRFs are conferred as the endpoints of cell signalling pathways that provide physiological context information distinct from that provided by non-covalently associated hormonal ligands. PTMs can confer allosteric transitions, create or inactivate protein interaction surfaces, and affect TRF protein localization, stability, DNA binding, ligand response and regulatory activity in a context-specific manner. GR can be modified by several PTMs at distinct sites65–69 (FIG. 5). Here, we summarize some of these PTMs and their effects on GR action.
Figure 5 |. Sites of glucocorticoid receptor post-translational modifications.
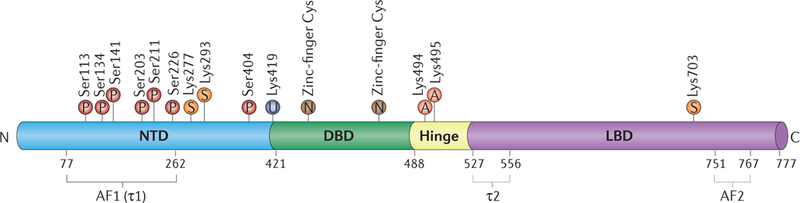
Major reported modifications, including phosphorylation (P), sumoylation (S), ubiguitylation (U), acetylation (A) and nitrosylation (N), are mapped onto the glucocorticoid receptor domain schematic. AF, activation function domain; DBD, DNA-binding domain; LBD, ligand-binding domain; NTD, amino-terminal domain.
Phosphorylation.
Phosphorylation (generally on Ser, Thr or Tyr) of nuclear receptors is involved in ligand binding, nuclear localization, DNA binding, and modulating interactions with co-regulators68,70. GR maintains a basal level of phosphorylation, but additional sites are phosphorylated on ligand treatment71–73. To date, there are seven experimentally confirmed phosphorylation sites on GR clustered within the NTD: Ser113, Ser134, Serl41, Ser203,Ser211, Ser226 and Ser404 (REF. 65). These residues are conserved among humans, mice and rats73. Although the enzymes responsible in vivo are uncertain, these sites can be modified in vitro by cyclin-dependent kinases (CDKs), mitogen protein kinases (MAPKs), JUN N-terminal kinases (JNKs) and glycogen synthase kinase 3 (GSK-3)74, implying that multiple signaling pathways communicate with GR. Mutational analysis of particular sites led to mixed reports of the effects of phosphorylation on the transcription regulatory activities of GR75,76, consistent with strong gene-, cell- and physiology-specific context dependence, as expected.
In general, phosphorylation of GR increases the protein half-life, and mutation to Ala at several different phosphorylation sites, one site at a time, each resulted in rapid protein degradation67,76. Phosphorylation of sites Ser203, Ser211 and Ser226, which are located within AF1, are predicted to affect exposure of protein surfaces critical for cofactor interactions77,78. Phosphorylation of Ser211 results in increased recruitment of GR to GORs and subsequent regulation of GR target genes79. Mutation of Ser203 prevents phosphorylation at Ser226, suggesting interdependence of those modifications72,80. Upon ligand binding, Ser203 is phosphorylated and GR is selectively partitioned to the nucleus. Mutation of that site precludes nuclear accumulation and subsequent GR-dependent gene regulation80, whereas Ser226 phosphorylation, presumably via JNK signalling pathways, results in increased nuclear export, thus decreasing gene regulation through GR68. Ser203 is phosphorylated in vitro by CDK and MAPK74,81. This apparent integration across different signalling pathways may contribute to the differences in transcriptional outcomes observed in different cellular and physiological contexts70,80. Another site, Ser404, phosphorylated in vitro by GSK-3, seems to be hypo-phosphorylated in nuclear fractions, but loss of Ser404 phosphorylation changes the conformation of GR, alters cofactor recruitment and transcriptional responses, and increases glucocorticoid-induced apoptosis82,83.
Ubiquitylation.
The 8.5-kDa ubiquitin polypeptide is covalently attached to Lys residues of a target protein and promotes protein turnover by targeting the protein to the proteasome for degradation. Cells treated with pro- teasome inhibitors showed enhanced GR regulatory activity67,84 and increased in vivo DNA occupancy time, indicating that ubiquitylation is involved in regulating GR stability11,12. Furthermore, ubiquitylation of GR at Lys419 (REF. 85) was shown to stimulate GR nuclear export and subsequent degradation86.
Sumoylation.
The small ubiquitin-related modifier 1 (SUMO-1) polypeptide, when covalently linked to Lys residues of target proteins, can alter their stability, localization or transcription regulatory activity67,87. GR can be sumoylated at three sites: Lys277 and Lys293 within the NTD and Lys703 within the LBD; these modifications have been shown to have effects on GR activity that are highly context dependent66,88. Genome-wide profiling of GR sumoylation mutants revealed enhanced GR recruitment to DNA, specifically at genes involved in cell growth, proliferation and survival89.
Although its functions are context dependent, sumoylation has most commonly been linked to transcription repression90,91. One recent example concerns the repression of inflammatory genes92,93 bearing IR-GBS- containing GORs. Specifically, sumoylation at Lys293 within the NTD is required for IR-GBS-mediated repression, but mutations of the other SUMO sites have no effect. Sumoylation of Lys293 also promotes the recruitment of co-regulators silencing mediator of retinoic and thyroid receptors (SMRT) and nuclear receptor co-repressor (NCoR) but does not affect GR-mediated activation from certain genes linked to canonical GBS- containing GORs92. It seems that GR sumoylation not only facilitates the assembly of repressive regulatory complexes but also assists in the binding of GR to weaker associated sites, at least at the IR-GBS-containing GORs examined. Furthermore, loss of this sumoylation site results in diminished GR-mediated repression at certain AP-1 and NF-κΒ tethering sites93, apparently owing to inhibition of regulatory complex assembly.
Acetylation.
GR is acetylated at Lys494 and Lys495 within the hinge region at a common acetylation motif, KXKK (where X is any amino acid). This motif is conserved among the 3-keto steroid receptors, suggesting that this acetylation may be important for certain general functions of these receptors. The HAT proteins circadian locomoter output cycles protein kaput (CLOCK) and BMAL1 (encoded by ARNTL) were inferred from cell-based assays to acetylate GR at these Lys residues94. KXKK acetylation reduced the affinity of GR binding to canonical GBSs in vitro and also reduced its ability to regulate transcription in transfection assays. The regulation of this modification by CLOCK is critical for circadian rhythm maintenance95. GR is deacetylated in vitro by histone deacetylase 2 (HDAC2), which seems to be important for repression of NF-κB-regulated genes96.
Nitrosylation.
S-Nitrosylation involves the covalent attachment of a nitric oxide to a thiol group on a Cys residue. Nuclear receptors contain two zinc-finger domains, with four Cys residues each. Nitric oxide can target these residues and cause the release of bound Zn2+. For GR, this modification has been shown to inhibit ligand binding69. Exposure of COS-7 cells to exogenous nitric oxide sources has been shown to inhibit DNA binding and dimerization of nuclear receptor complexes such as VDR-RXR97, but to date this has not been studied for GR.
