Epigenetic silencing of endogenous retroviruses (ERVs) is initiated by KRAB-ZFP-KAP1-SETDB1 repression complexes during early mammalian development. On the basis of biochemical evidence from histone H3.3-knockout embryonic stem (ES) cells, Elsässer et al.1 reported that histone H3.3 is deposited at KAP1-SETDBl-targeted ERVs by the chaperone DAXX-ATRX complex and that this deposition is required to repress ERV transcription and retrotransposition. However, our re-analysis of the published data revealed little evidence of genome-wide ERV upregulation in H3.3-knockout ES cells, and, more importantly, that the ES cells used for the analysis include polymorphic ERV insertions, which probably reflect a mixed genetic background and compromises their use for ERV expression and re-integration analysis. Thus, despite the strong evidence for H3.3 deposition at KAP1-SETDB1-targeted ERV elements, it remains to be determined whether this deposition plays a major role in preventing ERV reactivation. There is a Reply to this Comment by Elsässer, S. J. et al. Nature 548, http://dx.doi.org/10.1038/nature23278 (2017).
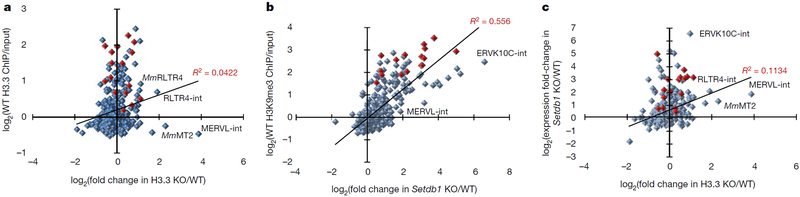
Elsässer et al.1 reported that H3.3 is deposited at ERVs by the H3.3 histone chaperone DAXX in mouse ES cells1. While we were able to confirm DAXX-dependent H3.3 deposition at ERVs in a newly generated conditional Daxx-knockout ES cell line (Extended Data Fig. 1a, b), we have concerns about the authors’ conclusion that H3.3 knockout leads to increased ERV expression1. Intracisternal A-type particle (IAP) ERVs were upregulated less than 1.5-fold in the two tested H3.3-knockout ES cell lines compared with a single wild-type line, and ERVK10C ERVs showed a modest upregulation (approximately 2-fold) in only one of these lines1. By contrast, IAPs and ERVK10C are upregulated 14- and 99-fold in Setdbl-knockout ES cells, respectively2, with comparable levels of ERV reactivation observed in Kapl-knockout ES cells3. As Elsässer et al.1 presented expression data of H3.3-knockout ES cells for only a small selection of ERVs1, we re-analysed the published H3.3-knockout RNA-sequencing (RNA-seq) datasets to obtain an expanded picture of ERV reactivation. Notably, we found that the number of annotated ERV families that are upregulated in H3.3-knockout ES cells approximately matches the number of ERV families that are downregulated in these cells (Extended Data Fig. 2a). Furthermore, comparing ERV expression in the two H3.3-knockout ES cell lines reveals that an up to 1.5-fold difference in expression is within the variation ‘noise’ between two ES cell lines of the same genotype (Extended Data Fig. 2b). If H3.3 directly represses ERVs, ERV upregulation in H3.3-knockout ES cells and H3.3 enrichment at these elements in wild-type ES cells should be positively correlated. However, in contrast to H3K9me3 enrichment and ERV upregulation in Setdbl-knockout ES cells, our re-analysis does not support such a correlation for H3.3 (Fig. 1a, b). Moreover, the vast majority of ERVs that are upregulated in Setdbl-knockout ES cells are not convincingly upregulated in H3.3-knockout ES cells (Fig. 1c and Extended Data Fig. 3), indicating that this histone variant is generally dispensable for KAP1-SETDB1-mediated ERV silencing. Curiously, the only robustly re-activated ERVs in H3.3-knockout ES cells, namely class III MERVL elements, are not enriched in H3K9me3 or H3.3 in wild-type ES cells (Fig. 1a, b), suggesting that the strongest effect of H3.3 knockout on ERV expression is indirect.
Figure 1 |. Comparative analysis of ERV expression and histone modifications at ERVs in knockout ES cell lines.
a, Mean fold change in ERV (long terminal repeat (LTR) elements annotated in UCSC RepeatMasker) expression in H3.3-knockout (KO) ES cells (two cell lines) over wild-type (WT) ES cells (one cell line) versus H3.3 enrichment in wild-type ES cells. ChIP, chromatin immunoprecipitation. b, Fold change in ERV expression in Setdb1-knockout ES cells over wild-type ES cells versus H3K9me3 enrichment in wild-type ES cells. c, Mean fold change in ERV expression in H3.3-knockout ES cells over wild-type ES cells versus fold change in ERV expression in Setdb1-knockout ES cells over wild-type ES cells. ERVs belonging to the IAP family are marked in red. Mm, mus musculus. Annotations including an ‘-int’ represent the internal regions, which are transcribed from the cognate 5′ LTR, of the annotated ERV. Only ERV groups with more than 100 family members were considered for analysis. Linear trend lines and corresponding R2 values are shown.
