Abstract
Germline mutations in the human SAMHD1 gene cause the development of Aicardi-Goutières Syndrome (AGS), with a dominant feature being increased systemic type I interferon(IFN) production. Here we tested the state of type I IFN induction and response to, in SAMHD1 knockout (KO) human monocytic cells. SAMHD1 KO cells exhibited spontaneous transcription and translation of IFN-β and subsequent interferon-stimulated genes (ISGs) as compared to parental wild-type cells. This elevation of IFN-β and ISGs was abrogated via inhibition of the TBK1-IRF3 pathway in the SAMHD1 KO cells. In agreement, we found that SAMHD1 KO cells present high levels of phosphorylated TBK1 when compared to control cells. Moreover, addition of blocking antibody against type I IFN also reversed elevation of ISGs. These experiments suggested that SAMHD1 KO cells are persistently auto-stimulating the TBK1-IRF3 pathway, leading to an enhanced production of type I IFN and subsequent self-induction of ISGs.
Keywords: SAMHD1, IFN, AGS, TBK1-IRF3, ISGs
1. Introduction
The sterile alpha motif HD domain 1 (SAMHD1) protein is best known as potently restricting HIV-1 infection of non-cycling cells such as macrophages, dendritic cells and resting CD4+ T cells (Arfi et al., 2008; Baldauf et al., 2012; Descours et al., 2012; Goujon et al., 2008; Goujon et al., 2003; Goujon et al., 2007; Hrecka et al., 2011; Laguette et al., 2011; St Gelais et al., 2012). The ability of SAMHD1 to block HIV-1 infection correlates with its dNTPase activity (Goldstone et al., 2011; Lahouassa et al., 2012; Powell et al., 2011), which is necessary to decrease levels of dNTPs in non-cycling cells. However, several known SAMHD1 mutations can decrease cellular levels of dNTPs without restricting HIV-1 infection, suggesting that the dNTPase activity of SAMHD1 is necessary but not sufficient to enable SAMHD1 to block HIV-1 infection (Cribier et al., 2013; Welbourn et al., 2013; Welbourn and Strebel, 2016; White et al., 2017; White et al., 2013b); rather the ability of SAMHD1 to block HIV-1 infection correlates with the phosphorylation state of T592 (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2017).
SAMHD1 is comprised of a sterile alpha motif (SAM) and a histidine-aspartic acid (HD) domain. SAM domains are protein interaction modules that mediate contact with other SAM domains (Qiao and Bowie, 2005) or non-SAM domain-containing proteins(Qiao and Bowie, 2005), or that bind to a specific DNA sequences of DNA, acting as transcription activators or repressors (Qiao and Bowie, 2005). In contrast, the HD domain is a dGTP-regulated dNTPase that decreases cellular dNTP levels (Goldstone et al., 2011; Kim et al., 2012; Lahouassa et al., 2012; Powell et al., 2011), and also is necessary for SAMHD1 to oligomerize and bind to RNA (Bhattacharya et al., 2016; Brandariz-Nunez et al., 2013; Goncalves et al., 2012; Koharudin et al., 2014; Seamon et al., 2015; Tungler et al., 2013; White et al., 2013a; Yan et al., 2013). Although the HD domain has been reported to have nuclease activity, this remains incompletely understood (Antonucci et al., 2016; Beloglazova et al., 2013; Choi et al., 2015; Ryoo et al., 2014; Seamon et al., 2015).
Germline mutations in the human SAMHD1 gene are responsible for the auto-inflammatory Aicardi-Goutieres syndrome (AGS), which mimics congenital infection and is manifested by increased production of type I interferon (IFN) (Crow et al., 2006a; Crow and Manel, 2015; Dale et al., 2010; du Moulin et al., 2011; Kretschmer and Lee-Kirsch, 2017; Leshinsky-Silver et al., 2011; Livingston and Crow, 2016; Ramantani et al., 2011; Rice et al., 2009; Thiele et al., 2010). If dysregulated IFN production is a dominant feature in the pathology, such a syndrome is now termed type I interferonopathy (Crow, 2014), where AGS is now classified, as are other AGS-like syndrome like ISG15 and USP18 deficiencies (Bogunovic et al., 2012; Hermann and Bogunovic, 2017; Meuwissen et al., 2016; Zhang et al., 2015). Besides SAMHD1, the AGS phenotype is associated with mutations in six additional genes, TREX1 (AGS1)(Crow et al., 2006a), RNASEH2A (AGS2)(Crow et al., 2006b), RNASEH2B (AGS3)(Crow et al., 2006b), RNASEH2C (AGS4)(Crow et al., 2006b), ADAR1 (AGS6)(Rice et al., 2012), and IFIH1 (AGS7)(Rice et al., 2014). As of now, all proteins affected in the AGS syndrome are involved in nucleic acid metabolism, leading to the hypothesis that AGS is caused by inappropriate activation of type I IFN, following recognition of endogenous DNA and/or RNA species that can activate nucleic-acid-sensing pattern-recognition receptors (Roers et al., 2016).
Perhaps, the best-studied protein associated with AGS development is the 3’ repair exonuclease 1 (TREX1) (Crow et al., 2006a; Hoss et al., 1999; Mazur and Perrino, 1999; Stetson et al., 2008). TREX1 is an essential negative regulator of the interferon-stimulatory DNA (ISD) response, an antiviral response triggered by innate sensing of intracellular DNA (Stetson et al., 2008). Loss-of-function mutations in TREX1 are associated with the development of AGS (Crow et al., 2006a; Lehtinen et al., 2008; O’Driscoll, 2008). TREX1 metabolizes reverse-transcribed DNA from endogenous retroelements, preventing their accumulation. The accumulation of reverse-transcribed DNAs from endogenous retroelements triggers the type I interferon response (Stetson et al., 2008). Overall, these experiments suggested that AGS observed in individuals with TREX1 mutations is triggered by the inability of TREX1 protein to degrade reverse-transcribed DNA from endogenous retroelements that are left in the cell, triggers the innate immune response. Moreover, it has recently been shown that SAMHD1 is involved in the restriction of endogenous retroelements in cycling cells and that this regulation is controlled by T592 phosphorylation, suggesting that the absence of SAMHD1 allows accumulation/replication of retroelements, which could trigger AGS or autoimmune diseases (Goodier, 2016; Herrmann et al., 2018; White et al., 2016; Zhao et al., 2013).
Inspired by the mechanism used by TREX1 to prevent type I IFN response and by the role of SAMHD1 in myeloid cells HIV biology in particular, we sought to test the hypothesis that absence of SAMHD1 in monocytes triggers spontaneous type I interferon response. In agreement with this hypothesis, here we demonstrate that stable KO of SAMHD1 expression indeed triggers the type I IFN response. Interestingly, SAMHD1 KO cells exhibited spontaneous transcription and translation of several interferon-stimulated genes (ISGs). By using an inhibitor of the catalytic activity of TBK1/IKKε (BX795)(Clark et al., 2009), which blocks its phosphorylation, and hence IRF3 activation and IFN-β production, we were able to prevent spontaneous transcription and translation of ISGs in SAMHD1 KOs. In agreement, we found that SAMHD1 KO cells present high levels of phosphorylated TBK1 when compared to control cells. These experiments suggested that these cells persistently secrete type I IFN, causing autocrine stimulation. To test whether spontaneous transcription and translation of ISGs in SAMHD1 KOs is triggered by secreted type I IFN, we used an antibody against IFNα/β that blocks the ability of IFNα/β to stimulate the interferon receptor. Remarkably, the use of this antibody blocked the spontaneous transcription and translation of ISGs in the knockouts. Taken together, the results described below showed that SAMHD1 KO monocytes spontaneously trigger the type I IFN response. More specifically, the results suggest that SAMHD1 is shielding innate immune sensors from interaction with nucleic acids, thereby preventing the activation of the type I IFN response.
2. Materials and Methods
2.1. Cell lines
Human THP-1 cells (ATCC TIB-202) were grown in growth medium consisting of RPMI 1640 supplemented with 10% fetal calf serum, 1× GlutaMAX (Gibco) and 1% penicillin-streptomycin (Gibco). All cells were cultured at 37 °C and 5% CO2.