Composite GRE-bound non-GR TRFs
The fourth class of allosteric effectors that alter GR activity are non-GR TRFs bound at composite GREs.Correlative studies suggest that composite GREs are 0.5–2-kb genomic segments containing one or more of the four classes of GR-binding motifs described above, clustered with binding sequences for non-GR TRFs36. In the first-described composite GRE, GR interacted both with GBS DNA and with an AP-1 factor bound contiguously, and either activated or repressed transcription, depending on the subunit composition of the AP-1 factor28,98,99. A simple interpretation is that particular combinations of non-GR TRFs bound at composite GREs could bias GR occupancy at those elements, and by directly interacting with GR confer allosteric effects, modulating GR activity. Occupancy of the non-GR TRF-binding sites at a given composite GRE will differ in different cell types, owing to cell context-specific differential absolute and relative expression and activities of the respective TRFs, thereby providing a context- dependent signal for transcription regulation mediated by GR.
Co-regulators as GR signalling readers
TRFs such as GR nucleate the assembly of large (~102 polypeptides and non-coding RNAs) transcription regulatory complexes, typically including other TRFs and distinct collections of co-regulatory factors, which confer structural or functional changes on the transcription machinery or chromatin, thereby positively or negatively modulating target gene mRNA production. Roughly 300 co-regulators have been identified, many of which are themselves multifactor complexes, and shown to exhibit a wide range of functions100,101. Which co-regulators interact with GR depends not only on co-regulator availability in a given cell type but also on the integrated effects of the four classes of signals that communicate context information to GR discussed above. Hence, co-regulators can be viewed as readers of GR-mediated signalling — it is the co-regulators that convert the integrated, signal-driven allosteric transitions at distinct receptor surfaces into context-specific transcription regulatory actions. Originally classified as co-activators and co-repressors, many co-regulators have been recognized to both activate and repress genes in a context-specific manner102. Here, we classify co-regulators based on their mechanisms of action rather than on the regulatory out-come in any particular context, and we discuss those classes shown to date to interact with GR (TABLE 1).
Table 1.
Five functional classes of co-regulators reported to interact with GR
| Functional class | Examples | GR-interaction surfaces | Targets and regulatory outcomes | Refs |
|---|---|---|---|---|
| Structural and enzyme- interacting |
pl60 SRC family: • SRC-1 (also known as NCoA-1) • SRC-2 (also known as NCoA-2, TIF2 and GRIP1) • SRC-3 (also known as NCoA-3, CIP, AIB1 and TRAMl) |
LXXLL motif interacts with AF2 and bHLH-PAS domain interacts with AF1 |
• Transcription activation best characterized; but at least SRC-2, at can also repress gene expression in appropriate contexts • Interaction increasesα-helical content of AF1, increasing its structural stability |
29,100, 109, 116–134 |
| Mediator | LXXLL motif within MED1 interacts with the LBD of GR, and MED14 interacts with AF1 |
• Recruitment and allosteric regulation of transcriptional machinery, including RNA polymerase II and TFIIH • Recruitment of multiple enzymatic activities, including phosphorylation and acetylation (at SerlO and Lys 14, respectively) of histone H3 through associated kinase CDK8 and HATGCN5L |
135–138, 140–143 |
|
| HIC-5 (also known asTGFB1|1) | HIC-5 binds to the tau2 domain within the GR hinge |
• Transcription complex assembly and Mediator recruitment • May block chromatin remodelling • Differently affects DNA binding and transcription regulatory activity of different gene classes |
112,113 | |
| COCOA | Binds indirectly via interaction with the amino terminus of pl60 |
• Acts synergistically with pl60 | 119,120 | |
| CCAR1 | Directly bindsto GR or is indirectly recruited by COCOA |
• Recruits Mediator and p160to response elements |
121 | |
| Chromatin- remodelling |
BRG1 (also known as SMARCA4) and BRM SWI/ SNF-related ATPases |
Multiple BAF subunits interact withthe DBD, LBD and AF1 in different contexts |
• ATP-dependent repositioning of nucleosomes • Relieves repressive state of chromatin • Multiple context-specific effects of BRM knockdown on GR-dependent genes |
146–153 |
| Met hyltransferases | CARM1 (also known as PRMT4) | Binds indirectly via interaction with carboxy terminus of pl60 |
• Methylates histone H3 and non-histone proteins in vitro • Activates SRC-2-dependent genes |
120,154 |
| Lys methyltransferase G9a (also known as EHMT2) |
N-terminal portion of G9a binds to the GR NTD |
• Positive regulation by enhanced recruitment of CARM1 and p300to GR target genes • Facilitates methylation of Lys9 on H3, which is associated with transcription repression |
155 | |
| HATs | CBP/p300 | Binds directly to AFl or indirectly through partner pl60-AF2 interations |
• Acetylation of histones • Cell-type-specific transcriptional outcomes |
100,117, 124,157, 158,160, 165 |
| PCAF (in ADA and SAGA complexes) |
Binds directly to AFl or indirectly through either partner pl60- AF2 or CBP/p300-pl60-AF2 interactions |
• Acetylation of histones • Less well defined, but contains TAF subunits, which have been postulated to aid in the recruitment of general transcription factors to DNA |
100,159, 166 |
|
| HDACs | NCoR, SMRT |
• (L/I)XX(I/V)I or LXXX(I/L)XXX(I/L) CoRNR motif interacts with AF2 when GR is bound to RU-486 • NCoR and SMRT interact with NTD in GR bound to IR-GBS or tethered GR when GR is bound to a standard ligand |
• Transcription repression correlated with histone deacetylation in most cases |
24,92,93, 128,163, 164,167, 168 |
ADA, Ada acetyltransferase; AF,activation function domain; BAF, BRG/BRM-associated factor; bHLH-PAS, basic helix-loop-Fielix-Per/ARNT/Sim; BRM, BraFima; CARM1, coactivator-associated Arg metFiyltransferase 1; CBP, CREB-binding protein; CCAR1, cell division cycle and apoptosis regulator 1; CDK8, cyclin-dependent kinase 8; COCOA, coiled-coil coactivator; CoRNR, co-repressor nuclear receptor; DBD, DNA-binding domain; GBS, GR-binding seguence; GR, glucocorticoid receptor; FIAT, hi stone acetyltansferase; HDAC, histone deacetylase; IR-GBS, in verted-repeat GBS; LBD, ligand-binding domain; MED, Mediator of RNA polymerase II transcription subunit; NCoR, nuclear receptor corepressor; NTD, N-terminal domain; PCAF, p300/CBP-associated factor; SAGA, Spt-Ada-Gcn5-acetyltransferase; SMRT, silencing mediator of retinoic and thyroid receptors; SRC, steroid receptor co-regulator; TAF, TBP-associated factor; TFIIH, transcription factor II human.
Basis of GR-co-regulator interactions
Structural analysis has provided deep insight into the interaction of co-regulators with regulatory domain AF2 near the GR C terminus, a highly conserved 12-α-helix and 4-β-sheet motif that folds into a 3-layer helical bundle103. AF2 of nuclear receptors is minimally comprised of helices 3, 4 and 12 (REFS 104,105), with which nuclear receptor co-regulators interact through highly conserved LXXLL motifs (so called nuclear receptor boxes)106·107 or through (L/I)XX(I/V)I or LXXX(I/L) XXX(I/L) motifs (referred to as co-repressor nuclear receptor (CoRNR) boxes)108 (FIG. 4c,d). By contrast, the entire NTD of GR, which includes the AF1 domain, is highly disordered. Interaction of steroid receptor co-regulator 2 (SRC-2; also known as NCoA-2, TIF2 and GRIP1) and TATA box-binding protein with AF1 stabilizes it and increases its α-helical content109·110 (similar disordered regions are commonly directed to fold into well-ordered functional domains in other TRFs and other proteins111); little else is known of the structure and dynamics of the functional domain or of the AF1- co-regulator interfaces. Still less is known structurally about the tau2 domain within the GR hinge domain (FIG. 1 a), which interacts with co-regulator HIC-5 (also known as TGFB1I1)112,113 (TABLE 1). Importantly, because systematic mutagenesis and mapping in multiple contexts has not been carried out, we are far from understanding even the number of functional surfaces that potentially might form within any given GR domain.
