Abstract
Patients with acute kidney injury (AKI) frequently require kidney transplantation and supportive therapies, such as rehydration and dialysis. Here, we show that radiolabelled DNA origami nanostructures (DONs) with rectangular, triangular and tubular shapes accumulate preferentially in the kidneys of healthy mice and mice with rhabdomyolysis-induced AKI, and that rectangular DONs have renal-protective properties, with efficacy similar to the antioxidant N-acetylcysteine—a clinically used drug that ameliorates contrast-induced AKI and protects kidney function from nephrotoxic agents. We evaluated the biodistribution of DONs non-invasively via positron emission tomography, and the efficacy of rectangular DONs in the treatment of AKI via dynamic positron emission tomography imaging with 68Ga-EDTA, blood tests and kidney tissue staining. DNA-based nanostructures could become a source of therapeutic agents for the treatment of AKI and other renal diseases.
Acute kidney injury (AKI), formerly named acute renal failure, is characterized by the accumulation of nitrogenous wastes and decreased urine output within hours or days of disease onset1,2. There are more than 13.3 million new cases of AKI, in addition to 1.7 million associated deaths, annually worldwide3. While kidney transplantation and supportive therapies, such as rehydration and dialysis, are available, there are currently no curative therapies4. N-acetylcysteine (NAC) showed positive effects in preventing contrast-induced AKI and preserving kidney function from nephrotoxic agents; yet, the poor bioavailability of NAC has hindered its clinical applicability to other types of AKI5,6.
Nanomedicine is the design and engineering of atoms or molecules at the submicrometre level to generate materials with fascinating physicochemical properties for biomedical applications7–9. The past decade has witnessed significant improvements in the development of new nanomedicines for imaging and therapy of various diseases, including AKI10–13. For example, carbon nanotubes have been employed as kidney-targeting vehicles for the delivery of small interfering RNA for the treatment of AKI14. However, there has been limited success in engineering nanomaterials for kidney targeting and treatment in vivo15–18.
DNA nanotechnology allows for the construction of predictable, programmable nanostructures by exploiting intra- and intermolecular Watson–Crick base pairing19, which enables the design of complex structures with distinct dimensions, shapes and sizes20,21. DNA nanostructures display excellent in vivo properties, including minimal toxicity, low immunogenicity and excellent biostability22–25. DNA origami nanostructures (DONs)—especially framework nucleic acids—have been used for the delivery of small interfering RNA, doxorubicin, gold nanorods and photosensitizers into tumour sites for imaging and treatment (for example, chemotherapy, photothermal therapy and photodynamic therapy) of cancer26–34. However, to date, no studies have explored the biodistribution of DONs in vivo using quantitative imaging strategies, which has limited further applications of DNA-based nanomaterials in the biomedical arena35,36.
In this study, we prepared and radiolabelled a series of DONs with Cu-64 (half-life: 12.7 h) for positron emission tomography (PET) imaging to investigate their biological behaviour in vivo. PET imaging revealed that DONs preferentially accumulate in the kidneys of healthy mice and mice with rhabdomyolysis-induced AKI. The rectangular DON (Rec-DON) was therapeutically efficacious in mice with AKI, as confirmed by quantitative renal PET imaging, blood tests and haematoxylin and eosin staining of kidney tissue sections. Intriguingly, Rec-DON (10 μg mouse−1) elicited excellent therapeutic effects comparable to those of NAC (4.2 mg mouse−1)—the current gold standard for the treatment of contrast-induced AKI, and the most commonly used dietary supplement for food and antioxidants for several clinical indications. Furthermore, DONs actively accumulated in both healthy and diseased kidneys, thus resulting in an increased therapeutic effect compared with traditional AKI therapy (Fig. 1a). We anticipate that these results will catalyse new avenues for the development of DNA nanotechnology and innovative therapies for various kidney-related diseases, and AKI treatment in this work only serves as an initial proof of principle.
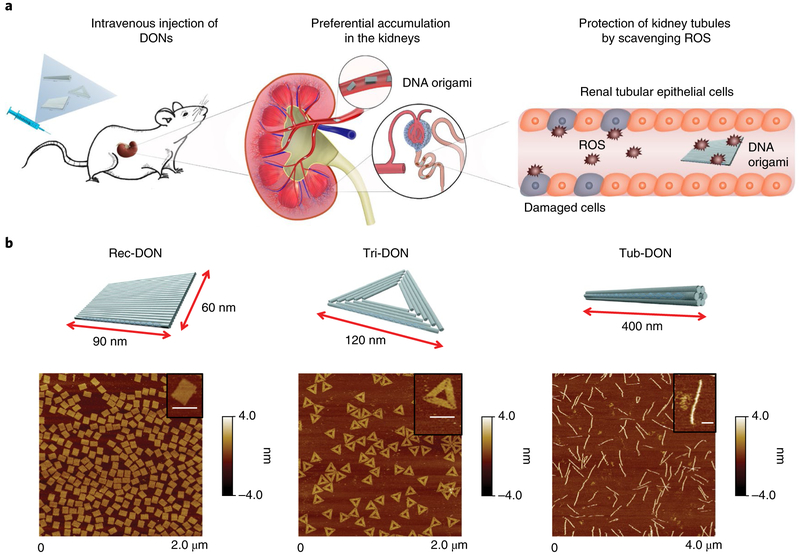
Fig. 1 ∣. Preferential renal accumulation of DONs enabled protection against AKI.
a, Three different DONs (Rec-DON, Tri-DON and Tub-DON) showed selective kidney accumulation after intravenous injection. When applied to murine models of rhabdomyolysis-induced AKI, DONs can scavenge ROS and alleviate oxidative stress locally, protecting kidney structures and alleviating AKI. b, Schematic structures of the three DONs (top) and corresponding AFM images (bottom). Insets: single DON structures. Scale bars, 100 nm.
Results
Preparation of DONs.
Using a single-step annealing procedure reported previously37,38, we constructed 3 different morphologies of DONs: Rec-DON (90 nm × 60 nm), triangular DON (Tri-DON; 120 nm per edge) and tubular DON (Tub-DON; 400 nm long), as shown in Fig. 1b. A Cy3/Cy5 dual-labelled Rec-DON (termed 'FRET Rec-DON', where FRET stands for fluorescence resonance energy transfer) was assembled to facilitate stability studies, where eight staple strands were functionalized either at their 5′-end with an acceptor Cy5 fluorophore or at their 3′-end with a donor Cy3 flu-orophore (Supplementary Fig. 4). We also assembled three partially folded DONs, in which a portion of staple strands was removed from the annealing process. Atomic force microscopy (AFM) images of all three partially folded DONs displayed structures with a significant part of the M13 (a circular single-stranded DNA (ssDNA) used as the scaffold for annealing) unfolded (Supplementary Figs. 13–15). Dynamic light scattering and gel electrophoresis revealed smaller sizes of partially folded DONs, compared with their intact counterparts (Supplementary Figs. 16–18). Eight overhangs were designed on the surface of each DON, allowing subsequent functionalization and 64Cu labelling of DONs for in vivo studies (Supplementary Figs. 1–18, Supplementary Table 1 and Supplementary Sheets 1–3).
Predominant renal uptake of DONs in healthy mice.
We radiolabelled ssDNA to generate 64Cu-ssDNA (with specific activity [SA] of at least 0.8–1.7 mCi nmol−1), which further provided 64Cu-DONs (specific activity of 6.4–13.6 mCi nmol−1) on hybridization to overhangs designed on different DONs (Supplementary Fig. 19). Ten ssDNAs complementary to different parts of M13 ssDNA were also radiolabelled to obtain 64Cu-M13 (specific activity of 8–17 mCi nmol−1). The radiolabelling efficiency of 64Cu-ssDNA was 84.8 ± 0.6% (n = 5; Supplementary Fig. 20). Longitudinal PET scans at different time points within 24 h were performed after intravenous injection of radiolabelled DONs or M13 into healthy mice.
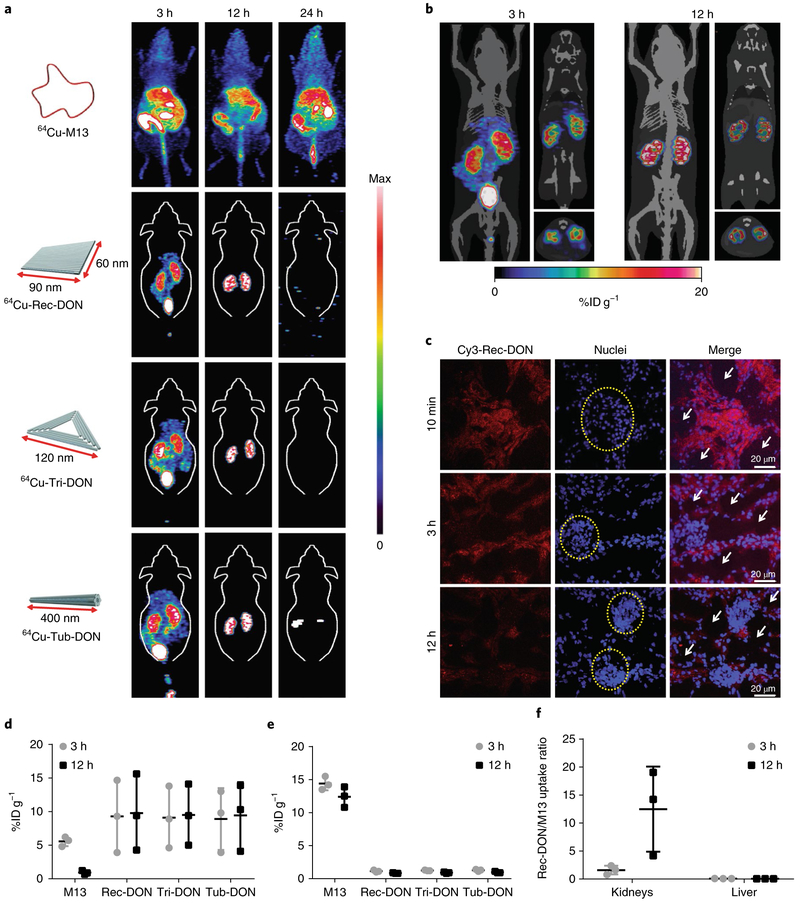
All 3 intact DONs showed preferential accumulation in the kidneys at 3 and 12 h post-injection. No retention of the radioactivity signal was observed at 24 h post-injection, indicative of complete clearance of intact DONs in vivo (Fig. 2a and Supplementary Videos 1–3). PET images of M13 presented high liver and gastrointestinal uptake, revealing its enterohepatic clearance from 3 to 24 h post-injection. PET imaging with three 64Cu-labelled, partially folded DONs showed increased liver sequestration compared with intact DONs (Supplementary Fig. 21). The similar biodistribution pattern of M13 ssDNA and partially folded DONs indicated a rapid response of the mononuclear phagocyte system (known to mediate the removal of foreign antigens from the circulation) to recognize and eliminate foreign DNA from the blood circulation25,39.
