Abstract
As a member of the retrovirus family, HIV-1 packages its RNA genome into particles and replicates through a DNA intermediate that integrates into the host cellular genome. The multiple genes encoded by HIV-1 are expressed from the same promoter and their expression is regulated by splicing and ribosomal frameshift. The full-length HIV-1 RNA plays a central role in viral replication as it serves as the genome in the progeny virus and is used as the template for Gag and GagPol translation. In this review, we summarize findings that contribute to our current understanding of how full-length RNA is expressed and transported, cis- and trans-acting elements important for RNA packaging, the locations and timing of RNA:RNA and RNA:Gag interactions, and the processes required for this RNA to be packaged into viral particles.
Keywords: HIV-1, retrovirus, RNA transcription and processing, full-length RNA, RNA export, Rev, RRE, RNA dimerization, RNA packaging, Gag, virus assembly
The Beginning: Biogenesis of HIV-1 RNA
HIV-1 packages its RNA genome into viral particles; upon entering the target cells, the RNA genome is reverse-transcribed into double-stranded DNA, which is then integrated into the cellular genome to form a provirus (reviewed in Coffin et al., 1997; Freed, E.O. and Martin, M.A., 2013). Integration of viral DNA affords retroviruses the ability to depend on host cell machinery for gene expression including transcription as well as post-transcriptional processing. The U3, R and U5 regions in the long terminal repeats (LTRs) of the provirus are the cis-elements important for the initiation of transcription by RNA polymerase II and for the modification of the RNA molecules (reviewed in Coffin et al., 1997; Freed, E.O. and Martin, M.A., 2013). Along with the use of host factors to mediate transcription, HIV-1 also encodes its own trans-activator of transcription (Tat) for efficient RNA synthesis (Gaynor, 1995a; Jones and Peterlin, 1994). Tat interacts with cyclin T1/CDK9 and binds to the stem-loop structure at the 5’end of the RNA transcripts called the trans-activating response element (TAR); this complex promotes the hyperphosphorylation of RNA polymerase II to ensure efficient elongation of viral transcripts (Fackler et al., 2001; Hauber and Cullen, 1988; Parada and Roeder, 1996; Rounseville and Kumar, 1992). Absence of Tat has been shown to lead to the accumulation of prematurely terminated transcripts (Adams et al., 1994; Gaynor, 1995b).
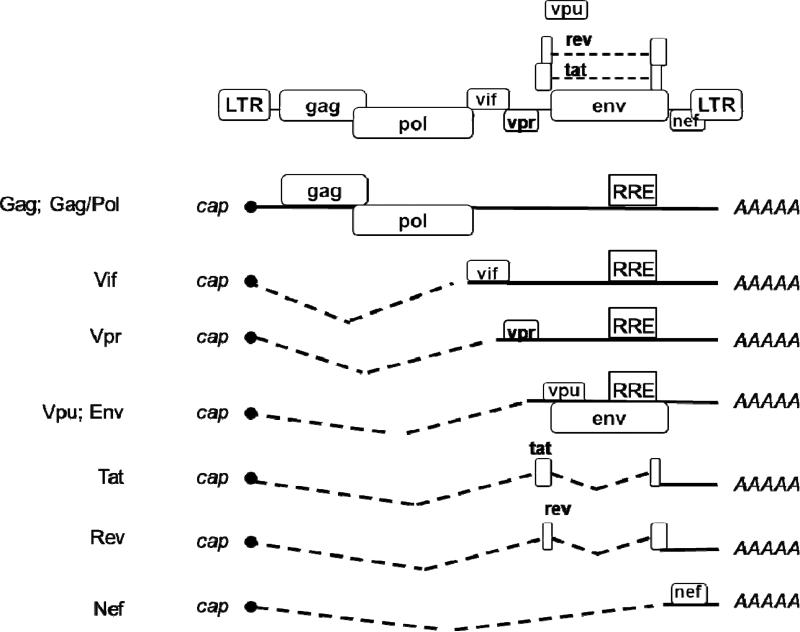
Following RNA transcription, HIV-1 transcripts undergo similar processing steps as cellular RNAs including the addition of the 5’ cap, cleavage and the polyadenylation of the 3’ end. As with transcription, HIV-1 relies on the components of the host spliceosome machinery for efficient splicing to generate various RNA molecules. Some of the HIV-1 RNA transcripts undergo splicing and generate several different RNA species depending on the selection of the splice sites and the number of splicing events that occur (Figure 1) (Coffin et al., 1997). Fully spliced HIV-1 RNAs accumulate early in the replication cycle and encode the viral regulatory proteins Tat, Rev and Nef (Figure 1). Singly spliced or partially spliced RNA species encode Env and the accessory proteins Vif, Vpr and Vpu (Figure 1) (Cullen, 1998). A portion of the HIV-1 RNA transcripts remains unspliced and these full-length HIV-1 RNAs serve two functions: they are used as a template for translation of Gag and GagPol polyproteins, and they are packaged into the virions as the RNA genome of the next generation (Cullen, 2003; Freed, E.O. and Martin, M.A., 2013). HIV replication is therefore dependent upon the efficient transcription, processing, and nuclear export of unspliced, singly spliced and fully spliced derivatives of the primary transcript.
Figure 1. HIV-1 generates multiple RNA species through splicing.
A single promoter from upstream LTR drives HIV-1 transcription. Along with the unspliced full-length transcript several alternatively spliced transcripts are generated depending on the number of splicing events that occur and which splice sites are selected. The dashed lines connect the major splice donor sites to the appropriate splice acceptor. The proteins expressed by these RNA species are indicated on the left.