For the THP-1 SAMHD1 KO generation, two single guide RNAs (gRNAs) targeting unique sequences targeting exon 1 of the SAMHD1 gene were designed (gRNA1: 66CTCAAACACCCCTTCCGCAG86 and gRNA 2: 88GGCAGACTGGTCCCCGGGCC108) and cloned into the lentiCRISPR v1 plasmid, which contains a puromycin resistance cassette and Cas9 nuclease. Both gRNA constructs were functionally tested for their ability to mediate genomic insertion, deletion or inversions by isolating the genomic DNA from stably transduced and selected polyclonal population of THP-1 cells and performing a Surveyor nuclease assay as per manufacturer’s instructions. Puromycin resistant cells were selected from each gRNA vector transduction, and then sorted by fluorescence-activated cell sorting (FACS) into 96-well plates for clonal expansion. PCR of the SAMHD1 gene was used to analyze the sequences of the KO cell lines. Clone THP-1 SAMHD1 KO 1 has a deletion of 4 nucleotides in exon 1 which gives a SAMHD1 protein with a STOP codon at the beginning of the open reading frame [CCCCTTCCG-Δ(82-87)-GGCAGACTGG], losing the first Met in the sequence. Clone SAMHD1 KO 1-3 has a 14 nucleotides deletion [AAACACCCCT-Δ(78-92)-CTGGTCCCCG], also losing the first Met in the sequence.
2.2. Cell priming with IFNα and BX795 treatment
THP-1, THP-1 Contrail orTHP-1 SAMHD1 KO cells were primed by incubation with 10, 100, 1000 or 5000 IFN Alpha A U/ml (Hu-IFN-αA; Hu-IFN-α2a, Millipore) in normal RPMI 10% FCS medium for 30 minutes, 24 h or as indicated. After incubation, cells were collected for transcript (RNA) and protein expression of ISGs.
The TBK1/IKKε catalytic inhibitor of phosphorylation BX795 (Invivogen), which prevents IRF3 activation and IFN-β production, was used at concentrations of 2 and 4 μM in non-PMA treated cells for 48 h at 37 °C in RPMI culture medium containing 10% FCS. After incubation, cells were collected for protein expression analysis.
2.3. ISG Expression Analysis by qRT-PCR
RNA was extracted from THP-1 cells (Qiagen RNeasy) and reverse-transcribed (SuperScrip III reverse transcriptase) according to the manufacturer’s instructions. Levels of ISG expression (IFN-β, IFIT1, MXA), relative to the ACTIN RNA housekeeping gene, were analyzed by Eppendorf quantitative real-time PCR using SYBR™ Green PCR Master Mix (Applied Biosystems). The primer sequences used were as follows: ACTIN Forward 5’AACACCCCAGCCATGTACGT’3 and Reverse 5’CGGTGAGGATCTTCATGAGGTAGT’3, IFN-β(NM002176.3) Forward 5’ACCTCCGAAACTGAAGATCTCCTA’3 and Reverse 5’TGCTGGTTGAAGAATGCTTGA’3, IFIT1 (NM001548.4) Forward 5’TTGCCTGGATGTATTACCAC’3 and Reverse 5’GCTTCTTGCAAATGTTCTCC’3 and MXA (NM001144925.1) Forward 5’AGGACCATCGGAATCTTGAC’3 and Reverse 5’TCAGGTGGAACACGAGGTTC’3. Relative expression of the ISGs were calculated by the ΔΔCt method, with comparison with the mean value for the mock-treated THP-1 Control 1 cells.
2.4. ISG Protein Expression
One million THP-1 cells, undifferentiated or treated for 24 h with PMA (40ng/mL), were grown in six-well dishes, and lysed in 0.1 ml of whole-cell extract (WCE) buffer [50 mM Tris pH 8.0, 280 mM NaCI, 0.5% IGEPAL, 10% glycerol, 5 mM MgCl2, 50 μg/ml ethidium bromide, 50 U/ml benzonase (Roche)]. The extract was then incubated for 1 h at 4 °C, and the cell debris removed by centrifugation at 20,000 g for 1 h at 4 °C. The supernatant was then mixed with 5× protein-loading buffer and the samples were analyzed by SDS-PAGE and Western blotting with anti-ISG antibodies [anti-MXA (1:1000; Abeam), anti-MXB (1:1000; Novus Biologicals), anti-ISG15 (1:500; Santa Cruz Biotechnology), anti-SAMHD1 (1:1000; house made rabbit anti-SAMHD1) and anti-GAPDH (1:5,000; Ambion)] in a buffer containing 5% non-fat Milk in PBS-Tween 1%. Secondary antibodies against rabbit and mouse conjugated to IRDye 680LT or IRDye 800CW were obtained from Li-Cor (1:10,000 diluted). Protein concentrations were quantified by optical densitometry of the gel band, using the program Image J and normalizing to the GAPDH band intensity.
In the case of STAT1, STAT2, pSTAT1, pSTAT2, USP18 and pTBK1, cells were lysed RIPA buffer (Thermo Scientific), and protease/phosphatase inhibitor cocktail (Cell signaling technologies) and analyzed by western blot. The following antibodies were used: USP18 (Cell Signaling Technology D4E7, 1:1,000); Stat1 (Santa Cruz C-111, 1:1,000); Stat2 (Santa Cruz C-20, 1:1,000); phospho-Stat1 (Cell Signaling Technology 9171 L, 1:1,000); phospho-Stat2 (Millipore 07224, 1:1000); pTBK1 (Cell Signaling Technology 5483 1:1,000); GAPDH (Millipore MAB374, 1:20,000). Antibody binding was detected by enhanced chemiluminescence (Western Lightning, Perkin Elmer).
2.5. IFN bioassay
To examine the production of type I IFN the reporter cell line HEK293-ISRE-Luc was used (Saito et al., 2016). This reporter cell line carries IFN-stimulated response element within the promoter region driving the expression of luciferase. One day prior to use, HEK293-ISRE-Luc cells were plated on a 96-well flat bottom plate at 30,000 cells per well with 100 μl of cell suspension. One day after, 100 μl of culture supernatant from THP-1 cells was harvested and transferred into the 96-well flat bottom plate containing HEK293-ISRE-Luc cells for 24 h. In each experiment, the IFN standard curve was generated using a stock solution of recombinant interferon alpha A (Millipore). A working solution of IFN at 10,000 units per mL was used to generate 10-fold serial dilutions. These standard dilutions were added at 10 μl per well in triplicates. The cells were placed in a tissue culture incubator after addition of supernatant samples and IFN standards. At the following day, the cells were lysed with 25 μl of 1× buffer prepared from Luciferase Cell Culture Lysis 5× Reagent (Promega) for 20 minutes at −80 °C. Twenty five μl of cells lysates was transferred to an opaque 96-well plate and 100 μl of the luciferase assay reagent (Promega) was added, followed by reading the relative luciferase units in a luminometer.
2.6. Statistical analysis
To compare the effects of each cell line in relation to its control, all data were analyzed using Student’s t-test. Differences were considered statistically significant at P < 0.05 (*), P < 0.01 (**) or P < 0.001 (***), not significant (ns).
3. Results
3.1. THP-1 cells stably knocked out for SAMHD1 expression show increased basal transcription and translation of IFN-stimulated genes (ISGs).
Mutations in the human SAMHD1 gene trigger AGS, in an autosomal recessive fashion with the phenotype being characterized by cerebral vasculopathy and early stroke. Interestingly, this phenotype correlates with high levels of type I IFN and ISGs in the peripheral blood of these patients (Dale et al., 2010; Rice et al., 2009). In an attempt to model this human in vivo phenotype in a traceable in vitro system, we stably knocked out SAMHD1 in a monocytic cell line.
