Abstract
Recent analysis of transcriptomes has revealed that RNA molecules perform a myriad of functions beyond coding for proteins. RNA molecules can fold into complex secondary and tertiary structures, which are critical for regulating their function. Selective Hydroxyl Acylation analyzed by Primer Extension, or SHAPE is a common method for probing RNA structure in and outside of cells. Recent developments in SHAPE include the design of acyl imidazole acylating electrophiles with alkyl azides to enrich the sites of SHAPE adduct formation. Enrichment is key for next-generation sequencing experiments as it dramatically improves the signal. In a recent comparison of different structures of such reagents, we realized that furoyl acylating reagents form hyper-stable ester adducts with hydroxyls. This prompted us to design, synthesize and test a novel dual-functioning SHAPE probe (FAI-N3), which has the stable furoyl scaffold and the alkyl azide for enrichment. Herein we present the results that show FAI-N3 is a suitable probe for RNA structure analysis by SHAPE and that it can be used for enrichment of SHAPE adducts. These results strongly demonstrate that FAI-N3 is an ideal probe for structure probing in cells and will be very useful for sequencing-based analysis of SHAPE.
Graphical Abstract
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.
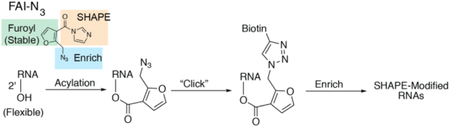
RNA molecules can be found at the heart of many normal biological pathways and are key players in the onset of many diseases.(1) For proper function, RNA molecules must fold into complex secondary and even tertiary structures.(2, 3)
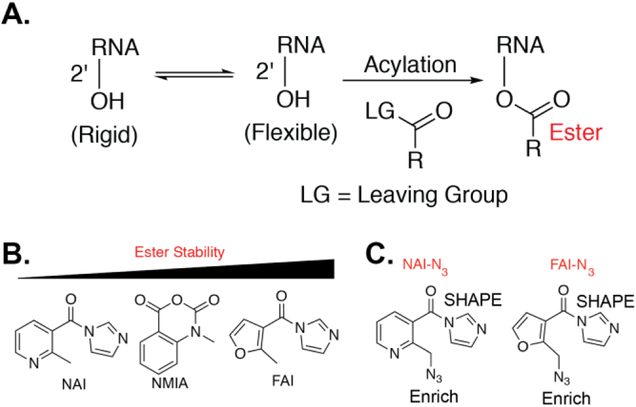
Several chemical methods have been developed for analyzing RNA structure. For example, dimethyl sulfate (DMS) can alkylate A and C residues not involved in Watson-Crick pairing.(4, 5) Selective hydroxyl acylation analyzed by primer extension, or SHAPE, reveals the internucleotide flexibility in RNA by 2’-OH acylation. SHAPE reactions form ester adducts on the 2’-OH (Figure 1)(6) which is accomplished by incubation with acyltion electrophiles such as anhydrides,(6) acyl cyanides,(7) and more recently acyl imidazole reagents.(8)
Figure 1. The SHAPE reaction and stability of SHAPE reagents.
A. The SHAPE reaction. B. Ester stability of different SHAPE electrophiles. C. Structure of NAI-N3 and FAI-N3.
A key aspect of SHAPE is the stability of the ester product on the 2’-OH, which is identified by reverse transcription. A recent analysis by our lab demonstrated that furoyl SHAPE reagents form hyper-stable ester products (Figure 1, B).(9) This critical observation suggests that the furoyl scaffold would be ideal for downstream in vivo SHAPE analysis.