Although even a subtle difference in ERV expression could be biologically important, it is possible that secondary effects of H3.3 knockout or clonal variation are responsible for the observed weak phenotype in H3.3-knockout ES cells. More importantly, the genomic copy number of active ERVs, including IAPs, differs widely in inbred mouse strains (described in detail below) and their expression is influenced by genetic background. Given the low level of upregulation reported previously1, and the fact that mice of mixed genetic background (C57BL/6 and 129) were used to generate the single wild-type and two knockout ES cell clones used in the study1, we conclude that the data as originally provided do not convincingly support a requirement for H3.3 in transcriptional silencing of ERVs.
The modest level of IAP derepression in H3.3-depleted ES cells1 was inconsistent with their observation of frequent de novo IAP insertions in this line. Notably, IAP elements are highly polymorphic among inbred mice4–7, that is, copies can be present in only one or a few strains. We were therefore concerned that the authors sequenced genomic DNA from wild-type and H3.3-knockout ES cell lines of mixed 129 and C57BL/6 origin, mapped the identified IAP elements to the reference C57BL/6 genome, and concluded that elements not mapping were novel insertions1. Furthermore, these experiments did not involve sequence-based elucidation of target site duplications, as is standard.
To determine whether the reported de novo IAP copies actually comprised pre-existing polymorphic IAPs, we intersected the genomic coordinates of these copies with the coordinates from two previous studies6,7. Indeed, all of the ‘wild-type only’ and ‘wild-type and H3.3-knockout’ IAPs reported1 were present in 129 mice in one or both datasets. For the 80 ‘H3.3-knockout only’ IAP copies, 16 were found in 129 strains and therefore excluded from further analysis.
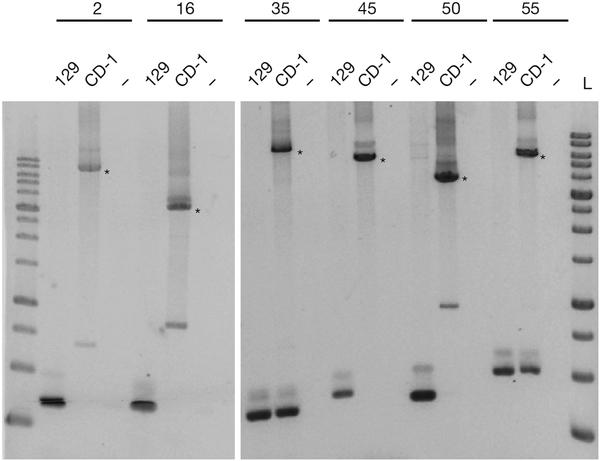
However, we found 47 of the remaining 64 ‘H3.3-knockout only’ copies in various other mouse strains, but not in 129 or C57BL/6, and only 17 were not present in the published datasets7 (Supplementary Table 1). This indicates that H3.3-knockout ES cells have a complex genetic background that includes additional laboratory mouse strains. To verify these insertions, we performed a more stringent re-analysis of the relevant whole-genome sequencing data using our own pipeline (described in the Supplementary Methods). We identified 37 out of the 47 polymorphic and 13 out of the 17 potentially new IAP copies (Supplementary Table 1). The majority of these IAP copies were found in multiple mouse strains, indicating that they were misannotated polymorphic IAPs and not de novo retrotransposition events. Notably, we also found 8 of the IAP copies reported as H3.3-knockout only1 to be present in both wild-type and H3.3-knockout ES cells, as well as in various other strains, including 129 (Supplementary Table 2). Moreover, 6 out of 10 randomly chosen polymorphic and putative new IAP copies could be readily detected in CD-1 feeder DNA (Fig. 2 and Supplementary Table 1), which share considerable genetic similarity to NOD/LtJ mice8. Altogether, these analyses show that the vast majority of the reported ‘new’ IAP insertions1 are polymorphic IAP elements already present in various laboratory mouse strains.
Figure 2 |. PCR analysis of polymorphic IAP insertions.
PCR assay of 129/SvJ and genomic DNA of the CD-1 feeders using primers flanking six ‘new’ copies described in H3.3-knockout ES cells in ref. 1. Asterisks indicate larger bands corresponding to the size expected for a typical IAP insertion. Non-template controls are shown (−) and 10-kb size ladders (L) are included on either side of the six copies. IAP copy indexes (2, 16, 35, 45, 50 and 55) are shown above the lanes and the coordinates can be found in Supplementary Table 1 along with the primer sequences.