Fig. 2 ∣. Biodistribution of DONs in healthy mice.
a, Maximum intensity projection PET images of 64Cu-labelled M13, Rec-DON, Tri-DON and Tub-DON in healthy mice at 3, 12 and 24 h after intravenous injection. One of three independent experiments is shown. DONs showed selective kidney uptake at 3 h and 12 h post-injection. The accumulation of DNA structures was denoted as the %IDg−1, and the maximum scale bar was 20%IDg−1 for PET images at 3 h and 12 h. At 24 h post-injection, maximum scale bars were 20, 0.1, 0.1 and 0.1%IDg−1 for 64Cu-M13, 64Cu-Rec-DON, 64Cu-Tri-DON and 64Cu-Tub-DON, respectively. b, PET imaging of 64Cu-Rec-DON merged with the computed tomography framework of a mouse at 3 h and 12 h post-injection. One of three independent experiments is shown. The renal cortex is clearly delineated at 3 h and 12 h, demonstrating prominent renal accumulation. The three images for each time point are maximum intensity projection (left), coronal (upper right) and axial slices (lower right). c, Confocal images of kidney sections after intravenous injection of Cy3-labelled Rec-DON (red signal; one of three independent experiments is shown). Nuclei were stained with 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI; blue). Yellow dashed circles denote glomeruli and white arrows denote renal tubular lumina. d, ROI analysis of kidney uptake at 3 h and 12 h post-injection. e, ROI analysis of liver uptake at 3 h and 12 h post-injection. M13 showed primarily liver uptake and minimal kidney accumulation, whereas all three DONs showed higher kidney accumulation and low liver uptake. f, Ratio of kidney and liver uptake between Rec-DON and M13 at 3 h and 12 h post-injection. In d–f, data represent means ± s.d. from three independent replicates.
A region-of-interest (ROI) analysis of PET data demonstrated a similar accumulation (approximately 10% injected dose per gram (%ID g−1)) of intact DONs in the kidneys at 12 h postinjection, with Rec-DON showing the highest renal uptake value (although not statistically significant) among all 3 (n = 3 for each DON; Fig. 2d). M13 ssDNA exhibited a 17.5-times-higher liver uptake (15.2 ± 1.5%ID g−1) and 11.7-times-lower renal uptake (0.8 ± 0.2%ID g−1) than Rec-DON (n = 3; Fig. 2d–f). Three partially folded DONs shared liver and kidney uptake values of approximately 10 and 4%ID g−1 at 24 h post-injection, respectively (n = 3 for each partially folded DON; Supplementary Figs. 22 and 23). Biodistribution studies at 24 h post-injection confirmed the whole-body clearance of Rec-DON, with all organ uptake values lower than 1%ID g−1 (n = 3; Supplementary Fig. 24). M13, instead, showed retained liver accumulation of higher than 10%ID g−1 (n = 3; Supplementary Fig. 25). For comparison purposes, we performed a PET imaging and biodistribution study of 64Cu-ssDNA (20 bases long) and found that short ssDNA underwent rapid renal filtration within 30 min post-injection, and the signal was detected primarily in the gallbladder and intestine from 3 to 24 h post-injection, with uptake values lower than 1%ID g−1 at 24 h post-injection (n = 3; Supplementary Figs. 26 and 27).
To better investigate the distribution of Rec-DON, the computed tomography framework of a mouse was merged with PET images of Rec-DON (Fig. 2b). We found minimal liver uptake of about 1%ID g−1 and prominent kidney retention of about 20%ID g−1 in the renal parenchyma at 12 h post-injection. Furthermore, we injected Cy3-labelled Rec-DON into healthy mice and harvested the kidneys at 10 min, 3 h and 12 h post-injection. Confocal laser scanning microscopy of the kidney sections demonstrated that Rec-DON initially accumulated in the glomerulus, and was later found primarily in the renal tubules (Fig. 2c and Supplementary Fig. 28), in agreement with PET images showing that the majority of Rec-DON stayed in the renal parenchyma. The fluorescence intensity of Rec-DON in the glomerulus decreased from 10 min to 12 h post-injection, suggesting that Rec-DON gradually passed through the glomeruli and was slowly excreted from the kidneys over time. A cell internalization study using human embryonic kidney 293 (HEK293) cells demonstrated that Rec-DON needed at least 6 h to be transported into cells, suggesting that the signal in the kidney tissue sections mostly represents the prolonged retention of Rec-DON (Supplementary Fig. 29). However, the basolateral uptake pathway (from the blood directly to tubular cells) may also be possible.
To assess the structural integrity of Rec-DON within the kidneys, we collected urine samples at 3 and 12 h post-injection of FRET Rec-DON and performed fluorescence imaging to detect the urinary elimination of Rec-DON. The donor–acceptor energy transfer demonstrated that some of the injected Rec-DON survived the renal excretion process (Supplementary Fig. 30). To fully test the stability of Rec-DON, we incubated FRET Rec-DON in 80% foetal bovine serum (FBS) and 50% mouse urine. The results indicated a decrease of FRET efficiency to approximately 50% after 12 h of incubation, suggesting that a significant portion of Rec-DON survived the challenge of enzymes/proteins in FBS and basic milieu of mouse urine (Supplementary Figs. 31–33).
From the PET imaging results, we discovered that DONs preferentially accumulated in the kidneys, while M13 ssDNA and partially folded DONs were sequestered by the liver. The different biodistribution patterns between intact DONs and other investigated DNA nanostructures emphasized that proper folding of M13 into intact DONs played an important role in downregulating hepatic sequestration of DNA nanomaterials, which may be of great advantage for designing DNA nanostructures towards lower liver accumulation and better bioavailability. To further verify the hypothesis, another compact DNA structure—DNA tetrahedron nanoparticle (DTN; edge length: 20 bases)40,41—was tested for its biodistribution (Supplementary Fig. 34 and Supplementary Table 4). We found that compared with three fully folded DONs, DTN presented a similarly low liver uptake and significantly higher renal excretion. We conclude that the compact structures and intact folding contributed to the decreased liver uptake and improved kidney excretion for DTN, and accumulation for DONs.
We hypothesized that active kidney accumulation of DONs maybe attributed to three factors. First, compared with M13 ssDNA and partially folded DONs, intact DONs are more compact structures without significant regions of DNA exposed to enzymes/proteins in the blood pool42. M13 and partially folded DONs expose virus DNA in the blood, which can trigger immune surveillance or enzyme/protein interaction and subsequent liver sequestration25. In contrast, fully folded DONs may elude unnecessary enzyme digestion due to their compact structures22. Second, negatively charged nanoparticles have a decreased chance of absorbing proteins in the blood to form a protein corona, resulting in reduced accumulation in the liver and spleen43–45. Third, the morphologies and sizes of DONs play pivotal roles in their localization to the kidneys46,47. Three biological filtration barriers stand before efficient renal targeting and delivery of DONs. The nanostructure, or any compound undertaking renal filtration, must pass through: (1) glomerulus endothelium containing pores (fenestra) of 80–100 nm; (2) the negatively charged glomerular basement membrane with pores of 5–8 nm; and (3) a slit diaphragm of podocytes with pores of around 15 nm12,47. Previous studies reported the glomerular filtration and urinary excretion of carbon nanotubes (average length of 300–500 nm and diameter of 20–30 nm)48,49. Tub-DON (400 nm in length and 7 nm in diameter) of a similar size and dimension may undergo the same process of renal excretion in vivo by aligning its rod axis with pores in kidney structures, as predicted using a mathematical model of the rotational diffusivity49. Other studies have reported that soft discoidal polymeric nanoconstructs (1,000 nm × 400 nm rectangular or circular discs) displayed resistance to macrophage uptake50, and a mesoscale organic nanoparticle (400 nm) with negative charges showed specific renal accumulation, as discovered by fluorescent imaging51. Integrating previous investigations and PET imaging results of DONs, we posited that compact structures and negative charges may contribute to the escape of hepatic sequestration, while the well-designed shapes and sizes of DONs result in the glomerular endothelial fenestrae filtration and ultimately lead to increased kidney accumulation in vivo. Overall, PET imaging provides a quantitative, reliable and real-time method with which to observe the circulation and biodistribution of DONs in vivo. Based on the PET data shown above, we selected Rec-DON as the best candidate for further investigation in AKI mice.