Recently, SHAPE has been extended from selected RNA studies to being used transcriptome-wide. This transition has been accompanied by the development of novel protocols to convert SHAPE adducts into sequencing reads. An important criterion of such protocol development is an enrichment step of SHAPE adducts for achieving high signal-to-noise ratios in sequencing experiments. As such, we have utilized dual-functioning SHAPE reagents. This is tackled through the incorporation of two moieties, an acyl imidazole for the acylation reaction, and an alkyl azide to permit biotin attachment and enrichment through Strain Promoted Azide Alkyne Cycloaddition (SPAAC) reactions (Figure 1, C).(10)
Our first-generation reagent, NAI-N3, was utilized to generate SHAPE data from the mouse embryonic stem cell transcriptome in both inside and outside cells.(10) However, we previously demonstrated that the nicotinoyl scaffold is highly prone to hydrolysis.(9) To retain the power of dual-functioning reagents coupled with our recent analysis of ester stability, we were prompted to design, synthesize, and test an optimized SHAPE reagent, FAI-N3 (Figure 1, C), for structural analysis of RNA.
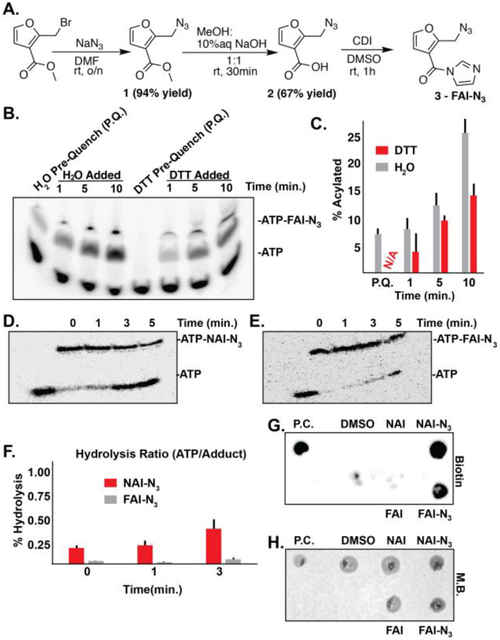
FAI-N3 probe was synthesized in three steps (Figure 2, A). First, a substitution reaction was done on 2-(bromomethyl)furan-3-carboxylate with sodium azide in dimethylformamide to yield 2-(azidomethyl)furan-3-carboxylate. Then the ester was hydrolyzed to the carboxylic acid using methanol and aqueous sodium hydroxide. Finally, 2-(azidomethyl)furan-3-carboxylic acid was treated with carbonyldiimidazole in dimethylsulfoxide to convert the carboxylic acid to the imidazole amide. The overall yield in three steps was 30%.
Figure 2. Synthesis and evaluation of FAI-N3 for hydroxyl acylation.
A). Synthesis of FAI-N3. B). DTT quenching of FAI-N3 acylation. Pre-quench (P.Q.) is designated for addition of either water or DTT to the ATP solution before the addition of FAI-N3 C). Bar-graph detailing the percent acylation (monoacylation) as a function of time in quenching conditions represented in Panel B. Black bars denote error from biological triplicates. N/A is abbreviated for no acylation observed. D). Stability of NAI-N3 at 95 °C. E). Stability of FAI-N3 at 95 °C. F). Bar-graph detailing the percent hydrolysis as represented in Panel D and E. Black bars denote error from biological duplicates. G). Dot blot demonstrating successful “click” for biotinylation of FAI-N3 acylated RNA. H). Methylene blue staining of G. Biotin = streptavidin dot blot. M.B. = methylene blue. P.C. = positive control bioinylated oligo.
A common requirement with these long lasting SHAPE probes is the ability for the probes to be quenched with dithiothreitol (DTT). Based on previous findings that FAI can be quenched by DTT, we tested if FAI-N3 could also be quenched with the same method. In Figure 2, B & C (Supplementary Information), we compared the rates by quenching with DTT as opposed to quenching with water. Preincubation of DTT with ATP followed by addition of FAI-N3 shows no reactivity, demonstrating that DTT is capable of quenching acylation. In subsequent lanes, the addition of DTT following the acylation reaction afforded better control in quenching compared to the addition of water. This shows that DTT is capable of quenching the acylation reactivity of furoyl SHAPE reagents
We recently reported that the furoyl scaffold on SHAPE reagents makes them stable to hydrolysis in conditions used in RNA-seq experiments.(9) We wanted to test whether FAI-N3 adducts were more stable in comparison to NAI-N3 adducts. We subjected isolated ATP-bound adducts with NAI-N3 and FAI-N3 to high-temperature hydrolysis conditions over time (95 °C; temperature used repeatedly for annealing of RNA over several steps including reverse transcription, primer binding, and also linker ligation. These results demonstrated that FAI-N3 esters are much more stable than those from NAI-N3 (Figure 2, D – F; Supplementary Information). This is consistent with our earlier observations comparing the two SHAPE scaffolds side by side, and demonstrates that the furoyl scaffold is a suitable reagent for RNA structure probing to form stable ester adducts for enrichment and opens the door for stringent washes at elevated temperatures.