The Journey: RNA Export and Transport
The export of cellular RNA molecules from the nucleus to the cytoplasm is a tightly regulated process that serves as a key step in the control of eukaryotic gene expression. Proper processing of mRNA is required for the nuclear export (Libri et al., 2002; Maniatis and Reed, 2002) as post-transcriptional processing events, including splicing facilitate the recruitment of protein factors necessary for export (Huang and Steitz, 2005; Lei and Silver, 2002; Moore and Proudfoot, 2009). This requirement for posttranscriptional processing poses a conundrum for retroviruses such as HIV-1 that need to export intron-containing mRNAs including the unspliced full-length RNA and several other partially spliced mRNAs. Therefore, HIV-1 uses a trans- acting viral protein, Rev, and a cis-acting viral Rev response element (RRE), to facilitate the export of intron-containing RNAs (Felber et al., 1989; Malim et al., 1989b; Rosen et al., 1988).
Temporal analysis showed that early after HIV-1 infection, only the fully spliced mRNAs encode for Tat, Rev, or Nef are detected in the cytoplasm (Kim et al., 1989). These fully spliced RNA transcripts use the NXF1 pathway for export, which is the pathway used for many cellular mRNAs. Rev is required for the intron-containing RNAs to be exported from the nucleus to the cytoplasm (Kim et al., 1989). In the absence of functional Rev, unspliced and singly spliced RNA transcripts can be detected in the nuclei but are present in the cytoplasm of infected cells at drastically reduced levels (Felber et al., 1989; Hadzopoulou-Cladaras et al., 1989; Kim et al., 1989; Malim et al., 1988).
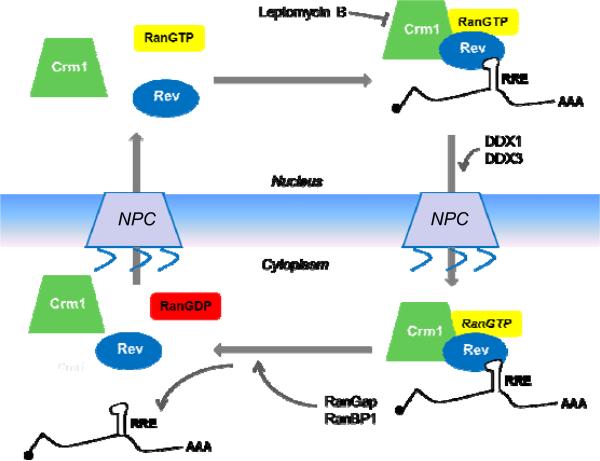
Rev allows the export of the intron-containing RNAs by acting as a bridge between the viral RNA and the host export machinery. A small nuclear shuttling protein, Rev contains a leucine-rich nuclear export signal (NES) and an arginine-rich RNA binding region (Kjems et al., 1991; Malim et al., 1990, 1989a; Zapp et al., 1991). Rev binds cooperatively to and multimerizes on RRE (Cook et al., 1991; Hadzopoulou-Cladaras et al., 1989; Malim et al., 1990, 1989b; Mann et al., 1994; Tiley et al., 1992). Located in the coding region of Env, RRE is present in all of the full-length and partially spliced HIV-1 transcripts. RRE is highly structured and folds into several stem-loops (Battiste et al., 1996; Daugherty et al., 2010a, 2010b; DiMattia et al., 2010; Hammarskjold and Rekosh, 2011); the structure of the HIV-1 RRE, which has recently been solved, forms a tree-dimensional “A” shape; the two legs and the distance between them all play a critical role in the Rev-RRE recognition (Fang et al., 2013). In addition to viral RNA, Rev directly interacts with a karyopherin Crm1 (chromosome region maintenance 1, also known as exportin 1 or Xpo1), which is a member of the importin β family of nuclear transport receptors (Fornerod et al., 1997; Nakielny and Dreyfuss, 1999; Neville et al., 1997). Leptomycin B, a metabolite of S. Pombe that specifically inhibits Crm1-mediated nuclear export, can inhibit Rev- dependent export of RNA transcripts (Daelemans et al., 2002; Kudo et al., 1999; Yashiroda and Yoshida, 2003). The Rev-RRE forms an export complex with Crm1 and RanGTP, a small GTPase (Ran) in a GTP-containing form; this export complex is then transported through the nuclear pore (Figure 2). Studies have shown that Crm1 interacts with several nucleoporins such as Nup358 and Nup214, which may facilitate the movement of the export complex through the pore (Bernad et al., 2004; Fornerod et al., 1997). Once in the cytoplasm, Ran GTPase-activating protein 1 (Ran GAP1) and Ran binding protein 1 (Ran BP1) modulate the activity of Ran by hydrolyzing the Ran-associated GTP into GDP, leading to the dissociation of the export complex (Figure 2).
Figure 2. Nuclear export mechanism of full-length and partially spliced HIV-1 RNA.
Viral protein Rev serves as a bridge that recruits Crm1and RanGTP to intron-containing HIV-1 RNAs by binding to the RRE. The Rev-RRE-Crm1-RanGTP complex moves through the nuclear pore complex (NPC). Once in cytoplasm, RanGap and RanBP1 lead to hydrolysis of Ran-associated GTP into GDP causing the dissociation of the export complex and the release of HIV-1 RNA. Although not shown, other host proteins, such as DDX3, may be involved in the nuclear export process (Figure not drawn to scale).
