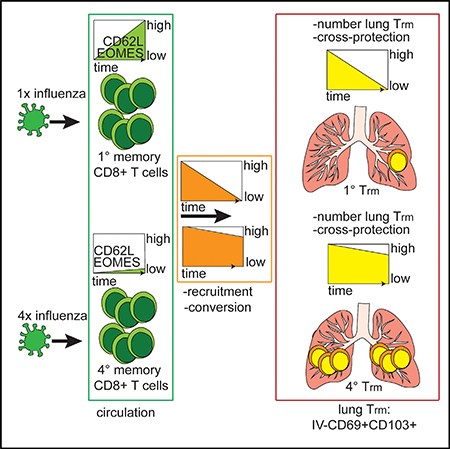
SUMMARY
Lung-resident primary memory CD8+ T cell populations (Trm) induced by a single influenza infection decline within months, rendering the host susceptible to new heterosubtypic influenza infections. Here, we demonstrate that, relative to single virus exposure, repeated antigen exposure dramatically alters the dynamics of influenza-specific lung Trm populations. Lung Trm derived from repeatedly stimulated circulating memory CD8+ T cells exhibit extended durability and protective heterosubtypic immunity relative to primary lung Trm. Parabiosis studies reveal that the enhanced durability of lung Trm after multiple antigen encounters resulted from the generation of long-lasting circulating effector memory (Tem) populations, which maintained the ability to be recruited to the lung parenchyma and converted to Trm, in combination with enhanced survival of these cells in the lung. Thus, generating a long-lasting Trm precursor pool through repeated intranasal immunizations might be a promising strategy to establish long-lasting lung Trm-mediated heterosubtypic immunity against influenza.
In Brief
Van Braeckel-Budimir et al. find that repeated antigen exposure by influenza virus infection in the lung enhances the durability of lung CD8+ T resident memory populations and extends the duration of heterosubtypic immunity against influenza virus.
Graphical Abstract
INTRODUCTION
Based on health and socioeconomic impact, influenza virus infections are a major global health burden (Kondrich and Rosenthal, 2017; Nicholson et al., 2003). This public health burden remains despite the approval of the first influenza vaccine almost 7 decades ago (Barberis et al., 2016). Current vaccine formulations aim for induction of neutralizing antibodies specific for the main surface antigen (hemagglutinin [HA]) of the influenza virus particle (Barberis et al., 2016). However, the HA protein undergoes high rates of mutation (antigenic drift) (Doherty et al., 2006) that enables successful escape from the immunological pressure of vaccination-induced antibodies and dramatically limits vaccine efficacy (Boni, 2008; de Jong et al., 2000). Additionally, reassortment of the segmented influenza virus genome in animal reservoirs can result in new HA sequences (antigenic shift) (Kim et al., 2018) that have not previously circulated in humans and have the potential for pandemic infections (Kim et al., 2018). It has been well documented in humans and rodent models that influenza-specific CD8+ T cells targeting conserved internal proteins of the virus can control virus titers and limit disease development in the absence of neutralizing antibodies (Altenburg et al., 2015; Kreijtz et al., 2007; McMichael et al., 1983; Sridhar et al., 2013). Recent research suggests that the population of lung-resident CD8+ T cells (Trm) induced by primary influenza infection plays a critical role in such heterosubtypic immunity (HI) (Hogan et al., 2001; Slütter et al., 2017; Wu et al., 2014). Thus, inducing a potent and long-lasting influenza-specific Trm population should be considered as a potentially useful vaccination target.
Waning of protection is one of the main limitations of T cellmediated heterosubtypic immunity after primary influenza infection (Liang et al., 1994; Slütter et al., 2017; Wu et al., 2014). We and others have shown that the gradual loss of protection closely correlates with the decrease in size of the influenza-specific lung Trm population (Slütter et al., 2017; Wu et al., 2014). Mechanistically, lung Trm cells undergo increased apoptosis that, in combination with time-dependent decreases in recruitment and conversion of circulating Trm precursors, limit the longevity of influenza-specific Trm in the lung (Slütter et al., 2017€ ). Of note, the vast majority of published studies addressing the formation and maintenance of influenza-specific lung Trm, are based on a “single-exposure” model (Slütter et al., 2017; Takamura et al.,€ 2016; Wu et al., 2014). This represents an important limitation given the repetitive, seasonal nature of influenza infections and the current approaches of yearly vaccine applications. Thus, it is pivotal to understand how repeated influenza antigen encounters impact the dynamics of lung Trm and, consequently, the longevity of heterosubtypic immunity.
RESULTS
Experimental Model
To study the impact of repeated influenza infections on lung Trm, we initially infected C57BL/6 mice with an antibody escape variant of the mouse adapted H1N1 A/PR/8, designated SEQ12 (Das et al., 2013; Van Braeckel-Budimir et al., 2017), followed at 60-day intervals by infection with X31 (H3N2), and then a high dose of the parental A/PR/8 (PR8) virus. Despite the careful selection of viruses, sequential infection with SEQ12 and X31 induced antibodies that neutralized PR8 and no virus was recovered after the tertiary challenge (not shown). Furthermore, repeated infections may lead to generation of memory populations with different antigen exposure history, by incomplete recruitment of existing memory CD8+ T cells together with de novo recruitment of naive CD8+ T cells (Masopust et al., 2006; Wirth et al., 2010). Thus, we sought to develop a model that would allow us to generate distinct populations of Trm with defined numbers of antigen encounters after lung infection.
To address these issues, we took advantage of a previously published model based on adoptive transfer (AT) of TCR transgenic CD8+ T cells (Wirth et al., 2010). Naive Thy1.2 C57BL/6 mice were seeded with low numbers (104) of naive spleenderived transgenic Thy1.1 P14 cells and subsequently intranasally (IN) infected with recombinant PR8 expressing the GP33 epitope of lymphocytic choriomeningitis virus (LCMV) recognized by P14 cells (PR8-GP33). Ninety days after infection, 105 spleen-derived primary memory (1°M) P14 CD8+ T cells were transferred into new naive congenic recipients followed by IN infection with PR8-GP33. Simultaneously, another group of naive recipients was seeded with naive P14 cells and infected IN with PR8-GP33. Adoptive transfer and viral infections were repeated until we simultaneously obtained 1°M and quaternary memory (4°M) P14 cells (Figure 1A). Of note, based on published data indicating that 3°M cells proliferate less than naive CD8 T cells in response to systemic infection (Wirth et al., 2010) and in order to achieve similar frequencies of circulating 1°M and 4°M P14 cells, mice received 105 3°M P14 cells. This model not only permitted isolation and transfer of P14 cells with a known antigen exposure history but also incorporated the recently established notion that circulating memory T cells can be recruited to tissues and generate Trm after repeated infections (Beura et al., 2018; Park et al., 2018). Figure S1A depicts selected phenotypic characteristics of naive and 3M spleenderived P14 which were the precursors of 1M and 4M lung Trm. Of note, the circulating precursor populations used for adoptive transfer lacked cells expressing both CD69 and CD103, which represent the canonical markers of lung Trm (Wakim et al., 2013; Wu et al., 2014). As expected, naive and 3°M populations exhibited differential expression of a number of cell surface markers, including CD44, CD11a, CD62L, and KLRG1. To verify that the different numbers of transferred cells did not impact the overall course of influenza infection and to confirm that both 1°M and 4°M cells developed in similar conditions, we assessed the lung PR8-GP33 titers at different days after infection. As depicted in Figure S1B, titers measured 4 and 6 days after infection were similar in recipients of naive or 3°M P14 and in the same range as titers measured in mice receiving no adoptive transfer prior to infection. This finding suggested that the input of circulating naive or 3°M P14 did not alter the course of infection and that 1°M and 4°M cells developed under similar infection conditions. For clarity, although all of the recipient mice were infected with influenza only once, we will refer to mice bearing 1°M P14 cells as 1°M mice, and those bearing 4°M P14 cells as 4M mice.