The virtue of the alkyl azide is to enrich for SHAPE modifications, which others and we have shown can greatly reduce background of unmodified RNA.(11, 12) We next investigated the possibility if FAI-N3 could be amenable for RNA enrichment with the same aproach. Dot blot analysis of the modified RNA demonstrated that biotin can be attached through SPAAC (Figure 2, G & H). Successful biotin attachment suggests that SHAPE adducts can be enriched before reverse transcription – an important aspect of applications of SHAPE for transcriptome-wide analysis.
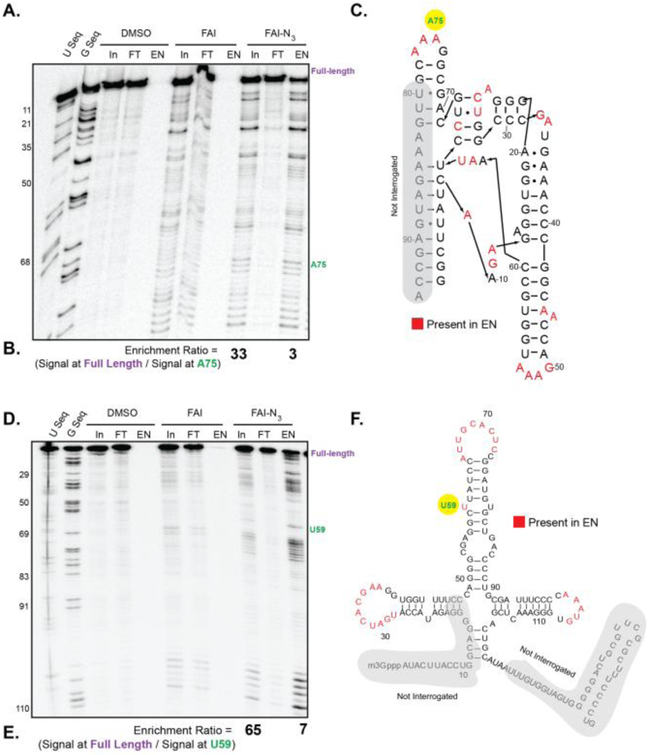
We tested if FAI-N3 would still recover viable RT stops for SHAPE on the SAM-I riboswitch and U1 snRNA.(13) We confirmed this by taking FAI-N3 through the protocol of SHAPE modification, biotinylation, enrichment, and then reverse transcription. Due to the low propensity for FAI-N3 adducts to be hydrolyzed, we added a high-temperature wash step. As shown in Figure 3 A & D, stringent wash steps with the FAI-N3 lane afforded clean RT stops and enrichment at single stranded sites. To determine enrichment, we compared the signal from full-length cDNA from RT to an enriched site from SHAPE (Full-length/SHAPE). We observed a marked reduction in full-length cDNA in the FAI-N3 enriched lane (IP, Figure 3, B & E) in contrast to observing majority of the full-length cDNA in the flowthrough (FT). The ratio of enriched cDNA bands at stops to full length increased almost 10-fold in the enriched samples. Enriched sites mapped to single-stranded regions of the RNA (Figure 3 C & F) Overall, these data nicely show that FAI-N3 is a viable probe for SHAPE and enrichment of RT stops with removal of full length cDNAs.
Figure 3. FAI-N3 can be used to enrich acylation sites in RNA structure modifications.