Recent studies suggest that an array of additional host factors may be involved in the regulation of full-length HIV-1 RNA expression. At this time, the mechanisms of actions of these proteins are not completely understood; some of these factors may act before or during RNA export whereas other factors may act in a more indirect manner such as RNA stabilization. Several DEAD-box RNA helicase proteins have been reported to be important to viral RNA expression and/or export. For example, DDX3 is thought to be required for the export of the Rev/RRE/Crm1/RanGTP complex (Yedavalli et al., 2004). Knockdown of DDX3 blocks the Rev-RRE-dependent Gag/Gag-Pol expression. Given that DDX3 can directly bind to Crm1, it may function by influencing the Crm1 export complex (Yedavalli et al., 2004). Other DEAD-box helicases may also play a role in HIV-1 RNA expression, including DDX1, DDX5, DDX17, and DDX24 although their mechanisms of actions are less understood (Fang et al., 2004; Ma et al., 2008; Naji et al., 2012; Yasuda-Inoue et al., 2013; Zhou et al., 2013). Among these, DDX1 has been reported to facilitate the binding and multimerization of Rev on RRE (Edgcomb et al., 2012; Robertson-Anderson et al., 2011). The nuclear retention of full-length HIV-1 RNA in the absence of Rev is mediated in part by hnRNP A2/B1 (Gordon et al., 2013). Although not directly involved in RNA export, other host proteins have been reported to play a role in full-length HIV-1 RNA expression. For example, analysis of the proteome associated with HIV-1 RNAs identified nuclear matrix protein Matrin 3 (MATR3) in Rev/RRE complexes (Kula et al., 2011), which is thought to stabilize unspliced and partially spliced HIV-1 RNAs (Yedavalli and Jeang, 2011). Peroxisome proliferator-activated receptor-interacting protein with methyltransferase domain (PIMT) can also interact with Rev and lead to hypermethylation the 5’ cap of full-length/partially spliced HIV-1 RNA to facilitate their expression (Yedavalli and Jeang, 2010).
Although HIV-1 uses the Crm1 pathway to export its intron-containing RNAs, other retroviruses may use different export pathways. For example, Mason-Pfizer Monkey Virus (MPMV) uses a structured RNA element termed constitutive transport element (CTE) that employs the cellular NXF1/Tap export pathway to avoid nuclear retention of intron-containing RNAs (Bray et al., 1994). This pathway is Crm1-independent, as treatment with Leptomycin B does not inhibit nuclear export of CTE-containing RNAs (Grüter et al., 1998; Otero et al., 1998; Pasquinelli et al., 1997; Saavedra et al., 1997). Though MPMV uses CTE to export its RNAs through a different pathway, CTE alone can functionally replace Rev/RRE during HIV-1 replication (Bray et al., 1994; McBride et al., 1997; Moore et al., 2009; Srinivasakumar et al., 1997; Ward et al., 2009; Wodrich et al., 2000).
Upon entering the cytoplasm, HIV-1 RNA must be transported to subcellular compartments to serve its functions. Interestingly, proper HIV-1 RNA export appears to tie to not only the RNA function in the cytoplasm but also the function of the protein encoded by that RNA. This phenomenon was first observed in modified mouse cells expressing the human cyclin T1 gene which allows for efficient HIV-1 RNA transcription elongation (Bieniasz et al., 1998; Swanson et al., 2004; Wimmer et al., 1999). HIV-1 RNA can be exported and translated in these cells, but the Gag proteins do not target to the plasma membrane properly resulting in an assembly defect (Mariani et al., 2000; Swanson et al., 2004). This defect can be rescued by rerouting the HIV-1 RNA export through the NXF1 pathway by replacing RRE with MPMV CTE (Swanson et al., 2004) or by supplementing rodent cells with human Crm1 protein (Elinav et al., 2012; Nagai-Fukataki et al., 2011; Okada et al., 2009; Sherer et al., 2011). A similar phenotype was also observed in human cells when HIV-1 RNA was exported using an unknown pathway (Jin et al., 2009). Therefore, RNA export is closely associated with RNA transport and RNA functions. At this time, the precise mechanism of HIV-1 RNA transport in the cytoplasm is not completely understood. For example, it was reported that later during replication after the expression of Gag, HIV-1 full-length RNA and Gag co-traffic using the microtubules to the virion assembly sites (Lehmann et al., 2009; Molle et al., 2009). However, disrupting microtubules by treating the cells with nocodazole does not inhibit HIV-1 virus production (Molle et al., 2009). Therefore, further studies are needed to understand how HIV-1 RNA is transported in the cytoplasm.
The Rendezvous: RNA Partner Selection / Dimerization and RNA-Gag Interaction
HIV-1 packages two copies of unspliced, single-stranded, full-length RNA genome into each viral particle (Chen et al., 2009). Both copies of genomic RNA are full-length, and each encodes all of the genetic information needed for viral replication. Packaging two complete RNA genomes provides the opportunity for frequent template switching events during reverse transcription, resulting in the generation of recombinant viruses that are genetically distinct from the two parental viruses (Hu and Temin, 1990a, 1990b; Rhodes et al., 2003). Strand transfer between the two RNA molecules may also act as a rescue mechanism to recover genetic information from an RNA molecule in which the integrity is compromised (Coffin, 1979; Hu and Temin, 1992).