We used human monocytic THP-1 cells and the CRISPR-Cas9 system to knockout SAMHD1. As shown in Figure 1A, three independent knockout clones were obtained (KO 1, KO 2, and KO 1-3). Expression of SAMHD1 was assayed by Western blotting using a specific antibody against SAMHD1. Two CRISPR control cell lines that retained normal SAMHD1 expression (THP-1 Control 1 and THP-1 Control 2) were also obtained (Fig. 1A). Next, we tested the basal transcription of different ISGs by qRT-PCR. For this purpose, we tested the basal transcription of MXA and IFIT1 (Fig. 1B). Remarkably, basal transcription of MXA increased ~20-, ~7-, and ~40-fold for THP-1 SAMHD1 KO 1, THP-1 SAMHD1 KO 2, and THP-1 SAMHD1 KO 1-3 cells, respectively, when compared to THP-1 Control 1 cells (Fig. 1B). Similarly, IFIT1 transcripts increased ~30-, ~6- and ~20-fold for THP-1 SAMHD1 KO 1, THP-1 SAMHD1 KO 2, and THP-1 SAMHD1 KO 1-3 cells, respectively (Fig. 1B). Expression of the different transcripts was normalized to actin (Fig. 1B). These experiments showed that stable SAMHD1 KO increased basal transcription of ISGs in THP-1 SAMHD1 KO clones.
Figure 1. THP-1 cells stably knockout for SAMHD1 expression showed increased basal transcription and translation of interferon-stimulated genes (ISGs).
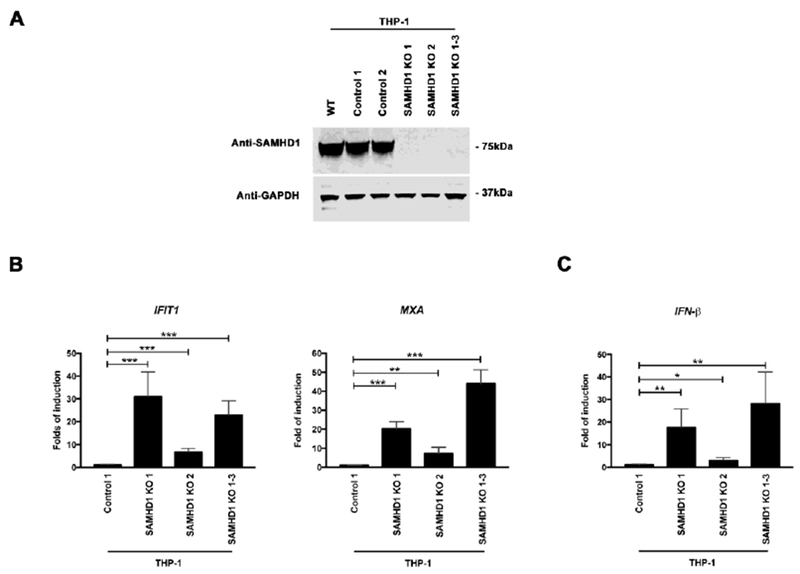
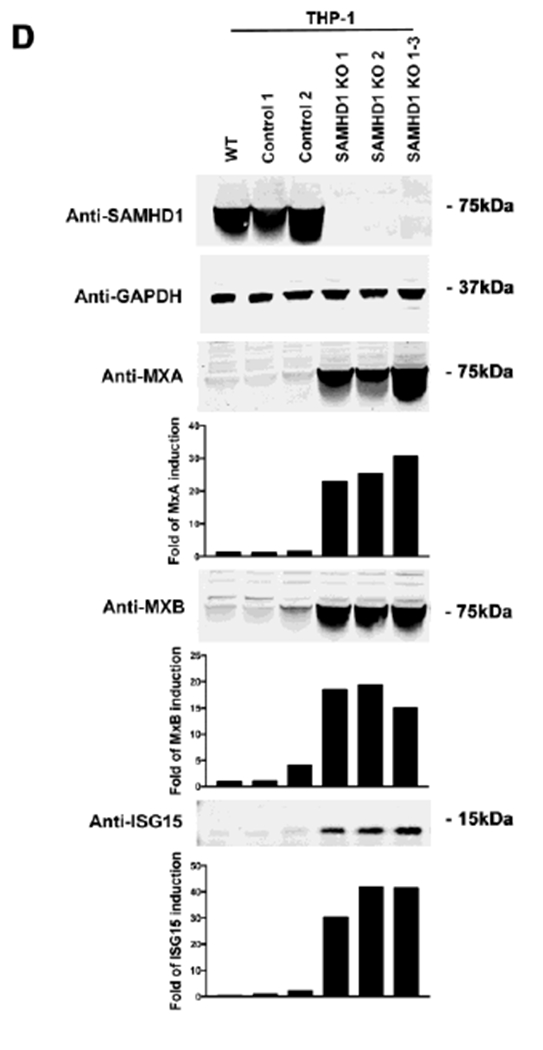
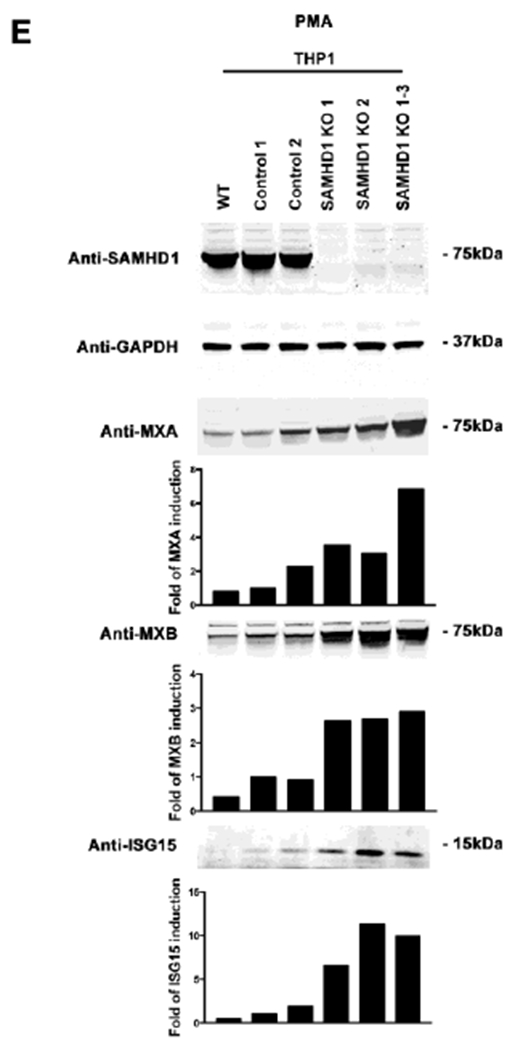
(A) SAMHD1 KO clones were prepared by using the CRISPR-Cas9 system. Three different THP-1 SAMHD1 KO clones (SAMHD1 KO 1, 2 and 1-3) were identified by Western blotting using antibodies against SAMHD1 (upper panel). THP-1 clones that underwent the CRISPR-Cas9 protocol but did not lose expression of SAMHD1 (Controls 1 and 2) were also identified (upper panel). In addition, a wild type THP-1 cell line (WT) was included. As a loading control, samples were also analyzed for GAPDH expression (upper panel). (B) Transcript levels of MXA and IFIT1 were analyzed by qRT-PCR in the indicated THP-1 SAMHD1 KO clones, and were compared to THP-1 control cells using a specific set of primers for each indicated gene (lower panel). Transcript levels were normalized to actin, and fold-induction relative to the THP-1 Control 1 is shown. P < 0.05 (*), P < 0.01 (**) or P < 0.001 (***) using two tailed Student’s t-test are shown. (C) Similarly the transcript levels of IFNβ were measured. Transcript levels were normalized to actin, and fold-induction relative to the THP-1 Control 1 is shown. (D) MXA, MXB and ISG15 protein levels were analyzed by Western blotting in the indicated THP-1 SAMHD1 KO clones. Expression levels for THP-1 Control clones are also shown. As a loading control, samples were also analyzed for GAPDH. Quantification of band intensities normalized to GAPDH is shown. Experiments were repeated at least three times and a representative example is shown. (E) Protein levels of MXA, MXB and ISG15 were also analyzed in PMA-treated THP-1 SAMHD1 KO clones. Quantification of band intensities normalized to GAPDH is shown. Experiments were repeated at least three times and a representative example is shown.
Because the increased basal transcription of ISGs may be due to production of type I IFNs, we tested whether production of IFN-β is increased in SAMHD1 KO clones (Fig. 1C). Interestingly, the basal transcription levels of IFN-β increased 15-, 3-, and 25-fold in THP-1 SAMHD1 KO 1, THP-1 SAMHD1 KO 2 and THP-1 SAMHD1 KO 1-3 cells, respectively, when compared to THP-1 CRISPR control cells (THP-1 Control 1) (Fig. 1C). This result indicated that IFN-β is up-regulated in SAMHD1 KO clones.