In summary, our data and the data from Elsässer et al.1 confirm the presence of H3.3-containing heterochromatin at ERV sequences in ES cells, but do not support a vital role for this H3 variant in suppressing ERV expression or mobilization. We speculate that in ES cells, SETDB1-dependent methylation of canonical H3 is generally sufficient to compensate for the loss of H3K9me3-marked H3.3 at ERVs.
Methods
Conditional Daxx-knockout ES cells were generated by CRISPR-Cas9-mediated loxP insertion into ES cells carrying a 4-hydroxytamoxifen-inducible Rosa cre-ERT allele. Native chromatin immunoprecipitation followed by sequencing (ChIP-seq) was performed with Daxx-knockout ES cells carrying a retrovirally delivered YFP-tagged H3.3 expression construct as described previously2. ERV expression and ChIP-seq analysis were performed as described previously2 and in the Supplementary Methods. For IAP integration analysis, genomic DNA from CD-1 feeders (StemCell Technologies) and 129/SvJ mice (Jackson Laboratory) was screened by PCR using primers shown in Supplementary Table 1. Full methods are available in the Supplementary Information.
Data availability.
Daxx-knockout ES cell RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE70967. All other data are available from the previously published accession numbers or from the corresponding author upon reasonable request.
Extended Data
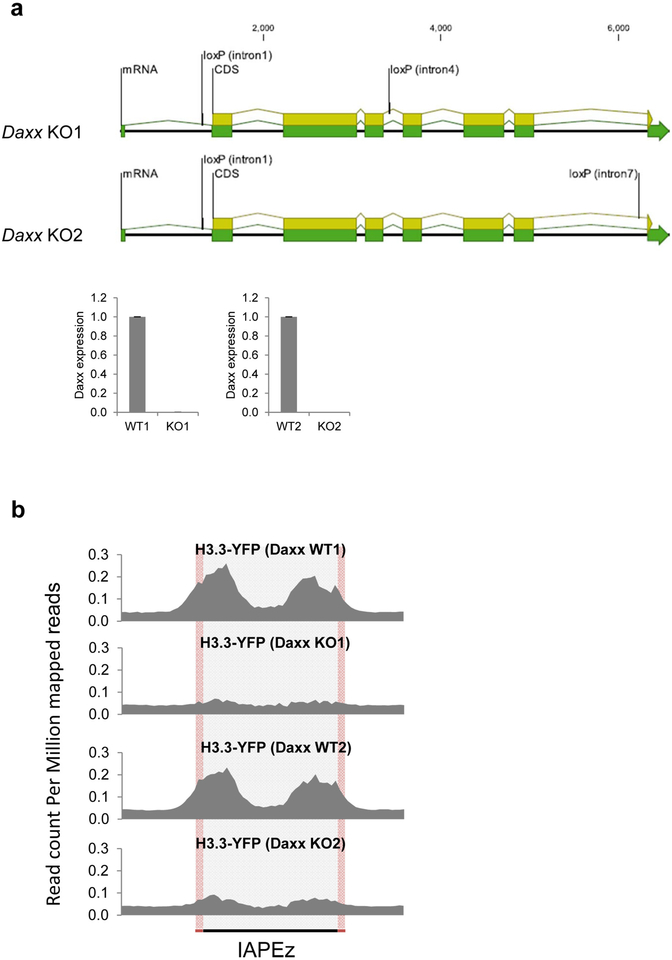
Extended Data Figure 1 |. H3.3 is incorporated into ERVs by the chaperone DAXX.
a, Top, representation of the two conditional Daxx-knockout alleles generated by CRISPR–Cas9-mediated loxP insertion at the indicated introns. Bottom, quantitative RT–PCR analysis of Daxx mRNA expression in Daxx-knockout ES cells. Data are mean ± s.d. expression (normalized to Gapdh) relative to the corresponding wildtype ES cells (n = 3, technical replicates). b, H3.3–YFP enrichment at 717 full-length IAP ERVs in two independently derived conditional Daxx-knockout ES cell lines.
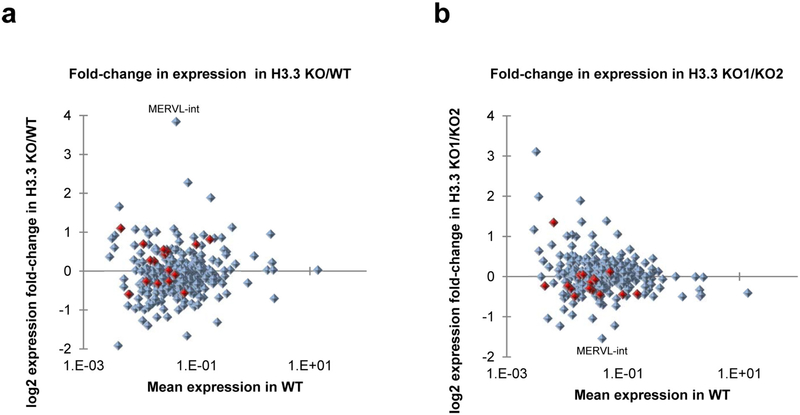
Extended Data Figure 2 |. ERV deregulation in H3.3-knockout ES cells.