PET imaging of 64Cu-Rec-DON in the AKI murine model.
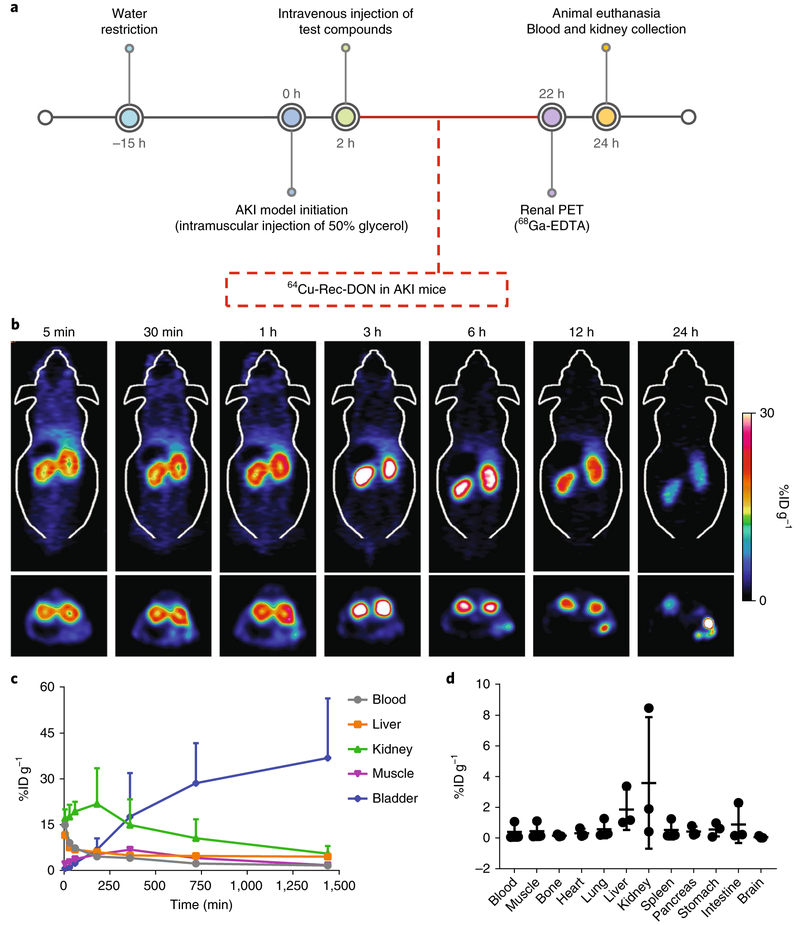
To investigate the treatment effect of DONs, we prepared a murine model of AKI through intramuscular injection of 50% glycerol into dehydrated healthy mice52–54. After the injection, massive myoglobin and haemoglobin were released from the dissolved muscle, and reactive oxygen species (ROS) were generated in forms of H2O2 and lipid hydroperoxides. In the absence of reductants (such as catalase, superoxide dismutase (SOD) and glutathione peroxidase), these extracellular ROS may undergo renal excretion, destroy renal tubules and eventually induce AKI52. We performed blood analysis and kidney tissue staining to validate the development of the model and found that the symptoms of AKI (increased nitrogenous wastes in the blood and tubular damage in the kidneys) developed as early as 2 h after glycerol injection and reached the end stage at 24 h (Supplementary Figs. 35 and 36). Further monitoring of AKI mice showed that all mice (n = 5) died at 3 d after AKI initiation and the average bodyweight declined to approximately 80% of the weight at day 0 (Supplementary Fig. 37).
Two hours after induction of AKI, we injected 64Cu-Rec-DON into AKI mice and performed longitudinal PET scanning to monitor its biodistribution (Fig. 3a). 64Cu-Rec-DON showed rapid accumulation in the kidneys as early as 5 min post-injection (Fig. 3b and Supplementary Video 4). The gradual renal accumulation of 64Cu-Rec-DON (green line in Fig. 3c) in AKI mice suggested that the injured kidneys may still extract and excrete DONs from the blood pool into the bladder, although at a decreased rate. The bladder signal of 64Cu-Rec-DON continued to rise for 24 h post-injection (blue line in Fig. 3c). While Rec-DON in healthy mice showed an overall clearance (calculated based on the whole-body ROI analysis) of nearly 100%ID at 24 h post-injection, the clearance of Rec-DON in AKI mice was approximately 28.3%ID. We conclude that the suppressed renal clearance and enhanced whole-body retention of Rec-DON in AKI mice was in good agreement with the clinical manifestation of the disease. These results also verify the preferential renal accumulation of Rec-DON in AKI mice. Furthermore, we injected Cy3-Rec-DON into AKI mice and harvested their kidneys at 10 min, 3 h and 12 h post-injection. Confocal laser scanning microscopy images of these kidney sections confirmed the accumulation of Rec-DON in the renal tubules (Supplementary Fig. 38). We also harvested organs and tissues at 24 h after initiation of the AKI model and measured the accumulation of Rec-DON in each tissue sample, with values denoted as %ID g−1. As shown in Fig. 3d, approximately 4%ID g−1 of Rec-DON was detected in the kidneys, and off-target accumulation was less than 5%ID g−1, emphasizing the low off-targeting potential of Rec-DON in AKI mice.
Fig. 3 ∣. Animal study design and PET imaging of 64Cu-Rec-DON in the murine model of AKI.
a, Preparation and treatment schedule of AKI mice. Dehydrated ICR mice were injected with 50% glycerol intramuscularly to induce muscle injury, which would further cause injury to the kidneys. Two hours after the initiation of AKI, different agents were administered intravenously. Three methods were employed to evaluate kidney function after treatment: dynamic PET imaging with 68Ga-EDTA, serum analysis of creatinine and blood urea nitrogen (BUN), and haematoxylin and eosin staining of kidney tissues. b, Longitudinal PET imaging of 64Cu-Rec-DON in mice with AKI. Images (top, coronal slices; bottom, axial slices) revealed that kidney injury does not change the biodistribution of Rec-DON in vivo, except that retention of 64Cu-Rec-DON in the kidneys is extended (n = 3 mice; one of 3 independent experiments is shown). c, ROI analysis of 64Cu-Rec-DON PET images in AKI mice showed low uptake of 64Cu-Rec-DON in the blood, liver and muscle. A gradual kidney accumulation process (green line) was observed, suggesting that injured kidneys slowly extracted Rec-DON from the blood pool, with the bladder signal continuing to rise over the 24 h time period. d, Biodistribution of 64Cu-Rec-DON in the murine model of AKI at 24 h post-injection. In c and d, data represent means ± s.d. from three independent replicates.
ROS scavenging effect in vitro.
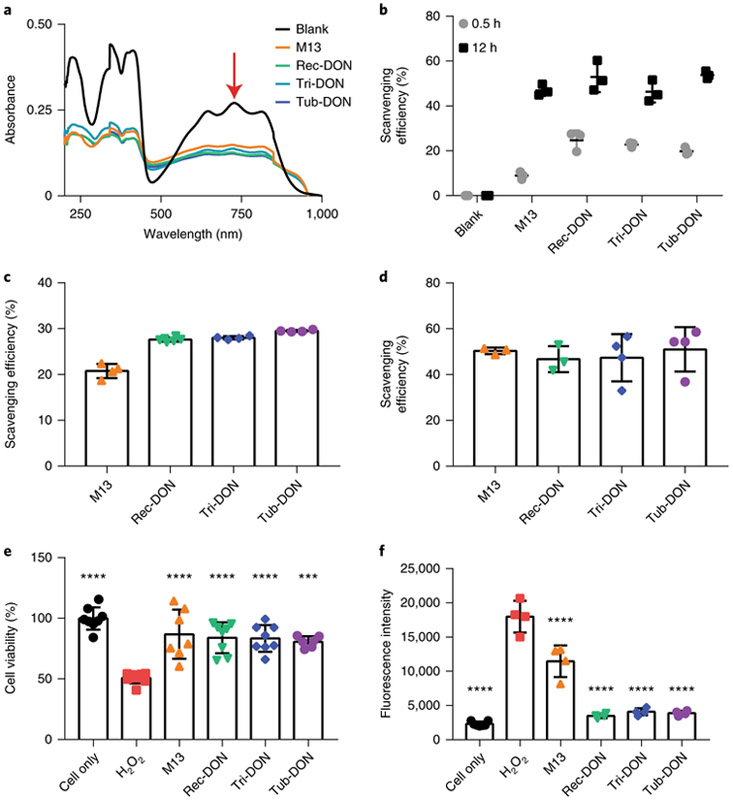
The development of AKI has been linked to cell damage caused by local ROS55,56. Since DNA is sensitive to ROS57, a variety of drugs have been developed to protect DNA from oxidative stress. Here, we asked whether DONs delivered to the kidneys can react with ROS in the kidneys, and eventually alleviate the clinical symptoms of AKI. To start off, ABTS radicals, obtained from the oxidation of 2,2’-azino-bis(3-ethylbenzothiazo-line-6-sulfonic acid) molecules (ABTS), were used as a representative ROS in vitro and mixed with M13 ssDNA (4 pmol) or each of the three DONs (2 pmol) with an equal number of DNA bases. As shown in Fig. 4a,b, all DNA constructs showed ROS eliminating effects when incubated with ABTS. Furthermore, we performed assays with hydroxyl radicals (•OH) and superoxide radicals (•O2−)—two typical types of ROS commonly seen in living organisms. We found that all DNA materials (M13 and three DONs) readily reacted with both types of ROS and lowered the level of oxidative stress in vitro (Fig. 4c,d).