To further determine whether the observed transcriptional activation translates into protein expression, we tested for MXA, MXB, and ISG15 production in the three THP-1 SAMHD1 KO clones 1, 2, and 1-3 by Western blotting (Fig. 1D). As a control, and to ensure that the observed phenotype was not an artifact of the CRISPR/Cas9 procedure, we utilized two different THP-1 CRISPR control clones (THP-1 Control 1 and Control 2). As shown in Figure 1D, all tested THP-1 SAMHD1 KO clones expressed higher protein levels of MXA, MXB and ISG15 when compared to THP-1 control clones or THP-1 wild-type (WT) cells. As a loading control, we measured expression of GAPDH. The results indicate that stable KO of SAMHD1 increases the basal levels of type I IFN, thereby increasing transcription and translation of ISGs. The bar graphs in Figure 1D show the fold-induction relative to THP-1 Control 1 cells normalized to GAPDH.
We next tested whether differentiation of THP-1 SAMHD1 KO cells to a non-cycling state with phorbol 12-myristate 13-acetate (PMA) changed the observed phenotype (Schwende et al., 1996). PMA-treated THP-1 SAMHD1 KO clones (Clones 1, 2, 1-3) still showed increased MXA, MXB and ISG15 expression when compared to THP-1 CRISPR control clones (THP-1 Control 1 and Control 2) (Fig. 1E). These experiments suggested that absence of SAMHD1 increases basal transcription levels of ISGs in cycling and non-cycling cells.
Next we tested whether knocking out the expression of SAMHD1 in epithelial cells resulted in the increased expression of ISGs. For this purpose, using the CRISPR/Cas9 methodology, we prepared HT-1080 cells that are KO for the expression of SAMHD1 (Data not shown). Surprisingly, none of the HT-1080 SAMHD1 KO cells exhibited increased expression of the protein MxB. These results indicate that this phenotype may only occur in cells that are derived from the immune system lineages.
To asses whether the phenotype observed in THP-1 SAMHD1 KO cell lines changes in the presence of human serum, we measured expression of MxA (Fig. 1S). Interestingly, we observed a stronger phenotype in human serum when compared to fetal bovine serum (Fig. 1S). The use of human serum, which is a condition closer to in vivo, increases the strength of this phenotype suggesting that human serum must contain agents that accentuate this phenotype.
Altogether our results suggested that ISGs are upregulated in THP-1 SAMHD1 KO clones and this may be due to the constant production of type I IFNs by KO clones.
3.2. THP-1 cells stably knocked out for SAMHD1 show intact responsiveness to type I IFN.
The type I interferon response needs to be tightly regulated for preservation of immune homeostasis (Chen et al., 2017). Dysregulation of IFN responses are linked to immune and autoimmune disorders (Kretschmer and Lee-Kirsch, 2017). Because AGS is an immune disorder, we tested whether response to type I IFN is intact in SAMHD1 KO cells. Thus, we stimulated THP-1 SAMHD1 KO clones with 1000 U/mlof IFNα, and measured MXA and IFIT1 transcription 24 h later. As shown in Figure 2A, stimulation of THP-1 SAMHD1 KO clones 1, 2, and 1-3 with 1000 U/ml of IFNα triggered equally robust MXA transcription response when compared to THP-1 Control 1 cells. Similarly, induction of IFIT1 transcription was similar when compared to THP-1 Control 1 cells (Fig. 2A).
Figure 2. THP-1 cells stably knocked out for SAMHD1 show intact responsiveness to type I IFN.
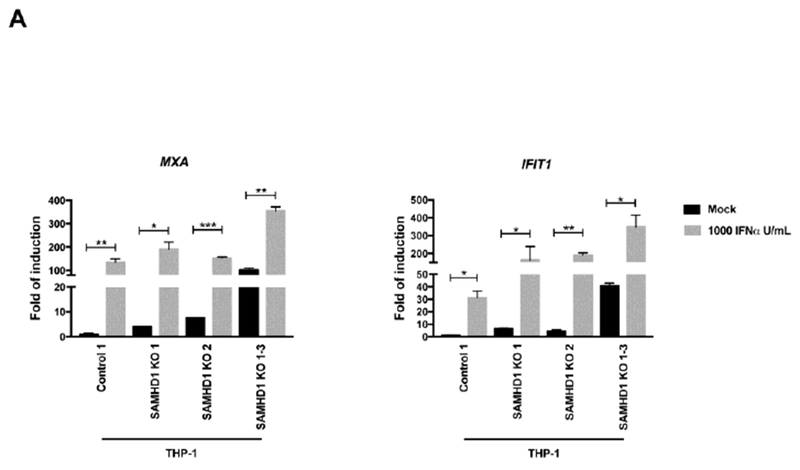
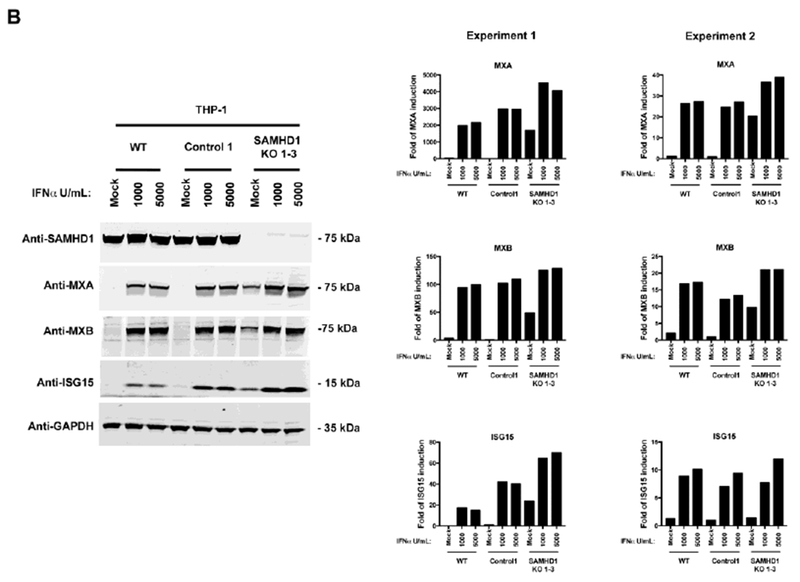
(A) Transcript levels for MXA and IFIT1 were determined in IFNα-treated THP-1 SAMHD1 KO clones. The indicated THP-1 SAMHD1 KO clones were treated with 1000 U/ml for 24 h, after which the cells were lysed and processed for RNA extraction. Transcription levels were determined by qRT-PCR using a set of specific primers for each indicated gene. Transcript levels were normalized to actin, and the fold-induction relative to THP-1 Control 1 is shown. P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) or not significant (ns), using two-tailed Student’s t-test are shown. (B) MxA, MxB and ISG15 levels were measured in THP-1 SAMHD1 KO 1-3 cells treated with the indicated concentrations of IFNα for 24 h (left panel). Protein levels were determined by Western blotting using the indicated antibodies (left panel). Expression of the different proteins was quantifying and normalized to GAPDH. Experiments were repeated at least three times and two independent experiments are shown (right panel). (C) Similar experiments to measure the levels of MxA, MxB and ISG15 were performed using lower concentrations of IFNα (left panel). Expression of the different proteins was quantifying and normalized to GAPDH. Experiments were repeated at least three times and a representative experiment is shown (right panel).
To test whether transcriptional activation in THP-1 SAMHD1 KO cells translates into increased protein expression, we performed the same experiment but measured protein expression by Western blotting. Thus, we stimulated THP-1 SAMHD1 KO cells with 1000 or 5000 U/ml of IFNα, and measured MXA, MXB and ISG15 protein expression 24 h later. As shown in Figure 2B, THP-1 SAMHD1 knockout cells (THP-1 SAMHD1 KO 1-3) showed similar levels of protein expression of MXA, MXB and ISG15 when compared to THP-1 Control 1 cells. In agreement with the transcription assay results, these experiments showed that basal translation of ISGs is likewise upregulated in THP-1 SAMHD1 KO cells. Similar results were obtained by using lower amounts of IFNα (Fig. 2C). Altogether these results indicated that SAMHD1 KO cells have normal responsiveness to type I IFN stimulation. This was important to document as type I IFN hypersensitivity was observed in patients with AGS-like disease, who lack USP18 or ISG15, a negative feedback regulators of type I IFN signaling (Meuwissen et al., 2016; Speer et al., 2016).