Functional classes of GR co-regulators
TABLE 1 displays five functional classes of co-regulators reported to interact with GR, together with specific examples from each class (for a more complete description of co-regulators, see REFS 102,114,115). As this is an active field of research, additional GR-associating co-regulator classes are probably yet to be discovered. These co-regulators typically interact with GR bound by one of the standard, cortisol-like glucocorticoid ligands, such as cortisol itself or dexamethasone. One class of co-regulators, the HDACs, binds GR at canonical GBS- containing sites only when bound by exogenous drug RU486, thus calling into question the physiological significance of these interactions. By contrast, at IR-GBS- containing sites, HDACs interact with GR bound by standard ligands. Below, we briefly consider examples from each functional class.
Structural and enzyme-interacting
pl60 SRC family.
The pl60 family contains three members: SRC-1 (also known as NCoA-1),SRC-2 (also known as NCoA-2, TIF2 and GRIP1) and SRC-3 (also known as NCoA-3, CIP, AIB1, ACTR and TRAM1)100. These proteins function as scaffolds and can associate with between six and ten other proteins116, including other coregulators, such as coiled-coil coactivator (COCOA), cell division cycle and apoptosis regulator 1 (CCAR1), histone acetyltransferases (CREB-binding protein (CBP)/p300 and CBP/p300-associated factor (PCAF; also known as KAT2B)) and histone methyltransferases (coactivator- associated Arg methyltransferase 1 (CARM1; also known as PRMT4) and protein Arg N-methyltransferase 1 (PRMT1))117–121. Knockdown of individual SRC proteins showed context-specific effects on GR-mediated transcription regulation122,123.
The SRC proteins contain three functional domains: the N-terminal basic helix-loop-helix-Per/ARNT/Sim (bHLH-PAS) domain, which binds to the AF1 domain of GR, increasing its stability and α-helical content109; the central receptor interaction domain (RID), which contains three LXXLL motifs and two transcription activation domains (AD1 and AD2) and associates with the AF2 domain of GR, within the LBD124,125 (see also FIG. 4C); and the C-terminal activation domains, which interact with HATs and histone methyltransferases118.
GR interacts preferentially with SRC-2 over other pl60 members126–128. SRC-2 can form distinct foci within the nucleus that also contain p300, PCAF and the nuclear receptors GR, AR and ER, as well as others. Dexamethasone-bound, but not RU486-bound, GR was observed to localize to these substructures129. SRC-2 can also be phosphorylated in a GR-interaction- dependent manner, and mutations to key phosphorylation sites result in reduced expression from selected target genes130. Also functional in repression, SRC-2 was the first co-regulator shown to deploy distinct regulatory domains131 for up- and downregulation of GR-dependent transcription in different contexts29,132–134.
Mediator and other structural and enzyme-interacting.
The ~30-protein, 1.2-MDa Mediator complex forms a physical link between TRFs, such as GR, and the general transcription machinery, and regulates transcription by affecting RNA polymerase II and TFIIH activity135. Structural studies suggest that Mediator activity is controlled allosterically136–139. For example, interaction with LBDs of nuclear receptors and other TRFs induces conformational changes resulting in the formation of a Mediator pocket domain, which enables Mediator-RNA polymerase II interaction137·140. Core Mediator components also stably interact with kinase modules, including CDK8 or CDK19, which in turn recruit other enzymes, such as the HAT GCN5L (also known as KAT2A)141. GR binds to two distinct Mediator subunits. Mediator of RNA polymerase II transcription subunit 1 (MED1) interacts with the GR LBD in a ligand-dependent manner via LXXLL motifs, whereas the MED14 mediator subunit, establishes interactions with the AF1 domain of GR independent of ligand binding142. GR target gene regulation seems to be differentially dependent on MED1 or MED14 (REF. 143). Other co-regulators, such as HIC-5, COCOA and CCAR1, also interact either directly or indirectly with GR, contributing to the formation of multi-subunit, context-specific transcription regulatory complexes112·113·119–121.
Chromatin-remodelling
Human cells contain an extensive family of evolutionarily conserved multi-protein SWI/SNF-related ATPases that catalyse various transformations of the regular nucleosome packaging of chromatin144,145. Predominant among them are the closely related Brahma (BRM) and BRG1 (also known as SMARCA4) complexes, which interact context-specifically with GR146,147, repositioning nucleosomes to facilitate the accessibility of DNA to TRFs and transcriptional machinery148–150. The recruitment of SWI/SNF complexes by GR to facilitate tran-scription151 served as the first published report of the existence of TRF co-regulators. GR has been shown to interact directly with multiple subunits of the BRM and BRG1 complexes152,153, with binding reported to the DBD, LBD and AF1 domains of GR, probably in a context-specific manner.
Methyltransferases
PRMTs, such as CARM1, and lysine methyltransferases, including G9a (also known as EHMT2), methylate histone and other proteins. They interact with GR directly or with GR-bound co-regulators p160 or p300, and either activate or repress GR target genes, in a context-dependent manner154,155.
HATs
HATs modify histones and other proteins, forming ε-Ν-acetyllysine at selected Lys residues156. Among the various HAT families, CBP, p300 and PCAF (as a component of ADA (Ada acetyltransferase) and SAGA (Spt-Ada-Gcn5-acetyltransferase) complexes) interact with GR directly, through interactions with the AF1 domain, or indirectly, through pl60 co-regulators associated with the AF2 domain of GR, to modulate transcription100,124,157–159. HATs target histones as well as non-histone proteins (including nuclear receptors) in a context-specific manner117,160 and, although they are generally associated with activation of transcription161, their mechanisms of action and regulatory outcomes seem also to be context-dependent162.
HDACs
NCoR and SMRT form multi-protein complexes that include HDACs163. NCoR and SMRT contain CoRNR boxes, which can interact with the LBD of GR. This binding occurs preferentially when GR is bound to RU-486 over when GR is bound to a standard glucocorticoid128, as helix 12 of the GR LBD in the RU-486-bound form is optimally positioned to permit CoRNR association164 (FIG. 4d). Although that GR interaction may not be physiologically significant, NCoR and SMRT co-occupy IR-GBS-containing or NF-κΒ- or AP- 1-tethered GORs at which standard glucocorticoid-bound GR is sumoylated within the NTD92,93.