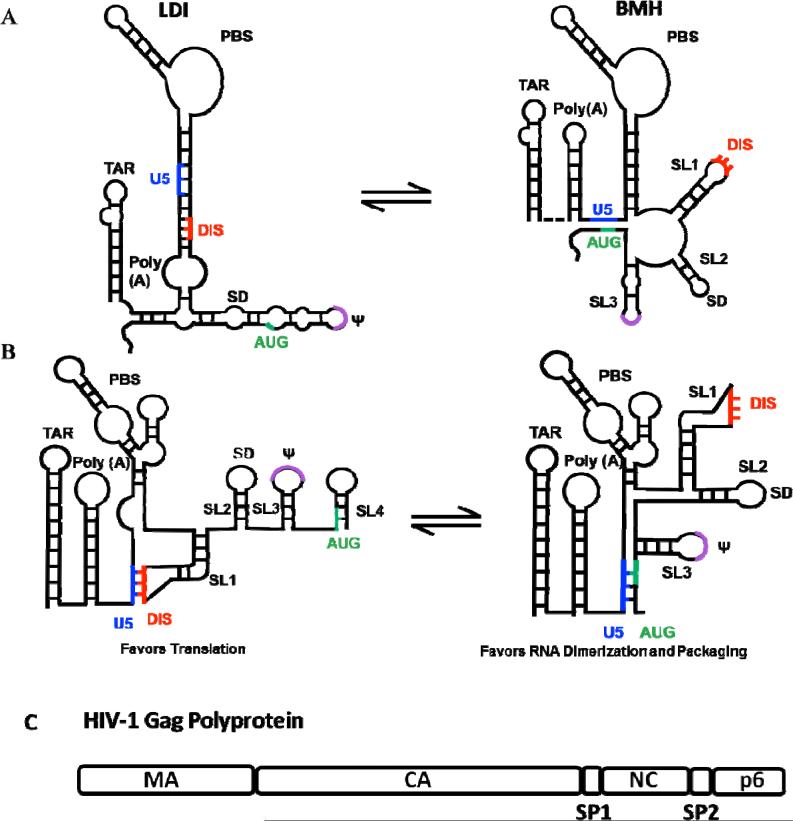
HIV-1 uses the cis-acting elements in the viral RNA and the trans-acting elements in Gag to achieve specific packaging of the viral RNA genome. The RNA elements important for packaging of HIV-1 genomic RNA have been mapped to the highly conserved and structured 5’ untranslated region (5’UTR) and extend into the 5’ end of the gag coding sequence (Aldovini and Young, 1990; Buchschacher and Panganiban, 1992; Clavel and Orenstein, 1990; Hayashi et al., 1992; Lever et al., 1989; Luban and Goff, 1994, 1991; McBride et al., 1997). The region required for RNA packaging and RNA dimerization overlap in HIV-1; multiple studies reported that the RNA fragments containing the aforementioned region can dimerize in vitro (Awang and Sen, 1993; Darlix et al., 1990; Marquet et al., 1991; Sundquist and Heaphy, 1993). The linkage of the 5’ ends of the two RNAs from several retroviruses can be visualized in electron micrographs (Bender and Davidson, 1976; Bender et al., 1978; Chien et al., 1980; Dube et al., 1976; Hoglund et al., 1997; Murti et al., 1981). Initially the entire 5’ packaging signal was refered to as Ψ (psi). Following extensive biochemical probing, mutagenesis, structural, phylogenetic and in silico analysis of this leader region, elements important for packaging became better defined (Baudin et al., 1993; Clever et al., 1995a; Harrison and Lever, 1992). Among the important elements in the HIV-1 leader that contribute to efficient RNA packaging are four stem loops (SL1, SL2, SL3, SL4) (Clever et al., 1995b; McBride et al., 1997). SL1 displays a GC-rich 6-nt palindromic sequence at the top of its loop called the dimer initiation signal (DIS). SL2 contains the major splice donor (SD) (Purcell and Martin, 1993). SL3, now denoted as psi (Ψ) is the most conserved region of the leader sequence and contains a GGAG RNA tetraloop that binds NC with high affinity (De Guzman et al., 1998). SL4 is downstream of SL3 and contains the AUG start codon of gag; although a documented structure, SL4 may not be present in the RNA competent to dimerize and be packaged (as discussed later). In addition to mediating RNA dimerization and packaging, the 336-nt 5’ leader sequence of the HIV-1 genome also is important for transcription (TAR), splicing (SD), and contains the reverse transcription primer binding (PBS), making it difficult to parse out what direct and indirect effects mutations in this region have on genome packaging. Many studies have linked the TAR hairpin to dimerization and packaging (Clever et al., 1999; Damgaard et al., 2004; Das et al., 1998; McBride et al., 1997). However, once the regulation of Tat/TAR on transcription was replaced with a tetracycline-inducible system, it became apparent that the TAR hairpin is not required for packaging (Das et al., 2007). Further studies confirm that the TAR hairpin as well as the Poly(A) signal are dispensable for RNA dimerization and packaging (Heng et al., 2012; Sakuragi et al., 2007).
Of the RNA elements described above, the DIS in SL1 is able to mediate in vitro dimerization of RNA fragments (Laughrea and Jetté, 1994; Muriaux et al., 1995; Paillart et al., 1994; Skripkin et al., 1994). Furthermore, the DIS sequence has proven to be the dominant factor driving RNA dimerization and RNA partner selection in vivo (Chen et al., 2009; Chin et al., 2005; Moore et al., 2007). The autocomplementary nature of the DIS supports the initiation of Watson-Crick base pairing between the two HIV-1 RNA molecules and the generation of a “kissing loop” (Clever et al., 1996; Kieken et al., 2006; Paillart et al., 1997). The most common DIS sequence is GCGCGC in subtype B, and GUGCAC in subtypes A, C and G HIV-1. The differences in the DIS primary sequences have proven to be a major barrier for recombination between two viral subtypes such as subtype B and subtype C HIV-1, reinforcing the role of the DIS in the initiation of RNA dimerization (Chin et al., 2005).
The influence of the DIS sequences on RNA copackaging is strong evidence that the viral RNAs select co-packaged RNA partners (or initiate RNA dimerization) before they are packaged into the viral particles (Chen et al., 2009; Moore et al., 2007). Furthermore, RNA partner selection occurs in the cytoplasm (Moore et al., 2009). Using cells harboring different proviruses and fusing cells at conditions that allow for the mixing of cytoplasmic but not nuclear content, it was shown that RNA dimerization occurs in the cytoplasm of the cell, not the nucleus (Moore et al., 2009). It is of note that RNAs using different export pathways are able to be copackaged, but to a lesser extent compared with RNAs using the same export pathways, suggesting that RNA partner selection in the cytoplasm occurs where the RNA molecules are still at least partially segregated (ibid).
A specific region of the murine leukemia virus (MLV) genome has been identified as necessary and sufficient for RNA packaging because insertion of this sequence can confer the ability of heterologous RNA to be packaged into viral particles (Adam and Miller, 1988; Hibbert et al., 2004). At this time, the necessary and sufficient HIV-1 packaging signal has not been defined. However, within the viral context, deletions have been made to trim down the sequences required for packaging of the HIV-1 genome. A minimal 159-nt RNA sequence that includes SL1-SL3 through the U5:AUG stem, but lacks TAR, Poly(A) and the upper PBS hairpin structure, can dimerize and is competent to bind NC in vitro (Heng et al., 2012). In addition, a 144-nt RNA sequence including a segment of sequence that forms the base of the PBS hairpin (but does not include the primer binding site), SL1, SL3 and SL4 proved to be sufficient to mediate intramolecular dimerization when inserted into an ectopic position in the HIV-1 genome, and was defined as the minimal element required for HIV-1 RNA dimerization in vivo (Sakuragi et al., 2007).