3.3. The small molecule BX795 inhibits spontaneous production of type I IFN in THP-1 SAMHD1 KO cells.
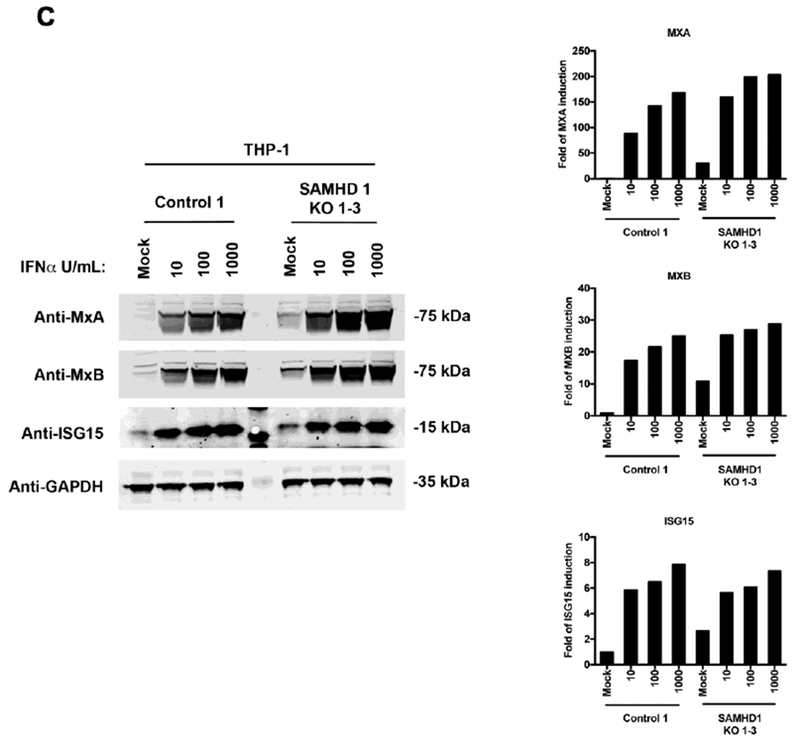
We sought to test whether spontaneous induction of ISGs in THP-1 SAMHD1 KO cells is due to persistent stimulation and production of type I IFN, which may auto-stimulate cells and expression of ISGs. To this end, we used BX795, a small-molecule inhibitor of the catalytic activity of TBK1/IKKε that blocks the latter’s phosphorylation, preventing IRF3 activation and IFN-β production (Clark et al., 2009); and ultimately shuts down the TBK1-IRF3 pathway, which is used by several intracellular sensors to induce type I IFN response. As shown in Figure 3A, use of increasing BX795 concentrations attenuated spontaneous expression of ISGs (MXA, MXB and ISG15) in all the three THP-1 SAMHD1 KO clones (1, 2, 1-3) when compared to THP-1 Control 1 cells or WT THP-1 cells. Interestingly, treatment with 2 μM BX795 for 48 h dramatically reduced expression of ISGs in THP-1 SAMHD1 KO clones. The bar graphs in Figure 3B represent fold-induction, normalized to GAPDH relative to the untreated KO clone (Mock). The results of these experiments suggested that spontaneous expression of ISGs in THP-1 SAMHD1 KO clones may be caused by persistent production of type I IFNs, which are responsible for the higher basal levels of ISGs in THP-1 SAMHD1 KO cells.
Figure 3. The TBK1 inhbitor BX795 prevents spontaneous expression of ISGs in THP-1 SAMHD1 KO cells.
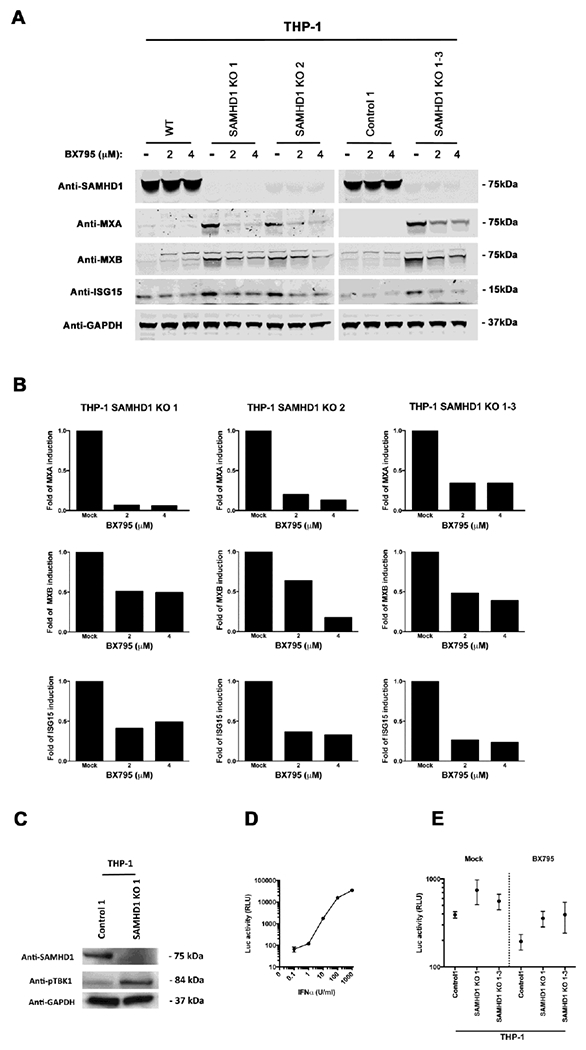
(A) Expression of MxA, MxB and ISG15 in BX795-treated THP-1 SAMHD1 KO clones. THP-1 SAMHD1 KO clones were treated with the indicated concentrations of BX795 for 48 hours, after which the cells were lysed, and protein expression was measured by Western blotting using specific antibodies against MxA, MxB, and ISG15, respectively. As a loading control, samples were also analyzed for GAPDH. (B) Band quantification normalized to GAPDH is shown. Experiments were repeated three times for each KO cell line, and a representative example is shown. (C) THP-1 SAMHD1 KO 1 cells were analyzed for the expression of phosphorylated TBK1 (pTBK1) by Western blotting. As a loading control, samples were also analyzed for GAPDH. (D) HEK293-ISRE-Luc cells were treated with the indicated concentrations of IFNαfor 24 h. Subsequently, cells were lysed and analyzed for luciferase activity. (E) Supernatant harvested from THP-1 control, THP-1 SAMHD1 KO 1 and KO 1-3, treated or not with 2μM of BX975 for 48h, were used to treat HEK293-ISRE-Luc cells for 24 h. Subsequently, cells were lysed and analyzed for luciferase activity.
Because BX795 concentrations attenuated spontaneous expression of ISGs (MXA, MXB and ISG15) in THP-1 SAMHD1 KO clones, we tested the phosphorylation state of TBK1 in THP-1 SAMHD1 KO 1 cells. As shown in Figure 3C, the levels of phosphorylated TBK1 (pTBK1) are upregulated in the THP-1 SAMHD1 KO 1 cells when compared with the THP-1 control cells. This result suggests that the element or elements triggering the expression of ISGs in the THP-1 SAMHD1 KO cell lines are promoted by the phosphorylation of TBK1.
To examine the production of IFN by THP-1 SAMHD1 KO cell lines, we used the HEK293T-ISRE-Luc reporter cell line (Saito et al., 2016). The HEK293T-ISRE-Luc reporter cell line contains the interferon stimulated response element (ISRE) fuse to the luciferase gene. To validate the assay, an IFNα standard curve was generated (Fig. 3D). To measure the levels of IFNα in the THP-1 SAMHD1 KO cells, we incubated supernatants from THP-1 SAMHD1 KO cell, treated or not with 2μM of BX795, with HEK293T-ISRE-Luc for 24 h. As shown in Figure 3E, luciferase activity was higher for THP-1 SAMHD1 KO cells when compared to control cells and is reduced when treated with BX795.