Precision and plasticity via allostery
Because it is expressed ubiquitously, GR provides a dramatic example of perhaps the most critical property of eukaryotic TRFs — their context specificity. That is, GR regulates networks of genes that are precisely determined in a given setting, yet differ dramatically as a function of cell type and physiological state. This context-driven plasticity likely reflects the integration of the four classes of context-specific signals outlined above — hormonal ligands, PTMs, GR-binding DNA sequences and adjacent binding sequence motifs for non-receptor TRFs. The hormones and PTMs provide physiological context information to GR; the two classes of DNA-binding sequences, which together make composite GREs, provide gene context; and the range of TRFs available to occupy non-GR-binding sequences provide cell context.
We and others have shown that certain GR signals, namely hormonal ligands and DNA-binding sequences, are allosteric effectors, conferring specific alterations on GR conformation. Our model assumes that all four classes of signals affecting GR regulatory activity in transcription operate allosterically, and that their effects integrate to produce context-specific patterns of GR surfaces. These patterns of surfaces are recognized and bound by specific co-regulator complexes, which typically possess enzymatic activities that confer structural and/or functional changes on the transcription machinery and/or the chromatin. Co-regulators are generally not cell specific; rather, we propose that they assemble in unique combinations at each GRE, based on the context-specific interactions established at these sites.
A provocative extension of these ideas is that GR and other TRFs may typically lack intrinsic regulatory activities, instead serving merely as molecular scaffolds, patterns of surfaces produced by signalling, to which co-regulators, the actual regulatory machinery, combi- natorially associate (FIG. 6). This idea accounts readily for the common observation that TRFs can activate transcription in one context and repress it in another. In this sense, the activities of GR reflect its molecular conformations, which emerge owing to context-specific cues, whereas the functions of GR are integrated regulatory outcomes of co-regulatory enzyme actions that associate with those various conformations. Thus, we suggest that GR structure, together with its determinants, is the key to understanding both its regulatory precision and plasticity. These ideas probably extend, at least conceptually, to other eukaryotic TRFs52, all of which face the same precision-plasticity challenge.
Figure 6 |. A model for transcription regulation: precision and plasticity of TRF function achieved via allostery.
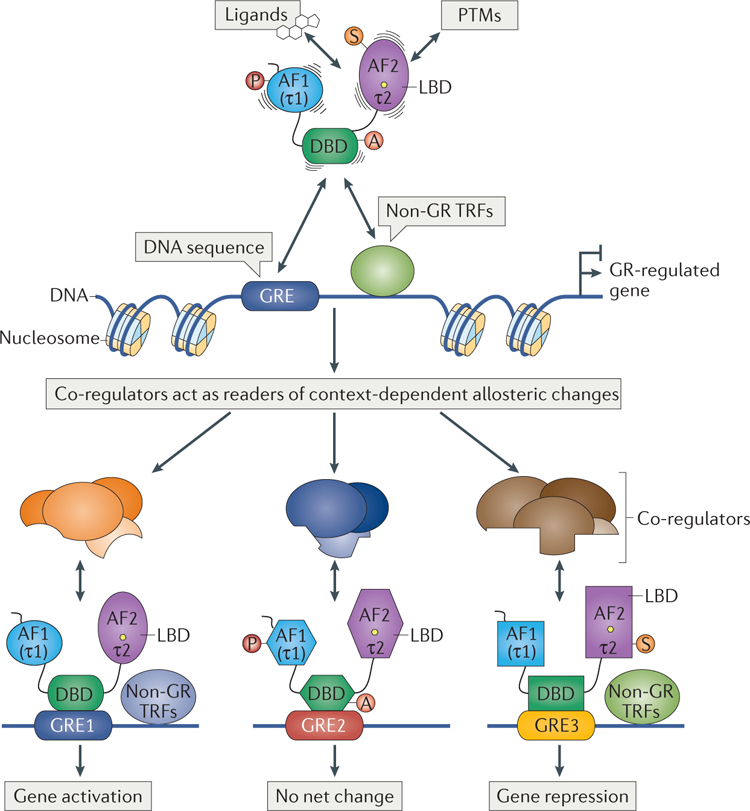
In our model for metazoan transcription regulation, transcriptional regulatory factors (TRFs), such as glucocorticoid receptor (GR), may lack intrinsic regulatory activities, serving instead as molecular scaffolds that can assume different conformations in response to modification by different combinations of specific signalling inputs. Four classes of signalling input are described: ligands and post-translational modifications (PTMs) provide physiological context, DNA-binding seguences provide gene context, and non-GRTRFs provide cell context. Each class of signal confers distinct allosteric effects, and their integrated actions can produce a vast range of GR conformations, which induce, expose or stabilize context-specific proteinsurfacesthat are read by co-regulators. These co-regulators, which are generally large multi-component enzyme-containing complexes, interact in distinct combinations with patterns of cognate GR surfaces and enzymatically modify the general transcriptional machinery and/or surrounding chromatin in and around glucocorticoid response elements (GREs) and target promoters and genes, resulting in the activation or repression of gene expression. Upper panel: lines around GR domains depict direct and allosteric conformational alterations imposed by the signalling inputs. Note that a lack of net regulation (that is, neither activation nor repression) could reflect balanced actions of one or more GREs acting on a single gene (not shown). AF, activation function domain; DBD, DNA-binding domain; LBD, ligand-binding domain. Circles with letters A, P,S indicate different PTMs.
Conclusions and perspectives
Biology is marked by astounding specificity of time and place, whether exemplified by butterfly migration, embryo patterning, neurite outgrowth or chromosome segregation. Here, we have considered such specificity from the perspective of a single polypeptide that regulates vertebrate gene expression. GR (which is ~90kDa) associates non-covalently with simple ligands (such as cortisol, which is only 362 Da) and controls transcription within networks comprising thousands of genes, distinct to cell type and condition. What explains such specificity?
We suggest here that the actions of GR are expanded, refined and directed through its interactions with multiple partner TRFs, dozens of signalling pathway-induced PTMs, hundreds of co-regulators, tens of thousands of potential genomic binding sites, hundreds of cell types (each with unique patterns of chromatin structure) and countless physiological states. This culminates in exquisitely complex regulation of gene transcription characterized by remarkable precision in any particlar contextual setting, yet facile plasticity to adapt when that context is altered. We propose that allosteric regulation of GR by these various inputs lies at the basis of the context specificity of gene expression.
This capacity of GR to integrate signalling inputs allosterically to produce distinct transcriptional outputs can be expanded further by the fact that multiple, distinct regulatory complexes established at individual GORs may collaborate to control any single GR-regulated gene. In addition, ligand chemistry and concentration readily alter GR-regulated gene network transcription, opening the door to speculation that endogenous ligands for GR may not be limited to cortisol. With appreciation of how daunting a task it is to understand this complexity sufficiently to render it predictive, what is a strategy to gain detailed insight into signal- and allostery-determined regulation of transcription? Undoubtedly, neither systems nor reductionist approaches alone will suffice41.
In our view, an essential step in obtaining holistic understanding of the regulation of metazoan gene transcription is to identify ‘causative primary-regulated genes’, which are defined as genes that are directly regulated by a given TRF and whose regulation by that factor is essential for a given phenotypic change. Next, functional response elements for such genes must be identified. Finally, allosteric, compositional and enzymatic changes in their corresponding regulatory complexes upon change of context could reveal properties and mechanisms (such as a distinct pattern of functional, allosterically specified surfaces) that correspond to the particular phenotype. Importantly, the identification of molecular features that can serve as surrogates for complex physiological or pathological outcomes could form a basis for predicting different combinations of contexts or signals that produce or preclude a given outcome, as well as frame a new approach for screening and assaying therapeutic candidates.