Fig. 4 ∣. ROS scavenging effect of DONs in vitro.
a, Ultraviolet–visible–near-infrared spectra of ABTS radicals incubated with various DONs of comparable concentrations (10.0 nM for M13 ssDNA and 5.0 nM for each DON) for 5 h. The characteristic absorbance of ABTS radicals at 734 nm (red arrow) decreased after incubation with DNA materials. One of three independent experiments is shown. b, Analysis of the scavenging efficiencies of ABTS radicals for different DONs. c,d, Analysis of the scavenging efficiencies of hydroxyl radicals (•OH; c) and superoxide radicals (•O2− d) for different DNA materials (10.0 nM for M13 ssDNA and 5.0 nM for each DON). e,f, Cell viability analysis by MTT assay (e) and ROS concentration (f) when incubating HEK293 cells with H2O2 (250 μM) and different DONs with equal numbers of DNA bases. All DONs are equally effective in vitro. Statistical significance compared with the H2O2 group is shown (***P < 0.001; ****P < 0.0001). Statistical analysis was performed by one-way ANOVA with Tukey's honest significant difference post-hoc test. Refer to Supplementary Table 5 for detailed P values for e, and Supplementary Table 6 for detailed P values for f. In b–f, data represent means ± s.d. from either three (b and d), four (c and f) or six (e) independent replicates.
Next, we used HEK293 cells (a human embryonic kidney cell line) to investigate the protective effect of DONs against oxidative stress produced by H2O2. After culturing HEK293 cells with 250 μM of H2O2, the cell survival rate decreased to approximately 50% at 24 h post-incubation (Supplementary Fig. 39). When incubated with M13 ssDNA or DONs, cell viability was significantly higher at about 80% (Fig. 4e and Supplementary Table 5). We also tested ROS levels in the cell culture medium and found that DONs alleviated oxidative stress within 2 h of incubation (Fig. 4f and Supplementary Table 6).
We further investigated the redox product of Rec-DON after its incubation with different concentrations of H2O2. AFM imaging and gel electrophoresis found no significant structural disassembly of the structure, as shown in Supplementary Figs. 41 and 42. Detailed measurement of the concentration of 8-hydroxy-2’-deoxyguanosine (one of the major oxidized derivatives of deoxyguanosine, and a biomarker of oxidative stress) in solutions after 30 min of incubation with H2O2 showed a more than fivefold increase for M13 and the 3 DONs (Supplementary Fig. 44).
Since the magnesium concentration is important to maintain the integrity of DONs24, we tested the tolerance of HEK293 cells with different concentrations of Mg2+ in the cell culture medium. We found that a high concentration of magnesium (as high as 12.5 mM) did not elicit any adverse effects for HEK293 cells in terms of viability (Supplementary Fig. 40), which assured us that intact DONs would not disassemble to M13 ssDNA in our ROS scavenging experiments with HEK293 cells. Our investigation of the interaction of DNA materials with ROS indicated that both M13 ssDNA and DONs may effectively neutralize ABTS radicals or the three types of ROS (•OH, •O2− and H2O2) via oxidation of DNA bases, and further protect cells from oxidative stress in vitro.
Treatment of AKI with Rec-DON.
Based on the in vitro ROS scavenging effect and in vivo PET imaging of 64Cu-DONs, we hypothesized that the accumulation of DONs in the kidneys may allow for local protection, and even effective treatment of AKI. For comparison, NAC—an antioxidant used clinically for the prevention of contrast-induced AKI5,58—was utilized as a positive control, while phosphate saline buffer (PBS) and M13 ssDNA were set as the negative controls.
We compared the therapeutic effect between five treatment groups, including Rec-DON (10 μg mouse−1), PBS buffer, M13 ssDNA (10 μg mouse−1), high-dose NAC (H-NAC; 4.2 mg mouse−1) and low-dose NAC (L-NAC; 10 μg mouse−1). These agents (PBS, M13 ssDNA, Rec-DON, H-NAC and L-NAC) were intravenously injected at 2 h after initiation of AKI. At 22 h after the creation of AKI models, a 30 min dynamic PET scan with 68Ga-EDTA was performed to evaluate the renal function of the mice. All mice were euthanized at 24 h after the model preparation, with kidney and blood samples collected and analysed for renal function (Fig. 3a).
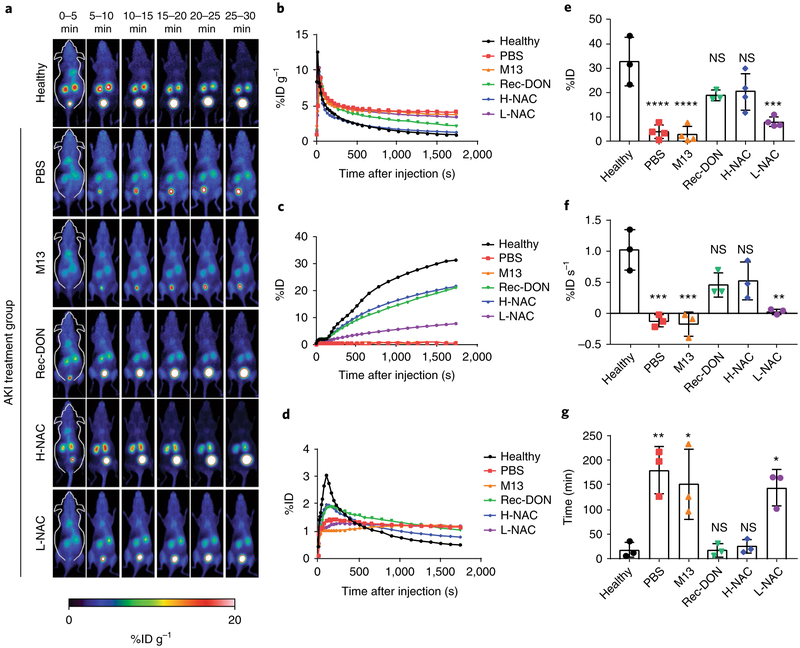
Dynamic PET imaging using 68Ga-EDTA (specific activity: 1–2 mCi nmol−1)—the traditional tracer used for renal imaging in the clinic59,60—allows for non-invasive and real-time evaluation of renal function. As shown in Fig. 5a and Supplementary Video 5, the 68Ga-EDTA signal in the kidneys of healthy mice peaked between 0 and 5 min and decreased from 10–30 min post-injection. Such short retention in the blood pool and high accumulation in the bladder provided baseline information for both 68Ga-EDTA in vivo and kidney function in healthy mice. In contrast, AKI mice treated with either PBS or M13 ssDNA showed sustained tracer retention in the blood pool and little excretion of 68Ga-EDTA to the bladder. Kidney accumulation of 68Ga-EDTA was rapid after injection, and the tracer remained in the kidneys, indicative of severe kidney impairment (that is, PBS and M13 ssDNA had no therapeutic effect). AKI mice treated with Rec-DON displayed enhanced kidney excretory function, and treatment of AKI with NAC showed a dose-dependent biological response.
Fig. 5 ∣. Dynamic PET imaging using 68Ga-EDTA for evaluation of kidney function.
a, Representative PET images of healthy mice from three independent experiments, alongside those of AKI mice treated with PBS, M13 (10 μg mouse−1), Rec-DON (10 μg mouse−1), H-NAC (4.2 mg mouse−1) or L-NAC (10 μg mouse−1). b, ROI analysis of signals in the blood pool. Lower signals indicate faster bladder clearance and better excretion of 68Ga-EDTA from the kidneys. c, ROI analysis of signals in the bladder. Higher signals indicate better function of the kidneys. d, ROI analysis of signals in the kidneys. A sharper peak indicates better excretory function of the kidneys. In b–d, one representative curve from three independent experiments is shown for each group. e, Clearance of 68Ga-EDTA into the bladder. Higher clearance defines better excretion, indicating better recovery of kidney function from AKI after treatment. Refer to Supplementary Table 7 for detailed P values. f, SIU of the kidney ROI curve between 1 and 2 min post-injection of 68Ga-EDTA. A positive SIU value indicates proper function of the kidney. Refer to Supplementary Table 8 for detailed P values. g, Washout half-time (t1/2) of the kidney ROI curve. A shorter time denotes better renal excretion, further suggesting better treatment efficacy of AKI. Refer to Supplementary Table 9 for detailed P values. In e–g, data represent means ± s.d. from three independent replicates; statistical significance compared with the healthy group is shown (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, non-significant); and statistical analysis was performed by one-way ANOVA with Tukey's honest significant difference post-hoc test.