A). FAI-N3 RNA adducts are capable of undergoing SPAAC for biotinylation and enrichment on SAM-I RNA. In = Input. FT = Flowthrough. EN = enriched. B). Calculation of enrichment of stops against full-length cDNA bands. C). SAM-I riboswitch with sites of FAI-N3 enrichment mapped on the secondary structure. Nucleotides that are denoted in red are enriched in the EN lane of Panel A. D). Same as in Panel A, but for U1 snRNA E). Calculation of enrichment of stops against full-length cDNA bands. F). U1 snRNA with sites of FAI-N3 enrichment mapped on the secondary structure. Nucleotides that are denoted in red are enriched in the EN lane of Panel D.
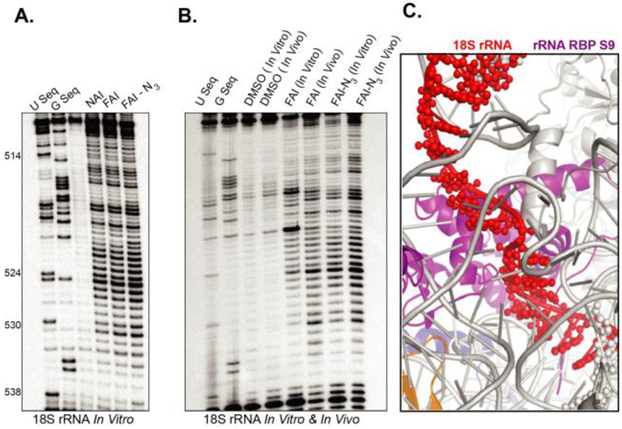
Lastly, we sought to demonstrate that FAI-N3 is a viable probe for in-cell analysis of RNA structure through SHAPE. In vitro analysis of RNA structure with SHAPE reagents does not always translate to robust signal for incell SHAPE.(14) We compared the in vitro SHAPE profiles of FAI and FAI-N3 on 18S rRNA isolated from cells. As shown in Figure 4, A, all reagents give similar robust SHAPE profiles in vitro.
Figure 4. FAI-N3 can be used to measure RNA structure inside living cells.
A. Comparison of NAI, FAI, and FAI-N3 for measuring RNA structure. B. Denaturing gel of adducts comparing in and outside of cell RNA acylation profiles. C. X-Ray structure model of the intact ribosome. (pdb 4V6X) The section in red is the single-stranded residues that have high acylation (SHAPE) in the denaturing gel in panel B.
Comparison of reagents in cells demonstrated that FAI-N3and NAI-N3 can both measure RNA structure robustly inside cells (Figure 4, B). Comparison of the SHAPE pattern to the published structure of the ribosome confirmed that residues with high SHAPE reactivity are not base paired and are in orientations that would severely weaken their base pairs to make them flexible, consistent with high SHAPE reactivity (Figure 4, C).(15)
RNA molecules are critical to the control of every biological pathway inside cells. Many RNA functions are influenced by unique RNA structures and developing methods to measure RNA structure inside cells is essential toward our understanding of RNA biology. Chemical probing is the go-to method for analyzing RNA structure. Extensions of chemical probing to transcriptomewide analyses have presented new opportunities for probe and protocol development. Herein we have presented the facile synthesis and evaluation of a dual-functioning furoyl probe for SHAPE, whose function is to measure RNA structure by hydroxyl acylation and also be amenable to enrichment through the attachment of a biotin handle.
We have demonstrated that our novel reagent, FAI-N3 is capable of measuring RNA structure, both inside and outside cells. FAI-N3 reactivity can be controlled through DTT quenching, affording complete experimental control over RNA probing. We have also demonstrated that FAI-N3 adducts can be enriched and such adducts are quite stable to conditions of hydrolysis. These two key observations suggest that FAI-N3 should be an ideal reagent for transcriptome-wide analysis by SHAPE. The recent surge in labs performing SHAPE in cells with acylimidazole reagents further underscores the importance of our findings.(16-19) The facile synthesis and utility of FAI-N3 should make it an ideal reagent for RNA structure probing in living cells.