Supplementary Material
Transcriptional regulatory factors
(TRFs). A general class of sequence-specific DNA-binding proteins that regulate transcription (for example, glucocorticoid receptor).
Nuclear receptor
A member of a superfamily of potentially ligand-gated DNA-binding transcriptional regulatory factors.
Glucocorticoid
A natural hormone that binds to glucocorticoid receptor, or a synthetic derivative with physiological effects similar to the natural hormone, cortisol.
Dexamethasone
A synthetic glucocorticoid receptor (GR) ligand, developed in 1957, which is OG specific, unlike cortisol (the natural ligand), which also binds to mineralocorticoid receptor with high affinity, Dexamethasone is universally used clinically as an anti-inflammatory agent and immunosuppressant.
Apo-GR
Inactive glucocorticoid receptor (DR) protein in a ligand-unbound state.
Glucocorticoid response elements
(GREs). Genomic DNA segments (typically 0,5–2 kb long) that confer a specific glucocorticoid receptor response in particular contexts in vivo. Tire term ‘response element’ is appropriately unbiased with respect to potential activation (‘enhancement’) or repression of target gene transcription.
Allostery
Conformational changes in one region of a molecule (usually a protein) that alter its function and are induced by binding of a modulator to a different, remote site on the target molecule.
GR-binding sequence
(GBS). A short DNA sequence motif bound specifically and with high affinity by glucocorticoid receptor in vitro.
Nuclear magnetic resonance
(NMR). A technique that uses the magnetic properties of atomic nuclei to probe chemical environments experienced by atoms, for example, within a small molecule, protein or protein-DNA complex. See Supplementary information S1 (table).
3-Keto steroid receptors
Members of nuclear receptor subfamily 5 (NR3), including the glucocorticoid receptor (encoded by NR3 group C member 1 (NR3C1)), mineralocorticoid receptor (encoded by NR3C2). progesterone receptor (encoded by NR3C3) and androgen receptor(encoded by NR3C4).
Epistatic mutations
Gene alterations that display a phenotype only in the context of another mutation.
Chromatin immunoprecipitation followed by sequencing
(ChIP-Seq). A technique to identify genomic segments occupied genome-wide in vivo by a particular antigen surface, such as a transcriptional regulatory factor epitope. See Supplementary information S1 (table).
GR-occupied regions
(GORs), Genomic DNA segments occupied by glucocorticoid receptor (GR) in particular contexts in vivo. COR terminology, typically identified by chromatin immunoprecipitation (ChIp), improves on the previously used GR-binding region (GBR or GRBR) nomenclature, which implies direct DNA binding rather than a broader proximity to DNA, the parameter measured by ChIp.
DNase I-hypersensitive sites
(DHSs), Short genomic regions that are cleaved by brief exposure to low concentrations of DNase I in permeabilized cells or isolated nuclei. See Supplementary information S1 (table).
Negative regulatory DNA sequence
(NRS). A short DNA sequence motif, under-represented at glucocorticoid receptor (GR)-occupied regions within the genome, that interferes with the ability of GR to functionally interact with DNA proximal to the motif.
Molecular dynamics simulations
A computer simulation method to model the physical movements of atoms within a macromolecule that occur over short, fixed time intervals, giving information about dynamics within a macromolecule. See Supplementary information S1 (table).
RU-486
A synthetic glucocorticoid receptor (GR) ligand, developed in 1980, which also has high affinity for progesterone receptor. As, a non-standard ligand, binding of RU-486 results, in both an altered GR conformation and a distinct pattern of transcription regulation compared to binding of standard glucocorticoids, such as, dexarnethasone and cortisol.
Selective GR modulators
(SCRMs). Glucocorticoid receptor (GR) ligands with a regulatory range distinct from that of the standard glucocorticoid ligands cortisol and dexarnethasone.
Acknowledgements
The authors thank the members of the Yamamoto laboratory for critical reading of the manuscript, with special note to Elaine Kirschke for insightful discussions, Samantha Cooper, Sheng-Hong Chen and Benjamin Schiller for use of unpublished data, and Kirk Ehmsen for use of unpublished data and assistance with Figure 3. E.R.W. is supported by US National Institutes of Health (NIH) predoctoral National Research Service Award (NRSA) 1 G31 GM 113397–01A1 from the National Institute of General Medical Sciences. M.T.K. is supported by NIH postdoctoral NRSA 5T32HL007731–20 from the National Heart, Lung, and Blood Institute and by NIH grant R01CA020535 from the National Cancer Institute. E.A.O. is supported by NIH grant R01DK095750 from the National Institute of Diabetes and Digestive and Kidney Diseases, by American Heart Association (AHA) grant 14GRNT20460124 and by a W.M. Keck Foundation Medical Research Grant. K.R.Y. is supported by NIH grants R01CA020535 from the National Cancer Institute and R21ES026068 from the National Institute of Environmental Health Sciences, and by grant MCB-161 5826 from the National Science Foundation.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Bridgham JT et al. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8, el000497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Revollo JR & Cidlowski JA Mechanisms generating diversity in glucocorticoid receptor signaling. Ann. NYAcad.Sci. 1179, 167–178(2009). [DOI] [PubMed] [Google Scholar]
- 3.Kadmiel M & Cidlowski JA Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 34, 518–530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis-Tuffin LJ, Jewell CM, Bienstock RJ, Collins JB & Cidlowski JA Human glucocorticoid receptor binds RU-486 and is transcriptionally active. Mol. Cell. Biol. 27, 2266–2282 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard D et al. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348, 166–168 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Chandler VL, Maler BA & Yamamoto KR DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell 33, 489–499 (1983). [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto KR Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 19, 209–252 (1985). [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto KR, Darimont BD, Wagner RL & Iniguez-Lluhí JA Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harb. Syrnp. Quant. Biol. 63, 587–598 (1998). Presents the idea that TRFs nucleate different multi-subunit regulatory complexes on chromatin that drive alternative transcriptional outcomes. [DOI] [PubMed] [Google Scholar]
- 9.McNally JG The glucocorticoid receptor: Rapid exchange rath regulatory sites in living cells. Science 287, 1262–1265 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Becker M Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3, 1188–1194 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavreva DA, Muller WG, Hager GL, Smith CL & McNally JG Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24, 2682–2697 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meijsing SH, Elbi C, Luecke HR, Hager GL & Yamamoto KR The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release. Mol. Cell. Biol. 27, 2442–2451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacta MA, Chinenov Y & Rogatsky I Glucocorticoid signaling: An update from a genomic perspective. Annu. Rev. Physiol. 78, 155–180 (2016). This review presents new insights into GR biology that have emerged with the development and refinement of systems approaches. [DOI] [PubMed] [Google Scholar]
- 14.Wyllie AH Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284, 555–556 (1980). [DOI] [PubMed] [Google Scholar]
- 15.Patel R, Williams-Dautovich J & Cummins CL Minireview: New molecular mediators of glucocorticoid receptor activity in metabolic tissues. Mol. Endocrinol. 28,999–1011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar R & Thompson ΗB The structure of the nuclear hormone receptors. Steroids 64, 310–319 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Luisi BF et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352, 497–505 (1991). Provides the first crystallographic analysis of the GR DBD-GBS complex and details how two GR DBDs dimerize on a canonical DNA-binding element. [DOI] [PubMed] [Google Scholar]