Three parameters are commonly used in the clinic to evaluate renal function based on the kidneys' time–activity curve: clearance (%ID), slope of initial uptake (SIU; min−1) and washout half-time (t1/2; min). Therefore, we performed ROI analysis of renal PET images to analyse these three parameters (Fig. 5b–d). 68Ga-EDTA clearance was denoted as the amount of tracer that was effectively cleared from the kidneys into the bladder by 30 min post-injection. We found that 3.7 ± 2.9, 2.7 ± 3.2, 17.0 ± 5.0, 20.0 ± 7.0 and 7.8 ± 2.1%ID of the tracer was cleared for the PBS, M13, Rec-DON, H-NAC and L-NAC treatment groups, respectively (n ≥ 3 for all imaging groups). In comparison, healthy mice displayed a clearance of 33.0 ± 10.0%ID (Fig. 5e and Supplementary Table 7; n ≥ 3). Next, we examined the SIU of the kidney ROI curves, which displayed a linear increase directly after injection (60 to 120 s) and represented the speed of kidney perfusion. Comparing the six groups, SIU values were significantly higher in the healthy, Rec-DON and H-NAC groups, indicative of better kidney function and efficient treatment of AKI (Fig. 5f and Supplementary Table 8). The washout half-times (t1/2; that is, the time interval between the peak tracer concentration and half of the peak value) in the healthy, Rec-DON and H-NAC groups were significantly shorter than those in the PBS, M13 and L-NAC groups, as shown in Fig. 5g and Supplementary Table 9.
Overall, PET imaging and ROI analysis of 68Ga-EDTA accumulation in the blood, bladder and kidneys confirmed that treatment of AKI with PBS or M13 was completely unsuccessful. Rec-DON (10 μg mouse−1) showed a similar effect to that of NAC at a therapeutic dose (H-NAC; 4.2 mg mouse−1)61, while L-NAC (10 μ g mouse−1) did not produce any therapeutic effect. Since all DNA nanostructures investigated in this study could scavenge ROS based on our in vitro studies, the fact that M13 did not cause any benefit to renal function excluded the possibility of potential secondary effects (vasodilatory or others) improving renal function. Different biodistribution behaviours of M13 (primarily in the liver) and Rec-DON (predominantly in the kidneys) greatly altered the amount of DNA in the vicinity of kidney structures and led to distinct results for AKI treatment. Rec-DON at a dose of 10 μg mouse−1 produced a comparable therapeutic effect in the AKI model to H-NAC.
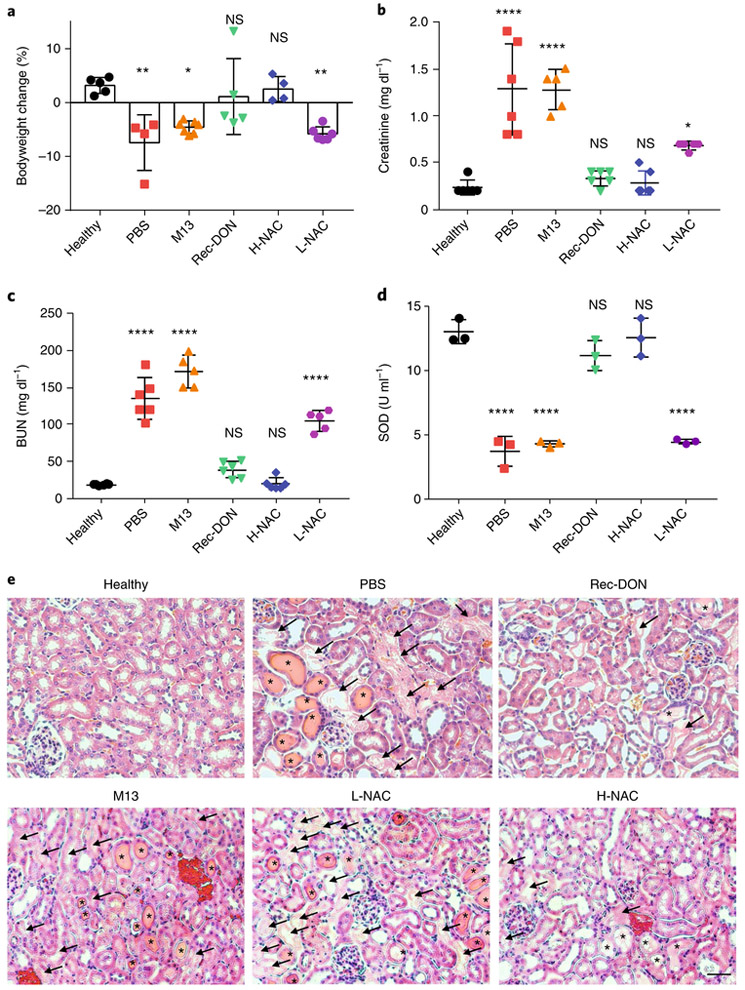
After PET imaging with 68Ga-EDTA, we measured the changes in bodyweight for all groups and found that healthy mice gained approximately 3% bodyweight in 24 h, while mice treated with PBS, M13 and L-NAC lost 4–8% bodyweight during the same time, suggesting the severity of AKI and lack of efficacy for PBS, M13 and L-NAC. However, treatment of AKI with Rec-DON and H-NAC prevented weight loss in AKI mice (Fig. 6a and Supplementary Table 10). Besides changes in bodyweight, high levels of serum creatinine and blood urea nitrogen (BUN) are clinical manifestations of the accumulation of nitrogenous wastes, and can serve as effective clinical indices of kidney excretory function62. Creatinine and BUN levels of mice confirmed our findings that Rec-DON and H-NAC could restore renal function in the AKI mice (Fig. 6b,c and Supplementary Tables 11 and 12).
Fig. 6 ∣. Bodyweight change, and blood and kidney tissue analysis after treatment for AKI.
a, Changes in bodyweight at 24 h post-treatment for AKI. A gain in bodyweight indicates treatment efficacy. Refer to Supplementary Table 10 for detailed P values. b, Analysis of creatinine levels in the blood. Lower levels indicate better kidney function. Refer to Supplementary Table 11 for detailed P values. c, Analysis of BUN levels in the blood. Lower levels indicate better kidney function. Refer to Supplementary Table 12 for detailed P values. d, Analysis of SOD levels in kidney homogenate from six groups. Reduced levels suggest enhanced vulnerability of kidney structures to ROS damage. Refer to Supplementary Table 13 for detailed P values. In a–d, data represent means ± s.d. from either four (a), five (b and c) or three (d) independent replicates; statistical significance compared with the healthy group is shown (*P < 0.05, **P < 0.01; ****P < 0.0001; NS, non-significant); and statistical analysis was performed by one-way ANOVA with Tukey's honest significant difference post-hoc test. e, Haematoxylin and eosin staining of kidney tissues from each group. One of three independent experiments from each group is shown. Arrows denote damaged tubules, and asterisks denote the formation of casts—structures formed via precipitation of denatured proteins in the tubules. Scale bar, 50 μm.
We also analysed the levels of SOD in the kidneys—an important defence to neutralize ROS for nearby renal cells. Reduced levels of SOD were found in the PBS, M13 and L-NAC treatment groups, while SOD levels in the Rec-DON and H-NAC groups were found to be similar to those in healthy mice (Fig. 6d and Supplementary Table 13), suggesting that Rec-DON and NAC both serve as reductants to neutralize ROS, restore SOD levels and further protect the renal cells of AKI mice. Finally, we analysed kidney sections from each group. Casts (a marker for kidney disease; asterisks in Fig. 6e) and damaged tubules (arrows in Fig. 6e) were observed in kidney sections from the PBS, M13 and L-NAC groups, while considerably fewer damaged structures were seen for the Rec-DON and H-NAC groups. Haematoxylin and eosin staining of the kidneys provided direct evidence that Rec-DON produces rapid and active protection of the kidneys.
Due to the dramatic difference between the molecular weight of Rec-DON and NAC (4.4 × 106 versus 163.2 g mol−1), simply comparing their doses in mass could not fully demonstrate the amount of required DON for AKI treatment. Since DNA bases would be oxidized after their interaction with various types of ROS, we estimated the amount of reductants in Rec-DON based on the number of DNA bases63,64. Details of the test materials are shown in Supplementary Tables 2 and 3. First, we performed in vitro ROS scavenging tests to compare the antioxidant effects of Rec-DON and NAC. As shown in Supplementary Fig. 43, Rec-DON and H-NAC can scavenge different types of ROS (ABTS, and hydroxyl and superoxide radicals) at a similar reaction rate, which leads to a similar protective effect for HEK293 cells. L-NAC was not sufficient to neutralize ROS in vitro or offset ROS damage to the cells. We further performed AKI treatment using different doses of Rec-DON and NAC, as well as ssDNA (20 bases long). Blood tests and kidney tissue staining results showed that NAC bearing the same number of reductant molecules (either 5 μg of NAC, equivalent to 10 μg of DNA bases in Rec-DON, or 10 μg of NAC, equivalent to 20 μg of DNA bases in Rec-DON) did not alleviate AKI symptoms. A therapeutic dose of NAC (4.2 mg) was needed to elicit similar therapeutic effects of Rec-DON at 10 μg mouse−1. As expected, a higher dose of Rec-DON (20 μg) proved efficacious in treating AKI, while ssDNA (10 μg) with a rapid renal filtration did not elicit any therapeutic effect (Fig. 6e and Supplementary Figs. 45 and 46).
To confirm the safety of Rec-DON, in addition to the above-mentioned therapeutic efficacy, we also tested the immunogenicity of DONs in vivo and measured blood levels of interleukin-6 and tumour necrosis factor-α after intravenous injection of M13 and DONs into healthy mice. The results indicated that these DNA nanostructures did not elicit an immune response in vivo (Supplementary Fig. 47). Furthermore, we examined the toxicity of DNA nanostructures and found that the kidney function (Supplementary Fig. 48), liver function (Supplementary Fig. 49) and haematology indices (Supplementary Figs. 50–52) of mice injected with M13 or Rec-DON were all within normal ranges, indicative of excellent biocompatibility and low toxicity of Rec-DON in vivo. Haematoxylin and eosin staining of primary organs (the heart, liver, spleen, lung and kidneys) showed no sign of toxicity by 24 h post-injection of M13 and Rec-DON—the time it takes to clear out from the mouse body based on PET imaging (Fig. 2a), as shown in Supplementary Fig. 53.