The HIV-1 RNA packaging signal is recognized by Gag, which is synthesized as a polyprotein that is later processed into six mature proteins: matrix (MA), capsid (CA), SP1, nucleocapsid (NC), SP2, and p6 (Figure 3C). Of these domains, NC plays the major role for the selective packaging of the HIV-1 RNA genome (Berkowitz et al., 1993; Luban and Goff, 1994). The NC protein contains two CCHC-type zinc knuckle motifs separated by a linker. Mutations of the NC domain including those in the CCHC sequences can cause severe defects in viral RNA packaging (Aldovini and Young, 1990; Dorfman et al., 1993; Gorelick et al., 1990). Furthermore, replacing the NC domain of HIV-1 Gag with the NC domain of MLV, allows this chimeric polyprotein to package MLV RNA genome, and vice versa (Berkowitz et al., 1995; Zhang and Barklis, 1995). However, the specificity of viral RNA packaging cannot always be changed by replacing the NC domain. For example, swapping the NC domains of HIV-1 and mouse mammary tumor virus (MMTV) does not alter the packaging preferences of these viruses (Poon et al., 1998). Containing a basic patch for membrane targeting, the MA domain has also been shown to have nucleic acid binding properties (Cai et al., 2010; Chukkapalli et al., 2010, 2008; Lochrie et al., 1997; Purohit et al., 2001). However, currently there is no evidence supporting that HIV-1 MA plays a role in specific packaging of viral genome.
Figure 3. Cis- and trans- factors of HIV-1 RNA packaging.
(A) and (B) Two models of the HIV-1 5’ leader RNA secondary structures favoring translation (left) or RNA dimerization and packaging (right). AUG start codon of gag (green), DIS (red), Ψ (purple), U5 (red). (A) Conformation switch of the HIV-1 5’ leader RNA proposed by the Berkhout group (Abbink and Berkhout, 2003). In the long distance interaction (LDI) structure (left) the DIS sequence is sequestered through base-pair interactions with the Poly(A). However, in the branched multiple hairpin (BMH) structure (right) the AUG start codon of gag binds the U5 region, exposing the DIS sequence and promoting RNA dimerization and packaging. (B) Conformation switch proposed by the Summers group (Lu et al., 2011). Alternative RNA secondary structures of the HIV-1 5’ leader RNA favoring translation (left) in which DIS is base-paired with the U5 region or dimerization and packaging (right) in which the AUG base-pairs with the U5 region the DIS sequence is exposed. (C) Domains of the HIV-1 Gag polyprotein. MA (Matrix), CA (Capsid), SP1 (Spacer 1), NC (Nucleocapsid), SP2 (Spacer 2), p6.
In addition to serving as the RNA genome, full-length HIV-1 RNA also serves as the mRNA template for Gag/GagPol translation (Butsch and Boris-Lawrie, 2000; Dorman and Lever, 2000). It is not well understood how these two functions of viral RNA are balanced. Multiple studies support a model in which two, or more, alternative structures for the HIV-1 RNA leader sequences exist, the two aforementioned RNA functions are carried out by RNA folded into different structures (Baudin et al., 1993; Berkhout and van Wamel, 2000; Berkhout et al., 2001; Huthoff and Berkhout, 2001a, 2001b; Huthoff et al., 2004; Ooms et al., 2004). Berkhout and colleagues have proposed that the 5’ end of HIV-1 RNA can fold into two alternative structures: the long distance interaction (LDI) structure or the branched multiple hairpin (BMH) structure (Figure 3A). In the LDI conformation, the DIS participates in a long-distance interaction with the upstream Poly(A) domain and therefore cannot engage in the “kissing loop” interactions that induce RNA dimerization (Huthoff and Berkhout, 2001a). However, in the BMH conformation, the DIS sequences are exposed, promoting RNA dimerization and packaging. Although, it has been shown that NC may disrupt the LDI conformation and promote RNA dimerization in vitro, the existence of the BMH and LDI structures have yet to be shown in vivo (Berkhout et al., 2001). A recent model proposed that the RNA conformation that favors translation sequesters the DIS sequence by interaction with part of the U5, not the Poly(A) region, whereas the AUG start codon participates in base-pairing in the stem of SL4 (Figure 3A) (Lu et al., 2011). In the alternative structure, the SL4 is lost as the AUG sequences base-pair with part of U5, displacing and exposing the DIS sequence and promoting dimerizations and packaging (Damgaard et al., 2004; Lu et al., 2011; Wilkinson et al., 2008).
It has been shown that MLV has two pools of RNAs, one serves as template for protein translation and the other serves as the RNA genome (Levin and Rosenak, 1976; Levin et al., 1974). Currently, there is no evidence that such division exists in the HIV-1 RNA population (Butsch and Boris-Lawrie, 2000; Dorman and Lever, 2000). In contrast, a cis packaging model was proposed in which the Gag polyprotein preferentially packages the RNA template from which it was translated (Poon et al., 2002). However, it was later shown that Gag efficiently packages RNA in trans and that trans-packaging is the primary mechanism used for HIV-1 RNA packaging (Nikolaitchik et al., 2006).
The precise location where the Gag and RNA genome interaction takes place is currently not known. It was proposed that HIV-1 full-length RNA and Gag first interact and colocalize in the perinuclear region and this complex subsequently traffics through the cytoplasm to the plasma membrane of the cell (Poole et al., 2005). This proposal was challenged later as HIV-1 RNA genome but not Gag was detected at the perinuclear region (Kemler et al., 2010). Using a co-immunoprecipitation assay coupled to membrane flotation, it was shown that HIV-1 RNA can be pulled down with Gag from the cytoplasmic fraction; additionally, the ability to co-precipitate viral RNA does not depend on the ability of Gag to target to the membrane but is stabilized by the Gag-Gag interaction. These experiments suggest that the initial Gag and RNA genome interaction may occur in the cytoplasm (Kutluay and Bieniasz, 2010).