- 18.Meijsing SH et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 (2009). Uses crystallographic analysis and functional assays done on multiple different GR DBD-GBS complexes to demonstrate that DNA binding acts as an allosteric effector of GR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson LC et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat. Struct. Mol. Biol. 20, 876–883 (2013). Uses biophysical analysis and NMR chemical-shift difference mapping to measure cooperative dimerization and to probe a potential allosteric pathway that extends from a GR DBD bound to one GBS half site, through specific regions within the bound GR DBD and the DBD dimerization domain, to the partner GR DBD bound to the other GBS half site. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heck S et al. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 13, 4087–4095 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiller BJ, Chodankar R, Watson LC, Stallcup MR & Yamamoto KR Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol. 15,418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reichardt ΗM et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93, 531–541 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Bledsoe RK et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 (2002). Provides the first crystal structure of ligand-bound GR LBD, which reveals the intricate network of protein-ligand interactions that define GR ligand selectivity [DOI] [PubMed] [Google Scholar]
- 24.Surjit M et al. Widespread negative response elements mediate direct repression by agonist- liganded glucocorticoid receptor. Cell 145, 224–241 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Hudson WH, Youn C & Ortlund EA The structural basis of direct glucocorticoid-mediated transrepression. Nat. Struct. Mol. Biol. 20, 53–58 (2013). Uses crystallographic analysis of the GR DBD- IR-GBS complex to reveal a new mode of GR-DNA recognition, in which two GR monomers bind opposite sides of the DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson WH et al. Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space. Proc. Natl Acad. Sci. USA 113, 326–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urn H et al. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res. 25, 836–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miner JN & Yamamoto KR The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev. 6, 2491–2501 (1992). [DOI] [PubMed] [Google Scholar]
- 29.De Bosscher K, Vanden Berghe W & Haegeman G Glucocorticoid repression of AP-1 is not mediated by competition for nuclear coactivators. Mol. Endocrinol. 15, 219–227 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Luecke HF & Yamamoto KR The glucocorticoid receptor blocks P-TEFb recruitment by NFkB to effect promoter-specific transcriptional repression. Genes Dev. 19, 1116–1127 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Bosscher K, Vanden Berghe W & Haegeman G The interplay between the glucocorticoid receptor and nuclear factor-κΒ or activator protein-1: Molecular mechanisms for gene repression. Endocr. Rev. 24, 488–522 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). Highlights the differences in experimental methodology and analysis of ChIP-Seq, which has become a mainstream method of analysing TRF- DNA interactions on a genome-wide scale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steger DJ et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 24, 1035–1044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John S et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grontved L et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBOJ. 32, 1568–1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.So AYL, Chaivorapol C, Bolton EC, Li H & Yamamoto KR Determinants of cell- and gene- specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 3, e94 (2007). Examines cell-type-specific GR occupancy on chromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy TE et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merkenschlager M & Nora EP CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17, 17–43 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Burd CJ. et al. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol. Cell. Biol. 32, 1805–1817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telorac J et al. Identification and characterization of DNA sequences that prevent glucocorticoid receptor binding to nearby response elements. Nucleic Acids Res. 44, 6142–6156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlenhaut NH et al. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol. Cell 49, 158–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Presman DM et al. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 12, e 1001813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starick SR et al. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 25, 825–835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.So AYL, Bernal TU, Pillsbury ML, Yamamoto KR & Feldman BJ Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl Acad. Sci. USA 106, 17582–17587 (2009). Identifies and characterizes the only gene-GRE pair confirmed to date, at its endogenous locus in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA et al. Genome engineering using the CRISPR- Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogatsky I et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc. Natl Acad. Sci. USA 100, 13845–13850 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas-Chollieir M et al. A naturally occuring insertion of a single amino acid rewires transcriptional regulation by glucocorticoid receptor isoforms. Proc. Natl Acad. Sci. USA 110, 17826–17831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen SH, Masuno K, Cooper SB & Yamamoto KR Incoherent feed-forward regulatory logic underpinning glucocorticoid receptor action. Proc. Natl Acad. Sci. USA 110, 1964–1969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangan S & Alon U Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA 100, 11980–11985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinenov Y, Coppo M, Gupte R, Sacta MA & Rogatsky I Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics 15, 656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hudson WH & Ortlund EA The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 15, 749–760 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefstin JA & Yamamoto KR Allosteric effects of DNA on transcriptional regulators. Nature 392, 885–888(1998). Introduces the concept of DNA as an allosteric regulator of DNA-binding proteins. [DOI] [PubMed] [Google Scholar]
- 53.Presman DM et al. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc. Natl Acad. Sci. USA 113, 8236–8241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gebhardt JCM et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robblee JP, Miura ΜT & Bain DL Glucocorticoid receptor-promoter interactions: Energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry 51,4463–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bain DL et al. Glucocorticoid receptor-DNA interactions: Binding energetics are the primary determinant of sequence-specific transcriptional activity. J. Mol. Biol. 422, 18–32 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Schӧne S et al. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nat. Commun. 7, 12621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat. Struct. Mol. Biol. 18, 556–563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thornton JW Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl Acad. Sci. USA 98, 5671–5676 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eick GN, Colucci JK, Harms MJ, Ortlund EA & Thornton JW Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 8, el003072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Y et al. Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 24, 713–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kauppi B et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J. Biol. Chem. 278, 22748–22754 (2003). Describes crystallographic analysis of the GR LBD bound to the non-standard ligand RU-486, which highlights the conformational malleability within GR to accommodate binding to ligands with disparate structures. [DOI] [PubMed] [Google Scholar]
- 63.Wang J-C et al. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 20, 689–699 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricketson D, Hostick U, Fang L, Yamamoto KR & Darimont BD A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J. Mol. Biol. 368, 729–741 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ismaili N & Garabedian MJ Modulation of glucocorticoid receptor function via phosphorylation. Ann. NY Acad. Sci. 1024, 86–101 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Tian S, Poukka H, Palvimo JJ & Jӓnne OA Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 367, 907–911 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace AD & Cidlowski JA Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J. Biol. Chem. 276, 42714–42721 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Itoh M et al. Nuclear export of glucocorticoid receptor is enhanced by c-Jun N-terminal kinase- mediated phosphorylation. Mol. Endocrinol. 16, 2382–2392 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Galigniana MD, Piwien-Pilipuk G & Assreuy J Inhibition of glucocorticoid receptor binding by nitric oxide. Mol. Pharmacol. 55, 317–323 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Ward RD & Weigel NL Steroid receptor phosphorylation: Assigning function to site-specific phosphorylation. BioFactors 35, 528–536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Housley PR & Pratt WB Direct demonstration of glucocorticoid receptor phosphorylation by intact L-cells. J. Biol. Chem. 258, 4630–4635 (1983). [PubMed] [Google Scholar]