Discussion
We investigated the biodistribution of three different DONs in mice using PET imaging, and found that all three DONs exhibited preferential kidney accumulation that was significantly different from short ssDNA (rapid renal clearance), M13 ssDNA (prolonged liver uptake) and partially folded DON (also prolonged liver uptake). The excellent biocompatibility and enhanced kidney-targeting efficiency of the DONs led to a rapid therapeutic response in AKI mice, suggesting that they are promising candidates for the treatment of clinical AKI and various other kidney diseases.
We conclude that there is preferential accumulation of DONs in the kidneys on the basis of PET imaging of various DNA materials and in vitro stability tests of Rec-DON. PET imaging of ssDNA (20 bases long) presented rapid urine clearance and minimal renal/liver uptake; M13 and three partially folded DONs showed predominant liver uptake; and DONs showed instant (as early as 5 min post-injection) and prolonged accumulation in the kidneys of healthy and AKI mice. Altogether, PET imaging suggests that the well-folded DONs remain intact before reaching the kidneys, as they did not break down and expose a significant amount of unfolded M13 to render themselves sensitive to liver sequestration. After accumulating in the kidneys, they were slowly excreted out of the body, as evidenced by the minimal signal observed in mice at 24 h post-injection. In vitro stability tests of Rec-DON were performed using FRET measurements, which are only observed when the FRET donors and acceptors (Cy3 and Cy5, respectively) are within a distance of ~5 nm. Although it is possible that the FRET signal was generated from degraded DNA strands in solution, it probably resulted from intact structures only, due to the close proximity of donors and acceptors that is required. Our results show that the structure was stable in 80% FBS and 50% mouse urine for the first few hours of incubation. FRET efficiencies of Rec-DON in both mediums were approximately 50% at 12 h post-incubation. The in vivo and in vitro findings combined suggest that well-folded DONs remain intact when reaching the kidneys and are later degraded and excreted.
Further studies are needed to determine optimum strategies for improving the kidney-targeting efficiencies of DNA origami frameworks. A recent study revealed that increasing the oligolysine polyethylene glycol coating of DNA nanostructures can lead to an increase in pharmacokinetic bioavailability, allowing for a longer time window for active targeting65. We expect that this inexpensive and effective protection could make DONs more resistant to denaturation and degradation. However, PEGylation of DONs could change their hydrodynamic radius and surface charge, leading to a different biodistribution pattern and altering the kidney accumulation. More experiments are thus required to determine the impact of size, morphology and surface modification on the renal retention of DONs. These nanostructures are easy to make at a reasonable cost with enhanced biocompatibility.
Considering the current kidney delivery efficiency (~20%ID g−1), a patient with AKI would require an injection of approximately 54 mg of DON using our delivery strategy, with a cost of approximately US$80 for the DNA materials only. This would result in a systemic administration of 1.1 ml of 100 nM Rec-DON (through %ID g−1 analysis of the mouse model) into a human patient to elicit a therapeutic effect. Recently, a study reported the production of ssDNA (32–205 nucleotides) for in vivo or in vitro DNA nanostructure assembly via Escherichia coli expression66. Later, it was reported that DNA origami could be biotechnologically mass produced using bacteriophages to generate single-stranded precursor DNA with arbitrary length and sequences67. With the recent development of scaled-up DNA self-assembly technologies, the cost estimate of DONs could be further reduced. We envision that our passive delivery strategy to the kidneys may find broad applications—both as an active drug (for example, in the treatment of AKI with Rec-DON) and a smart bioresponsive drug carrier (for NAC or other small-molecule drugs)—for the effective treatment of various kidney diseases, upon future optimization of the drug-loading capacity and systemic-delivery efficiency.
Methods
Materials.
All staple DNA strands (with lengths ranging from 20–60 bases) were purchased from Integrated DNA Technologies (www.IDTDNA.com) in the format of 96-well plates at 100 nmol synthesis scales with the concentration normalized to 100 μM. These strands were used directly for self-assembly without any purification. M13mp18 ssDNA scaffold (M13 ssDNA) was purchased from New England Biolabs (catalogue number: N4040S) and used without further purification. All other chemicals or materials were purchased from Sigma–Aldrich and used as received unless stated otherwise.
Self-assembly of DONs.
DONs with different shapes (rectangular, triangular and tubular) were assembled by annealing a 7,249-nucleotide viral genome M13mp18 (M13 ssDNA; the template strand) with various sets of synthetic oligonucleotides (that is, staple strands) of 20–60 bases. All DONs were prepared in 1× TAE-Mg2+ (12.5 mM magnesium acetate, 2 mM EDTA, 20 mM acetic acid and 40 mM Tris base, pH 8.0) using an established method1. For each DON, 20 nM circular M13 ssDNA was mixed with a set of tenfold molar excess of staple strands. The mixture was annealed from 90 to 4 °C using the temperature gradient shown in Supplementary Table 1. The annealed solution was filtered with 100 kDa Amicon Ultra filters in 1× TAE-Mg2+ buffer or phosphate-buffered saline-magnesium (PBS-Mg2+; 12.5 mM magnesium acetate, pH 7.2–7.5). The purity of the DONs was examined via agarose gel electrophoresis (AGE). The concentration of the DONs was measured by their absorbance at 260 nm, with an average extinction coefficient of 109,119,009 M−1 cm−1. DTN with four overhangs was assembled following previous reports2,3. In brief, four predesigned oligonucleotides (1 μM) were mixed in 1× TAE-Mg2+ buffer and annealed from 95 to 4 °C (Supplementary Table 4). Partially folded DONs were assembled with incomplete sets of staple strands and used as controls in this study. For design details and the sequences of the DONs, see Supplementary Figs. 1–18 and Supplementary Sheets 1–3.
AFM imaging of DONs.
DONs (2 μl 20 nM) were deposited onto a freshly cleaved mica cell and left to adsorb for 2–3 min. 1× TAE-Mg2+ buffer (400 μl was added to the liquid cell, and the sample was scanned in a tapping mode under fluid on a PicoPlus atomic force microscope (Agilent Technologies) with NP-S tips (Veeco).
Transmission electron microscopy (TEM) imaging of DONs.
TEM grids (400 mesh, copper grid coated with ultrathin carbon; Ted Pella) were glow discharged (Emitech K100X). Then, 2 μl concentrated samples were deposited onto the grids for 1 min, washed with 10 μl deionized water for 5 s, stained with 10 μl of 1% uranyl formate twice (2 s for the first time and 15 s for the second time) and imaged using a Philips CM12 transmission electron microscope. The grid was touched with a drop of 0.7% uranyl formate solution and excess solution was wicked away with filter paper. Again, the grid was touched with a drop of uranyl formate solution, this time for 15 s, and the excess solution was removed with filter paper. To evaporate any extra solution, the grid was kept at room temperature. Low-resolution TEM studies were conducted using a Philips CM12 transmission electron microscope operated at 80 kV in the bright field mode.
AGE.
AGE (0.8%) was performed in 1× TAE/Mg buffer (Tris-acetic acid 40 mM, pH 8.0, magnesium acetate 12.5 mM and EDTA 1 mM) at 80 V and 4 °C for 1.5 h. Then, DONs were stained with ethidium bromide for 5 min for further analysis on a chemiluminescence imaging system.
Dynamic light scattering measurements.
Measurements were made on a Zetasizer Nano ZS machine (Malvern Instruments). Dispersion technology software (Malvern Instruments) with a standard setting was used for data analysis.
Zeta potential measurements.
The zeta potential of DONs was determined with a Zetasizer Nano (Malvern Instruments) by taking the average of four samples at a final concentration of 20 nM in 1× TAE/Mg2+ buffer at pH 8.0 using a disposable zeta cell. Data were then analysed using Dispersion Technology Software (Malvern Instruments).
64Cu labelling of DONs.
Radiolabelling of the DONs was achieved by hybridizing 64Cu-labelled ssDNA with side-arms designed on DONs. Amine group-functionalized ssDNA was mixed with 2-S-(4-isothiocyanatobenzyl)-1,4,7-triazacyclononane-1,4,7-triaceticacid (p-SCN-Bn-NOTA) in dimethyl sulfoxide, and incubated at pH 8.5–9.0 at room temperature for 2 h. NOTA-ssDNA was purified using a PD-10 desalting column with 1× PBS as the elution buffer. 64Cu (decay half-life: 12.7 h) was produced by a PETtrace cyclotron (GE Healthcare) using the 64Ni(p,n)64Cu reaction. For the radiolabelling of NOTA-ssDNA, 64CuCl2 was diluted with sodium acetate (0.1 M, pH 5.5) and mixed with NOTA-ssDNA. Following 1 h incubation at 37 °C with constant shaking, the mixed solution was purified with a PD-10 column, and a fraction with the highest radioactivity was chosen to hybridize with the DONs.
PET imaging and biodistribution study of DONs in healthy mice.