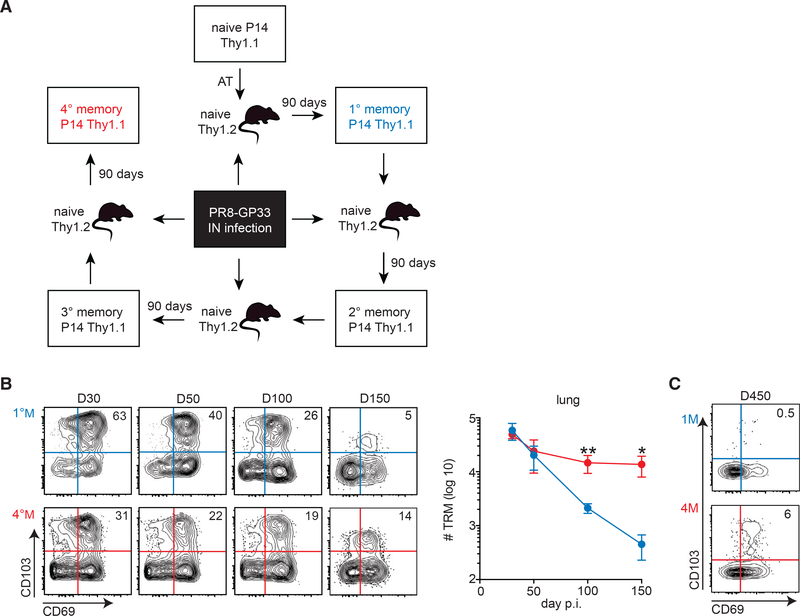
Figure 1. 4°M Lung Trm Exhibits Increased Durability Relative to 1M Lung Trm.
(A) Generation of memory CD8+ T cells. A sequential P14 adoptive transfer approach and IN infection with PR8-GP33 were used to obtain 1M (blue) and 4M (red) T cell responses that could be analyzed simultaneously.
(B) Groups of mice bearing 1M (blue) and 4°M (red) P14 cells were euthanized at days 30, 50, 100, and 150 p.i. Representative plots of % of CD69 and CD103 expression in IV−P14 at the indicated time points (left). Cumulative results depicting numbers of Trm (IV−CD69+CD103+) in the lung (right). n = 3–4 mice/group. Representative of three independent experiments. Error bars represent mean ± SD. **p = 0.0034, *p = 0.0162, unpaired t test.
(C) Representative plot of lung IV− P14 cells 450 days p.i. Percentages of CD69+CD103+ Trm are indicated.
Multiple Antigen Encounters Enhance the Durability of Influenza-Specific Lung Trm
To determine whether multiple antigen encounters altered the durability of lung Trm, lungs of 1°M and 4°M mice were analyzed at different time points after the PR8-GP33 infection. To discriminate circulating cells from those within the tissue parenchyma, we performed intravascular (IV) exclusion based on IV injection of a fluorescently labeled pan-lymphocyte marker (CD45.2) 3 min prior to euthanasia as described (Galkina et al., 2005; Teijaro et al., 2011). Trm were defined as CD8+IV CD69+CD103+ based on our previous results (Slütter et al., 2017). As described earlier (Slütter et al., 2017; Wu et al. 2014), 1°M lung Trm populations gradually waned over time, with only a few hundred cells detectable in the lung parenchyma 5 months after influenza exposure (Figure 1B). In sharp contrast, after an initial ~3-fold decline between day 30 and day 50 postinfection (p.i.), 4°M Trm numbers stabilized and were maintained at >10,000 cells/lung out to day 150 p.i. As a result, the 4°M Trm population was >50-fold larger than the 1°M Trm population at day 150 after infection (Figure 1B). Strikingly, 4°M Trm could be readily detected as late as day 450 p.i., a time point when 1°M Trm have essentially disappeared (Figure 1C). Thus, multiple antigen encounters after lung infection extended the durability of influenza-specific lung Trm.
Antigen Exposure History Defines the Tissue-Specific Distribution of Influenza-Specific CD8+ T Cells without Altering the Basic Trm Phenotype
Previous studies have emphasized the impact of repeated systemic infection on fundamental biological characteristics of memory CD8+ T cells and their distribution in tissues (Jabbari and Harty, 2006; Masopust et al., 2006; Rai et al., 2014; Vezys et al., 2009; Wirth et al., 2010). Thus, we assessed whether and how the history of influenza antigen encounter impacted the distribution of memory CD8+ T cells. To be able to study memory cell distribution while allowing for competition for distinct niches, we generated 1M and 4M cells in the same host (Figure S2A). For this purpose, naive Thy1.2/1.2 C57BL/6 recipients were seeded with mixture of 104 Thy1.1/1.2 naive P14 and 105 Thy1.1/1.1 3°M P14 cells and subsequently IN infected with PR8-GP33. Mice were analyzed at day 90 p.i., and the distribution of 1°M and 4°M cells was assessed in the spleen, blood, and lungs (Figure S2B). While the distribution of 1°M and 4°M cells was similar in the blood and spleen, 4°M cells were the dominant population in the lung, with slightly stronger preference for the parenchymal (IV−) niche compared to the vasculature (IV+) niche (Figure S2B).
Furthermore, we investigated whether repeated influenza antigen exposures altered some of the important phenotypic properties of lung Trm. For this purpose, we evaluated expression of CD103, CD69 (classical Trm markers) (Schenkel and Masopust, 2014; Wu et al., 2014), Eomes (a key transcription factor downregulated in Trm) (Mackay et al., 2015), CX3CR1 (expressed by Trm precursors) (Gerlach et al., 2016), and CXCR3 (facilitates homing to the lung/airway) (Slütter et al., 2013 ) on 1°M and 4°M P14 generated in the same host (Figures S2A and S2C). Overall, we observed no substantial difference in selected phenotypic characteristics between 1°M and 4°M lung Trm. Most 1°M and 4°M IVCD103+ cells expressed CD69, had low expression of Eomes and CX3CR1, and were CXCR3+. These data demonstrated that repeated influenza antigen exposure enhanced 4°M entry into the lung parenchyma, where some of the cells developed Trm phenotypic characteristics similar to those of their 1°M Trm counterparts. Thus, major changes in canonical Trm properties did not appear to account for the persistence of 4°M Trm in the lung parenchyma.