3.4. Use of anti-IFNα/β prevents spontaneous expression of ISGs in THP-1 SAMHD1 KO cells.
To further characterize the spontaneous induction IFN-β and activation of the type I IFN pathway, we measured the expression of phosphorylated STAT1 and STAT2 (pSTAT1 and pSTAT2) in THP-1 SAMHD1 KO cells upon IFNα stimulation. For this purpose, we incubated THP-1 SAMHD1 KO 1 cells with increasing concentrations of IFNα for 30 min, and measured phosphorylated STAT1 and STAT2. As shown in Figure 4A and B, levels of STAT1 and STAT2 expression were higher in THP-1 SAMHD1 KO 1 cells than in THP-1 Control 1 cells. Similarly, pSTAT2 levels and pSTAT1 levels were higher in THP-1 SAMHD1 KO 1 cells than in THP-1 Control cells.
Figure 4. Anti-IFNα/β antibody prevents spontaneous production of ISGs in THP-1 SAMHD1 KO cells.
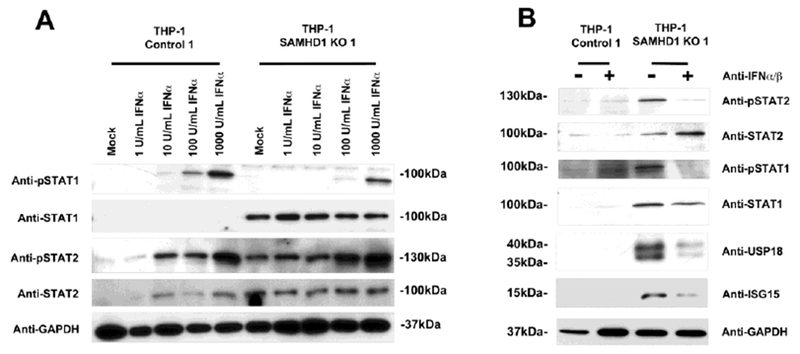
(A) THP-1 SAMHD1 KO 1 cells were treated with increasing amounts of IFNα. After 30 min, cells were lysed, and STAT1, STAT2, phosphorylated-STAT1 (pSTAT1), and phosphorylated-STAT2 (pSTAT2) expression was measured by Western blotting using specific antibodies. Similar analysis was performed in THP-1 Control 1 cells. As a loading control, samples were analyzed for GAPDH. (B) THP-1 SAMHD1 KO 1 cells were treated with IFNα for 30 min in the presence of anti-IFNα/β antibodies. The cells were then lysed and STAT1, STAT2, pSTAT1, pSTAT2, USP18 and ISG15 expression was measured by Western blotting using specific antibodies. As a loading control, samples were analyzed for expression of GAPDH. As a control, serum from sheep was used. Experiments were performed a least three times, and a representative is shown.
To test whether spontaneous transcription and translation of ISGs in SAMHD1 KO cells is triggered by secreted type I IFN, we used an antibody against IFNα/β that blocks the latter’s ability to stimulate the interferon receptor. For this purpose, THP-1 SAMHD1 KO 1 and control cells were treated with 5 ug/ml of anti-IFNα/β for 30 mins (Fig. 4B), and levels of ISG15, UPS18, STAT1, STAT2, pSTAT1, and pSTAT2 were measured by Western blotting. As shown in Figure 4B, anti-IFNα/β antibodies significantly reduced expression of the interferon-stimulated genes ISG15 and USP18. Similarly, levels of phosphorylated STAT1 and STAT2 were also reduced to those inTHP-1 Control 1 cells. These findings suggested that spontaneous expression of ISGs in THP-1 SAMHD1 KO clones is due to secreted type I IFN, which is persistently stimulating the type I IFN receptor.
4. Discussion
Compelling evidence in patients suggests that SAMHD1 is a negative regulator of the type I IFN response (Rice et al., 2009). AGS is a heritable disease linked to mutations of the SAMHD1 gene, causing afflicted individuals to exhibit increased production of type I IFN (Crow et al., 2015; Dale et al., 2010; du Moulin et al., 2011; Kretschmer and Lee-Kirsch, 2017; Leshinsky-Silver et al., 2011; Livingston and Crow, 2016; Rice et al., 2009; Thiele et al., 2010). However, the mechanism by which type I IFN is upregulated in patients with mutations or deletions of SAMHD1 is unknown. Consistently with these observations, SAMHD1 KO mice display increased levels of type I IFN (Maelfait et al., 2016a; Rehwinkel, 2014; Rehwinkel et al., 2013). In agreement with these observations, our experiments showed that stably knocking out SAMHD1 in THP-1 cells increased the basal levels of type I IFN, in a way replicating the phenotype seen in patients with AGS (Berger et al., 2011; Puigdomenech et al., 2013). The present work provided a model to study the spontaneous type I IFN stimulation that occurs in the absence of SAMHD1.
To demonstrate that THP-1 SAMHD1 KO cells spontaneously produce type I IFN, we measured transcript and protein levels of classic ISGs such as MXA and IFIT1. THP-1 SAMHD1 KO clones showed increased transcript and protein levels of the tested ISGs. This suggests that absence of SAMHD1 correlates with increased ISG transcription and translation. One possibility is that SAMHD1 degrades, or binds to, an as yet unidentified nucleic acid (RNA or DNA) with the ability to trigger innate immune sensors, e.g. RIG-I-like receptors (RLRs), cytosolic DNA sensors (CDSs) or the protein “stimulator of interferon“ (STING) (Habjan and Pichlmair, 2015; Mankan et al., 2014; Wu and Chen, 2014). RLRs such as RIG-I and MDA5 are cytoplasmic RNA helicases that sense double-stranded RNA, leading to activation of type I IFN through the TBK1-IRF3 pathway (Fujita et al., 2007; Gleason et al., 2011; Hiscott, 2007; Tu et al., 2013; Yoneyama and Fujita, 2007). Similarly, CDSs sense damaged or pathogenic DNA, leading again to type I IFN production through the TBK1-IRF3 pathway (Ma et al., 2015a). STING protein on the other hand is activated by cyclic dinucleotides, leading also to a potent type I IFN response mediated by the TBK1-IRF3 pathway (Ma et al., 2015b; Tanaka and Chen, 2012). In this scenario, SAMFIDTs role might be to degrade, or bind to, any of the aforementioned substrates (DNA and/or RNA), thereby preventing type I IFN activation. In the absence of SAMHD1, any or all of these intracellular substrates could conceivably activate type I IFN. (Maelfait et al., 2016b; Meuwissen et al., 2016).
AGS-like diseases are also developed by mutations that are involved in the type I IFN negative feedback regulation loop, as is the Ubiquitin-specific peptidase 18 (USP18) (Meuwissen et al., 2016; Taylor et al., 2018), which causes enhanced induction of ISGs after stimulation with type I IFN, a phenotype known as hypersensitivity response to IFN. To understand whether the absence of SAMHD1 triggers type I IFN hypersensitivity, we stimulated the THP1-SAMHD1 KO cells with IFNα and measure the levels of different ISGs, observing that the cells lacking SAMHD1 behaved like THP1 control cells, suggesting that a negative feedback loop may not exist.
Since most of the known pathways sensing cytosolic RNA and DNA lead to type I IFN via the TBK1-IRF3 pathway, we decided to test whether inhibition of this pathway by the small-molecule inhibitor BX795 can abrogate the spontaneous type I IFN phenotype (Pokatayev et al., 2016). Interestingly, BX795 restored the native phenotype in THP-1 SAMHD1 KO cells. In agreement, we found that the levels of phosphorylated TBK1 in THP-1 SAMHD1 KO cells are upregulated when compared to control cells, suggesting that the TBK1-IRF3 pathway is activated in these cells. Overall, these results suggested that absence of SAMHD1 leads to activation of type I IFN by a signaling mechanism initiated by a cytosolic nucleic acid. This is consistent with growing evidence that all known defective genes associated with AGS are somehow involved in the metabolism of nucleic acids (Roers et al., 2016).
In agreement with the conclusion that nucleic acids are activating innate immune sensors in THP-1 SAMHD1 KO cells, it is well established that SAMHD1 binds to RNA and DNA (Goncalves et al., 2012; Tungler et al., 2013). Triggering of the type I IFN response in THP-1 SAMHD1 KO cells by RNA or DNA might be of self or non-self origin. A similar situation is observed in the case of the DNAse protein TREX1 (O’Driscoll, 2008; Stetson et al., 2008), which binds to and subsequently degrades both self- and nonself- DNA species, thereby preventing activation of type I IFN response (Crow et al., 2015; Stetson et al., 2008; Yang et al., 2007). Future experiments will pursue the innate immune sensor that triggers the type I IFN response in THP-1 SAMHD1 KO cells.