We intravenously injected 10–50 μCi (0.37–1.85 MBq) of 64Cu-labelled DONs, including M13 ssDNA, fully folded DONs or partially folded DONs, into healthy Institute of Cancer Research (ICR) mice. Longitudinal PET scans were performed with an Inveon microPET/micro -computed tomography rodent model scanner (Siemens Medical Solutions) at 3, 12 and 24 h post-injection. For Rec-DON, a computed tomography model from the Inveon database was aligned with PET imaging at 3 and 12 h post-injection. ROI analysis of all PET images was performed to determine time–activity curves for different organs. To obtain ROI data, the manual delineations of major organs in animals were made on decay-corrected whole-body PET images using the in-house image analysis software (Inveon Research Workplace). The counts pixel−1 obtained from the raw PET data were converted to counts cm−3 based on a calibration coefficient obtained from phantom PET scans. By assuming the tissue density was approximately 1 g cm−3, counts cm−3 were converted to counts g−1, which were then divided by the injected dose to provide the final ROI in %ID g−1. By doing this, we were able to calculate and subtract any leftover dose in the tail or possible contamination on the skin to better quantify the actual dose in the organs or tissues of interest. After the scans at 24 h post-injection, mice were euthanized, and organs of interest were harvested to quantify the uptake of each DON using a gamma counter (PerkinElmer). The tracer accumulation in each organ or tissue was presented as the percentage of the injected dose per gram of tissue (%ID g–1; ≥3 mice in each group).
PET imaging and biodistribution study of 64Cu-labelled ssDNA in healthy mice.
Approximately 50 μCi (1.85 MBq) of 64Cu-labelled ssDNA was intravenously injected into healthy ICR mice, and serial PET scans were performed at 3, 12 and 24 h post-injection. ROI analysis was used to analyse the tracer uptake in major organs. At 24 h post-injection, mice were euthanized and organs of interest were harvested to quantify the accumulation of each DNA nanostructure using a gamma counter (PerkinElmer). The tracer accumulation in each organ or tissue was presented as %ID g−1 (n = 3).
Confocal imaging of fluorescently labelled Rec-DON in kidneys.
Cy3-labelled Rec-DON was injected intravenously into mice, and kidneys were harvested at 10 min, 3 h and 12 h post-injection and stored in optimum cutting temperature specimen matrix (VWR) for cryostat sectioning at −20 °C. Sectioning was performed by the Experimental Pathology Laboratory at the University of Wisconsin–Madison. Frozen kidney tissue slices of 5 μm thickness were washed with cold 1× PBS. Then, a cover glass was applied to each slide using VECTASHIELD Mounting Medium (Vector Laboratories), and confocal imaging was performed using a Nikon A1R confocal microscope (Nikon Instruments).
PET imaging of Rec-DON in AKI mice.
We intravenously injected 50 μCi (1.85 MBq) of 64Cu-labelled Rec-DON into murine models of AKI (n = 3), and serial PET scans were performed at 5 min, 30 min, 1 h, 3 h, 6 h, 12 h and 24 h postinjection. ROI analysis was used to analyse the tracer uptake in major organs.
At the 24 h time point, mice were euthanized, and the organs of interest were harvested to quantify the accumulation of Rec-DON using a gamma counter (PerkinElmer). The tracer accumulation in each organ or tissue was presented as %ID g−1 (n = 3).
Scavenging ABTS radicals with DONs.
The free radical scavenging capability of DONs was tested based on the reduction of •ABTS+ radicals using the ABTS radical cation decolourization assay. Some 7 mmol of ABTS was dissolved in deionized water and reacted with 2.45 mM potassium persulfate to produce ABTS radical cation (•ABTS+), and the mixture was kept in the dark at room temperature for 24 h before use. Then, the ultraviolet–visible spectra of the pure solution of •ABTS+ radicals and •ABTS+ radical solution with M13 ssDNA or different DONs were monitored to measure the absorbance at 734 nm. The inhibition rate of •ABTS+ radicals was calculated based on the ratio of neutralized •ABTS+ radicals to overall radicals. All measurements were carried out in triplicate.
Scavenging hydroxyl and superoxide radicals with DONs.
The hydroxyl radical scavenging efficiency was tested using a hydroxyl radical antioxidant capacity assay kit (catalogue number: STA-346-T; Cell Biolabs). The superoxide radical scavenging efficiency was assessed with a SOD assay kit (catalogue number: CS1000; Sigma–Aldrich). All tests were performed following the protocol provided by the manufacturers.
Protection of cells exposed to H2O2 with DONs.
HEK293 cells were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle medium supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were seeded into a 96-well cell culture plate at 106 cells well−1, then incubated for 24 h at 37 °C under 5% CO2.
An equal mass of DONs, including M13 ssDNA, Rec-DON, Tri-DON or Tub-DON, was added to the cells and they were incubated for 30 min. The cells were then incubated with H2O2 (final concentration in each well: 250 μM) for 24 h at 37 °C with 5% CO2. A colorimetric MTT assay was performed to examine the cell viability. To reliably measure ROS levels in live cells, a final concentration of 5 μM of fluorogenic probe (CellROX Deep Red Reagent) was added to the cells, which were incubated for 30 min at 37 °C after DONs incubation and H2O2 treatment. Afterwards, the cells were washed with 1× PBS three times, and the emission at 665 nm was measured with an excitation of 640 nm.
FRET assay for examining the stability of DONs.
Cy3/Cy5 dual-labelled Rec-DON was incubated in 80% serum or 50% urine, and the FRET spectra were taken at 10 min, 3 h and 12 h post-incubation. Since the adjacent Cy3/Cy5 fluorophores were placed within the estimated Forster distance of ~5.6 nm, a Cy5 emission signal in Rec-DON was expected when Cy3 excitation/Cy5 emission filters were used. If DONs were disassembled under the influence of enzymes in the FBS or the basic milieu of urine, the absence of FRET would cause a decrease in the Cy5 signal intensity. The FRET efficiency was calculated based on the Cy5 signal after Cy3 excitation (the FRET efficiency was considered to be 100% at time 0).
AKI model establishment.
All animal studies were conducted under a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee. Before initiation of the AKI model, all mice were deprived of water but had access to food for 15 h. At the end of the water restriction, 8 ml kg−1 of 50% glycerol was administrated into each hind limb of the mice intramuscularly. All mice were then given free access to water and food4,5.
Treatment of AKI.
Six groups of mice were studied: group 1 comprised healthy control mice (healthy; n ≥ 5) without any treatment; group 2 comprised AKI mice treated with 1× PBS (PBS; n ≥ 5); group 3 comprised AKI mice treated with 10 μg of M13 in 100 μl of 1× PBS (M13; n ≥ 5); group 4 comprised AKI mice treated with 10 μg of Rec-DON in 100 μl of 1× PBS (Rec-DON; n ≥5); group 5 comprised AKI mice treated with 4.2 mg of NAC in 100 μl of 1× PBS (H-NAC; n ≥ 5); and group 6 comprised AKI mice treated with 10 μg of NAC in 100 μl of 1× PBS (L-NAC; n ≥ 5). Two hours after the model induction, the agents were intravenously injected into AKI mice, and their kidney function was compared with that of healthy control mice. See Supplementary Tables 2 and 3 for more information on the doses of test materials for AKI treatment. To better illustrate the treatment of AKI using Rec-DON, we further performed treatment using different doses of ssDNA, Rec-DON and NAC (see Supplementary Fig. 45 for more details).
Dynamic 68Ga-EDTA PET imaging for kidney function evaluation.
68Ga (t1/2 = 68 min) was eluted from a 68Ge/68Ga radionuclide generator system (IDB Holland) with HCl (0.05 M) as the eluent. To prepare 68Ga-EDTA, 68GaCl3 was mixed with an equal volume of 10 mM EDTA in 2× PBS buffer, and the final pH was adjusted to ~7.5 with NaHCO3 (1 M)6. Radiochemical purity was determined using thin-layer chromatography silica gel plates developed with a solvent system of ammonium acetate buffer (1 M) and methanol (v/v = 50:50).
For dynamic PET imaging using 68Ga-EDTA, mice in the healthy control group or AKI treatment groups (n ≥ 3 for each group) were anaesthetized with 2% isoflurane and their tail veins were catheterized. Animals were placed on the PET bed in a prone position and sent into the scanner. Along with the intravenous injection of 68Ga-EDTA (200–500 μCi), a 30 min dynamic PET scan was acquired simultaneously. The histogram files were divided into 28 frames: 6 × 10 s, 6 × 30 s, 6 × 60 s and 10 × 120 s. Dynamic scans were reconstructed using an ordered subset expectation maximization three-dimensional/maximum a posteriori reconstruction algorithm. ROI analysis of the PET images was performed to determine the time–activity curve of the blood pool, kidneys and bladder. Tracer uptake was expressed as %ID g−1 for the blood pool, and %ID for the kidneys and bladder.
Blood tests.
We euthanized the mice in all groups 24 h after AKI initiation. Blood samples were collected into paediatric heparin tubes (BD Biosciences) and centrifuged at 2,000g for 15 min at 4 °C. Then, the plasma was sent to the Clinical Pathology Laboratory at the Veterinary Medical Teaching Hospital at the University of Wisconsin–Madison for analysis of the blood creatinine and BUN levels.
Haematoxylin and eosin staining of kidney sections.
Kidneys were collected at 24 h after the initiation of AKI, fixed with paraformaldehyde (4% in PBS), embedded in paraffin wax and sent to the University of Wisconsin Carbone Cancer Center Experimental Pathology Laboratory for sectioning and haematoxylin and eosin staining.
Toxicity assessment in vivo.