Repeated Influenza Antigen Exposures Extend the HalfLife of Trm in the Lung by Increasing Cell Survival
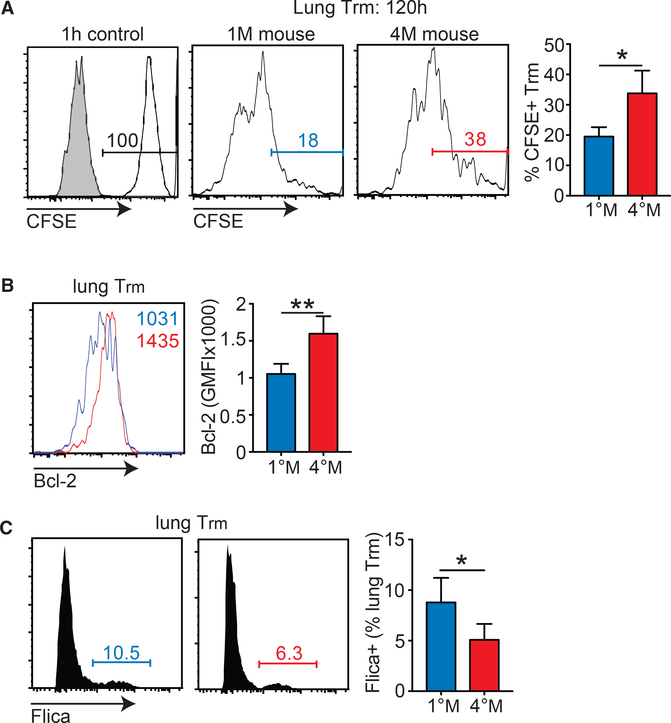
As shown previously, 1°M Trm have a relatively short half-life in the tissue parenchyma and exhibit higher frequencies of cells with active caspase 3/7 and decreased Bcl-2 expression compared to 1°M Trm in skin (Slutter et al., 2017 ). In the light of these findings, we hypothesized that the 4°M Trm population could exhibit prolonged survival in the lung relative to 1°M Trm, leading to their longer persistence. To probe this notion, we labeled lung Trm 90 days p.i. by IN inoculation of carboxyfluorescein diacetate succinimidyl ester (CFSE) (Legge and Braciale, 2003; Slütter et al., 2017 ). This approach labels all of the cells in the lung, with negligible labeling of cells in the other tissue compartments, and loss of CFSE signal with time relates to loss of labeled cells, not dilution due to proliferation (Slütter et al., 2017). To determine the initial level of labeling, we evaluated CFSE intensity in the lung Trm at 1 hr post-inoculation (Figure 2A). At this time point, 100% of lung Trm were CFSE+ (Figure 2A). When mice were analyzed 120 hr after labeling, only 20% of 1°M Trm were CFSE+, confirming the relatively short survival of this population in the lung. A significantly (p = 0.0127) larger fraction (>30%) of 4°M Trm were CFSE+ at the same time post-labeling (Figure 2A), suggesting prolonged survival of these cells in the lung. Additionally, 4°M Trm exhibited higher expression of Bcl-2, relative to 1°M Trm (Figure 2B), consistent with increased survival of this population in the tissue parenchyma. Finally, we observed a modest, but significant (p < 0.05) increase in the proportion of 1°M relative to 4°M Trm cells staining positive with Flica reagent, suggesting increased proapoptotic caspase 3/7 activity (Figure 2C). Together, these results suggested that enhanced durability of 4°M Trm in the lung parenchyma was due, at least in part, to increased survival. However, the differences in each “survival” assay were modest and are unlikely sufficient to entirely explain the enhanced durability of 4°M Trm.
Figure 2. 4°M Lung Trm Persists Longer and Exhibits Reduced Apoptosis Signature Compared to 1°M Trm.
(A) Mice bearing 1°M and 4°M P14 cells were IN inoculated with CFSE at day 90 p.i. and lung Trm cells (IVCD69+CD103+) were analyzed 1 and 120 hr later. Representative flow plots of CFSE labeling (left) and cumulative analysis of percentage of CFSE-labeled cells 120 hr post-labeling (right). n = 4 mice/group. Representative of three independent experiments. Error bars represent mean ± SD. *p = 0.0127, unpaired t test.
(B) Bcl-2 expression by 1°M and 4°M lung Trm measured 90 days p.i. Representative histograms (left); cumulative data (right). n = 4 mice/group. Representative of two independent experiments. Error bars represent mean ± SD. **p = 0.0072, unpaired t test. GMFI, geometric mean fluorescence intensity.
(C) Flica (caspase 3/7) staining of 1°M and 4°M lung Trm measured 90 days p.i. Representative histograms (left); cumulative data (right). n = 4 mice/group. Representative of two independent experiments. Error bars represent mean ± SD. *p = 0.0435, unpaired t test.