Supplementary Material
ACKNOWLEDGMENT
We thank members of the Spitale lab for their careful reading and critique of the manuscript. We thank the Hertel Lab for graciously donating the U1snRNA construct for our studies with FAI-N3. Spitale lab is supported by start up funds from the University of California, Irvine, and the NIH (1DP2GM119164 RCS; 1RO1MH109588 RCS). RCS is a Pew Biomedical Scholar.
Funding Sources
No competing financial interests have been declared.
ABBREVIATIONS
- SHAPE
Selective Hydroxyl Acylation analyzed by Primer Extension
- NAI
Nicotinoyl acylimidazole
- FAI
Furoyl acylimidazole
Footnotes
Supporting Information. Experimental methods, synthetic schemes and spectra, for all compounds are available free of charge via the Internet at http://pubs.acs.org.
The Supporting Information is available free of charge on the ACS Publications website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sharp PA (2009) The centrality of RNA, Cell 136, 577–580. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A, and Nudler E (2013) A decade of riboswitches, Cell 152, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevilacqua PC, Ritchey LE, Su Z, and Assmann SM (2016) Genome-Wide Analysis of RNA Secondary Structure, Annu. Rev. Genet 50, 235–266. [DOI] [PubMed] [Google Scholar]
- 4.Weeks KM (2010) Advances in RNA structure analysis by chemical probing, Curr. Opin. Struct. Biol 20, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunel C, and Romby P (2000) Probing RNA structure and RNA-ligand complexes with chemical probes, Methods Enzymol 318, 3–21. [DOI] [PubMed] [Google Scholar]
- 6.Merino EJ, Wilkinson KA, Coughlan JL, and Weeks KM (2005) RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE), J. Am. Chem. Soc 127, 4223–4231. [DOI] [PubMed] [Google Scholar]
- 7.Mortimer SA, and Weeks KM (2008) Time-resolved RNA SHAPE chemistry, J. Am. Chem. Soc 130, 16178–16180. [DOI] [PubMed] [Google Scholar]
- 8.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, and Chang HY (2013) RNA SHAPE analysis in living cells, Nat. Chem. Biol 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan D, Feng C, Zhen Y, Flynn RA, and Spitale RC (2017) Comparative Analysis Reveals Furoyl in Vivo Selective Hydroxyl Acylation Analyzed by Primer Extension Reagents Form Stable Ribosyl Ester Adducts, Biochemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, and Chang HY (2015) Structural imprints in vivo decode RNA regulatory mechanisms, Nature 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, and Chang HY (2015) Structural imprints in vivo decode RNA regulatory mechanisms, Nature 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulsen LD, Kielpinski LJ, Salama SR, Krogh A, and Vinther J (2015) SHAPE Selection (SHAPES) enrich for RNA structure signal in SHAPE sequencing-based probing data, RNA 21, 1042–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montange RK, and Batey RT (2006) Structure of the S-adenosylmethionine riboswitch regulatory mRNA element, Nature 441, 1172–1175. [DOI] [PubMed] [Google Scholar]
- 14.Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, and Chang HY (2017) Comparison of SHAPE reagents for mapping RNA structures inside living cells, RNA 23, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R (2013) Structures of the human and Drosophila 80S ribosome, Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- 16.Guo JU, and Bartel DP (2016) RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria, Science 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwok CK, Ding Y, Tang Y, Assmann SM, and Bevilacqua PC (2013) Determination of in vivo RNA structure in low-abundance transcripts, Nat. Commun 4, 2971. [DOI] [PubMed] [Google Scholar]
- 18.Hector RD, Burlacu E, Aitken S, Le Bihan T, Tuijtel M, Zaplatina A, Cook AG, and Granneman S (2014) Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution, Nucleic Acids Res. 42, 12138–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pirakitikulr N, Kohlway A, Lindenbach BD, and Pyle AM (2016) The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures, Mol. Cell 62, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.