- 72.Wang Z, Chen W, Kono E, Dang T & Garabedian MJ Modulation of glucocorticoid receptor phosphorylation and transcriptional activity by a C-terminal-associated protein phosphatase. Mol. Endocrinol. 21, 625–634 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Bodwell JE et al. Glucocorticoid receptor phosphorylation: Overview, function and cell cycle- dependence. J. Steroid Biochem. Mol. Biol. 65, 91–99 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Krstic MD, Rogatsky I, Yamamoto KR & Garabedian MJ Mitogen-activated and cyclin- dependent protein kinases selectively and differentially modulate transcriptional enhancement by the glucocorticoid receptor. Mol. Cell. Biol. 17, 3947–3954 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mason SA & Housley PR Site-directed mutagenesis of the phosphorylation sites in the mouse glucocorticoid receptor. J. Biol. Chem. 268, 21501–21504 (1993). [PubMed] [Google Scholar]
- 76.Jewell CM Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J. Biol. Chem. 272, 9287–9293 (1997). [DOI] [PubMed] [Google Scholar]
- 77.Chen W et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol. Endocrinol. 22, 1754–1766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garza AMS, Khan SH & Kumar R Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol. Cell. Biol. 30, 220–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller AL et al. p38 mitogen-activated protein kinase (MAPK) is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: Correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol. Endocrinol. 19, 1569–1583 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Wang Z, Frederick J & Garabedian MJ Deciphering the phosphorylation ‘code’ of the glucocorticoid receptor in vivo. J. Biol. Chem. 277, 26573–26580 (2002). [DOI] [PubMed] [Google Scholar]
- 81.King KL & Cidlowski JA Cell cycle regulation and apoptosis. Annu. Rev. Physiol. 60, 601–617 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Galliher-Beckley AJ, Williams JG, Collins JB & Cidlowski JA Glycogen synthase kinase 3-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol. Cell. Biol. 28, 7309–7322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galliher-Beckley AJ & Cidlowski JA Emerging roles of glucocorticoid receptor phosphorylation in modulating glucocorticoid hormone action in health and disease. IUBMB Life 61,979–986 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Deroo BJ et al. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 22, 4113–4123 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wallace AD, Cao Y, Chandramouleeswaran S & Cidlowski JA Lysine 419 targets human glucocorticoid receptor for proteasomal degradation. Steroids 75, 1016–1023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kino I, Liou SH, Charmandari E & Chrousos GP Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: Time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function. Mol. Med. 10, 80–88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gill G Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Le Drean Y, Mincheneau N, Le Goff P & Michel D Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology 143, 3482–3489 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Paakinaho V, Kaikkonen S, Makkonen H, Benes V & Palvimo JJ SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 42, 1575–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Treuter E & Venteclef N Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta 1812, 909–918 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Flotho A & Melchior F Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Hua G, Paulen L & Chambon P GR SUMOylation and formation of an SUMO-SMRT/NCoRl-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc. Natl Acad. Sci. USA 113, E626–E634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hua G, Ganti KP & Chambon P Glucocorticoid- induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/ NCoR 1-HDAC3 repressing complex. Proc. Natl Acad. Sci. USA 113, E635–E643 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nader N, Chrousos GP & Kino T Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEBJ. 23, 1572–1583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kino T & Chrousos GP Acetylation-mediated epigenetic regulation of glucocorticoid receptor activity: Circadian rhythm-associated alterations of glucocorticoid actions in target tissues. Mol. Ceil. Endocrinol. 336, 23–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ito K et al. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-κΒ suppression. J. Exp. Med. 203, 7–13 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krӧncke KD & Carlberg C Inactivation of zinc finger transcription factors provides a mechanism for a gene regulatory role of nitric oxide. FASEB J. 14, 166–173 (2000). [DOI] [PubMed] [Google Scholar]
- 98.Diamond M, Miner J, Yoshinaga S & Yamamoto K Transcription factor interactions: Selectors of positive or negative regulation from a single DNA element. Science 249, 1266–1272 (1990). Introduces differential context-specific regulation through the alternative interactions of GR with non-GR TRFs at composite elements. [DOI] [PubMed] [Google Scholar]
- 99.Miner JN, Diamond ΜI & Yamamoto KR Joints in the regulatory lattice: Composite regulation by steroid receptor-AP 1 complexes. Cell Growth Differ 2,525–530 (1991). [PubMed] [Google Scholar]
- 100.Jenkins BD, Pullen CB & Darimont BD Novel glucocorticoid receptor coactivator effector mechanisms. Trends Endocrinol. Metab. 12, 122–126 (2001). [DOI] [PubMed] [Google Scholar]
- 101.Bannister AJ & Kouzarides T Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Millard CJ, Watson PJ, Fairall L & Schwabe JWR An evolving understanding of nuclear receptor coregulator proteins. J. Mol. Endocrinol. 51, T23–T36 (2013). Provides a review of nuclear receptor co-regulator proteins, with a focus on the structural analysis of nuclear receptor-co-regulator interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vandevyveir; S, Dejager L & Libert C Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr. Rev. 35, 671–693 (2014). [DOI] [PubMed] [Google Scholar]
- 104.Parker MG & White R Nuclear receptors spring into action. Nat. Struct. Biol. 3, 113–115(1996). [DOI] [PubMed] [Google Scholar]
- 105.Wang Z et al. Structure and function of Nurr 1 identifies a class of ligand-independent nuclear receptors. Nature 423, 555–560 (2003). [DOI] [PubMed] [Google Scholar]
- 106.Torchia J et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387, 677–684 (1997). [DOI] [PubMed] [Google Scholar]
- 107.Heery DM, Kalkhoven E, Hoare S & Parker MG A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736 (1997). [DOI] [PubMed] [Google Scholar]
- 108.Hu X & Lazar MA The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402, 93–96 (1999). [DOI] [PubMed] [Google Scholar]
- 109.Khan SH et al. Binding of the N-terminal region of coactivator TIF2 to the intrinsically disordered AF1 domain of the glucocorticoid receptor is accompanied by conformational reorganizations. J. Biol. Chem. 287, 44546–44560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Khan SH, Ling J & Kumar R TBP binding-induced folding of the glucocorticoid receptor AF1 domain facilitates its interaction with steroid receptor coactivator-1. PLoS ONE 6, e21939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dahlman-Wright K, Almlӧf T, McEwan IJ, Gustafsson JA & Wright AP Delineation of a small region within the major transactivation domain of the human glucocorticoid receptor that mediates transactivation of gene expression. Proc. Natl Acad. Sci. USA 91, 1619–1623 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang L, Guerrero J, Hong H, DeFranco DB & Stallcup MR Interaction of the τ2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell 11,2007–2018 (2 000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chodankar R, Wu DY, Schiller BJ, Yamamoto KR & Stallcup MR Hic-5 is a transcription coregulator that acts before and/or after glucocorticoid receptor genome occupancy in a gene-selective manner. Proc. Natl Acad. Sci. USA 111, 4007–4012 (2 014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dasgupta S, Lonard DM & O’Malley BW Nuclear receptor coactivators: Master regulators of human health and disease. Annu. Rev. Med. 65, 279–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perissi V & Rosenfeld MG Controlling nuclear receptors: The circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6, 542–554 (2005). [DOI] [PubMed] [Google Scholar]