Repeated Influenza Antigen Exposures Stabilize the Circulating Trm Precursor Pool
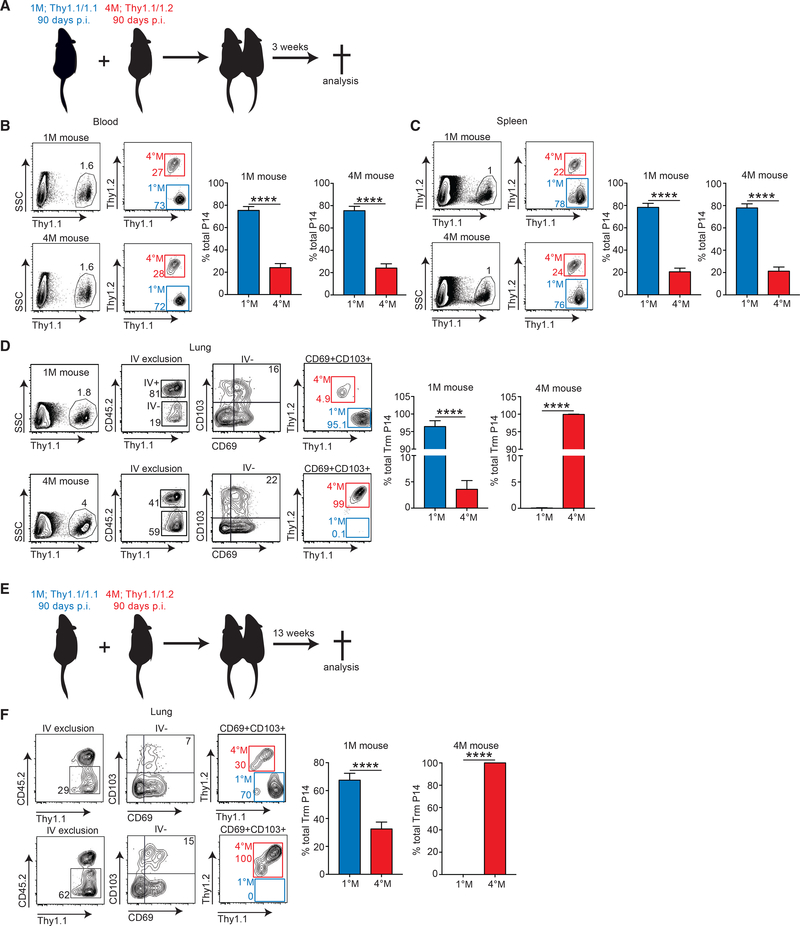
We previously showed that, despite their short half-life after CFSE labeling, 1°M Trm are sustained for some time by recruitment to the lung of circulating effector memory (Tem) precursors, some of which undergo phenotypic and functional conversion to Trm (Slütter et al., 2017 ). As a consequence of differentiation of circulating 1°M memory populations from Tem to primarily central memory (Tcm) with time, replenishment of lung Trm eventually ceases and the population declines. Thus, we hypothesized that, due to their increased antigen exposure history, circulating 4°M may exhibit a sustained Tem population allowing for prolonged recruitment and conversion to lung Trm. To probe this hypothesis, we took advantage of the mouse parabiosis model. 1°M mice, bearing Thy1.1/1.1 circulating memory and lung Trm cells, and 4°M mice, bearing Thy1.1/1.2 circulating memory and lung Trm cells, were surgically joined 90 days post-PR8GP33 infection (Figure 3A). Three weeks later, parabiotic pairs were analyzed for the distribution of 1°M and 4°M P14 cells in blood, spleens, and lungs (Figure 3A). Of note, similar frequencies of circulating P14 were found in the blood and spleen of both groups (Figures 3B and 3C), indicating that equilibrium had been reached. However, this equilibrium did not reflect a 1:1 distribution of 1°M and 4°M circulating cells. Instead, both groups of mice exhibited 75% 1°M P14 and 25% 4°M P14 ratio in the blood and spleen (Figures 3B and 3C). In contrast, a higher frequency of memory P14 was recovered from the total lung and IV− fraction of the lung in the 4°M parabiont (Figure 3D). As expected from prior work showing that late 1°M loses the ability to home to the lung parenchyma and convert to Trm (Slütter et al., 2017 ), and despite the dominance of 1°M in the circulation, Trm in the lungs of the 4°M parabiotic partner were exclusively 4°M Trm. In contrast, despite their reduced presence in the circulation, a clear population of 4°M Trm could be found within the Trm population of the 1°M parabiotic partner (Figure 3D). These data suggested that circulating 4M exhibited a prolonged capability to home to the lung parenchyma and convert to Trm.
Figure 3. Circulating 4°M Cells Home to the Lung and Convert to Trm for Extended Period of Time.
(A) 90 days post-PR8-GP33 infection, mice bearing Thy1.1/1.1 1°M P14 (blue; “1°M mice”) and Thy1.1/1.2 4M P14 (red; “4°M mice”) cells were joined by parabiosis. Three weeks later, parabionts were analyzed.
(B–D) Abundance of 1°M (blue) and 4°M (red) P14 cells in peripheral blood (B) and spleen (C) of parabiotic mice. Representative plots (left); cumulative data (right). n = 4 parabionts/experiment. Representative of two independent experiments. Error bars represent mean ± SD. ****p < 0.0001, unpaired t test.
(D) Abundance and Trm distribution of 1°M (blue) and 4°M (red) Trm P14 cells expressed as a percentage of the total Trm population (IVCD69+CD103+) in lungs of parabiotic mice. Representative plots (left); cumulative data (right).
(E) Parabionts were prepared as in (A) and were analyzed 13 weeks after the surgery.
(F) Abundance and Trm distribution of 1°M (blue) and 4M (red) Trm P14 cells expressed as a percentage of the total Trm population (IVCD69+CD103+) in lungs of parabiotic mice. Representative plots (left); cumulative data (right). n = 4 parabionts/experiment. Representative of two independent experiments. Error bars represent mean ± SD. ****p < 0.0001, unpaired t test.
The contribution of 4°M Trm P14 to the Trm compartment in 1M mice at 3 weeks of parabiosis was low (5%). Thus, we investigated whether representation of these cells would increase with additional time of parabiosis. For this purpose, 1°M and 4°M parabiotic partners were euthanized 13 weeks after joining, and distribution of P14 cells in lung tissue was assessed. Extended time of parabiosis did not change the contributions of 1°M and 4°M cells to blood and spleen compartments of parabiotic partners (not shown). Despite the added time of parabiosis, we still detected no 1 Trm in the lungs of the 4°M parabiotic partners. However, the representation of 4°M Trm P14 increased from 5% to 30% in the lungs of the 1°M mice with the additional parabiosis time. These data suggested sustained accumulation of 4°M-derived Trm from the circulating 4°M pool with time.
Consistent with this notion, splenic 1M progressed from 50% Tem (CD62Llo) at day 50 p.i. to <10% Tem at day 150 p.i., and this resulted in a decrease in total numbers of splenic Tem (Figure S3A). In sharp contrast, the splenic 4°M population remained CD62Llo and the Tem population was numerically stable in the same time frame (Figure S3A). This trend was extended well beyond day 150, as even 7 months p.i. circulating 4°M continued to express a Tem phenotype (CD62Llo) and a permissive transcription factor profile (Eomeslo), whereas the 1°M population was exclusively Tcm (CD62LhiEomeshi) at this late time point (Figure S3B). Although Tbet was modestly upregulated in 4°M, the Tbet regulated homing receptor CXCR3 was not differently expressed between splenic 1°M and 4°M (Figure S3C). These data were consistent with the notion that general impairment of lung-homing capacity was unlikely to underlie the failure to maintain 1°M Trm (Slutter et al., 2017€ ). In summary, circulating 4°M CD8+ T cells, due to their prolonged display of Tem properties were recruited to the lung and converted into Trm for an extended period of time, compared to circulating 1°M. Given the modest evidence for increased survival, sustained replenishment of lung Trm from the circulatory memory pool likely accounted for the enhanced durability of 4°M lung Trm.
Repeated Influenza Antigen Exposures Extend the Duration of the Heterosubtypic Protection
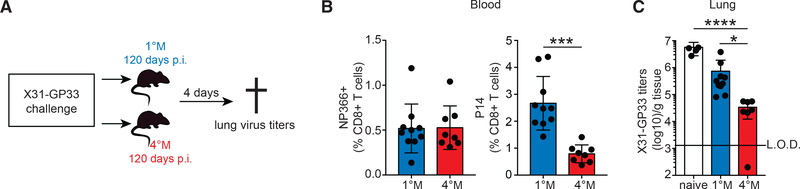
Waning of 1°M Trm correlates strongly with loss of heterosubtypic immunity (Slütter et al., 2017; Wu et al., 2014 ). Our results show that multiple exposures of circulating CD8+ T cell populations to pulmonary influenza infection extended the durability of lung Trm far beyond that observed with 1°M lung Trm. Thus, our next step was to investigate whether extending the durability of lung Trm through multiple antigen exposures resulted in prolonged heterosubtypic immunity. For this purpose, 1°M mice and 4°M mice, from the cohorts analyzed in Figure 1C, were challenged with a high dose of the heterologous X31-GP33 virus 120 days post-PR8-GP33 exposure (Figure 4A). Heterosubtypic immunity was evaluated by measuring X31-GP33 titers in the lung of challenged mice (Figure 4A).