Lastly this work explored whether the spontaneous induction of ISGs in THP-1 SAMHD1 KO cells is caused by IFNα/β autocrine stimulation. Interestingly, we observed that the use of anti-IFNα/β antibodies in the supernatant of THP-1 SAMHD1 KO cells prevents the induction of the type I IFN response. This result suggested that IFNα/β is produced and secreted by THP-1 SAMHD1 KO cells to subsequently stimulate the type I IFN receptor. Noteworthy was the fact that STAT1 and STAT2 levels are higher in THP-1 SAMHD1 KO cells than in control cells, also suggesting that the type I IFN pathway is active in THP-1 SAMHD1 KO cells.
The work reported here suggests that absence of SAMHD1 causes nucleic acids in the cytosol of THP-1 cells to trigger innate immune sensors that, in turn, use the TBK1-IRF3 pathway to activate the type I IFN response. These findings are a first step toward unraveling the complex molecular mechanism by which patients with SAMHD1 deficiencies exhibit high levels of type I IFN, and suggest the possibility of using the small-molecule inhibitor BX795 or an improved next-generation molecule to treat patients with early-stage AGS.
Supplementary Material
Highlights.
SAMHD1 KO human cells replicate the phenotype observed in patients with Aicardi-Goutières Syndrome.
SAMHD1 KO cells show spontaneous expression of type I IFNs and interferon-stimulated genes.
Inhibition of the TBK1-IRF3 pathway abrogated the production of type I IFNs in SAMHD1 KO cells.
Spontaneous expression of ISGs in SAMHD1 KO cells is due to secreted type I IFN.
ACKNOWLEDGEMENTS
We are grateful to the NIH/AIDS repository program for providing valuable antibodies and drugs. Dr. Klaus Strebel has generously provided the rabbit antibodies against SAMHD1. The work was funded by an NIH grant (R01 GM123540) to F.D.-G and March of Dimes, R01AI127372, R21AI134366, R21AI129827 to D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonucci JM, St Gelais C, de Silva S, Yount JS, Tang C, Ji X, Shepard C, Xiong Y, Kim B, Wu L, 2016. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nature medicine 22, 1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfi V, Riviere L, Jarrosson-Wuilleme L, Goujon C, Rigal D, Darlix JL, Cimarelli A, 2008. Characterization of the early steps of infection of primary blood monocytes by human immunodeficiency virus type 1. Journal of virology 82, 6557–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT, 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nature medicine 18, 1682–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF, 2013. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. The Journal of biological chemistry 288, 8101–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E, 2011. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS pathogens 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Wang Z, White T, Buffone C, Nguyen LA, Shepard CN, Kim B, Demeler B, Diaz-Griffero F, Ivanov DN, 2016. Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Scientific reports 6, 31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL, 2012. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandariz-Nunez A, Valle-Casuso JC, White TE, Nguyen L, Bhattacharya A, Wang Z, Demeler B, Amie S, Knowlton C, Kim B, Ivanov DN, Diaz-Griffero F, 2013. Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Liu J, Cao X, 2017. Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. Journal of autoimmunity 83, 1–11. [DOI] [PubMed] [Google Scholar]
- Choi J, Ryoo J, Oh C, Hwang S, Ahn K, 2015. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Plater L, Peggie M, Cohen P, 2009. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. The Journal of biological chemistry 284, 14136–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M, 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell reports 3, 1036–1043. [DOI] [PubMed] [Google Scholar]
- Crow YJ, 2014. Type I interferonopathies: Mendelian type I interferon up-regulation. Current opinion in immunology 32C, 7–12. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, Abdel-Hamid MS, Abdel-Salam GM, Ackroyd S, Aeby A, Agosta G, Albin C, Allon-Shalev S, Arellano M, Ariaudo G, Aswani V, Babul-Hirji R, Baildam EM, Bahi-Buisson N, Bailey KM, Barnerias C, Barth M, Battini R, Beresford MW, Bernard G, Bianchi M, Billette de Villemeur T, Blair EM, Bloom M, Burlina AB, Carpanelli ML, Carvalho DR, Castro-Gago M, Cavallini A, Cereda C, Chandler KE, Chitayat DA, Collins AE, Sierra Corcoles C, Cordeiro NJ, Crichiutti G, Dabydeen L, Dale RC, D’Arrigo S, De Goede CG, De Laet C, De Waele LM, Denzler I, Desguerre I, Devriendt K, Di Rocco M, Fahey MC, Fazzi E, Ferrie CD, Figueiredo A, Gener B, Goizet C, Gowrinathan NR, Gowrishankar K, Hanrahan D, Isidor B, Kara B, Khan N, King MD, Kirk EP, Kumar R, Lagae L, Landrieu P, Lauffer H, Laugel V, La Piana R, Lim MJ, Lin JP, Linnankivi T, Mackay MT, Marom DR, Marques Lourenco C, McKee SA, Moroni I, Morton JE, Moutard ML, Murray K, Nabbout R, Nampoothiri S, Nunez-Enamorado N, Oades PJ, Olivieri I, Ostergaard JR, Perez-Duenas B, Prendiville JS, Ramesh V, Rasmussen M, Regal L, Ricci F, Rio M, Rodriguez D, Roubertie A, Salvatici E, Segers KA, Sinha GP, Soler D, Spiegel R, Stodberg TI, Straussberg R, Swoboda KJ, Suri M, Tacke U, Tan TY, te Water Naude J, Wee Teik K, Thomas MM, Till M, Tonduti D, Valente EM, Van Coster RN, van der Knaap MS, Vassallo G, Vijzelaar R, Vogt J, Wallace GB, Wassmer E, Webb HJ, Whitehouse WP, Whitney RN, Zaki MS, Zuberi SM, Livingston JH, Rozenberg F, Lebon P, Vanderver A, Orcesi S, Rice GI, 2015. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American journal of medical genetics. Part A 167A, 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T, 2006a. Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature genetics 38, 917–920. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP, 2006b. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature genetics 38, 910–916. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N, 2015. Aicardi-Goutieres syndrome and the type I interferonopathies. Nature reviews. Immunology 15, 429–440. [DOI] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ, 2010. Familial Aicardi-Goutieres syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. American journal of medical genetics. Part A 152A, 938–942. [DOI] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M, 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Moulin M, Nurnberg P, Crow YJ, Rutsch F, 2011. Cerebral vasculopathy is a common feature in Aicardi-Goutieres syndrome associated with SAMHD1 mutations. Proceedings of the National Academy of Sciences of the United States of America 108, E232; author reply E233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Onoguchi K, Onomoto K, Hirai R, Yoneyama M, 2007. Triggering antiviral response by RIG-I-related RNA helicases. Biochimie 89, 754–760. [DOI] [PubMed] [Google Scholar]
- Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P, 2011. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon beta. The Journal of biological chemistry 286, 35663–35674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M, 2011. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T, 2012. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Human mutation 33, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Goodier JL, 2016. Restricting retrotransposons: a review. Mobile DNA 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Arfi V, Pertel T, Luban J, Lienard J, Rigal D, Darlix JL, Cimarelli A, 2008. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. Journal of virology 82, 12335–12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A, 2003. Heterologous human immunodeficiency virus type 1 lentiviral vectors packaging a simian immunodeficiency virus-derived genome display a specific postentry transduction defect in dendritic cells. Journal of virology 77, 9295–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A, 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A, 2015. Cytoplasmic sensing of viral nucleic acids. Current opinion in virology 11, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann M, Bogunovic D, 2017. ISG15: In Sickness and in Health. Trends Immunol 38, 79–93. [DOI] [PubMed] [Google Scholar]
- Herrmann A, Wittmann S, Thomas D, Shepard CN, Kim B, Ferreiros N, Gramberg T, 2018. The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mobile DNA 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, 2007. Triggering the innate antiviral response through IRF-3 activation. The Journal of biological chemistry 282, 15325–15329. [DOI] [PubMed] [Google Scholar]
- Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T, 1999. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. The EMBO journal 18, 3868–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J, 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA, 2012. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. The Journal of biological chemistry 287, 21570–21574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LM, Wu Y, DeLucia M, Mehrens J, Gronenborn AM, Ahn J, 2014. Structural basis of allosteric activation of sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. The Journal of biological chemistry 289, 32617–32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S, Lee-Kirsch MA, 2017. Type I interferon-mediated autoinflammation and autoimmunity. Current opinion in immunology 49, 96–102. [DOI] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M, 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F, 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature immunology 13, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW, 2008. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. The Journal of biological chemistry 283, 31649–31656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Malinger G, Ben-Sira L, Kidron D, Cohen S, Inbar S, Bezaleli T, Levine A, Vinkler C, Lev D, Lerman-Sagie T, 2011. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. European journal of human genetics : EJHG 19, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston JH, Crow YJ, 2016. Neurologic Phenotypes Associated with Mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1: Aicardi-Goutieres Syndrome and Beyond. Neuropediatrics 47, 355–360. [DOI] [PubMed] [Google Scholar]
- Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G, 2015a. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. Journal of immunology 194, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G, 2015b. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO reports 16, 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Bridgeman A, Benlahrech A, Cursi C, Rehwinkel J, 2016a. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell reports 16, 1492–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Bridgeman A, Benlahrech A, Cursi C, Rehwinkel J, 2016b. Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell reports 16, 1492–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V, 2014. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. The EMBO journal 33, 2937–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW, 1999. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′-->5′ exonucleases. The Journal of biological chemistry 274, 19655–19660. [DOI] [PubMed] [Google Scholar]
- Meuwissen ME, Schot R, Buta S, Oudesluijs G, Tinschert S, Speer SD, Li Z, van Unen L, Heijsman D, Goldmann T, Lequin MH, Kros JM, Stam W, Hermann M, Willemsen R, Brouwer RW, Van IWF, Martin-Fernandez M, de Coo I, Dudink J, de Vries FA, Bertoli Avella A, Prinz M, Crow YJ, Verheijen FW, Pellegrini S, Bogunovic D, Mancini GM, 2016. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. The Journal of experimental medicine 213, 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, 2008. TREX1 DNA exonuclease deficiency, accumulation of single stranded DNA and complex human genetic disorders. DNA repair 7, 997–1003. [DOI] [PubMed] [Google Scholar]
- Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Morris HD, Yan N, Crouch RJ, 2016. RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGAS-STING innate immune-sensing pathway in mice. The Journal of experimental medicine 213, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW, 2011. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. The Journal of biological chemistry 286, 43596–43600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomenech I, Casartelli N, Porrot F, Schwartz O, 2013. SAMHD1 restricts HIV-1 cell-to-cell transmission and limits immune detection in monocyte-derived dendritic cells. Journal of virology 87, 2846–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Bowie JU, 2005. The many faces of SAM. Science’s STKE : signal transduction knowledge environment 2005, re7. [DOI] [PubMed] [Google Scholar]
- Ramantani G, Hausler M, Niggemann P, Wessling B, Guttmann H, Mull M, Tenbrock K, Lee-Kirsch MA, 2011. Aicardi-Goutieres Syndrome and Systemic Lupus Erythematosus (SLE) in a 12-Year-Old Boy With SAMHD1 Mutations. J Child Neurol 26, 1425–1428. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, 2014. Mouse knockout models for HIV-1 restriction factors. Cellular and molecular life sciences : CMLS 71, 3749–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, Bieniasz PD, Towers GJ, Moita LF, Crow YJ, Bonthron DT, Reis e Sousa C, 2013. SAMHD1-dependent retroviral control and escape in mice. The EMBO journal 32, 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ, 2009. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature genetics 41, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Cordeiro NJ, Dale RC, Davidson JE, De Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenco CM, Mancardi MM, Masurel-Paulet A, McInnes IB, Menezes MP, Mignot C, O’Sullivan J, Orcesi S, Picco PP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ, 2014. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature genetics 46, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, De Laet C, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W Jr., Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O’Connell MA, Lovell SC, Crow YJ, 2012. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature genetics 44, 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Hiller B, Hornung V, 2016. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 44, 739–754. [DOI] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K, 2014. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nature medicine 20, 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Henning MS, Serrao E, Dubose BN, Teng S, Huang J, Li X, Saito N, Roy SP, Siddiqui MA, Ahn J, Tsuji M, Hatziioannou T, Engelman AN, Yamashita M, 2016. Capsid-CPSF6 Interaction Is Dispensable for HIV-1 Replication in Primary Cells but Is Selected during Virus Passage In Vivo. Journal of virology 90, 6918–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwende H, Fitzke E, Ambs P, Dieter P, 1996. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. Journal of leukocyte biology 59, 555–561. [PubMed] [Google Scholar]
- Seamon KJ, Sun Z, Shlyakhtenko LS, Lyubchenko YL, Stivers JT, 2015. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res 43, 6486–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F, Rubino E, Gardner TJ, Wedeking T, Hermann M, Duehr J, Sanal O, Tezcan I, Mansouri N, Tabarsi P, Mansouri D, Francois-Newton V, Daussy CF, Rodriguez MR, Lenschow DJ, Freiberg AN, Tortorella D, Piehler J, Lee B, Garcia-Sastre A, Pellegrini S, Bogunovic D, 2016. ISG15 deficiency and increased viral resistance in humans but not mice. Nature communications 7, 11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L, 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R, 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ, 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling 5, ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Cash MN, Santostefano KE, Nakanishi M, Terada N, Wallet MA, 2018. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV-1 infection in macrophages. Journal of leukocyte biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele H, du Moulin M, Barczyk K, George C, Schwindt W, Nurnberg G, Frosch M, Kurlemann G, Roth J, Nurnberg P, Rutsch F, 2010. Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Human mutation 31, E1836–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, Li Y, Dunn GP, Chan E, Thai T, Yang S, Ficarro SB, Marto JA, Jeon H, Hahn WC, Barbie DA, Eck MJ, 2013. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell reports 3, 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungler V, Staroske W, Kind B, Dobrick M, Kretschmer S, Schmidt F, Krug C, Lorenz M, Chara O, Schwille P, Lee-Kirsch MA, 2013. Single-stranded nucleic acids promote SAMHD1 complex formation. Journal of molecular medicine 91, 759–770. [DOI] [PubMed] [Google Scholar]
- Welbourn S, Dutta SM, Semmes OJ, Strebel K, 2013. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. Journal of virology 87, 11516–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Strebel K, 2016. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology 488, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Han K, Sawyer SL, Kim B, Diaz-Griffero F, 2016. Modulation of LINE-1 Retrotransposition by a Human SAMHD1 Polymorphism. Virology reports 6, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Martinez-Lopez A, Knowlton C, Lenzi G, Kim B, Ivanov D, Diaz-Griffero F, 2017. A SAMHD1 mutation associated with Aicardi-Goutieres Syndrome uncouples the ability of SAMHD1 to restrict HIV-1 from its ability to downmodulate type I interferon in humans. Human mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F, 2013a. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F, 2013b. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell host & microbe 13, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ, 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annual review of immunology 32, 461–488. [DOI] [PubMed] [Google Scholar]
- Yan J, Kaur S, DeLucia M, Hao C, Mehrens J, Wang C, Golczak M, Palczewski K, Gronenborn AM, Ahn J, Skowronski J, 2013. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. The Journal of biological chemistry 288, 10406–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE, 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131, 873–886. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T, 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine & growth factor reviews 18, 545–551. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, Rice GI, Chen C, Mansouri N, Mahdaviani SA, Itan Y, Boisson B, Okada S, Zeng L, Wang X, Jiang H, Liu W, Han T, Liu D, Ma T, Wang B, Liu M, Liu JY, Wang QK, Yalnizoglu D, Radoshevich L, Uze G, Gros P, Rozenberg F, Zhang SY, Jouanguy E, Bustamante J, Garcia-Sastre A, Abel L, Lebon P, Notarangelo LD, Crow YJ, Boisson-Dupuis S, Casanova JL, Pellegrini S, 2015. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature 517, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, Cheung LE, Wang G, Kazazian HH Jr., Yu XF, 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell reports 4, 1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