- 116.Lonard DM & O’Malley BW Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Mol. Cell 27, 691–700 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Fonte C et al. Involvement of β-catenin and unusual behavior of CBP and p300 in glucocorticosteroid signaling in Schwann cells. Proc. Natl Acad. Sci. USA 102, 14260–14265 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu J, Wu RC & O’Malley BW Normal and cancer- related functions of the p 160 steroid receptor co-activator (SRC) family. Nat. Rev. Cancer 9,615–630 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim JH, Li H & Stallcup MR CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 12, 1537–1549 (2003). [DOI] [PubMed] [Google Scholar]
- 120.Stallcup MR et al. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J. Steroid Biochem. Mol. Biol. 85, 139–145 (2003). [DOI] [PubMed] [Google Scholar]
- 121.Kim JH et al. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol. Cell 31,510–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Szapary D, Huang Y & Simons SS Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor-regulated gene expression. Mol. Endocrinol. 13,2108–2121 (1999). [DOI] [PubMed] [Google Scholar]
- 123.Trousson A et al. Recruitment of the p 160 coactivators by the glucocorticoid receptor: Dependence on the promoter context and cell type but not hypoxic conditions. J. Steroid Biochem. Mol. Biol. 104, 305-311 (2007). [DOI] [PubMed] [Google Scholar]
- 124.Voegel JJ et al. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17, 507–519 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Darimont BD et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12,3343–3356(1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li X, Wong J, Tsai SY, Tsai M, & O’Malley BW Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23, 3763–3773 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kurihara I et al. Expression and regulation of nuclear receptor coactivators in glucocorticoid action. Mol. Cell. Endocrinol. 189, 181–189 (2002). [DOI] [PubMed] [Google Scholar]
- 128.Ronacher K et al. Ligand-selective transactivation and transrepression via the glucocorticoid receptor: role of cofactor interaction. Mol. Cell. Endocrinol. 299, 219–231 (2009). [DOI] [PubMed] [Google Scholar]
- 129.Ogawa H et al. Nuclear structure-associated TIF2 recruits glucocorticoid receptor and its target DNA. Biochem. Biophys. Res. Commun. 320, 218–225 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Dobrovolna J, Chinenov Y, Kennedy MA, Liu B & Rogatsky I Glucocorticoid-dependent phosphorylation of the transcriptional coregulator GRIP 1. Mol. Cell. Biol. 32, 730–739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rogatsky I, Luecke HR, Leitman DC & Yamamoto KR Alternate surfaces of transcriptional coregulator GRIP 1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl Acad. Sci. USA 99, 16701–16706 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kamei Y et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85, 403–414(1996). [DOI] [PubMed] [Google Scholar]
- 133.Sheppard K-A et al. Nuclear integration of glucocorticoid receptor and nuclear factor-κΒ signaling by CRHB-binding protein and steroid receptor coactivator-1. J. Biol. Chem. 273, 29291–29294 (1998). [DOI] [PubMed] [Google Scholar]
- 134.De Bosscher K et al. Glucocorticoids repress NF-KB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl Acad. Sci. USA 97, 3919–3924 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allen BL & Taatjes DJ The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Knuesel ΜI & Taatjes DJ Mediator and post- recruitment regulation of RNA polymerase II. Transcription 2, 28–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meyer KD, Lin SC, Bernecky C, Gao Y & Taatjes DJ p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17,753–760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Taatjes DJ, Nӓӓr AM, Andel F, Nogales H & Tjian R Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 (2002). [DOI] [PubMed] [Google Scholar]
- 139.Knuesel ΜT, Meyer KD, Bernecky C & Taatjes DJ The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bernecky C, Grob P, Hbmeier CC, Nogales H & Taatjes DJ Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 9, e 1000603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meyer KD et al. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBOJ. 27, 1447–1457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP & Garabedian MJ Differential regulation of glucocorticoid receptor transcriptional activation via AF-1 -associated proteins. EMBO J. 18, 5380–5388 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen W, Rogatsky I & Garabedian MJ MED 14 and MED 1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol. 20, 560–572 (2006). [DOI] [PubMed] [Google Scholar]
- 144.Narlikar GJ, Sundaramoorthy R & Owen-Hughes T Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pazin MJ & Kadonaga JT SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88, 737–740 (1997). [DOI] [PubMed] [Google Scholar]
- 146.Fiyer CJ & Archer TK Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393, 88–91 (1998). [DOI] [PubMed] [Google Scholar]
- 147.Engel KB & Yamamoto KR The glucocorticoid receptor and the coregulator Brm selectively modulate each other’s occupancy and activity in a gene-specific manner. Mol. Cell. Biol. 31,3267–3276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ostlund Farrants AK, Blomquist P, Kwon H & Wrange O Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol. 17, 895–905 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Collingwood TN, Umov FD & Wolffe AP Nuclear receptors: Coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 23, 255–275 (1999). [DOI] [PubMed] [Google Scholar]
- 150.King HA, Trotter KW & Archer TK Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim. Biophys. Acta 1819, 716–726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yoshinaga S, Peterson C, Herskowitz I & Yamamoto KR Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258, 1598–1604 (1992). [DOI] [PubMed] [Google Scholar]
- 152.Wallberg AE et al. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ 1 activation domain. Mol. Cell. Biol. 20, 2004–2013 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Muratcioglu S et al. Structural modeling of GR interactions with the SWI/SNF chromatin remodeling complex and C/EBP. Biophys. J. 109, 1227–1239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen D et al. Regulation of transcription by a protein methyl transferase. Science 284, 2174–2177 (1999). [DOI] [PubMed] [Google Scholar]
- 155.Bittencourt D et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc. Natl Acad. Sci. USA 109, 19673–19678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee KK & Workman JL Histone acetyl transferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007). [DOI] [PubMed] [Google Scholar]
- 157.Almlӧf T, Wallberg AE, Gustafsson JÅ & Wright APH Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor τ 1 -core activation domain and target factors. Biochemistry 37, 9586–9594 (1998). [DOI] [PubMed] [Google Scholar]
- 158.Yao ΤP, Ku G, Zhou N, Scully R & Livingston DM The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl Acad. Sci. USA 93, 10626–10631 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wallberg AE et al. Histone acetyltransferase complexes can mediate transcriptional activation by the major glucocorticoid receptor activation domain. Mol. Cell. Biol. 19, 5952–5959 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fonte C, Trousson A, Grenier J, Schumacher M & Massaad C Opposite effects of CBP and p300 in glucocorticoid signaling in astrocytes. J. Steroid Biochem. Mol Biol. 104, 220–227 (2007). [DOI] [PubMed] [Google Scholar]
- 161.Verdin E & Ott M 50 yearn of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015). [DOI] [PubMed] [Google Scholar]
- 162.Shahbazian MD & Grunstein M Functions of site- specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007). [DOI] [PubMed] [Google Scholar]
- 163.Stewart MD & Wong J Nuclear receptor repression: Regulatory mechanisms and physiological implications. Prog. Mol. Biol. Transl Sci. 87, 235–259 (2009). [DOI] [PubMed] [Google Scholar]
- 164.Schoch GA et al. Molecular switch in the glucocorticoid receptor: Active and passive antagonist conformations. J. Mol. Biol. 395, 568–577 (2010). [DOI] [PubMed] [Google Scholar]
- 165.Kuznetsova T et al. Glucocorticoid receptor and nuclear factor kappa-b affect three-dimensional chromatin organization. Genome Biol. 16, 264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ogryzko VV et al. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94, 35–44 (1998). [DOI] [PubMed] [Google Scholar]
- 167.Guenther MG, Barak O & Lazar MA The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3.Mol. Cell. Biol. 21,6091–6101 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen JD & Evans RM A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