Figure 4. Durability of 4°M Trm Cells Enable Long-Lasting Heterosubtypic Protection.
(A) 120 days post-PR8-GP33 infection, mice bearing 1°M (blue) and 4°M (red) P14 cells were IN infected with 5 3 104 TCID50 X31-GP33. 4 days after the challenge, mice were euthanized, and lung virus titers were assessed.
(B) Prior to challenge influenza NP366-specific (left) and P14 (right) CD8+ T cell responses were evaluated in blood and expressed as percentage of total CD8+ T cells in peripheral blood. n = 8–10 mice/group. Representative of two independent experiments. Error bars represent mean ± SD. ***p = 0.0001, unpaired t test.
(C) Lung X31-GP33 virus titers measured in naive mice, and mice bearing 1°M (blue) or 4°M (red) P14 cells. n = 5–10 mice/group. Representative of two independent experiments. Error bars represent mean ± SD. *p = 0.0216, ****p < 0.0001, Kruskal-Wallis test. L.O.D., limit of detection.
It is important to note that both groups of mice were exposed to PR8-GP33 only once, both contained similar percentages of endogenous 1°M CD8+ T cells recognizing the NP366 influenza epitopes in their circulation, and challenge took place at a late time point when protection by endogenous lung Trm would have waned (Figure 4B). Thus, potential differences in protection would be due to differences in lung P14 Trm populations, which are lost in 1°M mice and sustained in 4°M mice (Figure 1C). While the magnitude of the endogenous influenza-specific 1°M responses were similar, in line with previous data (Figure 3) 1M mice had higher circulating P14 frequencies than 4M mice (Figure 4B) (Wirth et al., 2010). As expected at this late time point, 1°M memory mice showed only a modest reduction of X31-GP33 virus titers compared to naive infected mice, which did not reach statistical discernibility. In contrast, 4°M mice had 99% less virus than the naive infected group (Figure 4C). Thus, the extended durability of 4°M Trm in the lung enabled superior heterosubtypic immunity at time points after infection when protection mediated by 1°M Trm had waned.
DISCUSSION
Heterosubtypic immunity against influenza virus has been extensively studied in a single-infection mouse model of the disease (Budimir et al., 2012; Christensen et al., 2000; Furuya et al., 2010; Liang et al., 1994; Nguyen et al., 1999; Slutter et al., 2017;€ Wu et al., 2014). Recent studies show that heterosubtypic immunity mediated by 1°M CD8+ T cells wanes over time, as a consequence of gradual loss of lung Trm cells (Slutter et al.,€ 2017; Wu et al., 2014). In the context of annual influenza outbreaks and the potential for yearly immunizations, it has remained unknown how multiple antigen exposures influence the formation and maintenance of lung Trm and subsequent Trm-mediated heterosubtypic protection. To address these important knowledge gaps, we modified a multiple antigen exposure strategy previously used to assess responses to systemic infections (Wirth et al., 2010), by stimulating circulating memory CD8+ T cell populations with localized lung influenza infections. This approach allowed us to overcome limitations of repeated infections with influenza virus and to study homogeneous populations of memory CD8+ T cells with well-defined influenza exposure history (Masopust et al., 2006; Wirth et al., 2010). Our results revealed that 4°M Trm cells exhibit substantially increased durability in the tissue parenchyma compared to 1°M Trm. Mechanistically, the enhanced durability of 4M cells resulted from improved survival of this population in the lung and a dramatically extended capacity for the circulating 4M cells to enter the lung parenchyma and convert to Trm. These findings strongly suggest that, by inducing a long-lived Tem population, multiple influenza exposures license a circulating Trm precursor pool for sustained seeding and conversion in the lung, thus extending the duration of heterosubtypic immunity.
Our prior studies in systemic infection models (Jabbari and Harty, 2006; Nolz and Harty, 2011) showed that the history of antigen exposures defines fundamental biological properties of circulating memory CD8+ T cells, which jointly determine their main function—protection against subsequent infection. For example, 1°M and 2°M CD8+ T cells show differential capacity to protect against acute and chronic infection, with 2°M cells establishing better control of acute LCMV Armstrong or Listeria monocytogenes infection, but failing to protect against chronic LCMV clone 13 infection (Jabbari and Harty, 2006; Nolz and Harty, 2011). Importantly, this difference in protective capacity was driven by differential tissue preferences of 1°M and 2°M CD8+ T cells, determined by antigen exposure history. Furthermore, the previously described dramatic changes in gene expression signature between memory T cells with different antigen exposure histories, strongly suggests that a complex network of fundamental properties, defined by the number of antigen encounters, determines the protective capacity of memory CD8+ T cells. Here, we show that, similar to systemic infections, repeated exposures of circulating memory CD8+ T cells to influenza infection in the lungs determines their ability to persist as Trm and provide heterosubtypic protection against lung infection. Although further work will be needed to address possible differences in the per-cell quality of protection between 1°M and 4°M lung Trm, it is clear that 4°M Trm, on a population level, provide superior protection, as they can successfully control virus replication even very late after initial infection, when protection mediated by 1°M cells is lost (Figure 4C).
We have previously shown that maintenance of lung 1°M Trm is a dynamic process, defined by the short half-life and high apoptotic rate of these cells on one side and replenishment from circulating memory populations on the other (Slütter et al., 2017). Importantly, not all circulating memory populations exhibit the same capacity to home to the lung and convert to Trm (Slütter et al., 2017€ ). We have identified the population of circulating Tem cells (CD62Llo) as precursors for lung Trm cells, as their circulating Tcm counterparts were not able to successfully seed the tissue and convert to Trm. As 1°M CD8+ T cells differentiate toward a uniform Tcm phenotype with time (Martin et al., 2015), the number of potential Trm precursors decreases, resulting in a gradual loss of 1°M lung Trm. Of note, we previously showed that multiple encounters with systemic pathogens generate memory populations with sustained characteristics of Tem (Wirth et al., 2010). We now show that CD8+ T cells that undergo multiple encounters with influenza-derived antigens after lung infections exhibit similarly sustained Tem characteristics (Figure S3G), allowing for extended recruitment of these cells to the influenza-experienced lung and their conversion to Trm (Figures 3A–3F). Furthermore, we were able to find well-defined, albeit reduced populations of 4°M lung Trm as late as 450 days after infection (Figure 1C), suggesting that their lung persistence, although dramatically extended, may still be limited. Given that circulating 4M populations express exclusively a Tem phenotype as late as 210 days after influenza exposure, it is likely that a slow transition to Tcm is not the sole explanation for the eventual decline of 4°M lung Trm. At this point, we cannot rule out that possible time-dependent changes in the architecture of the lung tissue and immunological landscape after influenza infection have an important impact on long-lasting maintenance of the lung Trm population. These important knowledge gaps merit further in-depth studies in the realm of lung physiology in the context of influenza virus infection.