a, Mean fold change in ERV (LTR elements annotated in UCSC RepeatMasker) expression in H3.3-knockout ES cells (two cell lines) over wild-type ES cells (one cell line). b, Fold change in ERV expression comparing two H3.3-knockout ES cell lines. ERVs belonging to the IAP family are marked in red. Only ERV groups with more than 100 family members were considered for analysis.
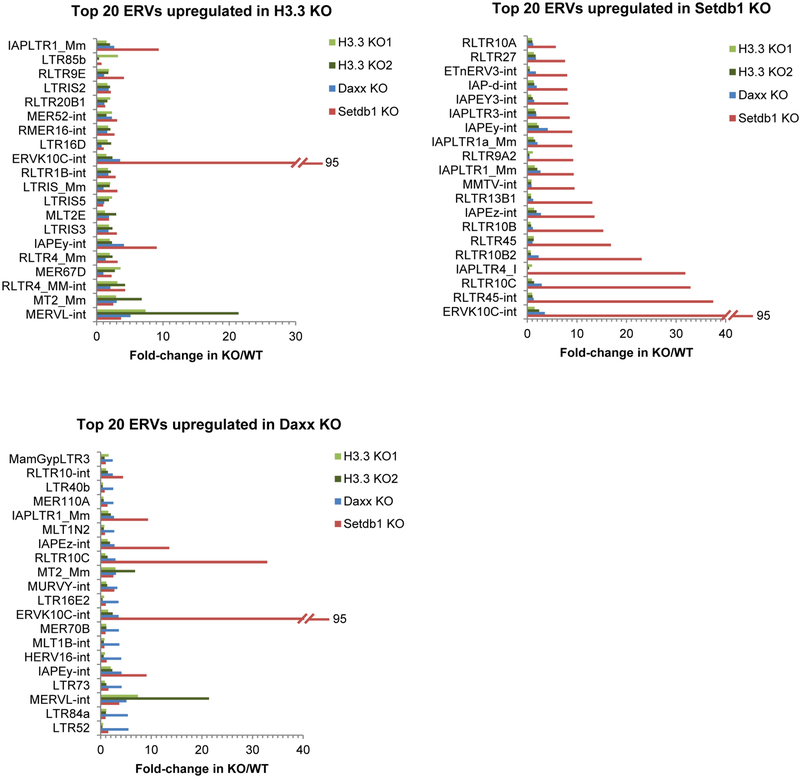
Extended Data Figure 3 |. ERV expression in Setdb1-, H3.3- and Daxx-knockout ES cells.
The fold change in expression over the corresponding wild-type control is shown for the top 20 upregulated ERV annotations in Setdb1-, H3.3- and Daxx-knockout ES cells. Annotations including –int represent the internal regions, which are transcribed from the cognate 5′ LTR, of the annotated ERV. For example, ERVK10C-int is the internal region of ERVK10C elements, with flanking LTRs: RLTR10A, RLTR10B and RLTR10C (depending on the specific genomic copy), which are also presented among the graphs. Similarly, IAPEz-int is the internal region of IAPEz elements with flanking cognate LTRs: IAPLTR1_Mm and IAPLTR1a_Mm, which are also represented. As the internal region is much longer and transcribed across its length, this is the most useful annotation to consider for expression analysis. The following published RNA-seq GEO data were re-analysed: GSM727424 (Setdb1-knockout ES cells); GSM1428580 and GSM1428581 (H3.3-knockout ES cells). Only ERV groups with more than 100 family members were considered for analysis.
Supplementary Material
Footnotes
Supplementary Information accompanies this Comment.
Competing Financial Interests Declared none.
References
- 1.Elsässer SJ, Noh KM, Diaz N, Allis CD & Banaszynski LA Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karimi MM et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowe HM et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Lueders KK, Frankel WN, Mietz JA & Kuff EL Genomic mapping of intracisternal A-particle proviral elements. Mamm. Genome 4, 69–77 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Li J et al. Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res. 22, 870–884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN & Mager DL Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 4, e1000007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nellåker C et al. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol. 13, R45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldinger KA, Sokoloff G, Rosenberg DM, Palmer AA & Millen KJ Genetic variation and population substructure in outbred CD-1 mice: implications for genome-wide association studies. PLoS ONE 4, e4729 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Daxx-knockout ES cell RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE70967. All other data are available from the previously published accession numbers or from the corresponding author upon reasonable request.