Toxicological analysis was carried out in five-week-old healthy female ICR mice (Envigo). Mice were randomized into three groups—control, M13 and Rec-DON (n = 10 for each group)—and injected with PBS (100 μl), M13 ssDNA (10 μg in 100 μl 1× PBS) or Rec-DON (10 μg in 100 μl 1× PBS), respectively. At 24 h post-injection, all mice were euthanized. Whole blood samples were collected for haematology analysis (Abaxis VetScan HM5 Hematology Analyzer). Major organs (including the heart, liver, spleen, lung and kidneys) from mice in the M13 and Rec-DON group were harvested for haematoxylin and eosin staining to evaluate morphological changes and/or damage caused by the administration of DNA materials.
Statistics.
Quantitative data are displayed as means ± s.d. Statistical differences were determined using a Student’s t-test for two groups or one-way analysis of variance (ANOVA) for three or more groups. Statistical analyses were performed using GraphPad Prism7.
Animal ethics.
All animal studies were conducted under a protocol approved by the University of Wisconsin Institutional Animal Care and Use Committee.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.
Supplementary Material
Acknowledgements
The authors thank J. J. Jeffery and A. M. Weichmann for help with the small-animal imaging studies. The authors are grateful for insightful input from B. Yu, R. Hernandez, S. Goel, L. Kang and E. B. Ehlerding. This work was supported in part by the University of Wisconsin–Madison, National Institutes of Health (1R01GM104960, P30CA014520 and T32CA009206), National Natural Science Foundation of China (31771036, 51703132 and 51573096), Guangdong Province Natural Science Foundation of Major Basic Research and Cultivation Project (2018B030308003), Fok Ying-Tong Education Foundation for Young Teachers in Higher Education Institutions of China (161032) and Basic Research Program of Shenzhen (JCYJ20170412111100742 and JCYJ20160422091238319). C.F. gratefully acknowledges the National Key R&D Program of China (2016YFA0201200), NSFC (21329501 and 21390414) and Chinese Academy of Sciences (QYZDJ-SSW-SLH031). This work is dedicated to the memory of Q. Huang, whose great insights inspired this project.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/ s41551–018-0317–8.
References
- 1.Bellomo R, Kellum JA & Ronco C Acute kidney injury. Lancet 380, 756–766 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Chawla LS, Eggers PW, Star RA & Kimmel PL Acute kidney injury and chronic kidney disease as interconnected syndromes. N. Engl. J. Med 371, 58–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewington AJP, Cerda J & Mehta RL Raising awareness of acute kidney injury: a global perspective of a silent killer. Kidney Int. 84, 457–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.VA/NIH Acute Renal Failure Trial Network Intensity of renal support in critically ill patients with acute kidney injury. N. Engl. J. Med 359, 7–20 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tepel M et al. Prevention of radiographic-contrast-agent–induced reductions in renal function by acetylcysteine. N. Engl. J. Med 343, 180–184 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Fishbane S N-acetylcysteine in the prevention of contrast-induced nephropathy. Clin. J. Am. Soc. Nephrol 3, 281–287 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Wagner V, Dullaart A, Bock A-K & Zweck A The emerging nanomedicine landscape. Nat. Biotechnol 24, 1211–1217 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Peer D et al. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotech 2, 751–760 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Sun WJ et al. Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J. Am. Chem. Soc 136, 14722–14725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min Y, Caster JM, Eblan MJ & Wang AZ Clinical translation of nanomedicine. Chem. Rev 115, 11147–11190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arai S et al. Apoptosis inhibitor of macrophage protein enhances intraluminal debris clearance and ameliorates acute kidney injury in mice. Nat. Med 22, 183–193 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kamaly N, He JC, Ausiello DA & Farokhzad OC Nanomedicines for renal disease: current status and future applications. Nat. Rev. Nephrol 12, 738–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelaz B et al. Diverse applications of nanomedicine. ACS Nano 11, 2313–2381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alidori S et al. Targeted fibrillar nanocarbon RNAi treatment of acute kidney injury. Sci. Transl. Med 8, 331ra339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun CJ et al. Controlling assembly of paired gold clusters within apoferritin nanoreactor for in vivo kidney targeting and biomedical imaging. J. Am. Chem. Soc 133, 8617–8624 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Choi CHJ, Zuckerman JE, Webster P & Davis ME Targeting kidney mesangium by nanoparticles of defined size. Proc. Natl Acad. Sci. USA 108, 6656–6661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manne N et al. Cerium oxide nanoparticles attenuate acute kidney injury induced by intra-abdominal infection in Sprague-Dawley rats. J. Nanobiotechnol 13, 75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H et al. Eupafolin nanoparticle improves acute renal injury induced by LPS through inhibiting ROS and inflammation. Biomed. Pharmacother 85, 704–711 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Pinheiro AV, Han D, Shih WM & Yan H Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotech 6, 763–772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seeman NC Nanomaterials based on DNA. Annu. Rev. Biochem 79, 65–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotech 10, 779–784 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Li J et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 5, 8783–8789 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Mei Q et al. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 11, 1477–1482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahn J, Wickham SF, Shih WM & Perrault SD Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 8, 8765–8775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Surana S, Shenoy AR & Krishnan Y Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotech 10, 741–747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotech 7, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu G et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl Acad. Sci. USA 110, 7998–8003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torring T, Helmig S, Ogilby PR & Gothelf KV Singlet oxygen in DNA nanotechnology. Acc. Chem. Res 47, 1799–1806 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Chen Y-J, Groves B, Muscat RA & Seelig G DNA nanotechnology from the test tube to the cell. Nat. Nanotech 10, 748–760 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Chao J et al. Hetero-assembly of gold nanoparticles on a DNA origami template. Sci. China Chem 59, 730–734 (2016). [Google Scholar]
- 31.Du Y et al. DNA-nanostructure–gold-nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy. Adv. Mater 28, 10000–10007 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Li J, Green AA, Yan H & Fan CH Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem 9, 1056–1067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Q, Li H, Wang L, Gu H & Fan C DNA nanotechnology-enabled drug delivery systems. Chem. Rev https://doi.org/10.1021/acs.chemrev.7b00663 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Li S et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol 36, 258–264 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 8, 6633–6643 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Jiang D, England CG & Cai W DNA nanomaterials for preclinical imaging and drug delivery. J. Control. Release 239, 27–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothemund PWK Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Fu J, Liu M, Liu Y, Woodbury NW & Yan H Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc 134, 5516–5519 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makarova KS, Wolf YI & Koonin EV Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 41, 4360–4377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin M et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. Engl 54, 2151–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Jiang D et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl. Mater. Interfaces 8, 4378–4384 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Keum J-W & Bermudez H Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun 0, 7036–7038 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto Y, Nagasaki Y, Kato Y, Sugiyama Y & Kataoka K Long-circulating poly(ethylene glycol)-poly(d,l-lactide) block copolymer micelles with modulated surface charge. J. Control. Release 77, 27–38 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Alexis F, Pridgen E, Molnar LK & Farokhzad OC Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm 5, 505–515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco E, Shen H & Ferrari M Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol 33, 941–951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albanese A, Tang PS & Chan WCW The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 14, 1–16 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Du BJ et al. Glomerular barrier behaves as an atomically precise bandpass filter in a sub-nanometre regime. Nat. Nanotech 12, 1096–1102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lacerda L et al. Dynamic imaging of functionalized multi-walled carbon nanotube systemic circulation and urinary excretion. Adv. Mater 20, 225–230 (2008). [Google Scholar]
- 49.Ruggiero A et al. Paradoxical glomerular filtration of carbon nanotubes. Proc. Natl Acad. Sci. USA 107, 12369–12374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Key J et al. Soft discoidal polymeric nanoconstructs resist macrophage uptake and enhance vascular targeting in tumors. ACS Nano 9, 11628–11641 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Williams RM et al. Mesoscale nanoparticles selectively target the renal proximal tubule epithelium. Nano Lett. 15, 2358–2364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutaud O et al. Acetaminophen inhibits hemoprotein-catalyzed lipid peroxidation and attenuates rhabdomyolysis-induced renal failure. Proc. Natl Acad. Sci. USA 107, 2699–2704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh AP et al. Animal models of acute renal failure. Pharmacol. Rep 64, 31–44 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Huang H et al. A porphyrin-PEG polymer with rapid renal clearance. Biomaterials 76, 25–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nath KA & Norby SM Reactive oxygen species and acute renal failure. Am. J. Med 109, 665–678 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Valko M et al. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol 39, 44–84 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Hemnani T & Parihar M Reactive oxygen species and oxidative DNA damage. Indian J. Physiol. Pharmacol 42, 440–452 (1998). [PubMed] [Google Scholar]
- 58.Shyu KG, Cheng JJ & Kuan P Acetylcysteine protects against acute renal damage in patients with abnormal renal function undergoing a coronary procedure. J. Am. Coll. Cardiol 40, 1383–1388 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Hofman M et al. 68Ga-EDTA PET/CT imaging and plasma clearance for glomerular filtration rate quantification: comparison to conventional 51Cr-EDTA. J. Nucl. Med 56, 405–409 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Hofman MS & Hicks RJ Gallium-68 EDTA PET/CT for renal imaging. Semin. Nucl. Med 46, 448–461 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Nair AB & Jacob S A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm 7, 27–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uchino S et al. Acute renal failure in critically ill patients—a multinational, multicenter study. J. Am. Med. Assoc 294, 813–818 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Cadet J & Wagner JR DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol 5, a012559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agnihotri N & Mishra PC Mechanism of scavenging action of N-acetylcysteine for the OH radical: a quantum computational study. J. Phys. Chem. B 113, 12096–12104 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Ponnuswamy N et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun 8, 15654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elbaz J, Yin P & Voigt CA Genetic encoding of DNA nanostructures and their self-assembly in living bacteria. Nat. Commun 7, 11179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Praetorius F et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.