It is important to acknowledge that our study did not compare lung Trm cells that encountered influenza once or multiple times due to repeated infection of the same host. Instead, we build on recent data describing the capacity of circulating memory CD8+ T cells to be recruited to the site of infection and convert to Trm (Beura et al., 2018; Park et al., 2018). Thus, we describe fundamental differences between Trm populations that were established by recruitment from the pool of circulating CD8+ T cells that were exposed to influenza lung infections once or multiple times, in an otherwise naive host. Understanding whether and how multiple influenza exposures shape the already existing lung Trm population remains an important knowledge gap and a major challenge, given the limitations of the available animal models and potential ethical and sampling concerns with multiple experimental challenges of humans.
Current influenza vaccines are highly purified subunit or splitvirion formulations (Soema et al., 2015; Wong and Webby, 2013), composed mainly of surface viral antigens (HA and neuraminidase [NA]), while conserved internal antigens (e.g., nucleoprotein [NP]) represent only a fraction of a split-virus vaccine (Soema et al., 2015). These vaccines aim at inducing neutralizing antibodies against surface antigens, such as HA. High mutation rate and antigenic plasticity of the main antibody targets can severely compromise the efficacy of seasonal influenza vaccines and preclude their efficacy against pandemic influenza (Boni, 2008; Kim et al., 2018). Thus, much effort is currently directed toward the concept of universal influenza vaccines. Although CD8+ T cells targeting conserved viral epitopes cannot provide sterilizing immunity to influenza infection, cumulative evidence points toward induction of lung-residing memory CD8+ T cells as a promising strategy to provide heterosubtypic immunity that could broadly limit influenza disease in humans (Kumar et al., 2017; Pizzolla et al., 2018). Our findings suggest that repeated lung immunizations with formulations containing conserved CD8+ T cell target epitopes, might generate durable lung Trm populations capable of sustained heterosubtypic immunity.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD8α (clone 53-6.7) | BioLegend | Cat# 100725 |
| Anti-mouse CD90.2 (clone 30-H12) | BuiLegend | Cat# 105306 |
| Anti-mouse CD90.1 (clone OX-7) | BioLegend | Cat# 202504 |
| Anti-mouse CD45.2 (clone 104) | BioLegend | Cat# 109832 |
| Anti-mouse CD103 (clone 2E7) | BioLegend | Cat# 121406 |
| Anti-mouse CD69 (cloneH1.2F3) | BioLegend | Cat# 104512 |
| Anti-mouse CD44 (clone IM7) | eBioscience | Cat# 11-0441-82 |
| Anti-mouse CD11a (clone M17/4) | eBioscience | Cat# 11-0111-85 |
| Anti-mouse CD62L(clone MEL-14) | eBioscience | Cat# 17-0621-83 |
| Anti-mouse Vb8.1/8.2 (clone KJ16-133) | eBiosciences | Cat# 11-5813-82 |
| Anti-mouse Va2 (clone B20.1) | eBiosciences | Cat# 47-5812-80 |
| Anti-mouse KLRG1 (clone 2F1) | eBioscience | Cat# 12-5893-82 |
| Anti-mouse CXCR3 (clone CXCR3-173) | eBioscience | Cat# 11-1831-82 |
| Anti-mouse CX3CR1 (clone SA011F11) | BioLegend | Cat# 149016 |
| Anti-mouse Bcl2 (clone BCL/10C4) | BioLegend | Cat# 633508 |
| Anti-mouse Eomes(clone Dam11Mag) | eBioscience | Cat# 12-4875-82 |
| Anti-mouse Tbet(clone 4B10) | BioLegend | Cat# 644810 |
| Tetramers | ||
| H-2Db-NP366–374 | Prepared In house | NA |
| Chemical and Peptides | ||
| LIVE/DEAD® Fixable Aqua | Thermo Fisher Scientific | Cat# L34957 |
| Brilliant Violet BV421 Streptavidin | BioLegend | Cat# 405225 |
| Vybrant FAM Caspase-3 and −7 Assay Kit | Thermo Fisher Scientific | Cat# V35118 |
| Chemical and Peptides | ||
| 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester | Sigma-Aldrich | 21888-25MG-F |
| Collagenase Type II | GIBCO | 17101-015 |
| DNase | Sigma-Aldrich | D4513-1VL |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| VitaLyse | CMDG | Cat# WBL0100 |
| Commercial Assays | ||
| Anti-PE MicroBeads | Miltenyi Biotec | Cat# 130-048-801 |
| Experimental models: Cell lines | ||
| Mouse: primary lymphocytes | This manuscript | NA |
| Flt-3L tumor cell line | (Mach et al., 2000) | B16-FLT3L (RRID:CVCL_IJ12) |
| Experimental models: Organisms/Strains | ||
| Mouse: C57BL/6J | National Cancer Institute | #556 |
| Mouse: LCMV P14 TCR Tg | Harty lab | NA |
| Recombinant influenza PR8-GP33 virus | (Mueller et al., 2010) | NA |
| Recombinant influenza X31-GP33 virus | (Mueller et al., 2010) | NA |
| Software and Algorithms | ||
| Diva 8 | BD Biosciences | http://www.bdbiosciences.com |
| FlowJo v10 | FlowJo, LLC | http://www.flowjo.com/solutions/flowjo/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John T. Harty (john-harty@uiowa.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Thy1.2/Thy1.2 C57BL/6 mice were purchased from the National Cancer Institute (Fredericksburg, MD). Thy1.1/Thy1.1 P14 TCR transgenic mice (on a C57BL/6 background) were originally acquired from Michael Bevan (University of Washington) and were maintained by brother sister mating in house. Thy1.1/Thy1.2 P14 TCR-tg mice were generated by crossing Thy1.1/Thy1.1 P14 TCR transgenic mice with Thy1.2 C57BL/6 mice in house. Mice used in all the experiments were female 6–10 weeks of age. All animal studies and procedures were approved by the University of Iowa Animal Care and Use Committee, under U.S. Public Health Service assurance, Office of Laboratory Animal Welfare guidelines.
METHOD DETAILS
Adoptive Transfer of P14 and Memory Generation
For generation of 1M CD8+ T cell response, peripheral blood cells from naive P14 TCR-transgenic mice were isolated, washed and characterized by flow cytometry for the frequency of Vb8.1,8.2+ Va2+ P14 TCR-tg T cells. Cell populations containing 104 naive Thy1.1. P14 cells were transferred iv in a 200 ul volume of sterile saline into naive C57BL/6 (Thy1.2) recipients. To transfer memory P14 cells, influenza PR8-GP33 immune mice were euthanized 90 days post infection, spleens were disrupted mechanically, made into a single cell suspension and stained with anti-Thy1.1-PE (clone Ox-7, Biolegend, San Diego, CA) in PBS with 5% FCS and subsequently purified using magnetic anti-PE beads (Mylteni, San Diego, CA) (Wirth et al., 2010). Purity of the population after enrichment was assessed by flow cytometry and ranged from 70%–85%. A cell mixture containing 105 memory Thy1.1 P14 cells was injected into naive Thy1.2 C57BL/6 mice. 2M, 3M and 4M memory P14 responses were initiated by IN infection of recipient mice with PR8GP33 24h after the adoptive transfer.