The Destination: HIV-1 Full-Length RNA Assembles into Virus Particles
To infect a new host, HIV-1 needs to generate new virions, which is a sophisticated process that involves the assembly of the appropriate numbers of viral proteins, viral RNA, host RNA including tRNA, and recruitment of host proteins to facilitate the exit of the newly generated particles from the cell. By directly visualizing the viral RNA content of individual particles, it was found that most HIV-1 particles contain viral genomes; furthermore, two copies of RNAs are packaged into one particle (Chen et al., 2009). Therefore, the packaging of the viral RNA is a tightly regulated process. As mentioned above, the interactions between Gag and RNA elements ensure the specificity of the RNA genome packaging; however, the mechanism by which HIV-1 regulates the packaging of the two copies of RNAs was unknown. Using RNA genomes of different lengths, it was shown that two copies of RNA were packaged regardless whether the genome was 3 kb, 8 kb, or 17 kb, indicating that viral genome packaging is not regulated by the mass of the RNA (Nikolaitchik et al., 2013). It was further shown that if two dimerization signals were present in a single viral RNA, it can form a self-dimer, and only one copy of the self-dimer is packaged (ibid). Taken together, these results indicate that HIV-1 RNA genome packaging is regulated by the recognition of one dimeric RNA.
Although controversial at one point, our current understanding is that the major assembly site for HIV-1 is the plasma membrane (Jouvenet et al., 2006; Ono, 2010). Furthermore, these assembly events preferentially take place in discrete domains of the plasma membrane enriched in cholesterol and sphingolipids, known as lipid rafts (Nguyen and Hildreth, 2000; Ono and Freed, 2001; Ono, 2009). The viral protein Gag orchestrates the assembly process including membrane targeting, Gag-Gag and Gag-GagPol multimerization, recruitment of Env, and the packaging of viral RNA genome (reviewed in Sundquist and Kräusslich, 2012).
The assembly and RNA packaging process have been visualized using total internal reflection fluorescence (TIRF) microscopy. By tagging Gag with a fluorescent protein and monitoring the signal emitted, the kinetics of virus assembly were examined. In these experiments, individual weak fluorescent signals first appeared on the plasma membrane and the intensity of the signals increased with time; eventually, the intensities of these signals reached a plateau and were in the range of those observed in viral particles. The measured assembly time varied but averaged between 5-6 minutes to 8-9 minutes (Ivanchenko et al., 2009; Jouvenet et al., 2008). Gradual increase of Gag signals on the plasma membrane is consistent with the biochemical studies indicating Gag multimerizes extensively on the membrane but not the in the cytoplasm (Kutluay and Bieniasz, 2010). The packaging of the viral RNA genome was studied by labeling the RNA with a fluorescently tagged bacteriophage MS2 coat protein which specifically interacts with the MS2 binding sites engineered into the HIV-1 genome. In these experiments, it was observed that viral RNA genomes can reach the plasma membrane even when Gag is not expressed; these RNA signals moved in a dynamic manner and only resided on the membrane for few seconds (Jouvenet et al., 2009). When Gag and RNA were both expressed, RNA signals appeared on the membrane first; weak Gag signals were detected and co-localized with the RNA approximately 4-5 minutes after the appearance of RNA signal. As the Gag signal intensities increased with time, the lateral mobility of the viral RNA decreased until the completion of the viral particle assembly (Jouvenet et al., 2009). It should be noted that although the RNA signals were detected prior to those of Gag in these experiments, it remains possible that at this time, a few Gag proteins were associated with RNA, but their signal intensity was below the limit of detection. Together these observations suggest that no more than a few Gag molecules bind RNA in the cytoplasm and together they are targeted to the membrane where Gag multimerization proceeds as Gag assembles around the viral RNA genome.
In summary, HIV-1 full-length RNA plays a central role in viral replication. Its complicated journey from the nucleus to the viral particles requires the sophisticated manipulation of cellular functions and dynamic interactions of viral components. There is still much to be learned about the various stages of the life of the full-length HIV-1 RNA, such as host proteins involved in nuclear RNA export, the mechanism of cytoplasmic RNA transport, the relationship between RNA structures and functions, the location(s) where Gag-RNA interaction takes place, and the molecular interactions between Gag and RNA that lead to specific packaging. Future studies will shed light on many of the currently unanswered questions.
Highlights.