Infection and Immunizations and Virus Titers
Mice were immunized by IN infection with a sublethal dose of recombinant influenza PR8-GP33 virus (2×104 TCID50) (Mueller et al., 2010), and protection was assessed by measuring lung virus titers defined as tissue culture infective dose 50 (TCID50) using MDCK cells after high-dose (8×104 TCID50) X31-GP33 (Mueller et al., 2010) IN challenge. Both viruses were grown on chicken eggs. Viruscontaining allantoic fluid was diluted in phosphate-buffered saline (PBS), and mice were inoculated with the specific viral dose after induction of anesthesia by ketamine + xylazine (80–100 mg/kg + 10–12.5 mg/kg) injection.
Tissue Preparation, T Cell Analysis, Flow Cytometry
For quantification of antigen-specific memory CD8+ T cells, influenza immune mice were euthanized at the indicated time points p.i., 3 min after IV administration of 2 mg fluorescently labeled anti-CD45.2 antibody (IV exclusion) (Slutter et al., 2017€ ). Spleen and PBL were harvested and processed into a single-cell suspension. Lungs were cut into small pieces and incubated in the presence of collagenase (125 U/ml) and deoxyribonuclease (0.1 mg/ml) for 1h at 37C. After incubation a single-cell suspension was obtained by physical homogenization through a 70-mm cell strainer. Leukocytes were enriched by centrifugation in 35% Percoll (GE Healthcare) diluted in HBSS and red blood cells were lysed using Vitalize (BioE). For flow cytometry analyses, single cell suspensions were incubated 30 min at 4C with the indicated antibodies and then washed extensively prior to analysis. Samples were stained for Thy1.1 to identify P14 cells or in house prepared MHC class I tetramers for Db-NP366–374 and analyzed by flow cytometry to identify transgenic P14 cells or endogenous influenza specific T cells and expression of the indicated markers detected with anti-CD8 (clone 53–6.7, eBioscience), anti-Thy1.2 (clone 30-H12, eBioscience), anti-Thy1.1 (clone OX-7, eBioscience), anti-CD45.2 (clone 104, BioLegend), anti-CD103 (clone 2E7, BioLegend), anti-CD69 (clone H1.2F3, BioLegend), anti-CD44 (clone 1M7, BioLegend), anti-CD11a (clone M17/4, eBioscience), anti-CD62L (clone MEL-14, BioLegend), anti-KLRG1 (clone 2F1, eBioscience), anti-CX3CR1 (clone SA011F11), anti-CXCR3 (clone CXCR3–173, eBioscience), anti-CX3CR1 (clone SA011F11, BioLegend), anti-Eomes (clone Dan11mag, eBioscience), Anti-mouse Bcl2 (clone BCL/10C4, eBioscience), Anti-mouse Tbet (clone 4B10 eBioscience). Active caspase3/7 (Flicka) staining was performed using Vybrant FAM Caspase-3 and 7 assay kit (Thermo Fischer Scientific, Waltham, MA) according to the manufacturer’s protocol. Lung residing cells were labeled by IN administration of 70 ug of 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester (CFSE) in 100 ul of PBS. Flow cytometry data were acquired using LSRFortessa (Becton Dickinson) and analyzed using the FlowJo software (FlowJo, LLC.).
Parabiotic Surgery
Mice bearing 1°M (Thy1.1/1.1) and 4°M (Thy1.1/1.2) P14 cells were surgically conjoined 90 days after PR8-GP33 infection. Parabionts were cohoused for two weeks prior to surgery. Two days prior to surgery skin preparation was performed. Under ketamine anesthesia hair was removed from one side of the first mouse by shaving, starting approximately 1 cm above the elbow to 1 cm below the knee. Hair was then removed from the opposite side of the second mouse.
On the day of the surgery mice were anesthetized using ketamine/xylazine and injected subcutaneously with Meloxicam to induce analgesia. Once anesthesia was induced, mice were transferred to the surgery area, and the shaved skin was aseptically cleaned with betadine and alcohol. Mice were placed back to back, on their sides, with adjacent shaved areas facing up. To avoid contamination of the surgical area, mice were covered with a sterile surgical drape exposing only the surgical area. To prevent hypothermia, mice were positioned on the heat pad for the duration of the surgery.
Longitudinal skin incisions were performed on the shaved side of each animal, starting at 0.5cm above the elbow to 0.5cm below the knee joint. Skin was gently detached from the subcutaneous tissue and the separation was performed along the entire incision. After attachment of knee and elbow joints with non-absorbable 3–0 suture the skin of the two animals was connected with a continuous absorbable 5–0 suture starting ventrally from the elbow toward the knee. Once ventral skin attachment was completed with a double surgical knot, the suture was continued dorsally ending with a double surgical knot. Bupivacaine was applied locally to both sutures and each mouse was subcutaneously injected with sterile saline to prevent dehydrataion.
To prevent bacterial infections, animals were fed an antibiotic-containing diet starting from day 2 prior to surgery until day 10 post surgery. Mouse recovery was followed daily for two weeks after the procedure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical differences between two study groups were evaluated using an unpaired, two-tailed t test. Statistical differences between more than two study groups (single factor) were evaluated using one-way ANOVA with Tukey’s multiple comparison post hoc test. Two-way ANOVA with Sidak’s multiple comparison post hoc test was used to assess comparison between more than two groups based on more than one parameter (multiple factors). Statistical significance was assigned as *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Statistical analyses were performed using Prism 7 software (GraphPad).
Supplementary Material
Highlights.
Influenza-specific quaternary (4°M) lung Trm persist longer than 1°M Trm
Circulating 4°M CD8+ T cells have prolonged Tem phenotype compared to 1°M CD8+ T cells
Circulating 4°M Tem exhibit sustained migration to the lung and conversion to Trm
Prolonged lung 4°M Trm maintenance extends heterosubtypic immunity to influenza
ACKNOWLEDGMENTS
We thank Lecia Epping and Lisa Hancox for technical assistance, Stacey Hartwig for virus preparations, and Scott Hensley (University of Pennsylvania) for the SEQ12 virus. This work was supported by NIH Grants AI42767 (to J.T.H.), GM113961 (to V.P.B.), AI114543 (to J.T.H. and V.P.B.), AI124093 (to S.M.V.), and T32AI007511 (to N.V.B.-B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.073.