Review steps affecting HIV-1 full-length RNA expression and transport
Discuss how HIV-1 exports introns-containing RNA species
Discuss where and how HIV-1 RNA:RNA and Gag:RNA interactions take place
Summarize the regulation of HIV-1 RNA genome packaging into viral particles
Acknowledgements
We thank Vinay K. Pathak for insightful discussions and suggestions; Olga Nikolaitchik, Krista Delviks-Frankenberry, and Yang Liu for critical reading of the manuscript. This work was funded by the Intramural Research Program of the Center for Cancer Research, NCI and the Intramural AIDS Targeted Antiviral Program, NIH. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- Adam MA, Miller AD. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J. Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M, Sharmeen L, Kimpton J, Romeo JM, Garcia JV, Peterlin BM, Groudine M, Emerman M. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3862–3866. doi: 10.1073/pnas.91.9.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–6. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry (Mosc.) 1993;32:11453–7. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5’ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–97. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- Bender W, Chien YH, Chattopadhyay S, Vogt PK, Gardner MB, Davidson N. High-molecular-weight RNAs of AKR, NZB, and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–96. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–95. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA N. Y. N. 2001;7:1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz RD, Luban J, Goff SP. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–56. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjöld ML. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchschacher GL, Jr., Panganiban AT. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–9. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Translation is not required To generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J Virol. 2000;74:11531–7. doi: 10.1128/jvi.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Huang Y, Craigie R, Clore GM. Structural basis of the association of HIV-1 matrix protein with DNA. PloS One. 2010;5:e15675. doi: 10.1371/journal.pone.0015675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U A. 2009;106:13535–40. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Deng CT, Chandler P, Davidson N. A method for the isolation of segments from the 5’ ends of retrovirus RNA. Anal Biochem. 1980;102:281–7. doi: 10.1016/0003-2697(80)90153-0. [DOI] [PubMed] [Google Scholar]
- Chin MP, Rhodes TD, Chen J, Fu W, Hu WS. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci U A. 2005;102:9002–7. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu W-S, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol- (4,5)-bisphosphate is essential for efficient gag membrane binding. J. Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F, Orenstein JM. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–4. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. J Virol. 1995a;69:2101–9. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. J Virol. 1995b;69:2101–9. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever JL, Eckstein DA, Parslow TG. Genetic dissociation of the encapsidation and reverse transcription functions in the 5’ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–9. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–8. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Daelemans D, Afonina E, Nilsson J, Werner G, Kjems J, De Clercq E, Pavlakis GN, Vandamme A-M. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14440–14445. doi: 10.1073/pnas.212285299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5’ region of the HIV-1 genome. J Mol Biol. 2004;336:369–79. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Gabus C, Nugeyre MT, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–99. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol. 2007;81:7742–8. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Klaver B, Berkhout B. The 5’ and 3’ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–23. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. U. S. A. 2010a;107:12481–12486. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010b;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–8. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–69. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J Virol. 2000;74:11413–7. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube S, Kung HJ, Bender W, Davidson N, Ostertag W. Size, subunit composition, and secondary structure of the Friend virus genome. J Virol. 1976;20:264–72. doi: 10.1128/jvi.20.1.264-272.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb SP, Carmel AB, Naji S, Ambrus-Aikelin G, Reyes JR, Saphire ACS, Gerace L, Williamson JR. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J. Mol. Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav H, Wu Y, Coskun A, Hryckiewicz K, Kemler I, Hu Y, Rogers H, Hao B, Ben Mamoun C, Poeschla E, Sutton R. Human CRM1 augments production of infectious human and feline immunodeficiency viruses from murine cells. J. Virol. 2012;86:12053–12068. doi: 10.1128/JVI.01970-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Peterlin BM, Weis K. Lessons from HIV: movement of macromolecules inside the cell. Curr. Mol. Med. 2001;1:1–7. doi: 10.2174/1566524013364167. [DOI] [PubMed] [Google Scholar]
- Fang J, Kubota S, Yang B, Zhou N, Zhang H, Godbout R, Pomerantz RJ. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Fang X, Wang J, O'Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SFJ, Rein A, Wang Y-X. An Unusual Topological Structure of the HIV-1 Rev Response Element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Field's Virology. Williams, and Wilkins; Lippincott: 2013. HIVs and their replication. pp. 1502–1560. [Google Scholar]
- Gaynor RB. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr. Top. Microbiol. Immunol. 1995a;193:51–77. doi: 10.1007/978-3-642-78929-8_3. [DOI] [PubMed] [Google Scholar]
- Gaynor RB. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr. Top. Microbiol. Immunol. 1995b;193:51–77. doi: 10.1007/978-3-642-78929-8_3. [DOI] [PubMed] [Google Scholar]
- Gordon H, Ajamian L, Valiente-Echeverrìa F, Lévesque K, Rigby WF, Mouland AJ. Depletion of hnRNP A2/B1 overrides the nuclear retention of the HIV-1 genomic RNA. RNA Biol. 2013:10. doi: 10.4161/rna.26542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick RJ, Nigida SM, Jr., Bess JW, Jr., Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–11. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M, Felber BK, Cladaras C, Athanassopoulos A, Tse A, Pavlakis GN. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold M-L, Rekosh D. A Long-Awaited Structure Is Rev-ealed. Viruses. 2011;3:484–492. doi: 10.3390/v3050484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GP, Lever AM. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J, Cullen BR. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J. Virol. 1988;62:673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–9. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–39. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert CS, Mirro J, Rein A. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 2004;78:10927–10938. doi: 10.1128/JVI.78.20.10927-10938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund S, Ohagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–9. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990a;250:1227–33. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Hu WS, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 1990b;87:1556–60. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Temin HM. Effect of gamma radiation on retroviral recombination. J Virol. 1992;66:4457–63. doi: 10.1128/jvi.66.7.4457-4463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a Messenger's Journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001a;7:143–57. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001b;29:2594–600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Girard F, Wijmenga SS, Berkhout B. Evidence for a base triple in the free HIV-1 TAR RNA. RNA N. Y. N. 2004;10:412–423. doi: 10.1261/rna.5161304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Kräusslich H-G, Eils R, Rohr K, Bräuchle C, Müller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Sturgeon T, Weisz OA, Mothes W, Montelaro RC. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PloS One. 2009;4:e6551. doi: 10.1371/journal.pone.0006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJD, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol. 2010;84:6352–66. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieken F, Paquet F, Brule F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–52. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J, Frankel AD, Sharp PA. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell. 1991;67:169–178. doi: 10.1016/0092-8674(91)90580-r. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula A, Guerra J, Knezevich A, Kleva D, Myers MP, Marcello A. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology. 2011;8:60. doi: 10.1186/1742-4690-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5’ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry (Mosc.) 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Milev MP, Abrahamyan L, Yao X-J, Pante N, Mouland AJ. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J. Biol. Chem. 2009;284:14572–14585. doi: 10.1074/jbc.M808531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Intron status and 3’-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–7. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Grimley PM, Ramseur JM, Berezesky IK. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Rosenak MJ. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. U. S. A. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3’-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochrie MA, Waugh S, Pratt DG, Jr, Clever J, Parslow TG, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5’-leader RNA that regulate genome packaging. Science. 2011;334:242–5. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Goff SP. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. J Virol. 1991;65:3203–12. doi: 10.1128/jvi.65.6.3203-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J, Goff SP. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–93. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Rong L, Zhou Y, Roy BB, Lu J, Abrahamyan L, Mouland AJ, Pan Q, Liang C. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology. 2008;375:253–264. doi: 10.1016/j.virol.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Malim MH, Böhnlein S, Hauber J, Cullen BR. Functional dissection of the HIV-1 Rev trans-activator--derivation of a trans-dominant repressor of Rev function. Cell. 1989a;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Fenrick R, Cullen BR. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989b;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mann DA, Mikaélian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler PJ, Gait MJ, Karn J. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- Mariani R, Rutter G, Harris ME, Hope TJ, Kräusslich HG, Landau NR. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 2000;74:3859–3870. doi: 10.1128/jvi.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–57. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle D, Segura-Morales C, Camus G, Berlioz-Torrent C, Kjems J, Basyuk E, Bertrand E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J. Biol. Chem. 2009;284:19727–19743. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–11. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5:e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA Processing Reaches Back toTranscription and Ahead to Translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Muriaux D, Girard PM, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5’ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–16. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- Murti KG, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–19. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai-Fukataki M, Ohashi T, Hashimoto I, Kimura T, Hakata Y, Shida H. Nuclear and cytoplasmic effects of human CRM1 on HIV-1 production in rat cells. Genes Cells Devoted Mol. Cell. Mech. 2011;16:203–216. doi: 10.1111/j.1365-2443.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- Naji S, Ambrus G, Cimermančič P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR, 3rd, Saphire AC, Gerace L. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteomics MCP. 2012;11:M111.015313. doi: 10.1074/mcp.M111.015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. CB. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaitchik O, Rhodes TD, Ott D, Hu W-S. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai S-HS, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu W-S. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Zhang X, Ben Fofana I, Nagai M, Suzuki H, Ohashi T, Shida H. Synergistic effect of human CycT1 and CRM1 on HIV-1 propagation in rat T cells and macrophages. Retrovirology. 2009;6:43. doi: 10.1186/1742-4690-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. HIV-1 Assembly at the Plasma Membrane: Gag Trafficking and Localization. Future Virol. 2009;4:241–257. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. HIV-1 assembly at the plasma membrane. Vaccine. 2010;28(Suppl 2):B55–59. doi: 10.1016/j.vaccine.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J Virol. 2004;78:10814–9. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero GC, Harris ME, Donello JE, Hope TJ. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Ernst RK, Lund E, Grimm C, Zapp ML, Rekosh D, Hammarskjöld ML, Dahlberg JE. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–55. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- Poon DT, Chertova EN, Ott DE. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology. 2002;293:368–78. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- Poon DT, Li G, Aldovini A. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J Virol. 1998;72:1983–93. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67:6365–78. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit P, Dupont S, Stevenson M, Green MR. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA N. Y. N. 2001;7:576–584. doi: 10.1017/s1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes T, Wargo H, Hu WS. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77:11193–200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson-Anderson RM, Wang J, Edgcomb SP, Carmel AB, Williamson JR, Millar DP. Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J. Mol. Biol. 2011;410:959–971. doi: 10.1016/j.jmb.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen CA, Terwilliger E, Dayton A, Sodroski JG, Haseltine WA. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounseville MP, Kumar A. Binding of a host cell nuclear protein to the stem region of human immunodeficiency virus type 1 trans-activation-responsive RNA. J. Virol. 1992;66:1688–1694. doi: 10.1128/jvi.66.3.1688-1694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. CB. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J Virol. 2007;81:7985–92. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Swanson CM, Hué S, Roberts RG, Bergeron JRC, Malim MH. Evolution of a species-specific determinant within human CRM1 that regulates the post-transcriptional phases of HIV-1 replication. PLoS Pathog. 2011;7:e1002395. doi: 10.1371/journal.ppat.1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci U A. 1994;91:4945–9. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjöld ML, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci U A. 1993;90:3393–7. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004;23:2632–2640. doi: 10.1038/sj.emboj.7600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen BR. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc. Natl. Acad. Sci. U. S. A. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AM, Rekosh D, Hammarskjold M-L. Trafficking through the Rev/RRE Pathway Is Essential for Efficient Inhibition of Human Immunodeficiency Virus Type 1 by an Antisense RNA Derived from the Envelope Gene. J. Virol. 2009;83:940–952. doi: 10.1128/JVI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer J, Fujinaga K, Taube R, Cujec TP, Zhu Y, Peng J, Price DH, Peterlin BM. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology. 1999;255:182–189. doi: 10.1006/viro.1998.9589. [DOI] [PubMed] [Google Scholar]
- Wodrich H, Schambach A, Kräusslich HG. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 2000;28:901–910. doi: 10.1093/nar/28.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda, Yoshida Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs. Curr Med Chem. 2003;10:741–748. doi: 10.2174/0929867033457791. [DOI] [PubMed] [Google Scholar]
- Yasuda-Inoue M, Kuroki M, Ariumi Y. Distinct DDX DEAD-box RNA helicases cooperate to modulate the HIV-1 Rev function. Biochem. Biophys. Res. Commun. 2013;434:803–808. doi: 10.1016/j.bbrc.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VSRK, Jeang K-T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VSRK, Jeang K-T. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology. 2011;8:61. doi: 10.1186/1742-4690-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VSRK, Neuveut C, Chi Y-H, Kleiman L, Jeang K-T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Zapp ML, Hope TJ, Parslow TG, Green MR. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Luo J, Mills L, Wu S, Pan T, Geng G, Zhang J, Luo H, Liu C, Zhang H. DDX5 facilitates HIV-1 replication as a cellular co-factor of Rev. PloS One. 2013;8:e65040. doi: 10.1371/journal.pone.0065040. [DOI] [PMC free article] [PubMed] [Google Scholar]