REFERENCES
- Altenburg AF, Rimmelzwaan GF, and de Vries RD (2015). Virus-specific T cells as correlate of (cross-)protective immunity against influenza. Vaccine 33, 500–506. [DOI] [PubMed] [Google Scholar]
- Barberis I, Myles P, Ault SK, Bragazzi NL, and Martini M (2016). History and evolution of influenza control through vaccination: from the first monovalent vaccine to universal vaccines. J. Prev. Med. Hyg 57, E115–E120. [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. (2018). Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol 19, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni MF (2008). Vaccination and antigenic drift in influenza. Vaccine 26 (Suppl 3), C8–C14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budimir N, Huckriede A, Meijerhof T, Boon L, Gostick E, Price DA, Wilschut J, and de Haan A (2012). Induction of heterosubtypic cross-protection against influenza by a whole inactivated virus vaccine: the role of viral membrane fusion activity. PLoS One 7, e30898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JP, Doherty PC, Branum KC, and Riberdy JM (2000). Profound protection against respiratory challenge with a lethal H7N7 influenza A virus by increasing the magnitude of CD8+ T-cell memory. J. Virol 74, 11690–11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Hensley SE, Ince WL, Brooke CB, Subba A, Delboy MG, Russ G, Gibbs JS, Bennink JR, and Yewdell JW (2013). Defining influenza A virus hemagglutinin antigenic drift by sequential monoclonal antibody selection. Cell Host Microbe 13, 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JC, Beyer WE, Palache AM, Rimmelzwaan GF, and Osterhaus AD (2000). Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol 61, 94–99. [PubMed] [Google Scholar]
- Doherty PC, Turner SJ, Webby RG, and Thomas PG (2006). Influenza and the challenge for immunology. Nat. Immunol 7, 449–455. [DOI] [PubMed] [Google Scholar]
- Furuya Y, Chan J, Regner M, Lobigs M, Koskinen A, Kok T, Manavis J, Li P, Mu€llbacher A, and Alsharifi M (2010). Cytotoxic T cells are the predominant players providing cross-protective immunity induced by gammairradiated influenza A viruses. J. Virol 84, 4212–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkina E, Thatte J, Dabak V, Williams MB, Ley K, and Braciale TJ (2005). Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J. Clin. Invest 115, 3473–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, and von Andrian UH (2016). The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan RJ, Usherwood EJ, Zhong W, Roberts AA, Dutton RW, Harmsen AG, and Woodland DL (2001). Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J. Immunol 166, 1813–1822. [DOI] [PubMed] [Google Scholar]
- Jabbari A, and Harty JT (2006). Secondary memory CD8+ T cells are more protective but slower to acquire a central-memory phenotype. J. Exp. Med 203, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Webster RG, and Webby RJ (2018). Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol. 31, 174–183. [DOI] [PubMed] [Google Scholar]
- Kondrich J, and Rosenthal M (2017). Influenza in children. Curr. Opin. Pediatr 29, 297–302. [DOI] [PubMed] [Google Scholar]
- Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, and Rimmelzwaan GF (2007). Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25, 612–620. [DOI] [PubMed] [Google Scholar]
- Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, et al. (2017). Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legge KL, and Braciale TJ (2003). Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity 18, 265–277. [DOI] [PubMed] [Google Scholar]
- Liang S, Mozdzanowska K, Palladino G, and Gerhard W (1994). Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol 152, 1653–1661. [PubMed] [Google Scholar]
- Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, and Dranoff G (2000). Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 60, 3239–3246. [PubMed] [Google Scholar]
- Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, and Carbone FR (2015). T-box transcription factors combine with the cytokines TGF-b and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Martin MD, Kim MT, Shan Q, Sompallae R, Xue HH, Harty JT, and Badovinac VP (2015). Phenotypic and functional alterations in circulating memory CD8 T cells with time after primary infection. PLoS Pathog. 11, e1005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Ha SJ, Vezys V, and Ahmed R (2006). Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J. Immunol 177, 831–839. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Gotch FM, Noble GR, and Beare PA (1983). Cytotoxic T-cell immunity to influenza. N. Engl. J. Med 309, 13–17. [DOI] [PubMed] [Google Scholar]
- Mueller SN, Langley WA, Li G, García-Sastre A, Webby RJ, and Ahmed R (2010). Qualitatively different memory CD8+ T cells are generated after lymphocytic choriomeningitis virus and influenza virus infections. J. Immunol 185, 2182–2190. [DOI] [PubMed] [Google Scholar]
- Nguyen HH, Moldoveanu Z, Novak MJ, van Ginkel FW, Ban E, Kiyono H, McGhee JR, and Mestecky J (1999). Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8+ cytotoxic T lymphocyte responses induced in mucosa-associated tissues. Virology 254, 50–60. [DOI] [PubMed] [Google Scholar]
- Nicholson KG, Wood JM, and Zambon M (2003). Influenza. Lancet 362, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolz JC, and Harty JT (2011). Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity 34, 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, et al. (2018). Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol 19, 183–191. [DOI] [PubMed] [Google Scholar]
- Pizzolla A, Nguyen TH, Sant S, Jaffar J, Loudovaris T, Mannering SI, Thomas PG, Westall GP, Kedzierska K, and Wakim LM (2018). Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J. Clin. Invest 128, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Martin MD, and Badovinac VP (2014). The longevity of memory CD8 T cell responses after repetitive antigen stimulations. J. Immunol 192, 5652–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, and Masopust D (2014). Tissue-resident memory T cells. Immunity 41, 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slu€tter B, Pewe LL, Kaech SM, and Harty JT (2013). Lung airway-surveilling CXCR3hi memory CD8+ T cells are critical for protection against influenza A virus. Immunity 39, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slu€tter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, and Harty JT (2017). Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol 2, eaag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soema PC, Kompier R, Amorij JP, and Kersten GF (2015). Current and next generation influenza vaccines: formulation and production strategies. Eur. J. Pharm. Biopharm 94, 251–263. [DOI] [PubMed] [Google Scholar]
- Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, and Lalvani A (2013). Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med 19, 1305–1312. [DOI] [PubMed] [Google Scholar]
- Takamura S, Yagi H, Hakata Y, Motozono C, McMaster SR, Masumoto T, Fujisawa M, Chikaishi T, Komeda J, Itoh J, et al. (2016). Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med 213, 3057–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefranc¸ ois L, and Farber DL (2011). Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol 187, 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Braeckel-Budimir N, Martin MD, Hartwig SM, Legge KL, Badovinac VP, and Harty JT (2017). Antigen exposure history defines CD8 T cell dynamics and protection during localized pulmonary infections. Front. Immunol 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, and Masopust D (2009). Memory CD8 T-cell compartment grows in size with immunological experience. Nature 457, 196–199. [DOI] [PubMed] [Google Scholar]
- Wakim LM, Gupta N, Mintern JD, and Villadangos JA (2013). Enhanced survival of lung tissue-resident memory CD8+ T cells during infection with influenza virus due to selective expression of IFITM3. Nat. Immunol 14, 238–245. [DOI] [PubMed] [Google Scholar]
- Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, and Badovinac VP (2010). Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity 33, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SS, and Webby RJ (2013). Traditional and new influenza vaccines. Clin. Microbiol. Rev 26, 476–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Hu Y, Lee YT, Bouchard KR, Benechet A, Khanna K, and Cauley LS (2014). Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol 95, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.